Abstract
PLAGL1/ZAC1 undergoes parental genomic imprinting, is paternally expressed, and is a member of the imprinted gene network (IGN). It encodes a zinc finger transcription factor with anti-proliferative activity and is a candidate tumor suppressor gene on 6q24 whose expression is frequently lost in various neoplasms. Conversely, gain of PLAGL1 function is responsible for transient neonatal diabetes mellitus, a rare genetic disease that results from defective pancreas development. In the present work, we showed that Plagl1 up-regulation was not associated with DNA damage-induced cell cycle arrest. It was rather associated with physiological cell cycle exit that occurred with contact inhibition, growth factor withdrawal, or cell differentiation. To gain insights into Plagl1 mechanism of action, we identified Plagl1 target genes by combining chromatin immunoprecipitation and genome-wide transcriptomics in transfected cell lines. Plagl1-elicited gene regulation correlated with multiple binding to the proximal promoter region through a GC-rich motif. Plagl1 target genes included numerous genes involved in signaling, cell adhesion, and extracellular matrix composition, including collagens. Plagl1 targets also included 22% of the 409 genes that make up the IGN. Altogether, this work identified Plagl1 as a transcription factor that coordinated the regulation of a subset of IGN genes and controlled extracellular matrix composition.
INTRODUCTION
Parental genomic imprinting is an epigenetic mechanism that was selected at the prototherian to metatherian transition during mammalian evolution (1–3). It results in mono-allelic, parent-of-origin-dependent expression of ∼150 genes in eutherian mammals. Although imprinted genes (IGs) appear to be mostly functionally unrelated, we recently showed that they are frequently co-regulated and modulate cell cycle withdrawal when cells enter quiescence or differentiate. This observation supports the hypothesis that IGs belong to a single gene network and that parental genomic imprinting targeted a distinct biological process (4). We also showed that the imprinted gene network (IGN) comprises non-imprinted genes, including a significant number of genes controlling the composition of the extracellular matrix (ECM). These observations raised several mechanistic questions, in particular with respect to the mechanisms that ensure the co-expression of IGN members. The majority of IGs is clustered at a limited number of loci, which are generally well conserved across eutherian genomes. This clustered organization suggests that IGs share regulatory elements, which may explain how IG expression is concerted within a given imprinted locus. However, at least 20 imprinted loci exist in eutherian genomes, some IGs are isolated, and non-imprinted IGN genes are mostly located outside imprinted loci; hence, additional mechanisms likely exist to coordinate IGN gene expression.
Zac1/Lot1/Plagl1 was originally identified as a gene encoding a zinc finger transcription factor with anti-proliferative activity (5) whose expression was lost upon cell transformation (6). The human orthologue is a candidate tumor suppressor gene on 6q24 whose expression is lost in mammary (7,8) and ovary tumors (9), and other types of neoplasm (10–12). Inversely, gain of PLAGL1 function is responsible for transient neonatal diabetes mellitus (13–15). The molecular mechanisms that underlie Plagl1 activity are not fully understood at the moment; the biological context that calls for Plagl1 activity is also unclear. We previously showed that Plagl1 is a member of the IGN and regulates the Igf2-H19 locus by direct binding to the enhancers located 3′ of H19 (4,16). We also showed that alteration of Plagl1 expression resulted in the modulation of the expression of other IGs, but the underlying mechanism was unclear. The relationship between Plagl1 activity and the expression of other IGN members was not known. In the present work, we identified Plagl1 target genes genome-wide and showed that Plagl1 regulated a subset of the IGN genes and ECM genes.
MATERIALS AND METHODS
Mouse tissues
All animal procedures were conducted according to the guidelines approved by the Institutional Animal Care and Use Committee. Different tissues were dissected out from C57BL/6J mouse E18.5 embryos and neonates and homogenized in RNA NOW (Ozyme, Saint Quentin en Yvelines). To study muscle regeneration, 10 μl of a 50 μg/ml notexin solution in 0.9% NaCl or saline alone were injected into the left tibialis anterior of adult male C57BL/6J mice (79.4 ± 5.0 days). Two mice were killed before injection and at 6 time points after injection in each condition. The injected tibialis anterior was removed and homogenized in RNA NOW. Saline-injected muscles were used to monitor the absence of significant gene regulation in the absence of notexin, not to normalize data.
Cell culture
Neuro-2a cells were grown in high-glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. AtT20 cells were grown in OptiMEM supplemented with 15% fetal bovine serum and 1% penicillin-streptomycin. For UV-C irradiation, cells were washed with PBS 1×, irradiated at 50 J/m2, and refed with culture medium. Mouse embryonic fibroblasts from wild-type and Plagl1+/-pat. C57BL/6J mice were prepared using standard protocols (17) and grown in high-glucose DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Min6 cells were grown in low-glucose DMEM supplemented with 15% fetal bovine serum and 50 mM β-Mercapto-ethanol. E14Tg2a (E14) mouse embryonic stem cells were maintained and differentiated to generate cortical cells in vitro as previously described (18).
Transfection
Plasmids encoding β-galactosidase, chloramphenicol acetyl-transferase (CAT), Plagl1/Zac1, CD2 and eGFP were described previously (4,5,8). 0.8–1 × 106 Neuro-2a cells were plated one day before transfection in 60-mm plates and transfected at 80% confluence with 30 μl of Lipofectamine 2000 (Thermo Fisher Scientific) and 10 μg of plasmid DNA including 4 μg of pRK5-mZac1 or pRK5-CAT as a control and 6 μg of carrier plasmid, according to the manufacturer's instructions. We quantified CAT and Plagl1 levels using RT-qPCR. The expression levels of CAT, respectively Plagl1, normalized to Tbp, Gus and Mrpl32 was 24.5 ± 5.4, respectively 12.5 ± 6.0. 3–4 × 106 Min6 cells were plated as above and transfected with 10 μl Lipofectamine 2000 and 5 μg of plasmid DNA including 2 μg of pRK5-mZac1 or pRK5-NLSeGFP as a control and 3 μg of carrier plasmid. The expression levels of eGFP, respectively Plagl1, normalized to Tbp, Gus and Mrpl32 was 789.4 ± 323.5, respectively 273.1 ± 91.0.
Immunological techniques
Western blots were performed on total cell lysates (40 μg total protein) using anti-Zac1 antibody (5) (1:4000) and anti-actin (Merck Cat. No. MAB1501; 1:2000). Anti-rabbit and anti-mouse secondary antibodies were from Cell Signaling (Cat. No. 7074) and Thermo Fisher Scientific (Cat. No. 32430), respectively. For immunofluorescence experiments, anti-Th antibody (Merck Cat. No. MAB318; 1:200) and anti-Zac1 antibody (1:4000) were incubated overnight at 4°C and secondary antibody (Thermo Fisher Scientific Cat. No. A11010, 1:500) for 45 min at room temperature. Nuclei were stained with DAPI (1 μg/ml). Coverslips were mounted with Mowiol and examined with an AxioimagerZ1 fluorescence microscope (Zeiss).
Cell cycle analysis
Neuro-2a cells were transfected as above with 1 μg of pRK5-CD2 together with 4 μg of pRK5-mZac1 or pRK5-CAT as a control and 5 μg of carrier plasmid. We selected CAT as a control for the cell cycle experiments because, among the three genes that we tested, i.e. CAT, GFP and β-galactosidase, CAT was the most neutral with respect to cell proliferation compared to untransfected Neuro-2a cells (data not shown). Thirty hours later, cells were prepared and stained with 0.5 μg FITC-conjugated anti-CD2 antibody and 50 μg/ml propidium iodide as previously described (7). Propidium iodide and FITC fluorescence intensities were determined using a FACScan flow cytometer (Becton–Dickinson) and the cell cycle distribution of the 10% most CD2-positive cells was analyzed using Modfit software (Verity Software House).
Promoter activity measurement
Nptx1 and Mpl promoters were PCR amplified from mouse genomic DNA with appropriate primers (Supplementary Table S1) and cloned upstream of the Luciferase/SV40 polyadenylation cassette in pGL3 (Promega). The genomic coordinates (mm9) were chr11(–): 119,409,017–119,409,960 for Nptx1_0.9kb and chr11(–): 119,409,017–119,410,530 for Nptx1_1.5kb constructs, and chr4(–): 118,130,034–118,131,876 for Mpl_1.8kb and chr4(–): 118,130,034–118,130,876 for Mpl_0.8kb constructs. The CMV-based pRK7-βgal plasmid was used to normalize for transfection efficiency. Plasmids were transfected into Neuro-2a cells and Luciferase and β-galactosidase activities were measured 24 hours later as previously described (15).
Digital gene expression
Sixteen hours after transfection, total RNA was extracted using the RNAqueous kit (Thermo Fisher Scientific, Cat. No. AM1912) and treated with DNase I (DNA-free, Thermo Fisher Scientific) according to the manufacturer's instructions. Libraries were prepared from two biological replicates of each condition (Plagl1- and CAT-transfected Neuro-2a) using the Digital Gene Expression-Tag Profiling Kit with DpnII (Illumina, Cat. No. FC-102-1007) according to the manufacturer's instruction. Briefly, 2 μg of total RNA were incubated with magnetic oligo(dT) beads to select poly(A)+ RNA. Reverse transcription was performed on captured RNA followed by cDNA second strand synthesis using random primers. Captured double stranded DNA was digested using DpnII. The fragments that remained attached to the beads were ligated to Illumina's GEX DpnII adapter 1 followed by digestion with MmeI resulting in the release of tags. The tags were ligated using Illumina's GEX Adapter 2, amplified by PCR (15 cycles), and purified on a polyacrylamide gel. Libraries were validated using a 2100 Bioanalyzer (Agilent), denatured using 0.1 M NaOH, diluted to 8 pM, and sequenced (single reads 36 nt, Illumina Genome Analyzer) at MGX facility following the manufacturer's instructions. Image analysis and base calling were performed using the Illumina pipeline version 1.3.
A tag library was made from a UCSC refFlat table of transcripts (refFlat.txt.gz; 15 January 2012) using an in-house Perl script available upon request. The 16-nucleotide sequences 3′ of all DpnII sites in every transcript were extracted. Unique tags that matched only one transcriptomic location were kept and annotated as canonic when the corresponding DpnII site was the last 3′ site on the cDNA, or transcriptomic when the corresponding DpnII site was not the last 3′ site. GeneIDs were retrieved from the NCBI (gene2refseq.gz; 19 January 2012).
Reads passing Illumina purity filters were kept for further analysis. After trimming the adapter sequences, the 16nt- and 17nt-trimmed reads were aligned to the DpnII-tag library without mismatch; counts for canonic and transcriptomic tags corresponding to the same GeneID were pooled using an in-house Perl script. Genes whose sum of tags in the four libraries was <10 were filtered out. Raw counts were normalized using the TMM (Trimmed Mean of M values) method (19) as implemented in the edgeR package. edgeR was used to identify differentially expressed genes with a generalized linear model likelihood ratio test using a paired design and a FDR set at 0.05 (20). DGE data from this study have been submitted to the NCBI GEO database under accession number GSE75942.
RNA-seq
The RNA-seq data regarding proliferation/quiescence/differentiation of the 3T3-L1 preadipocyte cell line have been described previously (4) and are available at the NCBI GEO web site under accession number GSE50612. Total RNA was prepared 16 hours after Min6 cell transfection as above. Strand-specific libraries were prepared using Illumina's TruSeq Stranded mRNA Sample Preparation kit from four independent replicates of Plagl1- and NLS-eGFP-transfected Min6 cells and sequenced on a HiSeq 2500 (single reads 50 nt) as described (4). Image analysis and base calling were performed using the HiSeq Control Software and Real-Time Analysis component. Demultiplexing was performed using Illumina's Conversion Software (bcl2fastq 2.17). The quality of the data was assessed using FastQC v0.11.5 from the Babraham Institute and the Sequence Analysis Viewer (Illumina). Potential contaminants were monitored with the FastQ Screen software from the Babraham Institute.
We aligned RNA-seq reads to the mouse genome (UCSC mm9) with the splice junction mapper TopHat 2.1.1, which used Bowtie 2.2.8. We downloaded gene model annotations from the UCSC database (genes.gtf 15 January 2012) and GeneIDs from the NCBI (gene2refseq.gz, 19 January 2012). Final read alignments having more than 3 mismatches were discarded. We performed gene counting using the union mode of HTSeq-count 0.6.1p1. Before statistical analysis, genes with less than 20 reads (cumulating the eight analyzed samples) were filtered out. Counts were normalized using the Relative Log Expression (RLE) method as implemented in edgeR (20), which was used to identify differentially expressed genes with a generalized linear model likelihood ratio test using a paired design. We corrected P-values for multiple testing using the Benjamini-Hochberg FDR method. Genes with adjusted P-value <0.01 were declared ‘differentially expressed’. RNA-seq data from this study have been submitted to the NCBI GEO under accession number GSE84914. We analyzed the transcriptome of two independent MEF preparations from Plagl1+/-pat. and control littermate E13.5 embryos. MEF RNA-seq libraries were prepared and sequenced as above. Data were processed as above with the following changes: bcl2fastq 2.18, Bowtie2 2.2.9, genes with <10 reads were filtered out. Data have been submitted to the NCBI GEO under accession number GSE99409.
RT-qPCR
Total RNAs from cell lines and tissues were prepared using RNA NOW, treated with DNase I (DNA-free, Thermo Fisher Scientific), and reverse-transcribed with MoMuLVRT (Thermo Fisher Scientific) according to the manufacturer's instructions. To quantify the expression of Plagl1 target genes, we designed and validated primer pairs (Qiagen-Operon, Supplementary Table S1) for real-time, quantitative PCR, which was performed with ABI Prism 7000 and the SybrGreen PCR Master Mix (Thermo Fisher Scientific). Primers used to quantify Plagl1 expression quantified all imprinted isoforms. We used geNorm (21) to select the most appropriate housekeeping genes for each experiment. The level of expression of each gene X was normalized to the geometric mean of the levels of expression of the selected housekeeping genes, e.g. R1, R2 and R3, according to the formula X/geometric mean (R1, R2, R3) = 2−(Cp(X) – arithmetic mean(Cp(R1), Cp(R2), Cp(R3))), where Cp is the threshold cycle. We normalized data with B2m in the comparison between transfected Neuro-2a cells and embryonic and neonate mouse tissues (Supplementary Figure S4), Mrpl32 in muscle regeneration experiments (Figure 2, Supplementary Figure S3), Tbp, Gus and Gapdh in in vitro corticogenesis experiments (Figure 2, Supplementary Figure S3), Tbp, Gus and Mrpl32 in MEF proliferation experiments (Figure 1) and in the comparison of Neuro-2a and Min6 cells (Figure 4).
Figure 2.
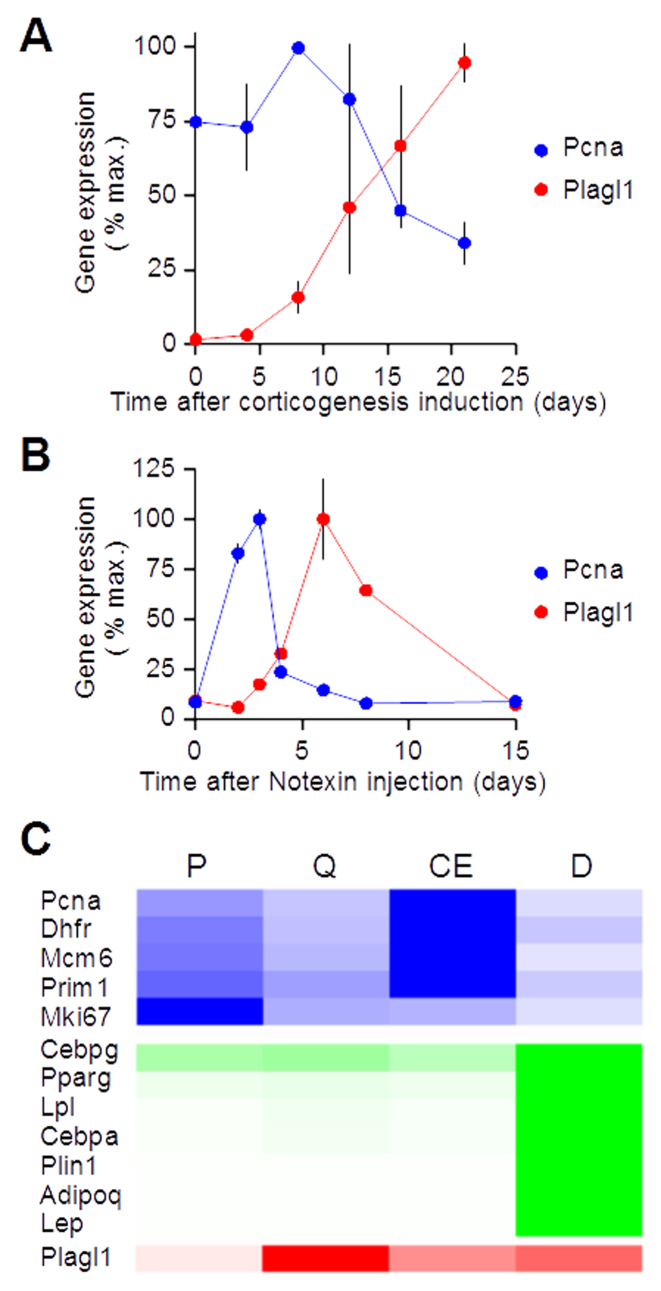
Plagl1 is induced during neurogenic (A), myogenic (B) and adipogenic differentiation (C). (A) Embryonic stem cells (ESCs) were grown in vitro and induced to cortical neural cells for the indicated period of time as in (18). ESCs initially lost their pluripotency and generated neural progenitors over the 12 first days. Neural progenitors progressively generated the different cortical neuron subtypes and ultimately glial cells; the expression pattern of proliferation, pluripotency and corticogenesis markers is displayed on Supplementary Figure S3A. Pcna (blue) and Plagl1 (red) expression levels were quantified by quantitative RT-PCR. Data are mean ± S.E.M. of three independent experiments reversed transcribed in duplicate and quantified in triplicate. (B) The tibialis anterior muscle of adult mice was injected with notexin, a snake venom toxin that is directly toxic to skeletal muscle. Satellite cells resumed proliferation, generated myoblasts at day 1–2 that exited cell cycle at day 3–4, and differentiated into mature myocytes over the following 10 days; the expression pattern of proliferation, inflammation, and myogenic markers is displayed on Supplementary Figure S3B. We collected muscles at different time points during muscle regeneration and quantified Pcna and Plagl1 expression by quantitative RT-PCR. Data are mean ± S.E.M. from one experiment on two animals per time point reverse transcribed in duplicate and quantified in octuplicate (Pcna) or quadruplicate (Plagl1). (C) Proliferating (P) 3T3-L1 preadipocytes were grown until confluence and remained quiescent (Q) for 2 days. Following addition of the adipogenic cocktail IDX (insulin, dexamethasone and IBMX), the cells resumed proliferation and entered the so-called clonal expansion (CE) phase. The cells exited the cell cycle and underwent adipogenic differentiation (D). Normalized RNAseq counts for proliferation markers (blue; Pcna, Dhfr, Mcm6, Prim1 and Mki67), adipogenic differentiation markers (green; Cebpg, Pparg, Lpl, Cebpa, Plin1, Adipoq, Lep) and Plagl1 (red) were displayed as a heat map.
Figure 1.
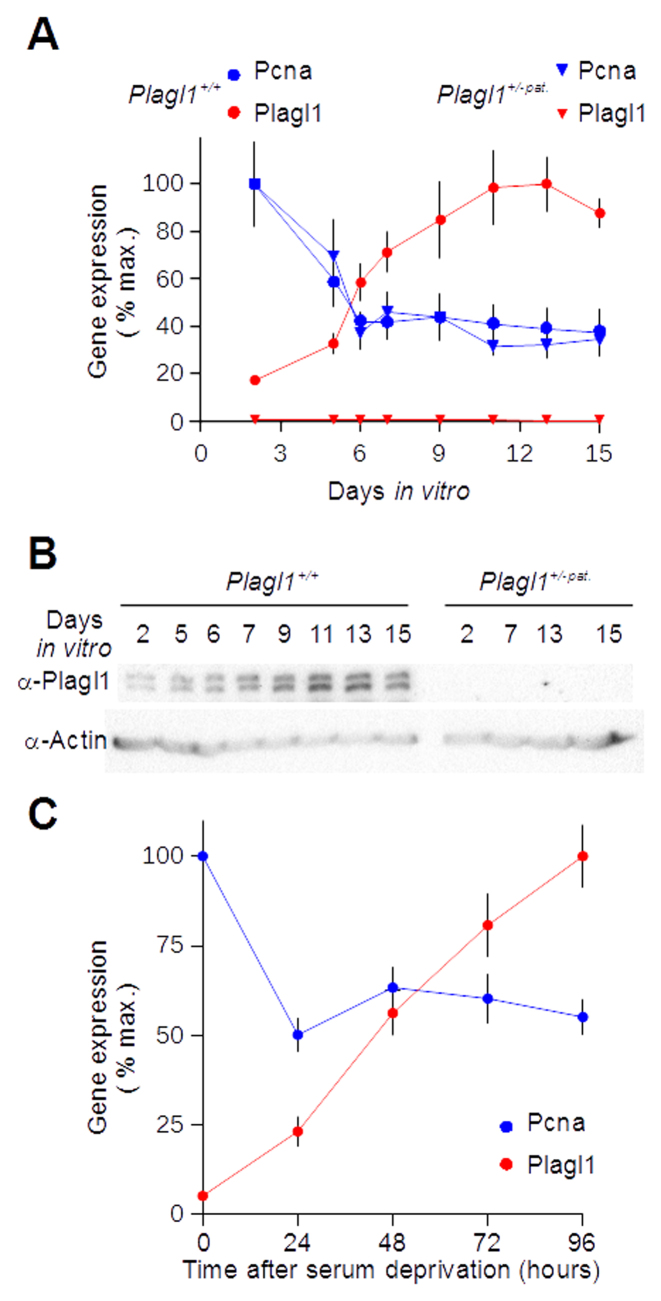
Plagl1 is induced during cell cycle exit. (A) Plagl1+/+ (circles) and Plagl1+/-pat. (triangles) mouse embryonic fibroblasts (MEFs) were grown in vitro until confluence (∼7 days in vitro), and kept confluent for 8 additional days with daily medium renewal. Pcna (blue) and Plagl1 (red) levels were quantified by quantitative RT-PCR. (B) Protein lysates were prepared from the same samples and probed with anti-Plagl1 and anti-actin antisera. (C) Wild-type, exponentially growing MEFs were deprived from fetal bovine serum for the indicated time period. Pcna (blue) and Plagl1 (red) expression levels were quantified as in A. One representative experiment of three independent experiments is shown in each panel. For real-time PCR experiments, data are mean ± S.D. of three independent reverse transcription reactions quantified in triplicate.
Figure 4.
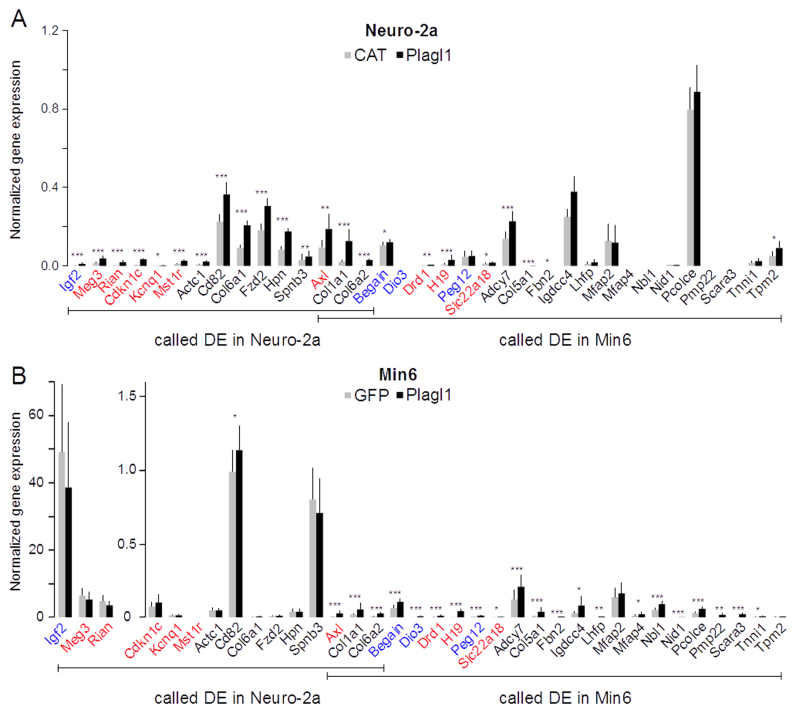
Validation of the DGE and RNA-seq data by quantitative RT-PCR. (A) Imprinted and IGN genes called ‘differentially expressed’ in Plagl1-transfected Neuro-2a or Min6 cells were quantified by qRT-PCR in CAT- or Plagl1-transfected transfected Neuro-2a cells. (B) Same as in A with GFP- or Plagl1-transfected Min6 cells. Paternally and maternally imprinted genes are labeled in blue and red, respectively. Bi-allelically expressed genes are labeled in black. Data are mean ± S.D. of four independent transfection experiments quantified in triplicate. The statistical significance of the difference between control and Plagl1-transfected cells was tested using the Mann–Whitney U test. ***P < 0.001, **P < 0.01; *P < 0.05.
ChIP-seq
Sixteen hours after transfection, we fixed Neuro-2a cells using 4% paraformaldehyde in PBS. We performed ChIP with an anti-Zac1 antibody (5) using the protocol described by A. Kouskouti and I. Kyrmizi (http://www.epigenome-noe.net/researchtools/protocol.php_protid=10.html). We used two biological replicates with input chromatins as controls. We prepared libraries according to Illumina's instructions using the ChIP-seq DNA sample prep kit (Cat. No. IP-102-1001). For each sample, 10 ng of DNA was repaired, resulting in blunt end fragments and adenylated on its 3′ ends. Illumina's adapters were ligated onto adenylated DNA. We sized the DNA fragments using an agarose gel (fragment size including adapters, 200 bp) followed by PCR (18 cycles) to amplify fragments with both adapters. We validated libraries on a 2100 Bioanalyzer (Agilent) and sequenced them (single reads 36 nt) using Illumina Genome Analyzer following the manufacturer's instructions. We performed image analysis, base calling and alignment to the mouse genome (mm9) using the Illumina pipeline version 1.3. Redundant reads that mapped at the exact same location were reduced to a single count. We called peaks using the two sample analysis method of Cisgenome v.1.2 (22) with input chromatin as control and a 0.001 FDR. Preliminary analysis of the two replicate experiments demonstrated a good overlap of the lists of peaks. The number of peaks was higher in one experiment but 75% of the peaks identified in the experiment which produced the lower peak number were also found in the replicate experiment. We performed further analysis on pooled reads from input and immuno-precipitated chromatins. Fifty one false-positive peaks located in amplified regions of the input genome were removed of the set of called peaks using Sole-Search (23). The remaining 4638 peaks were annotated with the three closest neighboring genes at a maximal distance of 20 kb using CisGenome. Enriched sequence patterns in Plagl1 binding regions were determined using Gibbs Motif Sampler provided by CisGenome that allowed calculating the Relative Enrichment Levels for these motifs. We assessed the abundance of the consensus motives in Plagl1 binding sites using the Motif Enrichment Analysis Framework (24). Representative illustrations were made using IGB (Integrated Genome Browser). ChIP-seq data from this study have been submitted to the NCBI GEO database under the accession number GSE75943.
ChIP-seq data for Nptx1 and Mpl promoters were confirmed by ChIP-qPCR using primers listed in Supplementary Table S1. Data were normalized using three loci L1, L2, L3, devoid of Plagl1 binding sites according to the formula: X/geometric mean (L1, L2, L3) = 2(Ct(X)-arithmetic mean (Ct(L1), Ct(L2), Ct(L3)), where Ct is the threshold cycle. Fold enrichments in anti-Plagl1 immuno-precipitated chromatin were calculated over input chromatin used as control (16).
Data contextualization
Functional annotation of gene lists with GO terms and KEGG pathways was performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.8; May 2016) tools (25) using the expressed genes as background list.
RESULTS
Plagl1 is induced during cell cycle exit
We first sought to identify the physiological context in which Plagl1 anti-proliferative activity was involved. Zac1/Plagl1 was originally isolated in a functional screen that also resulted in the isolation of a cDNA clone encoding p53 (5). Plagl1 and Trp53 encode structurally unrelated proteins but display similar functional properties as both induce cell cycle arrest and apoptosis upon transfection in different cell lines. Trp53 is best known for its physiological role in the cell cycle arrest and eventually apoptosis that DNA damage evokes. We thus tested whether the same experimental setting may also involve Plagl1. We irradiated AtT20 pituitary cells with UV light to induce DNA damage. We performed western blotting to detect p53 and Plagl1 at different time points after UV irradiation (Supplementary Figure S1). We observed the expected p53 induction 3 and 7 hours after UV irradiation. In contrast, Plagl1 was not induced at any time point. We concluded that, although Plagl1 and p53 share similar biological activities, these are used in different biological contexts.
We surveyed Plagl1 expression during physiological cell cycle arrest in various in vitro and in vivo models. Using real-time PCR, we monitored Plagl1 expression in primary mouse embryonic fibroblasts (MEFs) induced to quiescence following confluence and contact inhibition at ∼7 days in vitro (Figure 1A). We observed Plagl1 induction upon cell cycle exit, when the proliferation marker Pcna was down-regulated. Plagl1 was further induced upon prolonged quiescence. We confirmed these observations at the protein level as the two Plagl1 splice variants (5) were induced with very similar kinetics (Figure 1B). Using immunocytochemistry we also confirmed that Plagl1 expression was induced in the nucleus. (Supplementary Figure S2). We did not detect Plagl1 transcript and protein in Plagl1+/-pat. MEFs, no matter their proliferation status (Figure 1A, B and Supplementary Figure S2). This confirmed that reactivation of the methylated, silenced maternal allele was not involved in Plagl1 induction during cell cycle exit. We showed that Plagl1 was also induced upon serum withdrawal-induced quiescence (Figure 1C).
We used a differentiation paradigm in which mouse embryonic stem (ES) cells were induced to cortical neurons and glial cells in vitro (Figure 2A). This model was previously shown to recapitulate the major milestones of cortical development, including the generation of a diverse repertoire of cortical pyramidal neurons in a timely ordered fashion (18). Upon induction of corticogenesis, we confirmed the down-regulation of stem cell markers and the induction of cortical markers (Supplementary Figure S3A). We observed Plagl1 induction during the main period of neural progenitor cell cycle exit and differentiation into cortical neural cells (Figure 2A). We also monitored Plagl1 expression in vivo during muscle regeneration induced by injection of notexin, a snake venom toxin that kills myocytes and induces the proliferation and differentiation of satellite cells representing the muscle stem cell population (Figure 2B). We verified the transient expression of proliferation and inflammation markers and the progressive expression of myogenic factors until the restoration of myocyte markers (Supplementary Figure S3B). We observed Plagl1 induction when proliferating myoblasts exited the cell cycle to enter myogenic differentiation (Figure 2B). Finally, we studied an in vitro adipogenesis model in which a pre-adipocyte cell line was induced to adipogenic differentiation by addition of the IDX hormonal cocktail (26). Cells proliferated (Figure 2C, P) until they reached confluence and entered quiescence (Figure 2C, Q). Two days later, we added IDX; the cells resumed proliferation in the so-called clonal expansion phase (Figure 2C, CE), definitively exited the cell cycle 2–3 days later, and gradually differentiated into lipid-accumulating adipocytes (Figure 2C, D). We quantified gene expression using RNA-seq and draw a heat map with normalized gene counts for markers of proliferation (Pcna, Dhfr, Mcm6, Prim1, Mki67), early (Cebpg, Pparg) and late (Lpl, Cebpa, Plin1, Adipoq, Lep) adipogenic differentiation, and for Plagl1. We noticed that Plagl1 expression was maximal when cells were either quiescent or differentiated, i.e. they were not proliferating. All in all, these data showed that Plagl1 was up-regulated under physiological conditions when cells exited the cell cycle and entered quiescence or differentiated.
Identification of Plagl1-regulated genes
To gain insights into Plagl1 mechanism of action, we determined Plagl1 target genes using genome-wide transcriptomics. Constitutively Plagl1-deficient cells may adapt to loss of Plagl1 function by activating compensating regulatory mechanisms. We thus favored a cellular system in which we experimentally induced Plagl1 expression in a time-controlled fashion to focus on Plagl1 direct targets. We selected the Neuro-2a neuroblastoma cell line, which did not display detectable Plagl1 levels (data not shown). Neuroblastoma are tumors derived from sympathetic neuroblasts, which express Plagl1 in vivo (27), indicating loss of Plagl1 expression upon transformation (28). To avoid non-specific effects due to over-expression, we transfected Plagl1 at levels comparable to those observed in several tissues in vivo (Supplementary Figure S4A). Plagl1 mRNA levels in transfected Neuro-2a was 9.5 times higher than endogenous Plagl1 expression in whole P1 sympathetic ganglia (Supplementary Figure S4A). Immunohistochemistry using tyrosine hydroxylase (Th) and Plagl1 antibodies revealed that only a small subset of neural cells expressed Plagl1 in the superior cervical sympathetic ganglion (Supplementary Figure S4B). This indicated that Plagl1 expression levels in transfected Neuro-2a cells were comparable to that of Plagl1-positive sympathetic neural cells. We verified that Plagl1 displayed an anti-proliferative activity in Neuro-2a cells by measuring the distribution of transfected cells in the different phases of the cell cycle (Supplementary Figure S4C, D). Plagl1 transfection resulted in a G0/G1 arrest and a concomitant decrease of the number of cells in the S and G2/M phases. In contrast to previous observations in other cellular systems (5), we did not find evidence of apoptosis following Plagl1 transfection in Neuro-2a cells (data not shown).
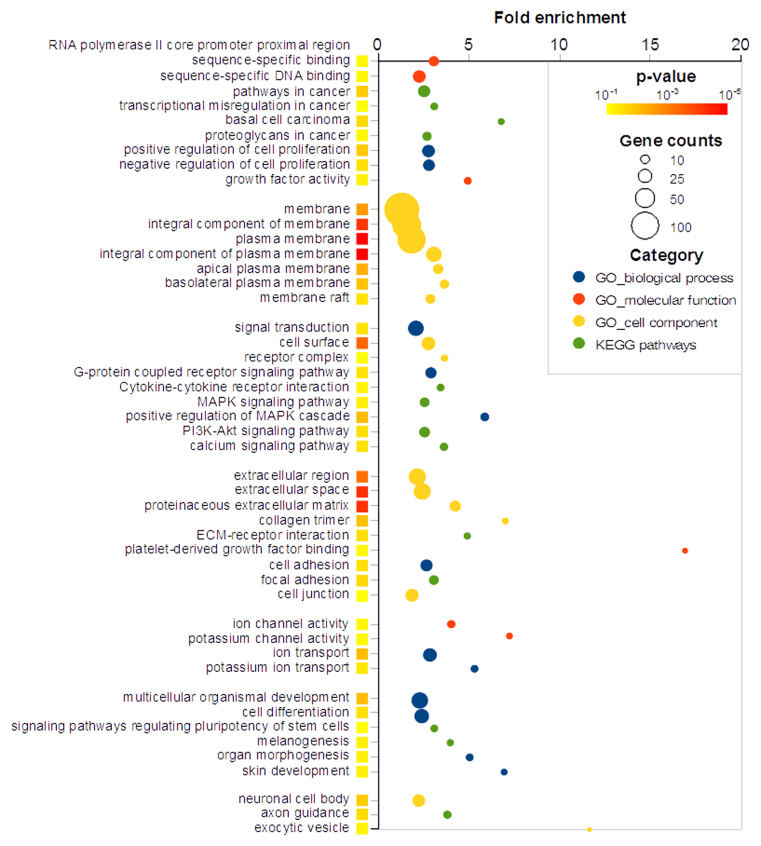
We transfected proliferating Neuro-2a cells with chloramphenicol acetyltransferase (CAT) as a control or Plagl1. We performed digital gene expression (DGE) profiling of 2 independent transfection experiments, and identified 351 DE genes (Table 1, Supplementary Table S2). We found only three genes significantly repressed by Plagl1 transfection, i.e. Rbm15 (x0.6), Hspa8 (x0.7) and Rps29 (x0.8). We tested the differential expression of 59 genes by quantitative RT-PCR (Supplementary Table S3), and 54 (92%) were confirmed as DE. To identify the biological processes that were regulated by Plagl1 in Neuro-2a cells, we performed enrichment analysis of GO terms and KEGG pathways (Supplementary Table S4). We grouped the terms from the three GO categories and KEGG pathways according to their semantic similarities (Figure 3). As expected for a putative tumor suppressor gene with proven anti-proliferative activity, we observed the enrichment of terms such as ‘regulation of cell proliferation’, ‘pathways in cancer’ and ‘basal cell carcinoma’. The most remarkable feature was the enrichment of genes encoding ECM proteins, genes involved in ECM-activated signaling, including cytoskeleton binding, and genes controlling cell adhesion and movement. We also noted the over-representation of terms linked to signaling, in particular cell–cell signaling, growth factor activity, PDGF signaling, steroid hormone receptor activity, receptor tyrosine kinase activity, and ion channels. The absence of enrichment of terms related to cell cycle control, despite Plagl1-induced cell cycle arrest (Supplementary Figure S4), was noteworthy.
Table 1. Enrichment analysis of different categories of genes among Plagl1-regulated genes and genes with at least one Plagl1 binding site in Neuro-2a neuroblastoma cells.
| DGE | ChIP-seq | |||||
|---|---|---|---|---|---|---|
| No. of ↓ among → | Plagl1-regulated genes | Expressed genes (12 861) | P-value | Genes with a Plagl1 binding site | All UCSC genes (23 100) | P-value |
| Plagl1-regulated genes | 351 | 217 | 351 | 1 × 10−81 | ||
| Genes with a Plagl1 binding site | 217 | 3082 | 5.4 × 10−53 | 3859 | ||
| Imprinted Genes | 7 | 61 | 0.001 | 25 | 115 | 0.03 |
| ECM genes | 17 | 177 | 5.3 × 10−6 | 92 | 369 | 1 × 10−5 |
| IGN members (4) | 15 | 297 | 0.009 | 88 | 407 | 0.002 |
| PLAG1 target genes (36) | 10 | 41 | 9.8 × 10−8 | 24 | 57 | 4 × 10−6 |
| Plagl2 target genes (37) | 8 | 20 | 2.6 × 10−8 | 17 | 23 | 2 × 10−9 |
| Plagl1 target genes (38) | 4 | 85 | ns | 19 | 108 | ns |
| Neuroblastoma signature genes (40) | 2 | 54 | ns | 16 | 59 | 0.02 |
IGN, PLAG1-, Plagl2-, Plagl1-target genes and the human neuroblastoma signature genes are from the indicated references. P-values were calculated using the hypergeometric distribution. ns, not significant.
Figure 3.
GO terms and KEGG pathways enrichment analysis. Genes regulated by Plagl1 transfection into Neuro-2a cells were analyzed using the DAVID bioinformatics resources (25). The figure displays the fold-enrichment of each statistically significant GO term/KEGG pathway. Circle color maps the GO term/KEGG pathway category. Circle size maps the number of genes in each GO term/KEGG pathway. The colored square next to each GO term/KEGG pathway maps the Benjamin–Hochberg-corrected P-value. The different GO terms/KEGG pathways were grouped according to their semantic similarities.
To test whether the genes regulated by Plagl1 in neuroblastoma cells were specific to this cell type, we transfected Min6 insulinoma cells with plasmids encoding eGFP or Plagl1. We performed RNA-seq analysis of the two transfected populations and identified 441 DE genes (Supplementary Table S5), including eight down-regulated. We performed GO terms and KEGG pathways enrichment analysis (Supplementary Table S6) as previously described, and identified terms very close to those associated with Plagl1-regulated genes in Neuro-2a cells (Supplementary Figure S5). Common enriched terms included ‘pathways in cancer’, ‘basal cell carcinoma’, terms related to ECM and ECM-activated signaling, cell/focal adhesion as well as terms related to other signaling pathways, e.g. Wnt, Hedgehog, adenylate cyclase, calcium etc. We found a large overlap between the genes regulated by Plagl1 in Neuro-2a and Min6 cells; 133 genes were regulated in the 2 cell lines, 218 specifically in Neuro-2a cells and 308 specifically in Min6 cells (Supplementary Figure S6).
The genes called ‘differentially expressed’ (DE) in Neuro-2a cells included 15 IGN genes, 5 of which were imprinted and 2 additional IGs (Begain, Dio3). Twenty-one IGN members were called DE in Min6 cells, including five IGs. Three IGN members, including the imprinted Axl gene, were called DE in both cell lines (Supplementary Figure S6). To confirm these findings, we assessed the expression levels of the 35 genes in control versus Plagl1-transfected Neuro-2a and Min6 cells using RT-qPCR (Figure 4). RT-qPCR confirmed the differential expression of all imprinted and IGN genes called DE in both Neuro-2a and Min6 cells. Four IGs and four IGN genes called DE in Min6 cells only were found differentially expressed in Neuro-2a cells using RT-qPCR.
Identification of Plagl1 binding sites
We performed ChIP-seq analysis of Plagl1-transfected Neuro-2a cells. Using a 0.001 FDR, we identified 4689 peaks in Plagl1 ChIP samples compared to the input chromatin; 51 peaks were located in amplified regions of the input genome and discarded from further analyses (Supplementary Table S7). We performed ChIP data quality control (Supplementary Table S8) according to the guidelines published by the ENCODE consortium (29). The average width of peaks was 125.9 ± 127.9 bp, indicating we identified mostly point-source peaks. The total number of uniquely mapped reads ranged from 4.8 to 6.5 M across the four samples we analyzed (duplicate input and immuno-precipitated chromatin preparations). This is less than the recommended 10 M uniquely mapped reads for ChIP samples consisting mostly of point-source peaks. However, the fraction of reads in peaks (FriPs), which is recommended to be >1%, was 3.2% and the cross-correlation of the number of reads assigned to the forward and reverse strands within peaks was 0.80 (Supplementary Figure S7, Supplementary Table S8). Therefore, our data fulfilled the three criteria for ChIP data quality assessment according to the ENCODE consortium. We also noted the presence of peaks at the H19 3′ enhancer, which we previously demonstrated to be bound and regulated by Plagl1 (16).
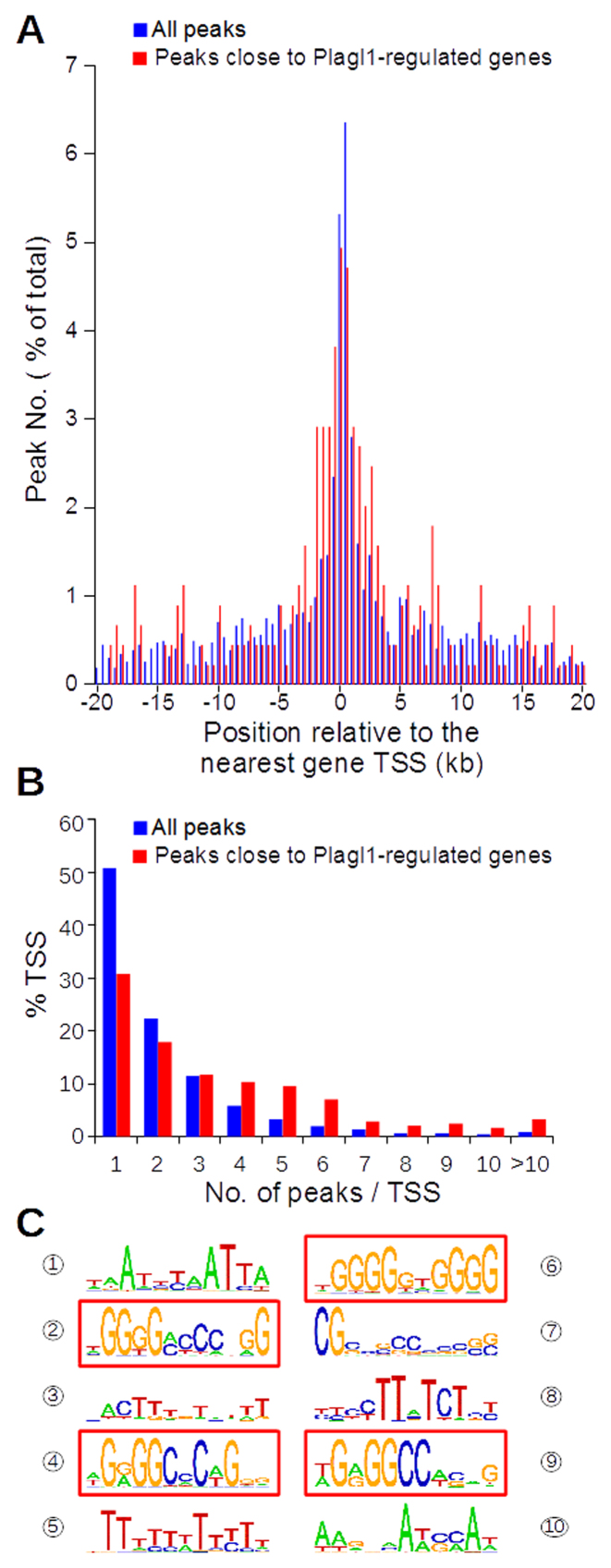
We annotated the ChIP peaks using CisGenome (Supplementary Table S9). A peak was annotated with a gene if it was located within ±20 kb of the gene body, i.e. from 20 kb upstream its most upstream transcription start site (TSS) to 20 kb downstream its most downstream transcription end site (TES). This region included the whole gene body, its proximal promoter(s) and a number of, but possibly not all, enhancers/silencers. Each peak was annotated with a maximum of three different genes. This strategy was a compromise to include as many potentially regulated genes as possible, but it had some obvious limitations as exemplified by the Igf2-H19 locus (see discussion). Of the 4638 peaks identified, 3876 were annotated by 3859 distinct genes, of which 3082 were expressed in Neuro-2a cells. This number was in vast excess compared to the number of Plagl1-regulated genes (Supplementary Figure S8) indicating that Plagl1 binding to a large number of loci did not result in the regulation of gene expression in Neuro-2a cells. The ChIP peaks were mostly located outside the gene body and in introns (Supplementary Figure S9). We looked at the distance between each peak and the TSS of the nearest gene and noticed that peak location was biased towards the proximal promoter region (Figure 5A) as expected for a conventional transcription factor. Plagl1 binding sites in Plagl1-regulated genes were also mostly located in the proximal promoter with a further peak enrichment from –2 kb to –0.5 kb relative to TSS (Figure 5A). We confirmed the higher Plagl1 binding site number in Plagl1-regulated genes when we looked at the distribution of peak number per TSS for all genes versus Plagl1-regulated genes; the vicinity of Plagl1-regulated TSS comprised more peaks than the population of TSS close to all peaks (Figure 5B).
Figure 5.
Identification of Plagl1 binding sites in Neuro-2a cells. (A) Comparison of the localization of Plagl1 binding sites (peaks) in Plagl1-regulated genes (red) versus all genes with at least one Plagl1 binding site (blue). (B) Comparison of the distributions of the number of Plagl1 binding sites in Plagl1-regulated genes (red) versus all genes with at least one Plagl1 binding site (blue). (C) Identification of a consensus Plagl1 binding site inferred from the sequences of all Plagl1 binding genomic regions.
Among the 351 genes regulated by Plagl1 in Neuro-2a cells, 217 (∼62%) had at least one Plagl1 binding site at <20 kb (Table 1, Supplementary Figure S8). We also compared the list of genes bound or regulated by Plagl1 to the genes of the IGN (Table 1, Supplementary Figure S8). Plagl1 was binding close to 88 of the 407 IGN genes present in the UCSC database (∼22%, P < 2 × 10−3, Fisher's exact test). Interestingly, among expressed genes, IGN genes were significantly more frequently regulated by Plagl1 than non-IGN genes (Supplementary Figure S8; 15/297 versus 336/12 564, P < 0.02, Chi-square test). To further illustrate how Plagl1 binding may contribute to IG regulation we provide maps of consensus and observed Plagl1 binding sites at imprinted loci (Supplementary Figure S10). Interestingly, Plagl1 bound in Neuro-2a 148 (48%) of the 308 genes regulated in Min6 cells only (Supplementary Figure S6).
Plagl1 consensus binding site
The genome-wide identification of Plagl1 binding regions offered the opportunity to deduce a Plagl1 consensus binding site in vivo. Using the de novo motif discovery module of CisGenome (22,30), we identified four GC-rich consensus motives that were significantly enriched among Plagl1 binding sites (Figure 5C, Supplementary Table S10). The two most statistically significant motives, nos. 4 and 9 on Figure 5C, were similar: G(g/a)GGC(c/a)C(a/t)G and G(g/a)GGCC(a/t) (g/c), respectively. The third statistically significant motif, no. 2, also included a stretch of G followed by a stretch of C. These consensus binding sites are reminiscent of the G4C4 motif we previously reported for the human PLAGL1 orthologue using an in vitro selection protocol (7) and of the primary Plagl1 GGG(C/G) (G/C)CCC motif determined by protein-binding microarray (31). The fourth significant binding site, G4N2G4, is also reminiscent of the second category of binding sites we identified in the same report, i.e. G4N6G4 (7). We also identified the Tn motif ubiquitously found in motif discovery experiments using ChIP-seq data (30) as well as other AT-rich motives that were not statistically significant.
We then assessed whether these consensus motives contributed to gene regulation by Plagl1. We used the Motif Enrichment Analysis framework (24) to compare the abundance of the consensus motives in 532 Plagl1 binding sites located in the vicinity of Plagl1-regulated genes versus 4106 binding sites located outside of Plagl1-regulated genes. We showed that the G4(c/a)C3 and the G4N2G4 motives were significantly over-represented in Plagl1 binding sites close to Plagl1-regulated genes (Supplementary Table S11).
Mpl and Nptx1 are direct Plagl1 targets
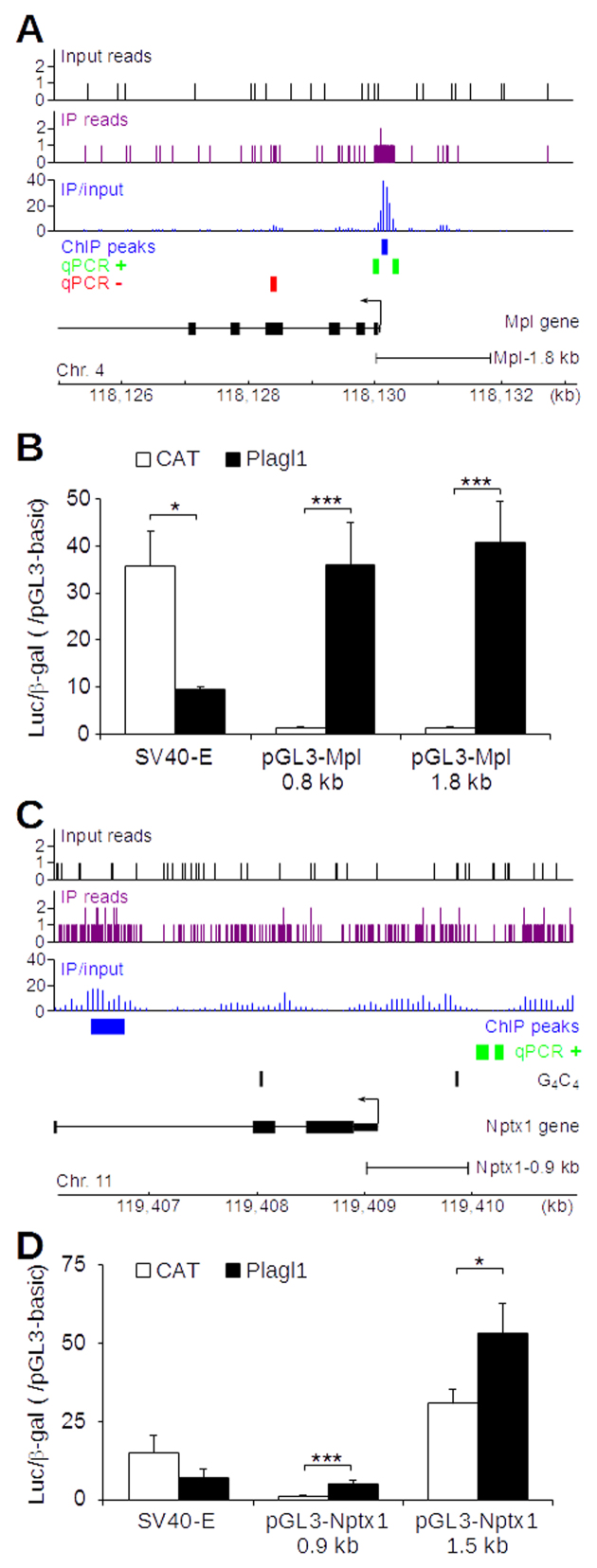
To confirm that some of the identified regulated genes were direct Plagl1 targets, we tested Mpl and Nptx1 promoter regulation by Plagl1 (Figure 6). Mpl was induced 1849-fold by Plagl1, mostly because it was barely detectable in CAT-transfected Neuro-2a. Mpl proximal promoter region displayed a ChIP-seq peak that was confirmed by ChIP-qPCR (Figure 6A). We cloned two fragments of the Mpl proximal promoter into the pGL3 luciferase reporter plasmid, and measured luciferase activity in CAT- and Plagl1-transfected Neuro-2a (Figure 6B). Plagl1 induced luciferase activity ∼25–30-fold from both constructs, confirming its ability to transactivate the Mpl promoter. On the other hand, the regulation of the Nptx1 gene was more complex. Nptx1 was induced ∼46-fold by Plagl1; in contrast to Mpl, Plagl1 binding occurred in intron 2 rather than in the proximal promoter (Figure 6C). We also noted the presence of ChIP-seq peaks that were not statistically significant in the vicinity of the Nptx1 promoter, as well as a G4C4 consensus binding site. ChIP-qPCR data close to this consensus site suggested that the promoter may indeed contain a weak Plagl1 binding site (Figure 6C). We cloned 2 fragments of the Nptx1 promoter into pGL3 and measured luciferase activity in CAT- and Plagl1-transfected Neuro-2a cells. We observed a 3-fold transactivation of the shortest fragment by Plagl1 (Figure 6D). The longer fragment displayed an elevated basal promoter activity, probably due to the presence of sequence from the neighboring Gm11762 gene promoter, and a weak 1.7-fold induction by Plagl1 (Figure 6D). These data confirmed that Plagl1 directly bound and regulated Mpl and Nptx1 promoters, possibly in combination with other regulatory elements in the case of the Nptx1 gene.
Figure 6.
Plagl1 regulates the thrombopoietin receptor (Mpl) and neuronal pentraxin 1 (Nptx1) genes by direct transactivation of their respective promoters. (A) ChIP-seq and ChIP-qPCR data were mapped to the Mpl locus. ‘IP reads’ designate reads from the anti-Plagl1 immuno-precipitated fraction. (B) Fragments of the SV40 early (SV40-E) or Mpl (Mpl 0.8 kb, Mpl 1.8 kb) promoters were subcloned into the pGL3 reporter plasmid encoding the Firefly Luciferase, and transfected into Neuro-2a cells. The pRK7-βGal plasmid containing the β-galactosidase cDNA under the control of a CMV promoter was co-transfected. The luciferase activity was normalized to the β-galactosidase activity of transfected cells. Data are displayed as ratio of the normalized luciferase activity in pGL3promoter- to pGL3basic-transfected cells. Plagl1 (closed bars) inhibited SV40 early promoter and stimulated Mpl promoter fragments compared to chloramphenicol acetyl transferase (CAT; open bars). (C) Same as A for the Nptx1 locus. The presence of two G4C4 sites in the Nptx1 gene is indicated. (D) Same as B for the Nptx1 locus. Data are mean ± S.D. of four independent experiments. ***P < 0.001; **P < 0.01; *P < 0.05.
Impact of loss of Plagl1 function in mouse embryonic fibroblasts
As we showed that Plagl1 is induced upon cell cycle exit in MEFs (Figure 1), we tested whether Plagl1 inactivation altered the gene regulation that occurs upon cell cycle exit. We plated MEFs at low density and let them grow until they reached confluence and entered quiescence (Supplementary Figure S11A). We observed no significant difference in the growth rate and saturation density in Plagl1+/-pat. versus wild type MEFs (Supplementary Figure S11A), indicating that Plagl1 inactivation was not sufficient to alter MEF proliferation in vitro. We performed RNA-seq analysis at different time points when MEFs were proliferating (2 days in vitro), confluent (8 days in vitro), and quiescent (10 and 12 days in vitro) (Supplementary Figure S11B). We observed a large number of DE genes when wild type MEFs exited the cell cycle; of the 15 788 genes expressed, 8799 were differentially expressed (FDR<0.01) upon confluence (2 versus 8 days in vitro, data not shown). It was noteworthy that several IGs, including Plagl1, and IGN members were significantly up-regulated upon confluence. In contrast, Plagl1 inactivation had a limited impact on the transcriptome at most time points (Table 2, Supplementary Table S12); 29 genes were called DE between wild type and Plagl1+/-pat. MEFs at day 2, 32 at day 8, 183 at day 10, and 9 at day 12 (Table 2). In contrast to Plagl1-transfected Neuro-2a and Min6 cells in which DE genes were mostly up-regulated, genes dysregulated in Plagl1+/-pat. MEFs included 99 up-regulated genes, 139 down-regulated genes, and three genes that were oppositely regulated at different time points. Of the 238 genes called DE at one time point at least, nine were called DE in transfected Neuro-2a or Min6 cells, 37 displayed a Plagl1 binding site in transfected Neuro-2a cells, 30 were part of the IGN, and 10 were IGs. Remarkably, 9 of the 10 IGs and 8 of the 9 genes called DE in Neuro-2a or Min6 were down-regulated in Plagl1+/-pat. MEFs. To identify the biological processes most affected by loss of Plagl1 function, we performed GO term enrichment analysis of the 238 DE genes (Supplementary Table S13). Over-represented terms were related to the ECM; ‘extracellular region’ (GO:0005576), ‘extracellular space’ (GO:0005615), ‘extracellular matrix’ (GO:0031012), ‘proteinaceous extracellular matrix’ (GO:0005578), ‘collagen binding’ (GO:0005518), ‘fibronectin binding’ (GO:0001968), and ‘insulin-like growth factor binding’ (GO:0005520). We noted that these terms were among those enriched in Plagl1-transfected Neuro-2a (Figure 3) and Min6 (Supplementary Figure S5).
Table 2. Enrichment analysis of different categories of genes among genes differentially expressed in wild-type versus Plagl1+/-pat. mouse embryonic fibroblasts. P-values were calculated using the hypergeometric distribution.
| Day 2 (proliferation) | Day 8 (confluence) | Day 10 (quiescence) | Day 12 (quiescence) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of ↓ among → | Genes called DE | Expressed genes (15 346) | P-value | Genes called DE | Expressed genes (15 945) | P-value | Genes called DE | Expressed genes (15 998) | P-value | Genes called DE | Expressed genes (15 788) | P-value |
| Genes called DE (down/up) | 29 (19/10) | 32 (6/26) | 183 (117/66) | 9 (3/6) | ||||||||
| IGN members | 5 | 383 | 6 × 10−4 | 8 | 387 | 6 × 10−7 | 20 | 390 | 2 × 10−8 | 3 | 384 | 1 × 10−3 |
| Imprinted genes | 0 | 101 | - | 2 | 106 | 2 × 10−2 | 7 | 107 | 2 × 10−4 | 2 | 108 | 1 × 10−3 |
| Plagl1-regulated genes in Neuro-2a cells | 1 | 269 | 3 × 10−1 | 1 | 291 | 3 × 10−1 | 4 | 295 | 2 × 10−1 | 1 | 281 | 1 × 10−1 |
| Genes with 1+ Plagl1 binding site in Neuro-2a cells | 5 | 3133 | 2 × 10−1 | 2 | 3234 | 3 × 10−2 | 31 | 3266 | 4 × 10−2 | 3 | 3266 | 2 × 10−1 |
| Plagl1-regulated genes in Min6 cells | 0 | 329 | - | 0 | 351 | - | 4 | 358 | 2 × 10−1 | 0 | 360 | 8 × 10−1 |
DISCUSSION
We showed that Plagl1 was up-regulated when cells from different lineages proceeded from a proliferative to a quiescent or differentiated state (Figure 1, Supplementary Figure S2, Figure 2). This cellular pattern of expression was in line with Plagl1 pattern of expression in the whole animal; Plagl1 was most abundantly expressed during late embryonic development and its expression decreased during post-natal growth deceleration in multiple organs (32). The induction of Plagl1 expression upon cell cycle exit was not limited to the murine gene as we previously showed that the human PLAGL1 gene was induced under similar circumstances, i.e. human keratinocyte differentiation (33). Furthermore, the context in which Plagl1/PLAGL1 induction occurred was similar to the one we recently observed for other imprinted genes (4), confirming that Plagl1 belongs to the IGN.
As anticipated from previous studies in other cell lines, Plagl1 expression led to cell cycle arrest in Neuro-2a neuroblastoma cells (Supplementary Figure S4). On the other hand, we did not observe apoptosis following Plagl1 transfection in this cell line. Only one term linked to apoptosis, i.e. ‘positive regulation of apoptotic process’, was over-represented among genes induced by Plagl1 in Neuro-2a cells. Of the 14 Plagl1-regulated genes that composed this GO category, only three were specific thereof: Sept4, Irf5 and Ctrb1. The 11 remaining genes were also part of other over-represented GO categories such as ‘regulation of cell proliferation’, ‘MAPK signaling pathway’ and other signaling pathways. This suggested that the main effect of Plagl1 induction in Neuro-2a was indeed the control of proliferation rather than apoptosis. This was further confirmed by the absence of terms related to apoptosis among Plagl1-regulated genes in Min6 cells. Although Plagl1 induced cell cycle arrest in Neuro-2a cells, we did not observe the regulation of components of the core cell cycle machinery, e.g. cyclins, cyclin-dependent kinases (CDKs), CDK inhibitors, pocket proteins, or E2F transcription factors. The only notable exception was the CDK inhibitor Cdkn1c/p57Kip2, which was induced ∼18-fold, in agreement with our observations that Cdkn1c was down-regulated in liver (16) and MEFs (this study) from Plagl1-deficient embryos. Interestingly, Cdkn1c is itself imprinted and displays functional properties not shared by other CDK inhibitors (34). This suggested that the regulation of Cdkn1c likely contributed to the cell cycle arrest induced by Plagl1, but may have fulfilled additional functions. In addition to Cdkn1c, other regulated genes likely contributed to the observed cell cycle arrest. Over-represented GO terms and KEGG pathways included several signaling pathways, which were shown to affect proliferation, e.g. ‘receptor protein tyrosine kinase activity’, ‘MAPK signaling pathway’, ‘growth factor activity’, ‘PDGF binding’. The most remarkable set of over-represented GO terms were related to the ECM. These included terms linked to ECM constituents (‘proteinaceous extracellular matrix’, ‘fibrillary collagen’), ECM-activated signaling (‘ECM-receptor interaction’, ‘collagen binding’, ‘focal adhesion’), and an ECM-mediated function (‘cell adhesion’). They also included terms related to the actin cytoskeleton, a major target of ECM-activated signaling: ‘actin binding’, ‘filamin binding’, ‘ankyrin binding’, ‘Rho GEF activity’, ‘cell–cell signaling’ and ‘cell movement’. We also examined the alteration of gene regulation induced by Plagl1 inactivation in mouse embryonic fibroblasts in vitro. We previously demonstrated that Plagl1 inactivation had profound impact on embryonic growth (16); however, Plagl1-deficient MEFs grown in vitro did not display a significant alteration of the proliferation rate and saturation density compared to wild type MEFs (Supplementary Figure S11). In agreement with this observation, Plagl1 inactivation did not extensively modify the transcriptome of proliferating, confluent, or quiescent MEFs (Table 2) suggesting that MEFs grown in vitro adapted to loss of Plagl1 function by activating compensatory mechanisms. Still, we found nine genes dysregulated by Plagl1 inactivation in MEFs that were identified as Plagl1 targets in Neuro-2a or Min6 cells. In addition, 37 dysregulated genes displayed a Plagl1 binding site in Neuro-2a cells, 30 were part of the IGN and 10 were IGs. A significant proportion of the genes regulated upon wild type MEF confluence were annotated by GO terms related to ECM structural constituents, ECM-activated signaling, and ECM-dependent processes. Interestingly, over-represented GO terms associated to genes dysregulated in Plagl1-deficient MEFs were all related to the ECM. We concluded that Plagl1-deficient MEF adapted to loss of Plagl1 function but for a limited number of Plagl1 targets, IGN members, and ECM genes. Altogether, the data from gain and loss of function Plagl1 mutants suggested that Plagl1 was not a modulator of the core cell cycle machinery; it rather impacted cell proliferation by modulating the composition of the ECM and the subsequent ECM-activated signaling. Retrospectively, the experimental setting we used to identify Plagl1 function and regulated genes may have been suboptimal; the rigid plastic vessels used for standard cell culture provide cells with adhesive cues that prevent the observation of the effects of the modulation of ECM composition. Future work will evaluate Plagl1 functional properties in a more permissive setting such as 3D culture systems.
Plagl1/PLAGL1 belongs to a family of three zinc finger transcription factors that share considerable homology in their zinc finger regions. In contrast to the candidate tumor suppressor ZAC1/PLAGL1, PLAG1 and PLAGL2 are proto-oncogenes that are not imprinted. Because we showed that the zinc finger region is involved in DNA binding (35), we tested whether Plagl1, Plag1 and Plagl2 shared some target genes. We showed a significant overlap between Plagl1 target genes identified in this study and the published PLAG1 (36) and Plagl2 (37) targets (Table 1). In contrast, we found only four genes in Neuro-2a cells, and three in Min6 cells, in common between our study and a previous one by Barz et al. (38) aimed at identifying Plagl1 target genes in the hippocampal cell line HW3–5 (Table 1). Although the observed difference might be attributed to the use of different cell lines, this is unlikely because we found a significant overlap between Plagl1 target genes in the neuroblastoma Neuro-2a cell line and the unrelated insulinoma Min6 cell line (Supplementary Figure S6). We also noted that 48% of the 351 Plagl1-regulated genes we identified in Neuro-2a cells were found in a recent study by Rraklli et al. using E14.5 cerebral cortices electroporated in vivo (39) (Supplementary Figure S12). Altogether, these data confirmed that we identified bona fide Plagl1 target genes, some of which were shared with other members of the Plagl1 family of transcription factors. Given Plagl1/PLAGL1 functional properties and chromosomal localization, we previously suggested that PLAGL1 might be the tumor suppressor gene on 6q24 (8). Interestingly, we noted that Plagl1 was binding close to a significant proportion of the 59-gene prognostic signature that sub-stratifies high-risk neuroblastoma patients (40) (Table 1). PLAGL1 is itself a member of this signature and our work suggested that it may indeed regulate a subset of the neuroblastoma signature genes.
We designed this study to identify direct Plagl1 target genes. Sixty-two percent of Plagl1-regulated genes in Neuro-2a cells displayed a Plagl1 binding site at ±20 kb of the gene body (Table 1). This percentage was an underestimate as the 20 kb arbitrary cut-off prevented us to identify some direct Plagl1 target as exemplified by the Igf2 gene. Igf2 and H19 are two neighboring, reciprocally imprinted genes on murine distal chromosome 7. They share an imprinting control region located in between the two genes as well as two enhancers located ∼8 kb downstream of H19 and ∼80 kb downstream Igf2. We previously showed (16), and confirmed in this study, that murine Plagl1 binds to these enhancers and directly regulates both genes. PLAGL1 binding to IGF2/H19 enhancers is conserved in humans (41). We have also shown here that Plagl1 bound and transactivated the promoters of two identified target genes, Mpl and Nptx1. This led us to conclude that at least ∼2/3 of the regulated genes we identified in this study were direct Plagl1 targets.
The number of Plagl1 binding sites was in vast excess to the number of Plagl1-regulated genes (Table 1, Supplementary Figure S8). This observation is not unusual in ChIP-seq experiments (42) and the number of Plagl1 binding sites we observed (4638) is average. ChIP-seq on MyoD in murine skeletal muscle cells identified approximately 30 000–60 000 MyoD binding sites (43). Similarly, the hematopoietic factor Gata1 was reported to have over 15 000 DNA binding sites in a mouse erythroblast line (44). Paradoxically, the calcium-response factor CaRF, which displayed 176 binding sites in cortical neurons from newborn mice (45), is rather exceptional. Plagl1 is comparable to transcription factors such as Tal1, which displayed 2994 binding sites in primary erythroid cells (46), the pregnane receptor Nr1i2/PXR, which displayed 3812 binding sites in mouse liver (47), or p53, which displayed 4785 binding sites in MEFs (48). One proposed explanation for large-scale genome-wide transcription factor binding is the presence of ‘non-functional’ binding sites that serve no biological purpose (49). Other hypotheses include the absence of appropriate transcriptional co-activators in Neuro-2a cells that may be expressed in other cells types. The Min6 data supported in part this hypothesis as 48% of the 308 Min6-specific, Plagl1-regulated genes included a Plagl1 binding site identified in Neuro-2a cells. Because we aimed at identifying mostly direct Plagl1 targets, including those that were transiently regulated, and because we wanted to restrict Plagl1 over-expression as much as possible, we looked at Plagl1-regulated genes early after Plagl1 induction. Some of the transcriptionally unproductive Plagl1 binding sites may therefore correspond to genes that will be regulated at a later stage. This argument may also explain why we identified very few genes repressed by Plagl1. We noted that Plagl1-regulated genes harbored more Plagl1 binding sites than genes annotating all Plagl1 binding sites (Figure 5B). This was suggestive of a mechanism in which occupancy of one Plagl1 binding site did not result in increased transcription but served as a nucleation event that permitted subsequent Plagl1 binding to neighboring loci to be transcriptionally productive. Models involving ‘pioneer’ transcription factors and homotypic cooperativity have been previously proposed (50,51). The Plagl1-regulated genes were generally enriched in Plagl1 binding sites compared to all genes located at ±20 kb of a Plagl1 binding site (Figure 5B). Many of these binding sites were located around the TSS of the nearest gene (Figure 5A). These sites most often comprised a GC-rich sequence (Figure 5C) in agreement with our previous results from SELEX experiments (7) and from electro-mobility shift assays using the H19 enhancer sequence (16). We concluded that Plagl1 is a transcription factor that binds most often a defined, G4C4-like sequence close to gene TSS, and that multiple Plagl1 binding at the same promoter resulted in transcriptional regulation.
We recently described the imprinted gene network, a set of imprinted and non-imprinted co-expressed genes that is involved in the control of the transition between proliferation and quiescence-differentiation (4). How exactly members of the IGN are co-regulated is unknown. Imprinted genes are mostly clustered at a few loci and share regulatory elements. Imprinting control regions (ICRs) are DNA elements that are differentially methylated on the paternal and maternal chromosome and govern the expression of the parental alleles of neighboring genes. We showed that co-regulation of imprinted genes occurs without remodeling of ICR methylation (4) suggesting that the co-regulation of imprinted genes does not involve their ICRs. Other shared regulatory elements include enhancers and silencers. We mentioned the enhancers downstream of H19 that regulate the maternal H19 and paternal Igf2 alleles. This mechanism was demonstrated only for this imprinted locus and cannot be generalized to all imprinted loci. Some imprinted genes and most non-imprinted genes that compose the IGN are not clustered; therefore, shared cis-regulatory elements cannot explain the co-expression of IGN genes. In the present work, we showed that imprinted genes and IGN genes were over-represented among Plagl1-regulated and Plagl1-bound genes (Table 1). In particular, ∼22% of the IGN genes were close to a Plagl1 binding site. As observed for all expressed genes, Plagl1 binding resulted in gene regulation for only a minority of IGN genes; interestingly, Plagl1 binding close to IGN genes resulted more frequently in gene regulation than binding close to non-IGN genes. All in all, our work identified the first transcription factor that contributed to the co-regulation of a significant proportion of the IGN genes, and gave further credit to the existence of the IGN as a novel identified biological machinery.
ACCESSION NUMBERS
Data from this study have been submitted to the NCBI GEO database under accession number GSE75942, GSE50612, GSE84914, GSE99409, and GSE75943.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aubin Thomas (Institut de Génétique Humaine, Montpellier) for help with bioinformatics, Hongkai Ji (Johns Hopkins University Bloomberg, Baltimore) for help with the CisGenome software, Anja Bellmann for help with quantitative PCR experiments, Isabelle Janoueix-Lerosey (Institut Curie, Paris) for providing P1 sympathetic ganglia RNA, Chrystel Lafont for help with imaging techniques, and Christophe Duperray (MRI-Montpellier Ressources Imagerie) for help in flow cytometry experiments.
Footnotes
Present addresses:
Christelle Dantec, Centre de Recherche de Biologie cellulaire de Montpellier, CNRS, Montpellier, France.
Laëtitia Chotard, ICON Plc Clinical Research, Saint-Laurent (Quebec), Canada.
Benoit Bilanges, UCL Cancer Institute, University College London, London, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Institut National de la Santé et de la Recherche Médicale; Université de Montpellier. Funding for open access charge, Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barlow D.P. Genomic imprinting: a mammalian epigenetic discovery model. Annu. Rev. Genet. 2011; 45:379–403. [DOI] [PubMed] [Google Scholar]
- 2. Ferguson-Smith A.C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011; 12:565–575. [DOI] [PubMed] [Google Scholar]
- 3. Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 2014; 15:517–530. [DOI] [PubMed] [Google Scholar]
- 4. Al Adhami H., Evano B., Le Digarcher A., Gueydan C., Dubois E., Parrinello H., Dantec C., Bouschet T., Varrault A., Journot L.. A systems-level approach to parental genomic imprinting: the imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 2015; 25:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spengler D., Villalba M., Hoffmann A., Pantaloni C., Houssami S., Bockaert J., Journot L.. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997; 16:2814–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdollahi A., Godwin A.K., Miller P.D., Getts L.A., Schultz D.C., Taguchi T., Testa J.R., Hamilton T.C.. Identification of a gene containing zinc-finger motifs based on lost expression in malignantly transformed rat ovarian surface epithelial cells. Cancer Res. 1997; 57:2029–2034. [PubMed] [Google Scholar]
- 7. Varrault A., Ciani E., Apiou F., Bilanges B., Hoffmann A., Pantaloni C., Bockaert J., Spengler D., Journot L.. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:8835–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bilanges B., Varrault A., Basyuk E., Rodriguez C., Mazumdar A., Pantaloni C., Bockaert J., Theillet C., Spengler D., Journot L.. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene. 1999; 18:3979–3988. [DOI] [PubMed] [Google Scholar]
- 9. Abdollahi A., Roberts D., Godwin A.K., Schultz D.C., Sonoda G., Testa J.R., Hamilton T.C.. Identification of a zinc-finger gene at 6q25: a chromosomal region implicated in development of many solid tumors. Oncogene. 1997; 14:1973–1979. [DOI] [PubMed] [Google Scholar]
- 10. Jarmalaite S., Laurinaviciene A., Tverkuviene J., Kalinauskaite N., Petroska D., Böhling T., Husgafvel-Pursiainen K.. Tumor suppressor gene ZAC/PLAGL1: altered expression and loss of the nonimprinted allele in pheochromocytomas. Cancer Genet. 2011; 204:398–404. [DOI] [PubMed] [Google Scholar]
- 11. Koy S., Hauses M., Appelt H., Friedrich K., Schackert H.K., Eckelt U.. Loss of expression of ZAC/LOT1 in squamous cell carcinomas of head and neck. Head Neck. 2004; 26:338–344. [DOI] [PubMed] [Google Scholar]
- 12. Pagotto U., Arzberger T., Theodoropoulou M., Grübler Y., Pantaloni C., Saeger W., Losa M., Journot L., Stalla G.K., Spengler D.. The expression of the antiproliferative gene ZAC is lost or highly reduced in nonfunctioning pituitary adenomas. Cancer Res. 2000; 60:6794–6799. [PubMed] [Google Scholar]
- 13. Gardner R.J., Mackay D.J., Mungall A.J., Polychronakos C., Siebert R., Shield J.P., Temple I.K., Robinson D.O.. An imprinted locus associated with transient neonatal diabetes mellitus. Hum. Mol. Genet. 2000; 9:589–596. [DOI] [PubMed] [Google Scholar]
- 14. Kamiya M., Judson H., Okazaki Y., Kusakabe M., Muramatsu M., Takada S., Takagi N., Arima T., Wake N., Kamimura K. et al. . The cell cycle control gene ZAC/PLAGL1 is imprinted–a strong candidate gene for transient neonatal diabetes. Hum. Mol. Genet. 2000; 9:453–460. [DOI] [PubMed] [Google Scholar]
- 15. Varrault A., Bilanges B., Mackay D.J., Basyuk E., Ahr B., Fernandez C., Robinson D.O., Bockaert J., Journot L.. Characterization of the methylation-sensitive promoter of the imprinted ZAC gene supports its role in transient neonatal diabetes mellitus. J. Biol. Chem. 2001; 276:18653–18656. [DOI] [PubMed] [Google Scholar]
- 16. Varrault A., Gueydan C., Delalbre A., Bellmann A., Houssami S., Aknin C., Severac D., Chotard L., Kahli M., Le Digarcher A. et al. . Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell. 2006; 11:711–722. [DOI] [PubMed] [Google Scholar]
- 17. Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. Ed. Frederick M Ausubel Al. 2005; doi:10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 18. Gaspard N., Bouschet T., Hourez R., Dimidschstein J., Naeije G., van den Ameele J., Espuny-Camacho I., Herpoel A., Passante L., Schiffmann S.N. et al. . An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008; 455:351–357. [DOI] [PubMed] [Google Scholar]
- 19. Robinson M.D., Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F.. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H.. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008; 26:1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blahnik K.R., Dou L., O’Geen H., McPhillips T., Xu X., Cao A.R., Iyengar S., Nicolet C.M., Ludäscher B., Korf I. et al. . Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010; 38:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLeay R.C., Bailey T.L.. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010; 11:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang D.W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 26. Green H., Kehinde O.. Sublines of 3T3 cells that accumulate lipids. Cell. 1974; 1:113–116. [Google Scholar]
- 27. Piras G., El Kharroubi A., Kozlov S., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L.. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 2000; 20:3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Preter K., Vandesompele J., Heimann P., Yigit N., Beckman S., Schramm A., Eggert A., Stallings R.L., Benoit Y., Renard M. et al. . Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol. 2006; 7:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012; 13:840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ji H., Vokes S.A., Wong W.H.. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Res. 2006; 34:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hume M.A., Barrera L.A., Gisselbrecht S.S., Bulyk M.L.. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015; 43:D117–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lui J.C., Finkielstain G.P., Barnes K.M., Baron J.. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008; 295:R189–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Basyuk E., Coulon V., Le Digarcher A., Coisy-Quivy M., Moles J.-P., Gandarillas A., Journot L.. The candidate tumor suppressor gene ZAC is involved in keratinocyte differentiation and its expression is lost in basal cell carcinomas. Mol. Cancer Res. 2005; 3:483–492. [DOI] [PubMed] [Google Scholar]
- 34. Pateras I.S., Apostolopoulou K., Niforou K., Kotsinas A., Gorgoulis V.G.. p57KIP2: ‘Kip’ing the cell under control. Mol. Cancer Res. MCR. 2009; 7:1902–1919. [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann A., Ciani E., Boeckardt J., Holsboer F., Journot L., Spengler D.. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol. Cell. Biol. 2003; 23:988–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Voz M.L., Mathys J., Hensen K., Pendeville H., Van Valckenborgh I., Van Huffel C., Chavez M., Van Damme B., De Moor B., Moreau Y. et al. . Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene. 2004; 23:179–191. [DOI] [PubMed] [Google Scholar]
- 37. Landrette S.F., Madera D., He F., Castilla L.H.. The transcription factor PlagL2 activates Mpl transcription and signaling in hematopoietic progenitor and leukemia cells. Leukemia. 2011; 25:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barz T., Hoffmann A., Panhuysen M., Spengler D.. Peroxisome proliferator-activated receptor gamma is a Zac target gene mediating Zac antiproliferation. Cancer Res. 2006; 66:11975–11982. [DOI] [PubMed] [Google Scholar]
- 39. Rraklli V., Södersten E., Nyman U., Hagey D.W., Holmberg J.. Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem Cell Res. 2016; 16:1–9. [DOI] [PubMed] [Google Scholar]
- 40. Vermeulen J., De Preter K., Naranjo A., Vercruysse L., Van Roy N., Hellemans J., Swerts K., Bravo S., Scaruffi P., Tonini G.P. et al. . Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009; 10:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iglesias-Platas I., Martin-Trujillo A., Petazzi P., Guillaumet-Adkins A., Esteller M., Monk D.. Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum. Mol. Genet. 2014; 23:6275–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. MacQuarrie K.L., Fong A.P., Morse R.H., Tapscott S.J.. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. TIG. 2011; 27:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L. et al. . Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010; 18:662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng Y., Wu W., Kumar S.A., Yu D., Deng W., Tripic T., King D.C., Chen K.-B., Zhang Y., Drautz D. et al. . Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009; 19:2172–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfenning A.R., Kim T.-K., Spotts J.M., Hemberg M., Su D., West A.E.. Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways. PLoS One. 2010; 5:e10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kassouf M.T., Hughes J.R., Taylor S., McGowan S.J., Soneji S., Green A.L., Vyas P., Porcher C.. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010; 20:1064–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cui J.Y., Gunewardena S.S., Rockwell C.E., Klaassen C.D.. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 2010; 38:7943–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kenzelmann Broz D., Spano Mello S., Bieging K.T., Jiang D., Dusek R.L., Brady C.A., Sidow A., Attardi L.D.. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013; 27:1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., MacArthur S., Bourgon R., Nix D., Pollard D.A., Iyer V.N., Hechmer A., Simirenko L., Stapleton M., Luengo Hendriks C.L. et al. . Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008; 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zaret K.S., Carroll J.S.. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011; 25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Todeschini A.-L., Georges A., Veitia R.A.. Transcription factors: specific DNA binding and specific gene regulation. Trends Genet. TIG. 2014; 30:211–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.