Abstract
Among nucleic acid–based delivery platforms, self-amplifying RNA (saRNA) vectors are of increasing interest for applications such as transient expression of recombinant proteins and vaccination. saRNA is safe and, due to its capability to amplify intracellularly, high protein levels can be produced from even minute amounts of transfected templates. However, it is an obstacle to full exploitation of this platform that saRNA induces a strong innate host immune response. In transfected cells, pattern recognition receptors sense double-stranded RNA intermediates and via activation of protein kinase R (PKR) and interferon signaling initiate host defense measures including a translational shutdown. To reduce pattern recognition receptor stimulation and unleash suppressed saRNA translation, this study co-delivered non-replicating mRNA encoding vaccinia virus immune evasion proteins E3, K3, and B18. It was shown that E3 is far superior to K3 or B18 as a highly potent blocker of PKR activation and of interferon (IFN)-β upregulation. B18, in contrast, is superior in controlling OAS1, a key IFN-inducible gene involved in viral RNA degradation. By combining all three vaccinia proteins, the study achieved significant suppression of PKR and IFN pathway activation in vitro and enhanced expression of saRNA-encoded genes of interest both in vitro and in vivo. This approach promises to overcome key hurdles of saRNA gene delivery. Its application may improve the bioavailability of the encoded protein, and reduce the effective dose and correspondingly the cost of goods of manufacture in the various fields where saRNA utilization is envisioned.
Keywords: : self-amplifying RNA, replicon, vaccinia virus E3, vaccinia virus K3, vaccinia virus B18, alphavirus
Introduction
Self-amplifying RNA (saRNA, also called “replicon RNA”) is engineered from genomes of plus-strand RNA viruses such as alphaviruses or flaviviruses. saRNA resembles mRNA; it is single-stranded, 5′-capped, and 3′-poly-adenylated and is of positive orientation. saRNA encodes an enzyme complex for self-amplification (replicase polyprotein) comprising an RNA-dependent RNA-polymerase function, helicase, capping, and poly-adenylating activity. The viral structural genes downstream of the replicase, which are under control of a subgenomic promoter, can be replaced by genes of interest (GOI). Upon transfection, the replicase is translated immediately, interacts with the 5′ and 3′ termini of the genomic RNA, and synthesizes complementary genomic RNA copies. Those act as templates for the synthesis of novel positive-stranded, capped, and poly-adenylated genomic copies, and subgenomic transcripts (Fig. 1). Amplification eventually leads to very high RNA copy numbers of up to 2 × 105 copies per cell.1 Thus, much lower amounts of saRNA compared to conventional mRNA suffice to achieve effective gene transfer and protective vaccination.2
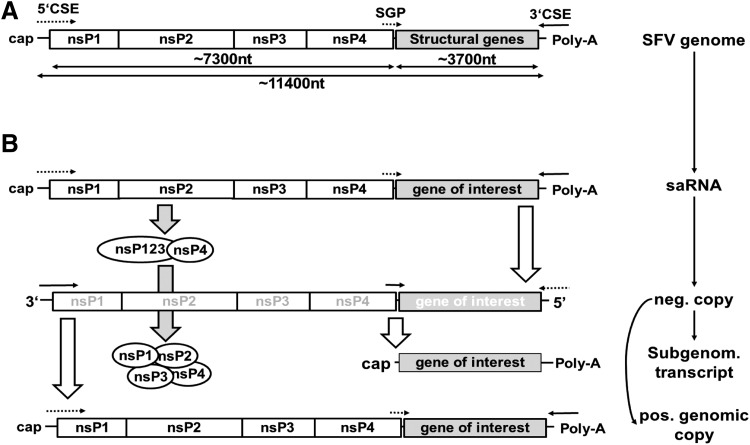
Figure 1.
Self-amplifying RNA vector structure and amplification. (A) Organization of the alphaviral genome of the Semliki Forest virus. The genomic RNA is positive-sensed, single-stranded RNA that encodes the non-structural polyproteins (nsP1–nsP4; replicase) at the 5′ end and structural genes (capsid and glycoproteins) at the 3′ end. The 3′ORF is replaced in saRNA with genes of interest under the transcriptional control of a subgenomic promoter (SGP). Conserved sequence elements (CSE) at the 5′ and 3′ end act as promoters for minus-strand and positive-strand RNA transcription. (B) Mechanism of self-replication. After transfection, the non-structural polyprotein precursor (nsP1234) is translated from in vitro transcribed saRNA. nsP1234 is at early stages auto-proteolytically processed to the fragments nsP123 and nsP4, which transcribes negative-stranded copies of the saRNA. Later, nsP123 is completely processed to single proteins, which assemble to the (+)strand replicase to transcribe new positive-stranded genomic copies, as well as (+)stranded subgenomic transcripts that code for the gene of interest. Subgenomic RNA as well as new genomic RNA is capped and poly-adenylated. This simplified scheme neglects that replication takes place at membrane invagination formed by nsP. Inactive promoters are dotted arrows; active promoters are lined arrows.
During saRNA translation, double-stranded RNA (dsRNA) intermediates are formed, which are natural ligands of cytoplasmic RNA sensors such as Rig-I, MDA5, and protein kinase R (PKR). This interaction initiates the release of interferons and activation of interferon response genes, and culminates in a cascade of innate immunity-related mechanisms.3 This strong intrinsic adjuvant activity of saRNA contributes to its higher immunogenicity at lower doses compared to not self-amplifying in vitro transcribed single-stranded mRNA.4,5 Activation of cytoplasmic RNA sensors, in particular of PKR, however, comes with the downside of a general inhibition of translation. Activated PKR phosphorylates the eukaryotic initiation factor 2 alpha subunit (eIF2α), thereby blocking cap-dependent translation,6 including that of saRNA.7 As a counter-mechanism to rescue translation, alphaviruses evolved an RNA stem-loop structure downstream of the capsid start codon (downstream loop, DLP) spanning the 5′-terminal 102 nucleotides (34 amino acids) of the capsid ORF, providing eIF2α independent translation.8,9 However, after replacement of the capsid ORF by a GOI, the resulting recombinant saRNA lacks a DLP and regains sensitivity toward activated PKR. Since a fusion of the DLP spanning part of the capsid to the GOI bears the risk of a functional alteration, alternative ways of rescuing the saRNA's translational efficiency are required.
In contrast to alphaviruses, most other viruses evade cellular immunity using a plethora of viral proteins counteracting cytoplasmic RNA sensors. Vaccinia virus (VACV) for instance expresses specialized immune evasion proteins, for example the PKR inhibitor and dsRNA binding protein E3, the PKR pseudo-substrate K3, and the interferon (IFN)-decoy receptor B18.10 It has been demonstrated previously that co-transfer of mRNA encoding VACV proteins E3, K3, and B18 (EKB) substantially improves the expression of synthetic non-replicating mRNA,11 and this approach has been commercialized for the generation of induced pluripotent stem cells. Recently, the influenza virus multifunctional immune evasion protein NS1 was used in a similar approach to enhance non-replicating synthetic mRNA expression.12
The objective of this study was to test EKB proteins for their capability to release dsRNA-mediated stalling of translation of saRNA-encoded GOI and to optimize this approach. For gene delivery of the VACV proteins, optimized non-replicating in vitro transcribed mRNA13,14 encoding each protein individually was used rather than co-expressing them from a second subgenomic transcript of the saRNA.15 This avoided inconveniently complex and long saRNA vectors, increased potential safety for clinical application, and was much more versatile.
This study shows that the co-transfer of synthetic mRNA encoding VACV PKR and IFN inhibitors improves saRNA expression by more than one order of magnitude and thereby helps overcome the major obstacles for translational application.
Material and Methods
Cell culture
All growth media, fetal calf serum (FCS), antibiotics, and other supplements were supplied by Life Technologies/Gibco, except when stated otherwise. Human foreskin fibroblasts obtained from System Bioscience (HFF, neonatal) or ATCC (CCD-1079Sk) were cultivated in minimum essential media (MEM) containing 15% FCS, 1 IU/mL of penicillin, 1 μg/mL of streptomycin, 1% non-essential amino acids, and 1 mM of sodium pyruvate at 37°C. Cells were grown at 37°C in a humidified atmosphere equilibrated to 5% CO2. BHK21 cells (ATCC; CCL10) were grown in Eagle's MEM supplemented with 10% FCS.
Animals
Balb/c_Rj mice, 6–8 weeks of age, were purchased from Janvier Labs and housed under normal laboratory conditions with circadian light/dark cycles and standard feeding. Animal experiments were approved by the Regional Council's Ethics Committee for Animal Experimentation (Koblenz, Rhineland-Palatinate, Germany).
RNA vectors and in vitro transcription
Template plasmid for in vitro transcription of mRNA was derived from pST1-2hBgUTR-A120 backbone14 that contain a tandem repeat of the human β-globin UTR, which stabilized mRNA followed by a poly-A stretch of 120 nucleotides. pST1 vectors encoding eGFP or firefly luciferase have been previously described.13,14 VACV genes E3, K3, and B18 were synthesized and codon-adapted to human cells (Geneart) and inserted into the multiple cloning site of the vector. The DLP–GFP fusion gene was constructed by fusing the 5′ terminal 102 nucleotides of the Semliki Forest virus capsid gene to GFP, separated by a T2A self-cleavage peptide. saRNA vector pSFV2gen derived from Semliki Forest virus isolate L10, clone SFV4 (accession number AJ251359) was kindly provided by Kenneth Lundström.16 SP6 was substituted by a T7 polymerase promoter using polymerase chain reaction (PCR)-based seamless cloning techniques (Cold Fusion; System Biosciences) and the 3′ poly-A stretch was extended to 120 nucleotides followed by a type-IIS restriction enzyme site (SapI) for linearization within the poly-A. In vitro RNA synthesis and purification were previously described using a synthetic cap analogue that provides superior translational efficiencies.13,14,17 Concentration, purity, and integrity of the RNA was assessed by spectrophotometry (NanoDrop 2000c; PeqLab) and on-chip electrophoresis (2100 BioAnalyzer; Agilent), respectively.
In vitro RNA transfection
RNA (mRNA and saRNA) was either electroporated or lipofected into cells. Electroporation was done at room temperature by applying defined pulses with a square-wave electroporator (BTX ECM 830; Harvard Apparatus); BHK21 (750 V/cm; one pulse of 16 ms), human fibroblasts (800 V/cm; one pulse of 24 ms). For lipofection, RNA was complexed with RNAiMAX or MessengerMax solution (Life Technologies) in a 1:4 ratio (w/v) and diluted with 200 μL of serum-free medium per microgram of RNA, following the manufacturer's instructions (5 μg/mL of final RNA concentration). To target cells cultured overnight at 2–4E + 04 cells/cm2 in 250 μL/cm2 of growth medium without antibiotics, 50 μL/cm2 of lipofection mixture was added and maintained until analysis.
Intramuscular injections and in vivo bioluminescence imaging
Balb/c_Rj mice were anesthetized by isoflurane inhalation (Abbott) before injecting 20 μL of premixed RNA in RNAse-free PBS (Life Technologies) into the musculus tibialis posterior or anterior. Bioluminescence imaging was performed following intraperitoneal injection of 100 mg/kg of body weight D-luciferin (PerkinElmer) into anesthetized mice using an IVIS® Spectrum imaging system (PerkinElmer), collecting emitted photons for 1 min. Total bioluminescence intensity from the muscular region of interest was quantified using Living Image software (PerkinElmer).
Luciferase assay
Luciferase assays with transfected cells were performed with the Bright-Glo Luciferase Assay System (Promega), according to the manufacturer's instructions. Bioluminescence (photons per second [p/s]) was captured using a microplate luminescence reader Infinite M200 (Tecan Group).
Flow cytometry
For flow cytometric analysis of fluorescent protein expression, the cells were harvested, washed with PBS, and fixed with PBS containing 2% formaldehyde. Expression of fluorescent proteins was assessed using FACS Canto II flow cytometer (BD Biosciences) and the companion Diva software.
Western blots
Cell lysates were prepared using Laemmli buffer18 and separated with pre-casted gels (4–12% NuPage Bis-Tris Gels; Life Technologies) and blotted onto nitrocellulose membranes using the XCell II Blot Module, solutions and procedure of the manufacturer (Life Technologies). Western blots were performed according to widely established protocols. The following antibodies were used: anti-eIF2α (9722), anti Phospho-eIF2α (9721; both Cell Signaling Technologies); anti PKR (ab45427), anti Phospho-PKR (ab32036), and rabbit anti-beta actin (ab75186; all Abcam).
Quantitative real-time reverse transcriptase PCR
Total cellular RNA was extracted using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions, and quantified by spectroscopy (NanoDrop 2000c; PeqLab). Total RNA (0.5–5 μg) was reverse transcribed with Superscript II reverse transcriptase (Invitrogen). The primer for first-strand synthesis was oligo-dT18 for subsequent quantification of endogenous transcripts. Quantitative real-time reverse transcriptase PCR was performed in triplicate using the ABI 7300 real-time PCR system, the companion SDS analysis software (Applied Biosystems), and the QuantiTect SYBR Green PCR Kit (Qiagen). Protocol followed the manufacturer's instruction, with 15 min at 95°C, and 40 cycles of 30 s at 94°C, 30 s at oligo-specific annealing temperature stated below, and 30 s at 72°C. Analysis was performed using the 2–ΔΔCT method,19 normalized to the housekeeping gene HPRT. The following specific primers and annealing temperatures were used for amplification: IFN-β, forward: 5′-AAGGCCAAGGAGTACAGTC-3′, reverse: 5′-ATCTTCAGTTTCGGAGGTAA-3′ (60°C); OAS1, forward: 5′ AGGTGGTAAAGGGTGGCTCC-3′, reverse: 5′-GGGTTAGGTTTATAGCCGCC-3′ (60°C); HPRT, forward: 5′-TGACACTGGCAAAACAATGCA-3′, reverse: 5′-GGTCCTTTTCACCAGCAAGCT-3′ (60°C); GFP, forward: 5′-ATCCGCCACAACATCGAGGAC-3′, reverse: 5′-CTCCAGCAGGACCATGTGATC-3′ (62°C).
Results
Co-transfection of mRNA-encoded VACV immune evasion proteins prevents saRNA-mediated PKR activation and IFN response
To assess the magnitude of PKR- and IFN-mediated translation inhibition, GFP-encoding saRNA was transfected into BHK21 cells, which are deficient of both pathways,8,20 and into primary human foreskin fibroblasts (HFF), which have an intact IFN response and express PKR.21 Twenty percent of HFF cells expressed GFP upon transfection with 2 μg GFP-encoding saRNA, while only 0.02 μg of saRNA (one-hundredth) resulted in the equivalent GFP expression in BHK21 cells (Fig. 2A). This finding confirmed the rationale of targeting PKR and IFN signaling for restoring translation efficiency of saRNA delivered GOI.
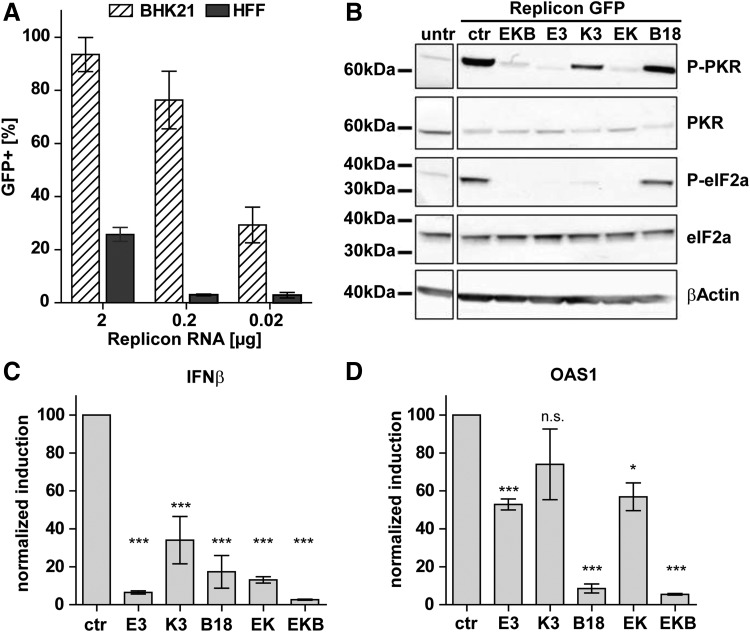
Figure 2.
Co-transfection of mRNA-encoded vaccinia virus (VACV) immune evasion protein E3 prevents saRNA-mediated protein kinase R (PKR) activation and interferon (IFN) response. (A) BHK21 cells and human foreskin fibroblasts (HFF) were lipofected with GFP-reporter-encoding saRNA. Total RNA amounts were adjusted to 2.5 μg using irrelevant infrared fluorescent protein (iRFP) and luciferase encoding mRNA. GFP expression was measured by flow cytometry 1 day after lipofection (mean of three experiments ± standard error of the mean [SEM]). (B) HFF were electroporated with 2.5 μg of GFP-saRNA RNA and in total 2 μg of in-vitro transcribed mRNA encoding VACV proteins E3, K3, or B18 or combinations thereof: E3 and K3 (EK) or all three (EKB). Each sample was in addition spiked with 2.5 μg of iRFP-mRNA to control success of electroporations. Controls (ctr) were GFP-saRNA RNA spiked with 2.5 μg of iRFP-mRNA. Cells were lysed 8 h after transfection, and Western blots were performed to detect phosphorylated (P-PKR) and total PKR, P-eIF2α and total eIF2α, and βactin. (C and D) HFF were co-lipofected with 0.75 μg of GFP-saRNA RNA, 0.5 μg of iRFP-mRNA, and a total amount of 1.25 μg of mRNAs encoding VACV proteins E3, K3, or B18 or combinations. Controls (ctr) were co-transfected with 1.25 μg of luciferase-mRNA instead of VACV proteins. Cells were harvested 24 h after transfection for cDNA extraction and quantitative real-time reverse transcriptase polymerase chain reaction analysis of IFNβ- and OAS1 transcript levels. Transcript levels were normalized to controls (mean of three independent experiments ± SEM). Statistical analysis was performed by one-way analysis of variance (ANOVA). Significance is given with respect to controls (n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.001).
The transfection of HFF cells with GFP saRNA resulted in the strong phosphorylation of PKR and consecutively of eIF2α, known to be associated with translational shutoff (Fig. 2B). However, co-transfection of VACV E3 encoding mRNA, either alone or combined with K3 and B18, prevented phosphorylation of both PKR and eIF2α. Co-transfected K3 mRNA alone reduced PKR auto-phosphorylation less effectively, but prevented eIF2α phosphorylation, while B18 had no detectable effect on the phosphorylation of PKR and eIF2α. Transfer of GFP saRNA into HFF cells strongly induced IFN-β and OAS1, the latter being a key IFN-response gene involved in viral RNA degradation (Fig. 2C and D). E3 mRNA co-transfection robustly reduced IFN-β induction by approximately 90%. K3 and B18 reduced IFN-β transcript levels by >60%. Co-transfection of all three mRNAs had the strongest effect in reducing IFN-β induction by 95% (Fig. 2C). E3 (alone or together with K3) reduced OAS1 induction by 50%. It was noteworthy that B18 inhibited OAS1 induction by >90% (Fig. 2D), showing that it is released in sufficient amounts from transfected cells to neutralize secreted IFN. Thus, B18 and E3 synergized to neutralize both of the most prominent pathways of the host cell's innate immune response. Despite a significant drop of IFN expression, no improvement of cell viability by E3 was observed, indicating that IFN response plays a minor role in saRNA-induced cell death (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/hum).
In conclusion, the data show that as a single agent, E3 mRNA, is most potent in inhibiting PKR activation and IFN induction, while combining it with B18 and K3 further strengthens the inhibition.
In vitro expression of genes transferred by saRNA is highly significantly augmented by co-transfection of mRNA encoding the VACV immune evasion protein E3
Next, the study tested the effect of VACV immune evasion proteins on transfection of saRNA and on expression of the encoded GOI in vitro. To assess how many transfected HFF cells expressed GFP encoded by saRNA, mRNA encoding infrared fluorescent protein (iRFP) was spiked into each sample. In the absence of VACV proteins, about 8% of iRFP-positive cells expressed GFP from saRNA at detectable levels (Fig. 3A). Co-transfection of E3 mRNA tripled the saRNA expression rate, while K3 and B18 mRNA had no or a small effect. Combining E3 with K3 and B18 had a synergistic effect, and transfection of all three mRNAs yielded the highest frequency of GFP-positive cells, significantly above controls without EKB. Another parameter of relevance is the mean fluorescence intensity (MFI) of GFP in saRNA-transfected cells, which reflects the GFP copy number and thus the translation rate of the encoding RNA. A 12-fold increase of the GFP MFI was achieved by E3 co-transfection compared to transfecting with saRNA alone. Again, co-transfection of K3 or B18 alone had no significant effect, and no further improvement was detectable after combining all three factors (Fig. 3B). A similar although less pronounced improvement was also found in mouse and human muscle cells (Supplementary Fig. S1B). These findings were also confirmed by fluorescence microscopy (Fig. 3C). In addition to an increased translation, a trend toward greater levels of plus-stranded vector RNA was also observed in the presence of VACV proteins except for B18 (Supplementary Fig. S1C). This indicates that saRNA replication increases as well, but the more robust benefit was found at the level of translation. However, expression of saRNA was not prolonged by E3 (data not shown), possibly related to the unchanged cytotoxicity of saRNA.
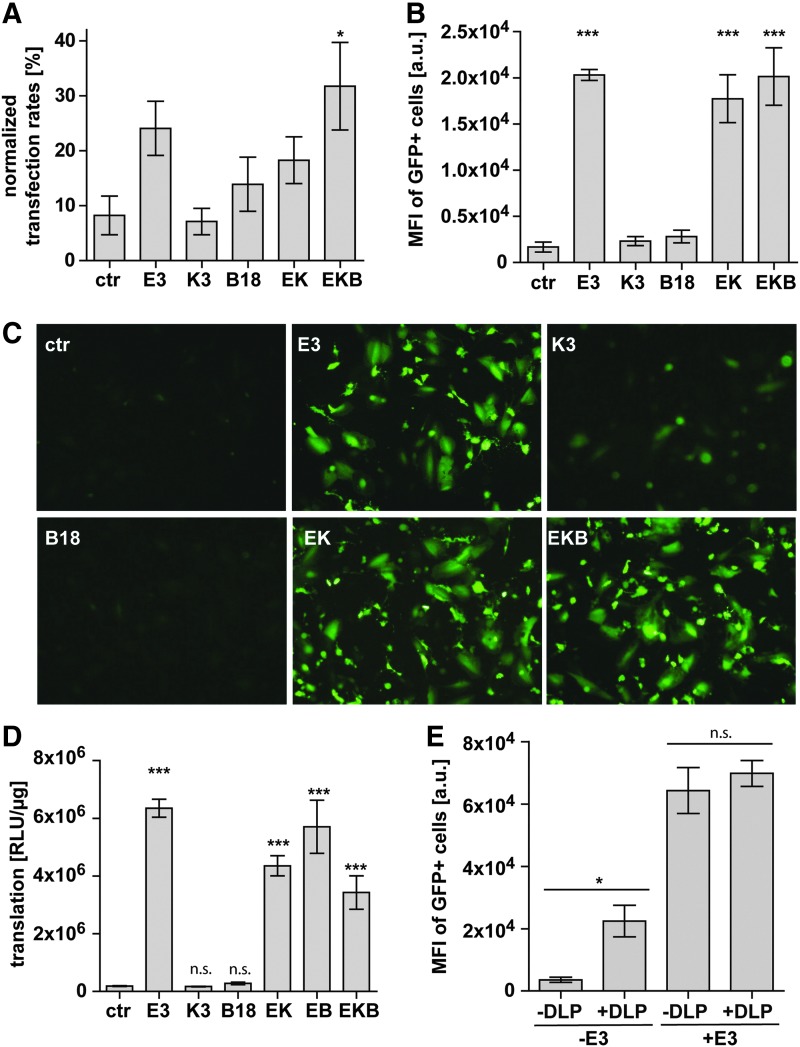
Figure 3.
In vitro expression of genes transferred by saRNA is highly significantly augmented by co-transfection of mRNA encoding the VACV PKR inhibitor E3. (A–C) Transfection efficiency of saRNA. HFF cells were co-lipofected with 0.75 μg of GFP-saRNA RNA, 0.5 μg of iRFP-mRNA, and a total of 1.25 μg of mRNA encoding single VACV proteins or combinations thereof. Controls (ctr) were co-transfected with 1.25 μg of luciferase-mRNA instead of VACV proteins, and iRFP and GFP expression were analyzed by flow cytometry 1 day later. Fraction (A) and mean fluorescence intensity (MFI) (B) of iRFP-positive cells that express GFP (mean of three experiments ± SEM) and representative examples of fluorescence microscopy of transfected cells (C). (D) Total protein translation from saRNA RNA. HFF were co-transfected with saRNA encoding secretable NanoLuc® and mRNA encoding VACV proteins. NanoLuc® accumulation in the cell culture supernatant was quantified 24 h after transfection to calculate cumulative protein translation per microgram of RNA (RLU/μg). (E) Comparison of E3 and DLP assisted saRNA expression. saRNA encoding either GFP as GOI or GFP fused to the viral DLP element was mixed with either E3 encoding or iRFP encoding mRNA in a 1:1 ratio and co-transfected in HFF (mean of three experiments ± SEM). All statistical significance calculated by One-way ANOVA with respect to controls (n.s., non-significant; *p < 0.05; **p < 0.01; ***p < 0.001).
To compare the total protein amount translated from saRNA with or without VACV-coding mRNA, saRNA encoding a secretable version of a deep sea shrimp luciferase (Nanoluc®) was used, with a half-life of several days. It accumulates in the culture supernatant and is therefore a good surrogate for total protein translation.22 Nanoluc® accumulation in the absence of VACV proteins was low, while co-transfection of E3 resulted in a highly significant 35-fold increase. Co-transfection of K3 and B18 alone had no effect on Nanoluc® accumulation and did not synergize with E3 (Fig. 3D). Thus, E3 was investigated more closely.
E3 counteracts translation arrest by ensuring eIF2α functionality, which appears to be an efficient target to inhibit translational shutoff. An alternative mechanism used by alphaviruses is overcoming of translation from eIF2α dependency via an RNA stem-loop structure downstream of the capsid start codon (DLP). The Semliki Forest virus DLP N-terminally fused to GFP was compared to the unmodified GFP saRNA in the context of E3 mRNA cotransfection. DLP enhanced GFP expression sixfold, whereas E3 co-expression alone resulted in an 18-fold higher GFP intensity. When E3 co-transfer prevented eIF2α phosphorylation, the DLP could no longer exert its benefit (Fig. 3E).
Taken together, these data indicate that E3 efficiently augments expression and protein production of saRNA-encoded GOI. Furthermore, the expression rate after transfection of saRNA benefits strongly from combining E3 with K3 and B18.
VACV immune evasion proteins substantially enhance expression of saRNA-encoded genes in mice in a dose-dependent manner
To test whether the improvement of saRNA-encoded GOI expression mediated by VACV immune evasion proteins in cell culture can be reproduced in vivo, immunocompetent BALB/c mice were used.
It has been shown in vitro that of the tested proteins, E3 had the highest potency of counteracting PKR and IFN signaling mediated translational arrest, and that the addition of B18 and K3 could incrementally improve this effect. Furthermore, it had been observed that for reversion of induction of the IFN response gene OAS1, B18 was more potent. Therefore, all three VACV proteins were combined for the in vivo setting.
RNAs were injected intramuscularly into the thigh either with firefly luciferase-encoding saRNA alone, or co-injected with an excess amount of the combination of E3, K3, and B18 mRNA (EKB) at two doses. In control mice, a signal was barely detectable by in vivo bioluminescence imaging. In mice co-injected with EKB, however, high-intensity luminescence signals were detectable at the injection sites and were sustained for >2 weeks (Fig. 4). The maximum intensity and measurable duration of expression were dependent on the dose level of EKB mRNA.
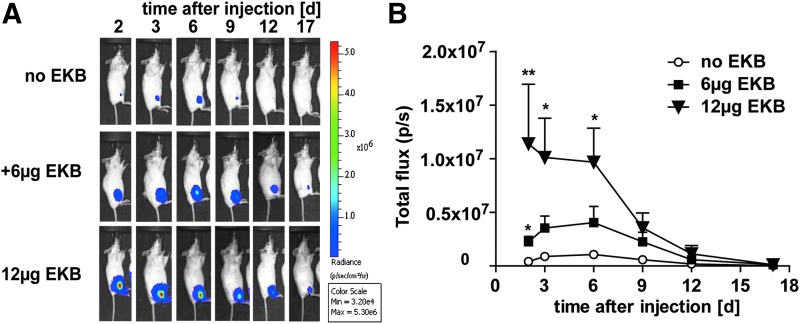
Figure 4.
VACV immune evasion proteins substantially enhance expression of saRNA-encoded genes in mice in a dose-dependent manner. Balb/c_Rj mice (five per group) were injected into the thigh muscle with 2 μg of luciferase-encoding saRNA in phosphate-buffered saline mixed with a total of either 6 μg or 12 μg of E3, K3, and B18 mRNAs (EKB) in equal amounts. Control mice received luciferase-encoding saRNA only (no EKB group). Bioluminescence was measured over 17 days at indicated time points. (A) Bioluminescence imaging of one representative animal of each group (n = 5). (B) Time course of mean luciferase expression per group ± SEM, total detected luminescence signal of each injection site was quantified (*p < 0.05; **p < 0.01 determined by two-way ANOVA with respect to control mice).
In conclusion, VACV immune evasion protein encoding mRNA is able to prevent suppression of translation in vivo upon intramuscular saRNA gene delivery.
Discussion
This study demonstrates that the expression of saRNA is substantially improved by co-transferring mRNA encoding VACV proteins inhibiting interferons and PKR. By comparison of saRNA translation in BHK21 and HFF cells, it is demonstrated that the impact of PKR and IFN activation on protein translation is in the range of two orders of magnitude and thus substantial, even if accounting for the different growth kinetics and physiology of the two cell lines.8,20
To counteract the host cell's innate immune defense mechanisms, the three VACV innate immune evasion proteins E3, K3, and B18 were chosen because they could potentially synergize by interfering with different pathways of the host cell's innate immune response. E3 alone was surprisingly effective at inhibiting PKR and did not act synergistically with K3, as has been previously described.23 E3 is known to sequester dsRNA, thereby eliminating an early trigger of the defense cascade not only for PKR but also for MDA5, another pivotal cytoplasmic sensor. Moreover, by complex formation, E3 affects PKR homodimerization,24 a prerequisite for its activation, and it was shown to inhibit OAS1 directly.25 K3 as an eIF2α mimic reduced the phosphorylation of eIF2α to some extent, but apparently not enough to unleash the full potential of saRNA translation. B18 is a type I IFN binding protein and, as expected, it did not alter PKR and eIF2α phosphorylation. It was found that B18 very effectively neutralized co-secreted IFNs and thus eliminated interferon receptor signaling and induction of IFN responsive genes such as OAS1. Even though the single-agent effects of K3 and B18 on reporter gene translation and protein production were marginal in vitro, as compared to E3, it was found that when all three were combined, K3 and B18 added an incremental improvement of saRNA translation. Therefore, the three VACV immune evasion proteins were combined for the subsequent in vivo setting.
By adding E3, a tripling of the expression rate and a 10- (GFP) to 35-fold (NanoLuc®) increase in translation was achieved. Secreted NanoLuc® accumulates over time in the cell culture supernatant and thus reflects total translation, explaining the more pronounced increase of Nanoluc® compared to GFP. Thus, the data are in line with a previous study, which described a 20-fold increase of translation for dominant-negative PKR co-expressed from a second subgenomic promoter.9 Whether the maximum achievable expression has been reached with the approach presented here remains to be studied. Providing E3 alone appears to inhibit PKR completely, as this measure surpassed the effect of the alphaviral DLP, supposedly because cap-dependent translation of the replicase also benefits from E3. The enhancement of subgenomic RNA levels also suggests that replicase translation benefits from E3 co-transfection.
The use of EKB in this study is meant to be rather exemplary than exclusive. Recently, it was shown that non-replicating mRNA translation was enhanced by 2 logs using Influenza virus NS1.12 Viruses have developed a plethora of different immune evasion proteins and mechanisms to hijack the host translation machinery, which may be worth testing in similar settings.
The observation that the blend of three VACV proteins enhanced expression in vivo is a good rationale to study the effect of VACV proteins on immune response in upcoming experiments. Increased expression of antigens should result in stronger immune responses, while a reduced innate immune response may counteract it. Thus, to prepare for a possible clinical application, a set of complex in vivo experiments are required to assess beneficial dose ranges and ratios of the different VACV proteins.
In summary, this study demonstrates that mRNA based co-delivery of viral immune evasion proteins results in a log scale increase of saRNA translation. This RNA-based approach can ease the tasks of producing sufficient amounts of vaccines for large cohorts of patients, for example in the case of recurrent viral infections such as influenza or emerging pathogens with pandemic potential such as Ebola or Zika.
Supplementary Material
Acknowledgments
We thank Anna-Lena Baumgarten, Tina Hempel, Silke Brill, Natalie Krimmel, and Alexandra König for excellent technical support, and Annette Vogel and Richard Rae for critically reviewing the manuscript. We thank Kenneth Lundström for providing the SFV vector backbone and his advice for handling saRNA.
Author Disclosure
U.S., T.B., L.K., M.D., S.K., and Ö.T. are inventors on patents and patent applications, which cover parts of this article. S.E., U.S., and K.C.W. are employees at BioNTech AG (Mainz, Germany). M.P. was an employee at BioNTech AG (Mainz, Germany). Ö.T. was employed at and U.S. was consultant at Ganymed Pharmaceuticals. U.S. and Ö.T. were stock owners of Ganymed Pharmaceuticals AG. A.S. has no potential conflicts of interest.
References
- 1.Lundstrom K. Biology and application of alphaviruses in gene therapy. Gene Ther 2005;12:S92–S97 [DOI] [PubMed] [Google Scholar]
- 2.Geall AJ, Verma A, Otten GR, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A 2012;109:14604–14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNab F, Mayer-Barber K, Sher A, et al. Type I interferons in infectious disease. Nat Rev Immunol 2015;15:87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leitner WW, Hwang LN, deVeer MJ, et al. Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat Med 2003;9:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brito LA, Chan M, Shaw CA, et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol Ther 2014;22:2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res 2011;31:59–70 [DOI] [PubMed] [Google Scholar]
- 7.Ventoso I, Sanz MA, Molina S, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 2006;20:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventoso I. Adaptive changes in alphavirus mRNA translation allowed colonization of vertebrate hosts. J Virol 2012;86:9484–9494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorchakov R, Frolova E, Williams BR, et al. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J Virol 2004;78:8455–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res 2009;29:581–598 [DOI] [PubMed] [Google Scholar]
- 11.Poleganov MA, Eminli S, Beissert T, et al. Efficient reprogramming of human fibroblasts and blood-derived endothelial progenitor cells using nonmodified RNA for reprogramming and immune evasion. Hum Gene Ther 2015;26:751–766 [DOI] [PubMed] [Google Scholar]
- 12.Phua KKL, Liu Y, Sim SH. Non-linear enhancement of mRNA delivery efficiencies by influenza A derived NS1 protein engendering host gene inhibition property. Biomaterials 2017;133:29–36 [DOI] [PubMed] [Google Scholar]
- 13.Kuhn AN, Diken M, Kreiter S, et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 2010;17:961–971 [DOI] [PubMed] [Google Scholar]
- 14.Holtkamp S, Kreiter S, Selmi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006;108:4009–4017 [DOI] [PubMed] [Google Scholar]
- 15.Domingo-Gil E, Toribio R, Najera JL, et al. Diversity in viral anti-PKR mechanisms: a remarkable case of evolutionary convergence. PLoS One 2011;6:e16711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrengruber MU, Lundstrom K. Semliki Forest virus and Sindbis virus vectors. Curr Protoc Hum Genet 2002;Chapter 12, Unit 12.2 [DOI] [PubMed]
- 17.Kreiter S, Konrad T, Sester M, et al. Simultaneous ex vivo quantification of antigen-specific CD4+ and CD8+ T cell responses using in vitro transcribed RNA. Cancer Immunol Immunother 2007;56:1577–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson JO, Ostwald K, Kabjorn C, et al. A method for protein assay in Laemmli buffer. Anal Biochem 1994;219:144–146 [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 20.Chinsangaram J, Piccone ME, Grubman MJ. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J Virol 1999;73:9891–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010;7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall MP, Unch J, Binkowski BF, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 2012;7:1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies MV, Chang HW, Jacobs BL, et al. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol 1993;67:1688–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romano PR, Zhang F, Tan SL, et al. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol 1998;18:7304–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas C, Gil J, M[ebreve]lková Z, et al. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 1998;243:406–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.