Abstract
Spliced leader (SL) trans-splicing is a critical element of gene expression in a number of eukaryotic groups. This process is arguably best understood in nematodes, where biochemical and molecular studies in Caenorhabditis elegans and Ascaris suum have identified key steps and factors involved. Despite this, the precise details of SL trans-splicing have yet to be elucidated. In part, this is because the systematic identification of the molecules involved has not previously been possible due to the lack of a specific phenotype associated with defects in this process. We present here a novel GFP-based reporter assay that can monitor SL1 trans-splicing in living C. elegans. Using this assay, we have identified mutants in sna-1 that are defective in SL trans-splicing, and demonstrate that reducing function of SNA-1, SNA-2 and SUT-1, proteins that associate with SL1 RNA and related SmY RNAs, impairs SL trans-splicing. We further demonstrate that the Sm proteins and pICln, SMN and Gemin5, which are involved in small nuclear ribonucleoprotein assembly, have an important role in SL trans-splicing. Taken together these results provide the first in vivo evidence for proteins involved in SL trans-splicing, and indicate that continuous replacement of SL ribonucleoproteins consumed during trans-splicing reactions is essential for effective trans-splicing.
INTRODUCTION
A wide range of eukaryotes engage in the trans-splicing of their pre-mRNAs, a process which results in the replacement of the 5′-end of the transcript with a short, ‘spliced leader’ (SL) (1,2). The essential mechanism of SL trans-splicing involves an intermolecular splicing event between the donor molecule, the SL RNA and the acceptor pre-mRNA, which possesses an intron-like 5′ untranslated region (5′ UTR), containing an unpaired 3′ splice site, termed the ‘outron’. However, most organisms that possess SL trans-splicing also contain a sub-set of genes organized into operons, consisting of several genes under the control of a shared promoter. Operons thus produce polycistronic transcripts, which are resolved into monocistronic mRNAs through SL trans-splicing coupled to 3′ end formation.
SL trans-splicing occurs on approximately 70% of mRNAs in Caenorhabditis elegans, including those that are derived from polycistronic transcripts (3). C. elegans uses two classes of SLs, SL1 and SL2, the former being trans-spliced to pre-mRNAs produced by monocistronic genes and to the pre-mRNAs derived from the first genes in operons (4). In contrast, SL2 is spliced exclusively to pre-mRNAs that are derived from downstream genes in operons (5). SL1 is essential, but lack of SL1 can at least in part be complemented by overexpression of SL2 RNA (6), presumably because they participate in fundamentally similar splicing events and the end products of these events are functionally similar. However, features that distinguish SL1 and SL2 trans-splicing have been defined, making it clear that these are distinct events (7–9).
SL RNAs form SL ribonucleoprotein particles (RNPs) and share some properties with the small nuclear RNPs (snRNPs) involved in cis-splicing reactions, in that they adopt similar secondary structures and are bound by proteins recognized by anti-Sm antibodies (10–13).
Additional proteins associated with SL RNAs were isolated from Ascaris splicing extracts, and homologs of these were identified in C. elegans (14,15). C. elegans SNA-2 was identified as the ortholog of the 95 kDa Ascaris protein, and was found to interact with SL1, but not SL2. The smaller, SL-30p Ascaris protein, was shown to have two C. elegans homologs, SNA-1 and SUT-1, with the former also interacting with SL1, while the latter was found to interact with a novel, nematode-specific class of snRNA termed SmY RNA (15,16). Subsequent work has shown that SNA-1 and SUT-1 are conserved throughout the Rhabditida (17). RNAi knockdown of sna-1 or sut-1 causes cold-sensitive sterility, and loss of both sna-1 and sut-1 function results in synthetic defects in viability, despite the fact that they show distinct biochemical properties (15).
To account for this MacMorris and colleagues have proposed that SNA-1 and SUT-1, together with SNA-2, associate with SL1 and SmY RNP complexes, respectively, and may function to recycle Sm proteins following trans-splicing (15). This hypothesis is based on the fact that SL trans-splicing consumes SL RNPs by transfer of the 5′ terminal spliced leader sequence onto mRNAs. According to this model, SNA-1 is involved in the recycling of Sm proteins following SL1 trans-splicing and thus loss of SNA-1 is compensated for by the presence of the other SUT-1/SmY-dependent Sm protein recycling pathway. Similarly, loss of SUT-1 would be tolerated because of the presence of the SNA-1/SL1 Sm recycling pathway. Loss of both SNA-1 and SUT-1 would compromise both pathways and thus results in a lethal loss of Sm protein recycling. Consistent with this model, loss of SNA-2, an essential component of both complexes, is lethal (15).
While this model explains the available molecular and genetic evidence, these novel proteins have not been directly shown to be required for SL trans-splicing in vivo. To better characterize SL trans-splicing we have designed a novel GFP reporter-based assay allowing us to genetically identify factors involved in this process in living C. elegans. We show that depletion of sna-1, sna-2 and sut-1 impairs SL trans-splicing in vivo and have identified new loss-of-function alleles of sna-1 from a genetic screen using our assay. We also show that depletion of Sm protein expression, and factors involved in snRNP assembly, leads to reduced SL trans-splicing. A role for these factors in SL trans-splicing was confirmed by analyzing SL trans-splicing of the GFP reporter gene transcripts, and of an endogenous trans-spliced RNA, by qPCR. To our knowledge, our molecular analysis demonstrates for the first time in a living system a role for these factors in SL trans-splicing. SL RNPs consumed during trans-splicing need to be replaced and our observations imply that the synthesis of new Sm proteins makes a significant and crucial contribution to the pool of Sm proteins available for the assembly of new SL RNPs.
MATERIALS AND METHODS
Strains
Nematodes were maintained using standard protocols at 20°C on Escherichia coli B strain OP50 unless otherwise stated (18,19). The following strains were used: N2 (Bristol); PE720 [fex308]; PE774 [sna-1(fe45) V; feEx308]; PE766 [sna-1(fe47) V; feEx308]; PE793 [feIs11 X]; FX3079 [sut-1(tm3079) II]. Transgenic genotypes are as follows: feEx308 is an extrachromosomal array consisting of [Pvit-2::outron::gfpM1APmyo-3::mCherry] (‘gfpM1A’ indicates that the initiator methionine of GFP has been changed to an alanine codon); feIs11 was generated by γ-ray-mediated integration of the feEx308 array into the X chromosome.
Construction of SL trans-splicing reporter transgene and generation of transgenic strains
We amplified the vit-2 promoter (Pvit-2) from wild type (wt) genomic DNA and inserted the resulting amplicon into BamHI-XbaI cut pPD95.75 (gift from A. Fire, Stanford School of Medicine; available from Addgene; plasmid #1494) to give pAWF29B. A synthetic ‘OU141’ outron based on previous studies (20) was assembled from oligonucleotides with overlaps filled in by polymerase chain reaction (PCR), and inserted into pGEM-T Easy (Promega). The resultant plasmid was digested with AgeI and BamHI and the OU145 outron insert was cloned into AgeI-BamHI cut pAWF29B to produce pAWF30. The GFP initiation codon was changed to an alanine (GCT) by site directed mutagenesis using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs), generating pLP6. The plasmid was sequenced to ensure no additional mutations were induced during the various cloning steps. This revealed that the OU141 outron sequence, which contained four copies of a 34 bp synthetic sequence (20) had undergone a truncation in pLP6 leaving only three copies (Supplementary Figure S2).
pLP6 was co-injected with pCFJ104, which expresses mCherry in body wall muscles (21). Five transgenic lines were generated, and all behaved similarly. One line, containing the extrachromosomal array feEx308, was selected for further study. The extrachromosomal array was integrated by subjecting transgenic animals to 3800 rad γ irradiation for 20 min (22). Four independent strains (feIs11-14) were recovered and backcrossed to wt six times. feIs11 was mapped to the X chromosome and chosen for use in this study.
Spliced leader trans-splicing reporter construct genetic screen
A population of PE720 worms were subject to ethyl methanesulfonate (EMS) mutagenesis (23) and allowed to become gravid adults. F1 progeny were picked singly to separate plates and their progeny screened using a stereomicroscope equipped with an epifluorescent light source for the presence of intestinal GFP expression. Individual fluorescent animals were then picked to fresh plates and their progeny scored to confirm the intestinal GFP expression phenotype.
Each mutant line was outcrossed with wt males and non-transgenic cross-progeny males crossed with unmutagenized PE720 hermaphrodites to ensure that the phenotype was not associated with changes to the transgenic array, and to determine the inheritance pattern of the mutation. Mutations in sna-1 were identified by PCR amplification of exons followed by Sanger sequencing (DNA Sequencing and Services, University of Dundee).
Epifluorescence microscopy
To determine the proportion of animals expressing GFP, ∼50 eggs were seeded on 3 cm RNAi plates with E. coli K-12 derived HT115(DE3) transformed with the appropriate feeding plasmid and allowed to grow at 20°C for 3 days to L4/adult stage. The total number of hatched animals and the number of GFP fluorescent animals were determined using a Leica MZ16F fluorescent stereomicroscope, and used to calculate the proportion of GFP expressing animals. To measure the intensity of GFP fluorescence, 10 animals were immobilized on slides with 1% agarose cushions, using 0.5% l-phenoxy-2-propanol as an anaesthetic, covered with a cover slip and sealed with Vaseline. A Zeiss Axioplan 2 fluorescence microscope was used to capture GFP and mCherry fluorescence for each animal. ImageJ software (24) was used to analyze the intensity of GFP and mCherry fluorescence. The GFP fluorescence intensity of each animal was normalized with respect to its mCherry fluorescence intensity. Statistical analysis by One-way ANOVA on ranks followed by Dunn's post-test was done using GraphPad Prism version 5.00 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com.
RNA Interference
To prepare the RNAi feeding vectors, DNA was PCR amplified from genomic DNA or cDNA and inserted by In-Fusion cloning (Takara Bio Inc.) into the plasmid pPD129.36 cut with SpeI. The targeted genes and the primers used for PCR amplification are shown in the Supplementary Table S1. The mixtures were directly used to transform E. coli HT115. DNA sequences were verified by Sanger sequencing (DNA Sequencing and Services, University of Dundee). The unc-22 RNAi construct was described earlier (25). RNAi-by-feeding was carried out as previously described (26), by seeding eggs, isolated by alkali hypochlorite treatment of gravid adults, onto RNAi plates. RNA was then prepared from L4/young adult animals.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was prepared from C. elegans strains grown on E. coli using the PureLink RNA Mini kit (Life Technologies) with modifications for TRIzol treated samples and DNAse treatment as described by the manufacturer. For cDNA synthesis, total RNA was reverse transcribed using oligo(dT) Primers and M-MLV Reverse Transcriptase, according to the manufacturer's instructions. qPCR assays were designed using the Universal Probe Library Assay Design Centre (Roche) and tested for efficiency (Supplementary Table S2). qPCR was done in three technical replicates on a Roche Lightcycler 480 using standard settings. The Minimum Information for Quantitative Real-Time PCR Experiments (MIQE) is included as Supplementary Data. Data was analyzed using the comparative CT method, assuming a primer efficiency of 2 (27). Outron levels were standardized with respect to internal RNA. Outron levels detected in unc-22(RNAi) treated animals were defined as 1.
RESULTS AND DISCUSSION
An in vivo assay to detect loss of SL1 trans-splicing
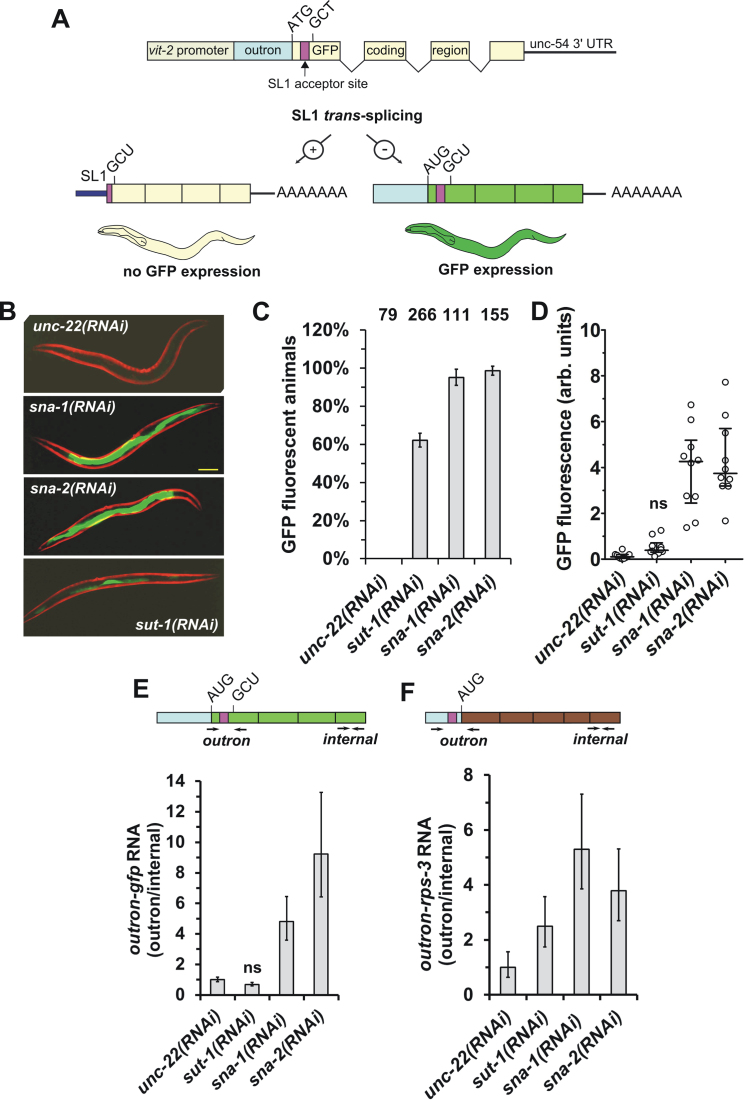
We generated a transgenic strain that would display GFP fluorescence dependent on suppression of SL trans-splicing (Figure 1A). The transgene consists of a modified GFP gene that is under the transcriptional control of the vit-2 promoter (20). The GFP gene was modified such that the original ATG initiation codon was changed into an alanine codon, GCT (GFPM1A). Upstream of this we engineered a synthetic outron sequence, which has previously been shown to confer on a pre-mRNA the ability to act as SL1 trans-splicing substrate (20). We placed a new ATG codon in-frame with the GFP coding region upstream of the 3′ splice site. Thus, in wild type worms, the removal of the synthetic outron from the pre-mRNA during SL trans-splicing would be expected to produce an mRNA lacking an AUG, and so no GFP fluorescence would be observed, whereas in animals with reduced SL trans-splicing, mRNA retaining the outron and its associated in-frame AUG would be predicted to accumulate, and thus allow GFP expression (Figure 1A). As predicted, transgenic lines established with this Pvit-2::outron::GFPM1A construct displayed only sporadic, weak fluorescence (Figure 1B–D: unc-22(RNAi)).
Figure 1.
A novel reporter gene assay for the in vivo detection of reduced SL1 trans-splicing. (A) The Pvit-2::outron::gfpM1A transgene engineered to monitor SL1 trans-splicing. The original ATG of the GFP open reading frame (yellow shading) was changed to a GCT alanine codon. Note the gfp gene contains three introns. The two mRNA products produced in the presence, or absence of SL1 trans-splicing are shown below, and a cartoon of the predicted GFP expression in wild-type (wt) worms (green shading indicates intestinal GFP fluorescence). (B) Detection of SL1 trans-splicing inhibition by fluorescence microscopy. Micrographs of GFP expression in animals carrying the Pvit-2::outron::GFPM1A transgene subjected to sna-1, sna-2, sut-1 or unc-22(RNAi). Exposure times were identical for all micrographs (5 ms). The scale bar corresponds to 100 μm. (C and D) Quantitation of GFP expression in animals carrying the Pvit-2::outron::GFPM1A transgene subjected to sna-1, sna-2, sut-1 or unc-22(RNAi). (C) The proportion of GFP fluorescent animals was determined by fluorescence microscopy. Error bars represent the standard deviation based on between two and five independent experiments (unc-22(RNAi): 2; sna-1(RNAi), sna-2(RNAi): 3; sut-1(RNAi): 5). The total number of animals analyzed is indicated at the top of the graph. (D) The intensity of GFP fluorescence (in arbitrary units) was determined from micrographs as shown in (B), and standardized with respect to mCherry fluorescence. The graph plots the GFP fluorescence of 10 animals analyzed for each RNAi treatment; shown are the median and the first and third quartile. ‘ns’ indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05; ANOVA). (E and F) Detection of SL1 trans-splicing inhibition by RT-qPCR. Animals carrying the Pvit-2::outron::GFPM1A transgene were subjected to sna-1, sna-2, sut-1 or unc-22(RNAi), and trans-splicing of gfp reporter gene and rps-3 transcripts was analyzed by reverse transcription followed by qPCR. The schematic diagrams indicate the position of the primers used (drawings are not to scale). Outron sequences are shown in blue and the SL1 acceptor sites in purple. Outron-gfp and outron-rps-3 RNA levels were standardized with respect to an internal part of the mRNA (internal) and levels in unc-22(RNAi) animals were defined as 1. Note that elevated outron-gfp or outron-rps-3 levels indicate inhibition of SL trans-splicing. ‘ns’ indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05, t-test). Error bars show standard deviation from three technical replicates. Similar results were obtained in two independent experiments.
To test the response of the transgenic strain to loss of SL trans-splicing, we carried out RNA interference (RNAi) targeted against sna-1 and sna-2, since these genes encode known components of the SL RNP, and sut-1, because of its genetic interaction with sna-1 and proposed role in Sm protein recycling (15). We observed a strong GFP expression response in animals subjected to sna-1(RNAi) and sna-2(RNAi), indicating that our assay is responding as expected (Figure 1B–D). We observed GFP expression in more than 60% of sut-1(RNAi) treated animals. While fluorescence was clearly detectable, it was weak and not significantly above the background values obtained in unc-22(RNAi) controls (Figure 1B–D), largely because typically relatively few intestinal cells displayed fluorescence in sut-1(RNAi) animals. To confirm that GFP expression correlates with retention of the outron in the GFP mRNA we used qRT-PCR to measure the levels of non-trans-spliced outron sequences, and of an internal part of the reporter gene transcript not involved in SL trans-splicing. Relative trans-splicing efficiencies were calculated by standardizing the levels of outron containing transcripts with respect to the internal fragments, which allows a direct comparison between different RNAi treatments (Figure 1E). This was complemented by the analysis of the SL trans-splicing of endogenous rps-3 transcripts, which are known to be trans-spliced by SL1 (3) (Figure 1F). In animals treated with sna-1(RNAi) and sna-2(RNAi), the levels of non-trans-spliced GFP reporter transcripts (from here on referred to as outron-gfp RNA) were significantly higher than in the unc-22(RNAi) treated control animals, reflecting the increase in GFP expression observed (Figure 1B–D). Furthermore, the levels of non-trans-spliced rps-3 RNA (from here on referred to as outron-rps-3 RNA) were elevated in sna-1 and sna-2 (RNAi) treated animals (Figure 1F), indicating that the knockdown of these genes also interferes with the trans-splicing of endogenous transcripts. In animals subjected to sut-1(RNAi) we were not able to detect a significant increase of outron-gfp transcripts, reflecting the weak activation of the reporter observed by microscopy (Figure 1B–D). However, the level of outron-rps-3 transcripts was elevated, consistent with a role of sut-1 in SL trans-splicing.
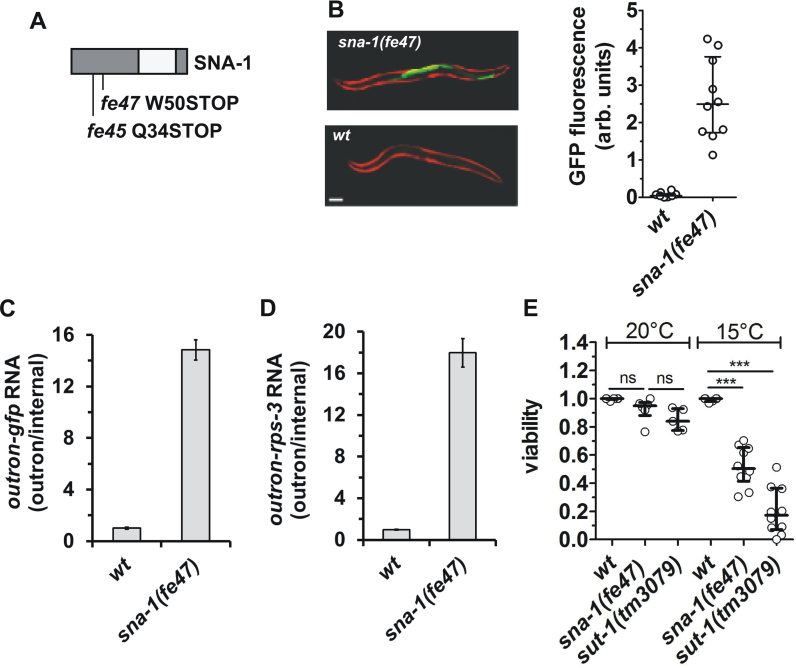
As further support for the role of SNA-1 in SL trans-splicing, we identified two loss-of-function sna-1 mutations from a genetic screen for PE720 worms that displayed elevated levels of GFP fluorescence. The fe45 allele is a C to T transition on chromosome V at position 6,710,253, creating a premature stop at codon 34 (CAA > TAA, Q34STOP) and fe47 is a C to T transition on chromosome V at position 6,710,203, creating a premature stop codon at codon 50 (GGA > TGA; W50STOP) (Figure 2). Detailed analysis of GFP fluorescence was carried out for sna-1(fe47) homozygotes (Figure 2B). In addition, the levels of outron-gfp and outron-rps-3 transcripts were more than 15-fold higher than in wild type animals (Figure 2C and D), confirming the inhibition of SL trans-splicing in these animals.
Figure 2.
Loss-of-function mutations in sna-1 recovered from a mutagenesis screen for genes involved in SL1 trans-splicing. (A) Schematic representations of the location of the residues affected by the mutations in the SNA-1 protein. The regions in dark gray are conserved in nematode SNA-1 orthologues (Supplementary Figure S1). (B) Representative images of wild type (wt) and sna-1(fe47) animals transgenic for Pvit-2::outron::gfpM1A, showing intestinal GFP (green) expression and constitutive mCherry (red) expression in body wall muscle cells. Scale bar represents 100 μm. The graph plots the GFP fluorescence of 10 animals analyzed; shown are the median and the first and third quartile. (C and D) RT-qPCR analysis of SL1 trans-splicing in wt and sna-1(fe47) animals carrying the Pvit-2::outron::gfpM1A transgene. Trans-splicing of gfp reporter gene (C) and rps-3 transcripts (D) was analyzed as described in the legend of Figure 1, with levels in wt animals set as 1. Error bars indicate standard deviation from three technical replicates. (E) Viability of wt, sna-1(fe47) and sut-1(tm3079) animals at 20 and 15°C, as expressed by the proportion of eggs that develop to L4/adult stage in 5–10 independent experiments. (***P ≤ 0.001; ‘ns’ not significant; ANOVA). Note that while at 20°C there is a slight reduction in viability in sut-1(tm3079) animals, this is not significantly different from the viability of sna-1(fe47) animals.
Although the two sna-1 alleles that we isolated are viable, previous studies have shown that sna-1(RNAi) results in cold-sensitive sterility defects and that combined loss of sna-1 and sut-1 function results in embryonic lethality (15). We replicated these observations with the sna-1(fe47) allele, demonstrating that it confers cold-sensitive defects in viability, similar to the sut-1(tm3079) allele (Figure 2E). Furthermore, sut-1; sna-1 double mutants are sickly, with the majority (92.5%; n = 40) being completely sterile.
In conclusion, these experiments clearly demonstrate that reduced expression of sna-1 or sna-2 interferes with SL trans-splicing. The genetic interaction of sna-1 with sut-1, combined with the weak disturbance of SL trans-splicing by sut-1 depletion supports a function of sut-1 in SL trans-splicing. It has been demonstrated that SUT-1 interacts with SmY RNAs (15), and this provides further support for a functional role of SmY RNAs in SL trans-splicing. Taken together, our data demonstrate a role for these molecules in SL trans-splicing, as proposed in earlier molecular studies.
Previous biochemical analyses of SL RNPs using anti-Sm antisera imply that SL1 RNA is associated with Sm proteins (10,13,15). However, since the post-translational modification of Sm proteins, symmetrical dimethylation of arginines, is the major epitope for anti-Sm antibodies (28), it is possible that other proteins containing this epitope, such as Lsm4 or piRNA associated Mili protein, might explain the ability of anti-Sm antisera to interact with SL RNPs (29,30). It was therefore important to directly test the role of Sm proteins in SL trans-splicing.
Sm proteins form heptameric rings that bind to a variety of snRNAs and are found in snRNPs acting in, for example, mRNA splicing. The major U1, U2, U4 and U5 snRNAs associate with a ring formed by the Sm proteins B, D1, D2, D3, F, E and G. In C. elegans, these proteins are encoded by snr-1, snr-2, snr-3, snr-4, snr-5, snr-6 and snr-7, respectively (31). In addition, the related LSm proteins form heptameric rings that interact with other RNAs (32). U6 snRNA, involved in cis-splicing is bound by a ring formed from LSm2, 3, 4, 5, 6, 7, 8 proteins, which in C. elegans are encoded by the gut-2, lsm-3, lsm-4, lsm-5, lsm-6, lsm-7 and lsm-8 genes, respectively. A similar ring where mammalian LSm8 is replaced by LSm1 (encoded by lsm-1) is involved in the degradation of mRNA (32). Using similarity searches we also identified the genes K07A1.15, C49H3.4 and M142.5 as encoding proteins with Sm-like domains (33). M142.5 was identified by others as an orthologue of human LSm12 (34), while K07A1.15 is annotated in Wormbase as the orthologue of human LSm10. LSm10 protein associates with LSm11 and Sm proteins B, D3, G, E and F to form the U7 snRNP particle involved in histone RNA processing (35,36). Drosophila Lsm10 and Lsm11 mutants die as pupae (37), indicating that these functions are essential. Surprisingly, no phenotype has been attributed to K07A1.15 loss-of-function (33,38,39). We thus wanted to determine the effect of knocking down the expression of Sm and LSm protein function on GFP fluorescence in our reporter strain, as well as on the levels of outron-gfp and outron-rps-3 transcripts.
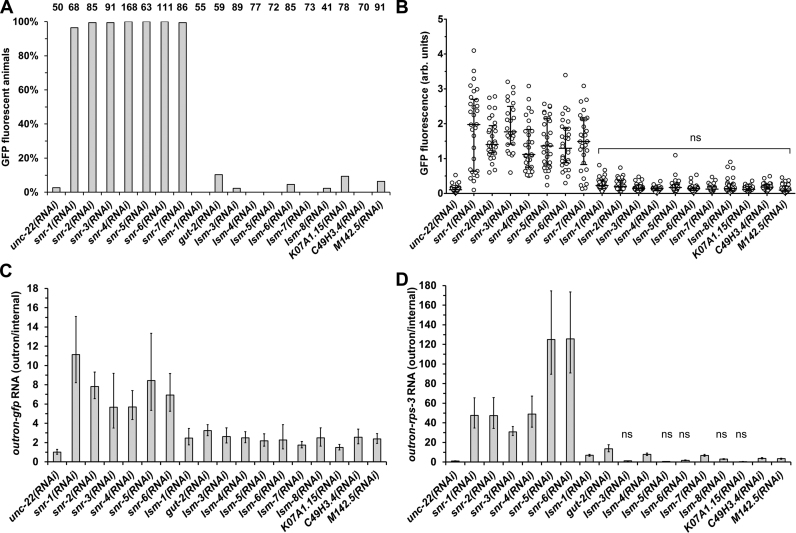
Figure 3 shows that the knockdown of snr gene transcripts led to GFP fluorescence in all, or nearly all animals tested, with a more than seven-fold increase of fluorescence compared to the unc-22(RNAi) treated control animals (Figure 3A and B). This was accompanied by five-fold or higher increase of outron-gfp transcripts in animals subjected to snr(RNAi) (Figure 3C). Importantly, snr(RNAi) also impaired trans-splicing of rps-3 transcripts (Figure 3D). The effect on these transcripts appeared more severe than the effect on the reporter gene transcripts, with up to more than 100-fold higher levels of non-trans-spliced transcripts detected in snr-5 and snr-6(RNAi) treated animals compared to the control treatment.
Figure 3.
SL1 spliced leader trans-splicing is sensitive to the knockdown of snr, but not lsm, gene transcripts. Detection of SL1 trans-splicing inhibition by fluorescence microscopy. Quantitation of GFP expression in animals carrying the Pvit-2::outron::GFPM1A transgene subjected to RNAi was done as described for Figure 1. (A) The proportion of GFP fluorescent animals. The number of animals analyzed are indicated above each dataset. (B) GFP fluorescence intensity. The graph summarizes individual measurements (n = 30) from three independent experiments. The distribution of measurements for each of the experiments is similar to the distribution of the pooled data. Median and the first and third quartile are indicated. ‘ns’ indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05; ANOVA). (C and D) Detection of SL1 trans-splicing inhibition by RT-qPCR. Animals carrying the Pvit-2::outron::GFPM1A transgene were subjected to RNAi, and trans-splicing of the gfp reporter gene (C) and rps-3 transcripts (D) was analyzed by reverse transcription followed by qPCR. Outron-gfp and outron-rps-3 transcript levels were standardized as described in the Figure 1 legend, and levels in unc-22(RNAi) animals were defined as 1. Note that snr-7(RNAi) was not analyzed by RT-qPCR. ‘ns’ indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05; t-test). Error bars show standard deviation from three technical replicates.
In contrast to the knockdown of Sm protein transcripts, knockdown of each of the eleven LSm protein transcripts failed to cause notable GFP fluorescence (Figure 3A and B). A low proportion of animals, not exceeding 10%, expressed GFP at a detectable level. This is distinct from the effect of sna-1 and sna-2(RNAi), and the knockdown of snr RNAs, which results in clearly detectable GFP fluorescence in all or most animals subjected to the RNAi treatments (Figures 1C and D, 3A and B). It is also distinct from sut-1(RNAi), which resulted in 62% of animals expressing GFP, albeit at a low level (Figure 1C and D). In addition, the levels of non-trans-spliced transcripts in these animals were generally low. Outron-gfp transcript levels varied between 1.5- and 3.3-fold of levels observed in the unc-22(RNAi) control treated animals, while levels of outron-rps-3 transcripts varied between 0.5- and 13.6-fold of those observed in the control treated animals, and were in many cases not significantly different from the control (Figure 3D).
The analysis of GFP fluorescence and that of trans-splicing by qRT-PCR were done independently. The difference between fluorescence and the detection of non-trans-spliced transcripts by qPCR in, for example, many of the lsm(RNAi) treatments may be caused by different sensitivities of the two methods. Importantly, the combination of the two approaches, with the inclusion of the analysis of trans-splicing of an additional endogenous gene, provides a coherent picture of the effect of the treatments.
Our findings indicate that SL1 trans-splicing is highly sensitive to the knockdown of the genes encoding Sm proteins: the knockdown of snr genes, similar to the knockdown of sna-1 and sna-2, results in strong GFP fluorescence in the majority or all animals subjected to the RNAi treatments. This is supported by the significant increase of non-processed outron-gfp and outron-rps3 RNA in these animals, which also indicates that trans-splicing is impaired. In contrast, lsm gene knockdown only results in weak GFP fluorescence in a small number of the treated animals. The molecular analysis of trans-splicing confirms that any GFP fluorescence detected is due to a defect in trans-splicing. Together these data indicate that SL trans-splicing is highly sensitive to the knockdown of the genes encoding Sm proteins, while the knockdown of genes encoding LSm proteins has no consistent detectable effect.
Since the Sm proteins are also required for assembly of cis-splicing U snRNPs, one possible interpretation is that the loss of these factors, all of which apart from U1 are required for SL trans-splicing (40,41), could contribute to the impaired SL trans-splicing that we observe. However, two observations suggest that the depletion of Sm proteins primarily impacts upon SL RNP function in our experiments. First, the removal, by cis-splicing, of the three GFP transgene introns, two of which contain stop codons, is required for fluorescence, implying that cis-splicing factors are not significantly impaired by Sm protein depletion. Second, depletion of multiple LSm proteins, which are required for U6 snRNP formation, has minimal effects on SL trans-splicing, despite the fact that U6 is required for trans-splicing (42). Thus, the most likely explanation for the impaired SL trans-splicing in animals depleted for Sm proteins is due to the effect of this loss on the formation of the SL RNP. This raises the question: why would cis-splicing U snRNPs be less sensitive to Sm protein depletion than SL RNPs? One explanation may be that the latter are consumed by the splicing reaction in which they participate, and thus SL trans-splicing is dependent on the continuous formation of new SL RNPs. As described earlier, it has been proposed that Sm protein recycling contributes to the formation of new SL RNPs (15). Our findings indicate that this is not sufficient, and that the formation of new SL RNPs is heavily reliant on the synthesis of new Sm proteins. An alternative, but not mutually exclusive, explanation is that SL RNA is stabilized by the integration into RNPs and is therefore sensitive to a reduction in RNP components.
Recycling of the snRNPs after completion of cis-splicing is an important, but poorly understood process. Investigations have mainly focused on the U6 snRNP, which undergoes major changes during the cis-splicing reaction and is associated with LSm protein (43,44). Our findings are compatible with a model where U6 snRNA and associated LSm proteins participating in SL trans-splicing are recycled, and do not require continuous replacement.
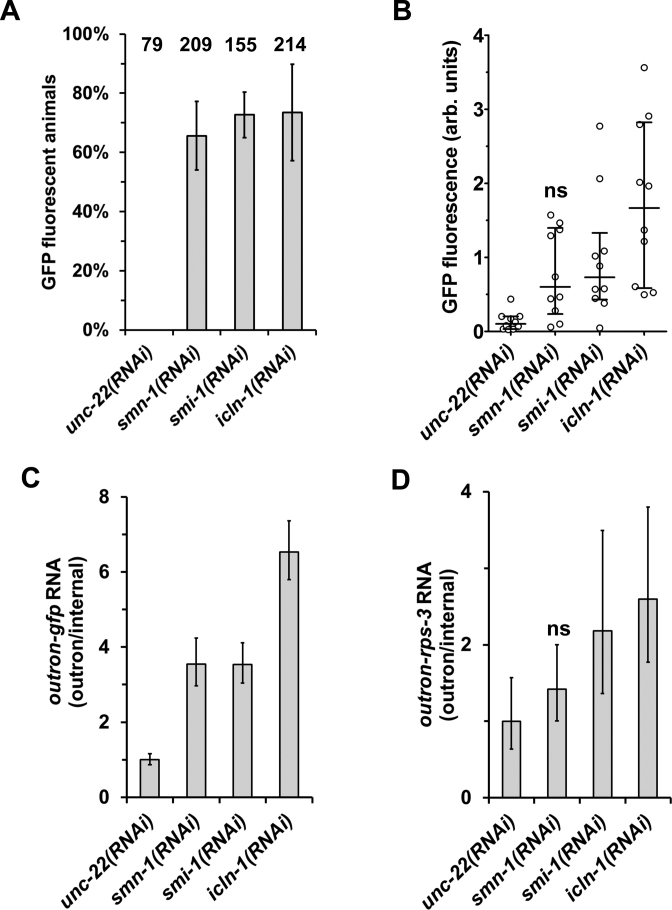
To determine whether factors involved in snRNP assembly also play a role in SL trans-splicing, we knocked down the expression of smn-1, smi-1, icln-1 by RNAi. smn-1 encodes a homolog of human SMN and smi-1 an ortholog of human Gemin2 that interacts with SMN, and, together with SMN, is a component of the SMN complex (45). This complex mediates the specific assembly of the heptameric Sm protein ring onto snRNAs and interacts with both Sm proteins and snRNAs (46). icln-1 encodes a homolog of the human methylosome subunit, pICln. Sm proteins, B/B’, D1 and D3 are modified by symmetrical dimethylation of arginine residues by a complex containing pICln, which promotes the interaction with the SMN complex for snRNP assembly (29,47,48).
Knockdown of smn-1, smi-1 and icln-1 led to GFP fluorescence in most of the animals subjected to RNAi, similar to the knockdown of sna-1, sna-2 and the snr genes described above (Figures 1C and D, 3A and B), with icln-1(RNAi) giving the strongest effect (Figure 4A and B). The inhibition of SL trans-splicing by smi-1 and icln-1 knockdown was confirmed by the increased levels of outron-gfp and outron-rps-3 transcripts (Figure 4C and D), indicating that the knockdown of these genes inhibits SL trans-splicing. smn-1 knockdown caused a weaker effect: while reporter gene activation was detected in the majority of animals, the fluorescence was not strong enough to produce a significant difference in the quantitative comparison to unc22(RNAi) animals (Figure 4A and B). However, the increase of non-processed outron-gfp RNA in smn-1(RNAi) animals indicates that trans-splicing is impaired. These data suggest that the same biogenesis factors involved in snRNP assembly are required for these processes during the lifecycle of SL RNPs, further reinforcing the similarities between the U snRNPs and SL RNPs.
Figure 4.
Knockdown of snRNP assembly factors inhibits SL1 spliced leader trans-splicing. Detection of SL1 trans-splicing inhibition by fluorescence microscopy. Quantitation of GFP expression in animals with the Pvit-2::outron::GFPM1A transgene subjected to RNAi was done as described for Figure 1. (A) The proportion of GFP fluorescent animals. The graph summarizes results from two or five independent experiments (unc-22(RNAi): 2; smn-1(RNAi), smi-1(RNAi), icln-1(RNAi): 5). The total number of animals analyzed is indicated. (B) GFP fluorescence intensity. The graph shows measurements of fluorescence intensity of 10 animals analyzed for each RNAi treatment. ns indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05; ANOVA). (C and D) Detection of SL1 trans-splicing inhibition by RT-qPCR. Animals carrying the Pvit-2::outron::GFPM1A transgene were subjected to RNAi, and trans-splicing of the gfp reporter gene (C) and rps-3 transcripts (D) was analyzed by reverse transcription followed by qPCR. Outron-gfp and outron-rps-3 transcript levels were standardized as described in the Figure 1 legend, and levels in unc-22(RNAi) animals were defined as 1. ‘ns’ indicates values not significantly higher than in unc-22(RNAi) animals (P ≥ 0.05, t-test). Error bars show standard deviation from three technical replicates.
In conclusion, we have developed an in vivo assay that allows us to monitor the levels of SL trans-splicing in C. elegans, and have confirmed that there is a strong correlation between GFP fluorescence and outron-retention in SL trans-spliced mRNAs. This has allowed us to demonstrate physiological roles in SL trans-splicing for SNA-1, SNA-2 and SUT-1, and for Sm proteins and snRNP assembly factors. Our findings indicate that Sm protein recycling is not sufficient to replace the SL RNPs used up during trans-splicing, but that the assembly of new SL RNPs is heavily reliant on the synthesis of new Sm proteins. Furthermore, we have shown that this assay system can be employed to identify mutations in genes specifically involved in SL trans-splicing. Further application of such screens, including searches for lethal mutations, should allow the comprehensive identification of molecules involved in this fundamental nematode process. Finally, we envisage that this will be useful as a bioassay in the search for compounds that inhibit SL trans-splicing.
Supplementary Material
ACKNOWLEDGEMENTS
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We would also like to thank Prof. Shohei Mitani, at the National Bioresource Project for the Experimental Animal ‘Nematode C. elegans’, Japan, for FX3079. We are grateful to Prof. Tom Blumenthal (University of Colorado, Boulder) for suggestions and support of this work; and to Kathrine Wood for her contribution to the initial stages of part of this work.
Author contributions. L.P., G.P., R.F., N.H., J.P. and B.M. performed experiments; B.M., J.P. and B.C. designed and lead the study; B.M. and J.P. drafted the manuscript. All authors reviewed the manuscript.
Footnotes
Present addresses:
Lucas Philippe, Department of Cellular and Molecular Physiology, Yale University School of Medicine, 333 Cedar Street, New Haven, CT 06510, USA.
George Cherian Pandarakalam, European Screening Centre, University of Dundee, Biocity Scotland, Bo’ness Road, Newhouse ML1 5UH, Scotland, UK.
Neale Harrison, School of Biosciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [Project grant BB/J007137/1]; Medical Research Council (MRC) Confidence in Concept 2014 - University of Aberdeen Award(MC_PC_14114v.2); University of Aberdeen Elphinstone Scholarship (to R.F.) and TETFund support through Adekunle Ajasin University, Nigeria (to R.F.). Funding for open access charge: Biotechnology and Biological Sciences Research Council and Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pettitt J., Harrison N., Stansfield I., Connolly B., Müller B.. The evolution of spliced leader trans-splicing in nematodes. Biochem. Soc. Trans. 2010; 38:1125–1130. [DOI] [PubMed] [Google Scholar]
- 2. Lasda E.L., Blumenthal T.. Trans-splicing. Wiley Interdiscip. Rev. RNA. 2011; 2:417–434. [DOI] [PubMed] [Google Scholar]
- 3. Allen M.A., Hillier L.W., Waterston R.H., Blumenthal T.. A global analysis of C. elegans trans-splicing. Genome Res. 2011; 21:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blumenthal T. Trans-Splicing and Operons in C. elegans. WormBook. 2012; The C. elegans Research Community. [Google Scholar]
- 5. Blumenthal T., Evans D., Link C.D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W.L., Duke K., Kiraly M. et al. . A global analysis of Caenorhabditis elegans operons. Nature. 2002; 417:851–854. [DOI] [PubMed] [Google Scholar]
- 6. Ferguson K.C., Heid P.J., Rothman J.H.. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 1996; 10:1543–1556. [DOI] [PubMed] [Google Scholar]
- 7. Evans D., Blumenthal T.. Trans splicing of polycistronic Caenorhabditis elegans pre-mRNAs: analysis of the SL2 RNA. Mol. Cell. Biol. 2000; 20:6659–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans D., Perez I., MacMorris M., Leake D., Wilusz C.J., Blumenthal T.. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3′ end formation and trans-splicing in C. elegans operons. Genes Dev. 2001; 15:2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lasda E.L., Kuersten S., Blumenthal T.. SL trans-splicing in a C. elegans in vitro extract. Cold Spring Harb. Protoc. 2011; 2011, pdb.prot5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Doren K., Hirsh D.. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature. 1988; 335:556–559. [DOI] [PubMed] [Google Scholar]
- 11. Van Doren K., Hirsh D.. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol. Cell. Biol. 1990; 10:1769–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruzik J.P., Maniatis T.. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992; 360:692–695. [DOI] [PubMed] [Google Scholar]
- 13. Thomas J.D., Conrad R.C., Blumenthal T.. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 1988; 54:533–539. [DOI] [PubMed] [Google Scholar]
- 14. Denker J.A., Maroney P.A., Yu Y.T., Kanost R.A., Nilsen T.W.. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA. 1996; 2:746–755. [PMC free article] [PubMed] [Google Scholar]
- 15. MacMorris M., Kumar M., Lasda E., Larsen A., Kraemer B., Blumenthal T.. A novel family of C. elegans snRNPs contains proteins associated with trans-splicing. RNA. 2007; 13:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones T.A., Otto W., Marz M., Eddy S.R., Stadler P.F.. A survey of nematode SmY RNAs. RNA Biol. 2009; 6:5–8. [DOI] [PubMed] [Google Scholar]
- 17. Pettitt J., Philippe L., Sarkar D., Johnston C., Gothe H.J., Massie D., Connolly B., Müller B.. Operons are a conserved feature of nematode genomes. Genetics. 2014; 197:1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stiernagle T. Maintenance of C. elegans. WormBook. 2006; The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conrad R., Lea K., Blumenthal T.. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995; 1:164–170. [PMC free article] [PubMed] [Google Scholar]
- 21. Fr⊘kjær-Jensen C., Davis M.W., Hopkins C.E., Newman B., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M.. Single copy insertion of transgenes in C. elegans. Nat. Genet. 2008; 40:1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans T. C. Transformation and microinjection. WormBook. 2006; The C. elegans Research Community. [Google Scholar]
- 23. Kutscher L. M., Shaham S.. Forward and reverse mutagenesis in C. elegans. WormBook. 2014; The C. elegans Research Community. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Timmons L., Court DL, Fire A.. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001; 263:103–112. [DOI] [PubMed] [Google Scholar]
- 26. Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., Ahringer J.. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001; 2:doi:10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 28. Brahms H., Raymackers J., Union A., de Keyser F., Meheus L., Lührmann R.. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 2000; 275:17122–17129. [DOI] [PubMed] [Google Scholar]
- 29. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R.. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001; 7:1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reuter M., Chuma S., Tanaka T., Franz T., Stark A., Pillai R.S.. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009; 16:639–646. [DOI] [PubMed] [Google Scholar]
- 31. Barbee S.A., Lublin A.L., Evans T.C.. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr. Biol. 2002; 12:1502–1506. [DOI] [PubMed] [Google Scholar]
- 32. Li D.K., Tisdale S., Lotti F., Pellizzoni L.. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 2014; 32:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whitelaw S. An investigation of the mechanism of histone mRNA 3′ end formation in the nematode Caenorhabditis elegans. 2006; University of Aberdeen, Ph.D. Thesis. [Google Scholar]
- 34. Albrecht M., Lengauer T.. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 2004; 569:18–26. [DOI] [PubMed] [Google Scholar]
- 35. Pillai R.S., Grimmler M., Meister G., Will C.L., Lührmann R., Fischer U., Schümperli D.. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003; 17:2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B.. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001; 20:5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Godfrey A.C., White A.E., Tatomer D.C., Marzluff W.F., Duronio R.J.. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre-mRNA processing and play an essential role in development. RNA. 2009; 15:1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al. . Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003; 421:231–237. [DOI] [PubMed] [Google Scholar]
- 39. Sonnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E. et al. . Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005; 434:462–469. [DOI] [PubMed] [Google Scholar]
- 40. Maroney P.A., Yu Y.T., Jankowska M., Nilsen T.W.. Direct analysis of nematode cis- and trans-spliceosomes: a functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA. 1996; 2:735–745. [PMC free article] [PubMed] [Google Scholar]
- 41. Denker J.A., Zuckerman D.M., Maroney P.A., Nilsen T.W.. New components of the spliced leader RNP required for nematode trans-splicing. Nature. 2002; 417:667–670. [DOI] [PubMed] [Google Scholar]
- 42. Hannon G.J., Maroney P.A., Nilsen T.W.. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J. Biol. Chem. 1991; 266:22792–22795. [PubMed] [Google Scholar]
- 43. Ruegger S., Miki T.S., Hess D., Grosshans H.. The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Res. 2015; 43:3344–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A.. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002; 21:2724–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paushkin S., Gubitz A.K., Massenet S., Dreyfuss G.. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002; 14:305–312. [DOI] [PubMed] [Google Scholar]
- 46. Pellizzoni L., Yong J., Dreyfuss G.. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002; 298:1775–1779. [DOI] [PubMed] [Google Scholar]
- 47. Meister G., Eggert C., Bühler D., Brahms H., Kambach C., Fischer U.. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 2001; 11:1990–1994. [DOI] [PubMed] [Google Scholar]
- 48. Friesen W.J., Massenet S., Paushkin S., Wyce A., Dreyfuss G.. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell. 2001; 7:1111–1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.