Abstract
RNA serves many functions in biology such as splicing, temperature sensing, and innate immunity. These functions are often determined by the structure of RNA. There is thus a pressing need to understand RNA structure and how it changes during diverse biological processes both in vivo and genome-wide. Here, we present Structure-seq2, which provides nucleotide-resolution RNA structural information in vivo and genome-wide. This optimized version of our original Structure-seq method increases sensitivity by at least 4-fold and improves data quality by minimizing formation of a deleterious by-product, reducing ligation bias, and improving read coverage. We also present a variation of Structure-seq2 in which a biotinylated nucleotide is incorporated during reverse transcription, which greatly facilitates the protocol by eliminating two PAGE purification steps. We benchmark Structure-seq2 on both mRNA and rRNA structure in rice (Oryza sativa). We demonstrate that Structure-seq2 can lead to new biological insights. Our Structure-seq2 datasets uncover hidden breaks in chloroplast rRNA and identify a previously unreported N1-methyladenosine (m1A) in a nuclear-encoded Oryza sativa rRNA. Overall, Structure-seq2 is a rapid, sensitive, and unbiased method to probe RNA in vivo and genome-wide that facilitates new insights into RNA biology.
INTRODUCTION
RNA structure influences numerous biological processes (1). Many of these can be informed via a global RNA structurome and thus genome-wide information on RNA structure is highly valuable. High-throughput methods provide an efficient, cost-effective alternative to classical one-off gene-specific, typically gel-based studies of RNA structure. Recently, several high-throughput RNA structural methods have been developed (1–4). Among these methods, Structure-seq, the method we developed (5,6), has some advantages in experimental and computational pipelines. Most importantly, because Structure-seq relies on chemical modification rather than nuclease cleavage, it can be performed in vivo, which is significant as in vivo and in vitro structures often differ (7). The experimental approach of Structure-seq has an advantage over other protocols in that reverse transcription (RT) is conducted immediately after RNA purification to minimize RNA degradation. Structure-seq also provides a powerful, user-friendly computational pipeline called StructureFold (8).
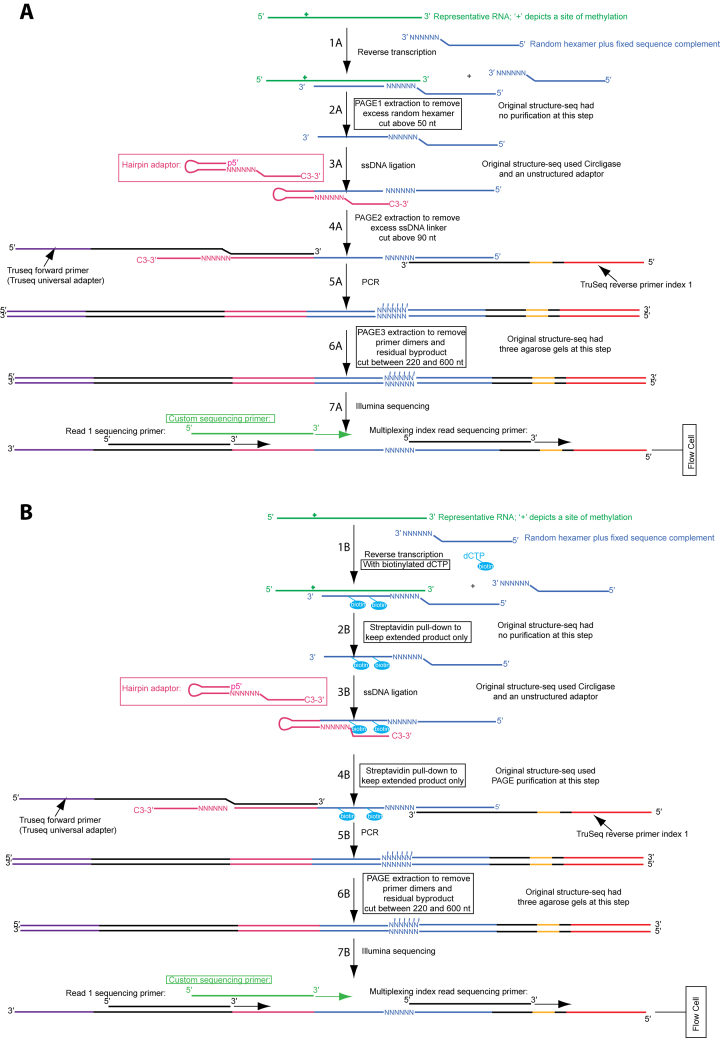
In our original Structure-seq method (6), we probe RNA in vivo with dimethyl sulfate (DMS), which covalently modifies unprotected adenines and cytosines. After RNA extraction and mRNA enrichment, reverse transcription (RT) with a random hexamer-containing primer is performed, which stops at the nucleotide before the modified nucleotide. After adaptor ligation to the cDNA 3′ end, the product is PCR-amplified and sequenced. The RT stop signal of a minus DMS sample is subtracted from that of the plus DMS sample and reactivities are calculated which can be used as restraints to predict RNA structures genome-wide (9). While Structure-seq is powerful, we identified steps that could be improved. Herein, we describe Structure-seq2 (Figure 1, Supplementary Figure S1), and demonstrate its applicability using a new species, rice (Oryza sativa). In Structure-seq2, the amount of starting material needed is reduced from 2000 to 300–500 ng poly(A)-selected RNA, a different ligation method is used, and two additional denaturing PAGE gels are introduced (Figure 1). To circumvent the time requirement and cost of these gels, we also developed a variation that utilizes streptavidin pulldown of biotinylated dCTP incorporated during RT, which streamlines the protocol.
Figure 1.
Two versions of Structure-seq2 produce high quality data. In Structure-seq2, RNA (kelly green) is first modified by DMS or another chemical that can be read-out through reverse transcription. The RNA is then prepared for Illumina NGS sequencing by conversion to cDNA (Step 1A/1B, blue), ligating an adaptor (Step 3A/3B), and amplifying the products while incorporating TruSeq primer sequences (Step 5A/5B). In order to increase library quality, numerous improvements were made to the original Structure-seq protocol (boxed). These include performing the ligation with a hairpin adaptor and T4 DNA ligase (Step 3A/3B; pink) (10), and adding various purification steps to remove a deleterious by-product (Figure 2A). We present two options for purification: PAGE purification (A) or a biotin–streptavidin pull down (B). In the PAGE purification method, an additional PAGE purification step is added after reverse transcription (Step 2A). In the biotin–streptavidin pull down method, biotinylated dNTPs (cyan) are incorporated into the extended product during reverse transcription (Step 1B) and are purified via a magnetic streptavidin pull down after reverse transcription (Step 2B) and after ligation (Step 4B). There is also a common, final PAGE purification step following amplification (Step 5A/5B). Finally, a custom sequencing primer (light green) is used during sequencing (Step 7A/7B) to further provide high quality data. Supplementary Figure S1 is a version of this figure with all the nucleotides shown explicitly.
MATERIALS AND METHODS
Plant growth
Wild-type rice (O. sativa ssp. japonica cv. Nipponbare) was used in this study. Rice seeds were sown on wet filter paper in a petri dish for germination in a greenhouse with a 16 h/8 h day/night photoperiod. Light intensity was 500 μmol m−2 s−1 with daytime temperatures of 28–32°C and nighttime temperatures of 25–28°C. After 4–5 days, the rice seedlings were transferred to 6 × 6 in. nursery pots with water saturated soil (Metro Mix 360 growing medium, Sun Gro Horticulture, Bellevue, WA, USA). Five plants were grown per pot. The plants were watered one additional time, a week after transferring to pots. The shoot tissue of two-week-old plants was used for in vivo DMS probing.
In vivo DMS treatment
All experiments involving DMS were conducted with double gloves and in a chemical fume hood. All disposables that came into contact with DMS were disposed of as hazardous waste.
Rice shoots (1 g total) were excised at the soil line and immersed in 20 ml DMS reaction buffer (100 mM KCl, 40 mM HEPES (pH 7.5), and 0.5 mM MgCl2) in a 50 ml Falcon centrifuge tube. For DMS treatment, 150 μl DMS was added (final concentration 0.75% or ∼75 mM) to the solution, and the DMS reaction was allowed to proceed for 10 min with intermittent inversion and mixing. To quench the reaction, 1.5 g of DTT was added to the solution (final concentration of 0.5 M). Vigorous vortexing was applied for 2 min. The solution was decanted from the centrifuge tube, and 50 ml of distilled deionized water was added to wash the samples. The wash step was repeated once, then the material was patted dry and immediately frozen in liquid nitrogen. A control treatment (–DMS) was performed as described, but without the addition of DMS.
RNA extraction and purification
All RNA extraction steps were done in a chemical fume hood with strong airflow (>250 fpm). Total RNA was extracted using the NucleoSpin RNA Plant kit (Macherey-Nagel, Germany) following the manufacturer's protocol. For each library, 500 micrograms total RNA comprised the starting material for one-round of poly(A) selection using the Poly(A) purist Kit (Thermo Fisher Scientific). To obtain proportionally more reads from mRNA, an additional round of poly(A) selection can be included.
Library construction
Fifteen different libraries were prepared to determine the outcomes of various modifications to the original Structure-seq method. Supplementary Table S1 highlights these changes. Two biological replicates each of Structure-seq2 –/+DMS without (Libraries 1–4) and with (Libraries 6–9) the biotin variation were made. Each of the other libraries converted one step of the protocol (Figure 1) back to what was performed in the original version of Structure-seq (5,6).
Reverse transcription
For each sample, two 20 μl reverse transcription (RT) (Figure 1, Step 1A) reactions were performed in two separate tubes each containing 250 ng (half of the total amount) of poly(A)-selected RNA. To increase coverage of primer annealing, the denaturation and annealing steps of the SuperScript III First-Strand Synthesis kit (Invitrogen) were adjusted. Namely, in the Structure-seq2 samples, the mRNA, random hexamer fused with an Illumina TruSeq Adapter, the 10× RT buffer, and the dNTP mix, were denatured at 90°C for 1 min then cooled on ice for 1 min before adding MgCl2 and DTT to a final concentration of 5 mM each. The samples were then preheated to 55°C for 1 min and the SuperScript III was added and the reaction allowed to proceed for 50 min. Each reaction contained 250 ng poly(A) RNA, 5 μM RT primer, 20 mM Tris–HCl (pH 8.4), 50 mM KCl, 0.5 mM dNTP (each), 5 mM MgCl2, 5 mM DTT and 200 U SuperScript III. The reaction was terminated by heating to 85°C for 5 min. Residual RNA was cleaved by adding 5 U of RNase H and incubating at 37°C for 20 min. Library 12 used the RT denaturation conditions from the original Structure-seq method; the RNA, and the dNTP mix were denatured at 65°C for 5 min then cooled on ice for 1 min before adding the 10× RT buffer, MgCl2 and DTT to the same final concentrations as in Structure-seq2. Library 13 tested the RT reaction temperature of the original Structure-seq method in which the RT reaction was conducted at 50°C rather than 55°C to monitor mutation rates during RT.
For the biotin variation of Structure-seq2 (libraries 6–9) and library 5, which was a control library to test the addition of biotin only (without streptavidin purification), RT was performed as in Structure-seq2, except with biotin-16-aminoallyl-2′-deoxycytidine-5′-triphosphate (TriLink BioTechnologies) doped into the reaction mixture (Figure 1, Step 1B). The final reactions contained 20 mM Tris–HCl (pH 8.0), 50 mM KCl, 5% DMSO, 0.5 mM dNTP (each), 0.125 mM biotin-dCTP, 5 mM MgCl2, 5 mM DTT, and 200 U SuperScript III.
PAGE purification
The two separate reaction tubes of each sample were combined for all samples and fractionated on a denaturing PAGE gel containing 10% acrylamide and 8.3 M urea. The gel containing the product was excised above 50 nt, according to a GeneRuler Low Range size ladder (ThermoFisher), to avoid excess RT primer (27 nt) (Figure 1, Step 2A). The excised gel piece was placed in a 50 ml Falcon tube, crushed to fine pieces, and weighed. A volume of TEN250 at least twice as much in ml as the mass of the gel piece in grams was used to submerge the gel pieces. The tube was then placed in a shaker/incubator at 37°C overnight. Ethanol precipitation was performed by first using a 0.2 μm syringe filter (PALL Scientific) to remove gel fragments and expel the buffer into a new 50 ml Falcon tube, then adding 2.5–3× the volume of 100% ice cold ethanol and 0.5 μl of GlycoBlue, and placing the tube on dry ice for at least 1 h. The sample was spun down at 12 000 g for 30 min before decanting the liquid and re-suspending the pellet with 1–2 ml 70% ice cold ethanol. The sample was spun down at 12 000 g for 5 min, the liquid was decanted, and the sample spun down for 1 min before removing the last bit of liquid with a pipette. The pellet was dried to completion in a 37°C incubator and then dissolved in 100 μl of water and transferred to a 1.7 ml Eppendorf tube. The sample was then concentrated to the proper volume for the subsequent reactions. The above RT-PAGE purification step was excluded for library 15, which tested the necessity of this gel.
Streptavidin purification
For the biotin variation, the two separate RT reaction tubes of each sample were combined and diluted to 100 μl. Phenol:chloroform extraction was performed as described in the original Structure-seq method (5). The final extraction product was purified with an illustra MicroSpinG-50 column (GE Healthcare) to remove excess dNTP and biotin-dCTP. Ethanol precipitation was performed as described previously (5) and the cDNA was dissolved in 50 μl of 1× wash/binding buffer (0.5 M NaCl, 20 mM Tris–HCl (pH 7.5), 1 mM EDTA).
During the final ethanol precipitation step, 25 μl of magnetic hydrophilic streptavidin beads were washed with 50 μl of 1× wash/binding buffer in a 1.7 ml microcentrifuge tube. A magnet was applied to pull the beads to the side of the tube, and the supernatant was pipetted off. The beads were washed two more times with 50 μl of 1× wash/binding buffer. After the final wash was discarded, the cDNA in 50 μl of 1× wash/binding buffer was added to the beads, and the beads were suspended by vortexing. The sample was incubated at room temperature for 10 min with occasional agitation by hand. A magnet was applied, and the supernatant was discarded. The beads were washed twice with 100 μl of 1× wash/binding buffer, and twice with 100 μl warm (40°C) low salt buffer (0.15 M NaCl, 20 mM Tris–HCl (pH 7.5), 1 mM EDTA). Each wash included vortexing to suspend the beads, pulse spinning to pull the solution to the bottom of the tube, applying a magnet, and pipetting off the supernatant. To elute the product from the beads, 25 μl of formamide buffer (95% formamide, 10 mM EDTA) was added to the beads, the tubes were vortexed and incubated at 95°C for 2 min, a magnet was applied and the supernatant was transferred to a clean 1.7 microcentrifuge tube. The elution was repeated with another 25 μl of formamide buffer, and the supernatant added to the first elution. The solution was diluted to 200 μl with RNase-free H2O, and ethanol precipitation was performed (Figure 1, Step 2B).
T4 DNA ligation
The ligation method was performed with T4 DNA ligase (Figure 1, Step 3A/3B) (10). After renaturing the purified cDNA with betaine, polyethylene glycol 8000 (PEG 8000) and hairpin donor (5′-pTGAAGAGCCTAGTCGCTGTTCANNNNNNCTGCCCATAGAG-3′-Spacer, where ‘5′-p’ is a 5′ phosphate and ‘3′-Spacer’ is a 3-carbon linker), 10× T4 DNA ligase buffer and T4 DNA ligase (NEB) were added to give a final 10 μl reaction mixture containing 500 mM Betaine, 20% PEG 8000, 10 μM hairpin donor, 1× T4 DNA ligase buffer, and 400 U T4 DNA ligase. The reaction proceeded at 16°C for 6 h, followed by 30°C h for 6 h, and was stopped by incubating at 65°C for 15 min. Library 11 tested the ligation method of the original Structure-seq. A 20 μl reaction containing the cDNA, 5 μM ssDNA unstructured linker (5′-pNNNAGATCGGAAGAGCGTCGTGTAG-3′-Spacer), 1× Circligase reaction buffer, 50 μM ATP, 2.5 mM MnCl2 and 100 U Circligase was incubated at 65°C for 12 h and was stopped by incubating at 85°C for 15 min.
The ligated cDNA was fractionated on a denaturing PAGE gel containing 10% acrylamide and 8.3M urea. The gel containing the product was excised above 90 nt to avoid excess hairpin donor (40 nt) and by-product (67 nt), according to GeneRuler low range DNA size ladder and custom ssDNA oligonucleotides of 67 nt and 91 nt (Figure 1, Step 4A). For the biotin variation, streptavidin purification was performed as described above (Figure 1, Step 4B).
Library amplification by PCR
PCR amplification (Figure 1, Step 5A/5B) was performed using Q5 High Fidelity DNA polymerase (NEB) and Illumina TruSeq primers (Illumina TruSeq forward primer, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGAACAGCGACTAGGCTCTTCA-3′; Illumina TruSeq reverse primers, 5′-CAAGCAGAAGACGGCATACGAGAT-barcode-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-3′ where ‘barcode’ refers to the unique 6–8 nt Illumina barcode for each sample). Reactions (25 μl) contained 1× Q5 reaction buffer, 0.2 mM dNTPs (each), 0.4 μM forward primer and 0.4 μM reverse primer and 0.5 U Q5 DNA polymerase. The samples were initially denatured at 98°C for 1 min, cycled through a denaturation step of 98°C for 8 s and an extension step of 72°C for 45 s, then subjected to a final extension step at 72°C for 10 min. Library 10 used the original Structure-seq protocol for amplification; the 25 μl reaction contained 1× Ex Taq buffer, 0.2 mM dNTPs (each), 0.2 μM forward primer and 0.2 μM reverse primer and 0.1 U Ex Taq DNA polymerase. After a PCR cycle test to determine the minimum number of cycles needed to obtain sufficient product, the amplification was completed at the selected cycle number, and the PCR product was purified via a 10% acrylamide 8.3 M urea denaturing PAGE gel to remove the by-product and obtain products between 220 and 600 nt according to a ss100 DNA ladder from Simplex Sciences (Figure 1, Step 6A/6B). Note that it is important that this gel have even heating across the entire glass plate to avoid slower migration of the DNA at the outer edges of the plate, often referred to as ‘smiling’, as this can lead to imprecise excision of the desired DNA and carry over of the by-product into sequencing (see Results). Library 14 tested this final purification using the original version of Structure-seq; the sample was extracted from three successive agarose gels instead of extracting from a PAGE gel.
Illumina sequencing
The quality of the purified libraries was evaluated by analysis on an Agilent Bioanalyzer system to evaluate the relative amounts of desired product vs. by-product, and by qPCR to quantify the concentration of each library and balance between them in order to achieve even sequencing output from the libraries. Libraries were sequenced using a MiSeq desktop sequencer (Illumina) with single-end reads of 150 bp. Approximately 20 nt are the minimum needed for accurate read mapping to the rice transcriptome, although this value may vary for other organisms, and this is the basis for cutting no closer than 20 nt above the primer.
Sequence generation, processing and mapping
Sequenced reads (150 nt) were obtained with an Illumina MiSeq. For Structure-seq2, adapters were removed computationally and reads were filtered for a quality score of >30 and a length of >20 using cutdapt (11), whereas Structure-seq used iterative mapping. Filtered reads were mapped to the rice reference cDNA and rRNA libraries (http://plants.ensembl.org/info/website/ftp/index.html) using Bowtie2 (12) (as compared to iterative Bowtie mapping in Structure-seq). Reads with a mismatch on the first 5′ nucleotide were discarded in Structure-seq2. Biological replicates were combined after validating RT stop correlation on rRNA (PAGE –DMS libraries r = 0.999; PAGE +DMS libraries r = 0.983; biotin –DMS libraries r = 0.923; biotin +DMS libraries r = 0.992) (Supplementary Figure S2A–D, respectively). When analyzing biological aspects rather than technical improvements, PAGE and biotin libraries were combined (–DMS r = 0.891; +DMS r = 0.990) (Supplementary Figure S2E and F). Raw DMS reactivities were derived using the same computational pipeline as for Structure-seq, except that 2–8% normalization was performed at the transcript level rather than at the global level as in Structure-seq (8).
RESULTS
Preparation and sequencing of optimized Structure-seq2 libraries
In this section we discuss how the libraries of Structure-seq2 are prepared and sequenced. The Structure-seq2 method is summarized in Figure 1 (nucleotide-level detail is shown in Supplementary Figure S1). Key improvements of Structure-seq2 are removal of a by-product, reduction of ligation bias, leveling out of read depth, lowering of mutation rate and improvement of sequencing quality. We then benchmark Structure-seq2 with rRNA and mRNA structure.
Removal of the deleterious by-product
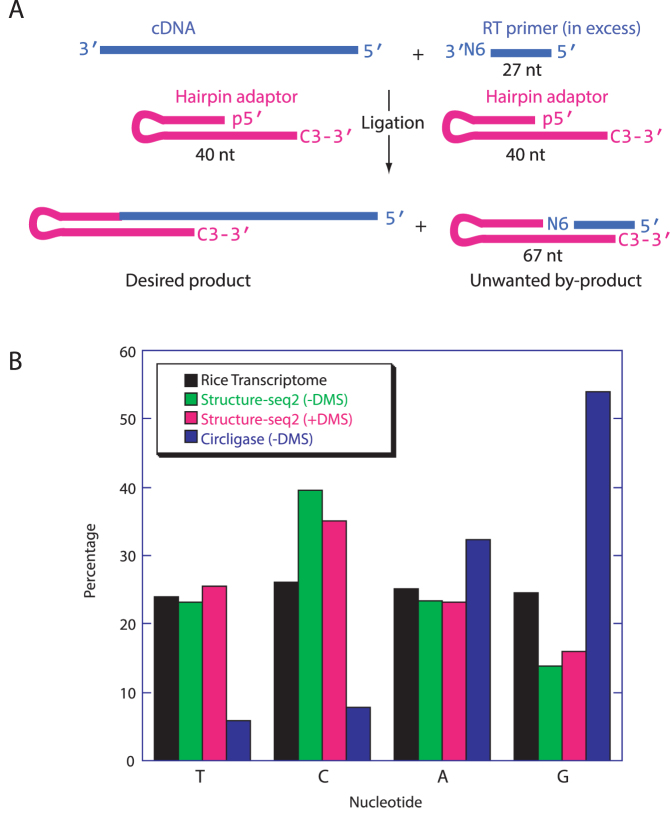
We discovered that Structure-seq leads to an undesired by-product between the RT primer and ligation adaptor (Figure 2A, Supplementary Figures S3 and S4). Because the by-product is shorter than a ligated extension product, it amplifies readily in PCR making it especially problematic. Presence of the by-product in the libraries reduces the proportion of useful reads. Previous runs with the original Structure-seq often became poisoned with the by-product such that either the desired library could not be prepared at all or the effective read rates were as low as 10–50% (data not shown). Structure-seq2 now produces results with effective read rates ∼90% (Supplementary Table S1). To minimize formation of this by-product, we perform three single nucleotide resolution PAGE purifications.
Figure 2.
Structure-seq2 leads to a lower ligation bias. (A) After RT (Figure 1, step 1A/1B), excess of the 27 nt primer (blue, top, right) is still present in the solution. During ligation (Figure 1, step 3A/3B), this primer can also ligate to the 40 nt hairpin adaptor (pink) to form an unwanted 67 nt by-product which has no insert and so results in sequencing reads with no utility. (B) The complement of the first nucleotide after the adaptor sequence read during sequencing is the nucleotide that ligated to the adaptor. Our new T4 DNA ligase-based method (green, –DMS and pink, +DMS) substantially decreases ligation bias as compared to the previous Circligase-based method (blue). Percentages equaling the transcriptomic distribution of the four nucleotides (black) are ideal.
In the first gel (Figure 1, Step 2A), excess RT primer is removed. The RT product smear is fractionated by denaturing PAGE and the gel is excised above 50 nt, which is ∼20 nt above the 27 nt RT primer. This significantly reduces by-product formation. Without the reduction in by-product afforded by this new Step 2A, the lower amount of starting RNA yields insufficient PCR product for library preparation and sequencing (Supplementary Figure S5). The next PAGE gel (Figure 1, Step 4A), which was also present in the original Structure-seq, removes excess ligation adaptor as well as any residual by-product by excising above 90 nt, which is ∼20 nt above the by-product (67 nt from the 27 nt RT primer plus the 40 nt ligation adaptor). The third PAGE gel, representing the second new PAGE gel, removes any residual by-product amplified during PCR, as well as PCR primers and any primer dimers (Figure 1, Steps 6A and 6B). This PAGE gel replaces three consecutive native agarose gels used in Structure-seq. Native agarose gels are potentially problematic because they do not offer single nucleotide resolution; moreover, we found that single-stranded nucleic acids in this protocol do not migrate true to size on lower-resolution native agarose gels (Supplementary Figure S6). Given these limitations, native agarose gel purifications have been entirely removed from Structure-seq2. Proper size selection on the third PAGE gel is 220–600 nt, which avoids the 149/151 bp by-product (Supplementary Figures S3 and S7). Notably, while PAGE gels do offer better separation of nucleic acids than agarose gels, we found that imprecise cutting at this third PAGE gel step will result in a lower effective sequencing rate. This is due to the fact that PCR has already occurred, and so any carryover of by-product has been amplified (Supplementary Figure S7, Supplementary Table S1).
While Structure-seq2 removes the by-product, running three PAGE gels is labor intensive. In practice, it takes approximately a day for each PAGE gel step in the protocol. Accordingly, we devised a facile variation that incorporates biotinylated dNTPs into the RT extension product (13) (Figure 1, Step 1B), allowing the extension product to be separated from the RT primer and by-product by two pull-downs with streptavidin-coated magnetic beads (Figure 1, Steps 2B and 4B). Each of these steps takes only ∼30 min. This variation of Structure-seq2 supplants two PAGE gels (Steps 2A and 4A) and thus is more efficient, reducing the library preparation time from over a week to 2.5 days. Importantly, adding biotin-dCTP during RT does not alter the distribution of nucleotide reads or read depth (Supplementary Figure S8), increase the overall mutation rate during RT or PCR (Table 1), or change the read profiles (Supplementary Figure S9).
Table 1. Higher mismatch rate with Ex Taq DNA polymerase and a lower reverse transcription reaction temperature.
| Library | RT reaction temperature | PCR polymerase | Mismatch rate per nucleotidea |
|---|---|---|---|
| Structure-seq2 (–DMS) | 55°C | Q5 | 0.89% |
| Structure-seq2 biotin variation (–DMS) | 55°C | Q5 | 0.83% |
| Ex Taq DNA polymerase | 55°C | Ex Taq | 1.15% |
| Lower RT reaction temperature | 50°C | Q5 | 0.97% |
aReads with more than three mismatches are not included as they cannot be confidently mapped.
Ligation-bias reduction
The original Structure-seq used Circligase to ligate an adaptor onto the 3′ end of the cDNA, but Circligase has a known nucleotide bias (10,14). We recently developed a ssDNA ligation method that overcomes this bias (10). A hairpin adaptor is used that base pairs with the 3′ end of the cDNA, which is then ligated by T4 DNA ligase. When comparing libraries prepared using T4 DNA ligase and the hairpin adaptor to a library prepared using the Circligase ligation, nucleotide ratios are much closer to transcriptome ratios, demonstrating reduced bias (Figure 2B). For example, when using Circligase the percentage of T nucleotides at the ligation junction is 6%, while the percentage of G nucleotides is 54%. However, when using T4 DNA ligation, the percentages of T and G residues improve to 23% and 14%, respectively, much closer to the transcriptome values of 24% and 25%, respectively (Figure2B).
More even read depth
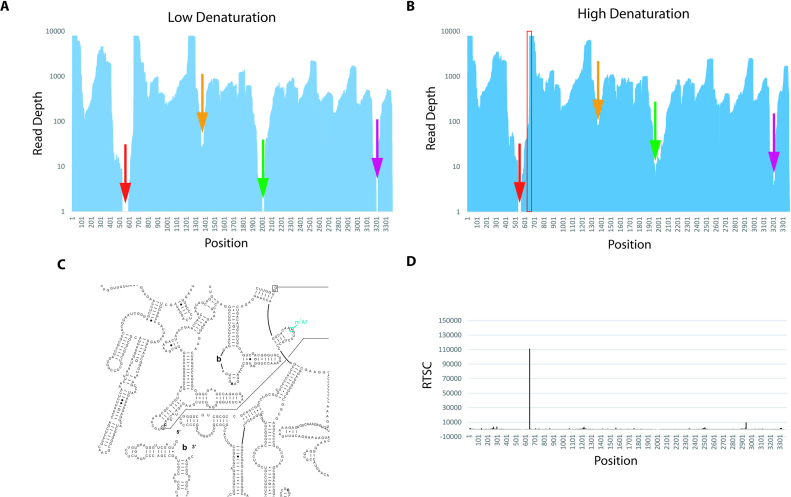
Structure-seq uses a random hexamer during RT to allow hybridization along the entire length of each RNA. Although each transcript should be covered evenly, certain regions are not read as deeply as others and some regions have no reads (Figure 3A). Regions of low/no coverage could be due to RNA structure interfering with RT primer binding. To address this possibility, we altered two features of the original Structure-seq method. The temperature of the RT annealing step was increased to favor RNA denaturation, and 50 mM KCl was added to favor DNA-RNA annealing.
Figure 3.
Structure-seq2 identifies a previously unreported m1A in 25S rRNA. (A) Using the original Structure-seq method for RT denaturation (65°C with no monovalent salt), there are regions that receive no reads (denoted with arrows). (B) Increasing the denaturation conditions (90°C with monovalent salt) allows these regions to be read (denoted with color-matched arrows) and narrows regions of low read depth. Total number of reads is similar in panels a and b. Location of the large drop in reads downstream of the single region in 25S that remains absent of reads (red arrow) corresponds to a site known to contain a m1A in yeast, human, and H. marismortui (C, Supplementary Figure S13) (16,18). Reads continue to decrease until they go to zero at nucleotide 539. The region between nucleotides 432 and 644 is 79% GC-rich with a read depth <100 on each nucleotide. (D) This site corresponds to a high RT stop count at the precise location in the –DMS data.
These changes increased read depth at sites of low or no reads. For example, regions in 25S rRNA that had just 27, 1 and 0 reads improved to 83, 6 and 4 reads (Figure 3A and B); moreover, the width of these three poor read regions narrowed almost 2-fold. Certain other positions still had no reads but these also narrowed. For example, there were no reads between 533 and 582, but this region narrowed to 534–539. The cause of these low read regions is likely in vitro RNA self-structure. Specifically, the three regions in 25S rRNA that have <10 reads (Figure 3B, red, green and purple arrows) have GC contents of 83%, 77% and 94%, compared to an overall GC content of 59% for 25S rRNA.
Lower mutation rate and higher quality sequencing rates
Mutations lower the number of reads that can be reliably mapped to the transcriptome. We reasoned that increasing the RT temperature and changing to a higher fidelity polymerase during PCR might decrease the number of mismatches (Table 1). Upon increasing the RT temperature from 50°C to 55°C, the mismatch rate per nucleotide decreased from 0.97% to 0.89% (an 8% decrease). When comparing Ex Taq DNA polymerase to the higher fidelity Q5 DNA polymerase, the mismatch rate per nucleotide decreased from 1.15% to 0.89% (a 23% decrease). We thus use both elevated RT temperature and high fidelity Q5 polymerase in Structure-seq2.
In Structure-seq2, the first 22 nt sequenced are identical for all reads (Figure 1, pink). Such low diversity can lead to poor sequencing quality by reducing the fidelity of cluster identification during Illumina sequencing (15). To address this, we designed a custom sequencing primer that abuts the unique region (Figure 1, light green). Using this custom primer, the mapping rate of effective reads in Structure-seq2, averaged over all libraries, increased sharply from 75% to 94% (Supplementary Table S1). We thus use this custom primer in Structure-seq2.
Benchmarking Structure-seq2
To assure that Structure-seq2 reliably reports on RNA structure, we benchmarked it in three different ways. First, we compared reactivity between Structure-seq2 and gel-based probing, which was done on 5.8S rRNA. As shown in Supplementary Figure S10, there is excellent agreement between the two methods. Second, we mapped reactivity data onto 25S rRNA. As shown in Supplementary Figure S11A, the reactivities agree with 25S rRNA secondary structure known from comparative analysis, confirming the ability of Structure-seq2 to report on the structure of the rRNA (16). Third, we compared Structure-seq2 to the original Structure-seq performed on Arabidopsis by assessing the continuous reactivity on the completely conserved ancient peptidyl transferase center in rice and Arabidopsis (Supplementary Figure S11A). There is a strong correlation (r = 0.7738) between continuous reactivity values in the two methods. We also compared reactivity between a region of the orthologous transcripts of RUBISCO SMALL SUBUNIT 2B in OS12T0274700-02 (rice) and AT5G38420.1 (Arabidopsis) (149/196 bp, identity 76%) (Supplementary Figure S12) (17). Our result shows a similar pattern of continuous reactivity (r = 0.4239; P-value = 3.9e−05) between rice and Arabidopsis on this mRNA, implying both fidelity between both Structure-seq methods and partial conservation of RNA secondary structure.
Using Structure-seq2 to identify novel biological features
We wanted to evaluate whether Structure-seq2 could lead to novel insights into biological systems. Ribosomal RNAs are known to be methylated at the N1 position (m1A) of A648 (rice numbering) of the large ribosomal subunit in human, Saccharomyces cerevisiae, and Haloarcula marismortui (18). This region is likely to be methylated in rice given the conserved secondary structures and sequences in this region (Figure 3C, Supplementary Figure S13). In fact, we find that the –DMS data in Structure-seq2 provide a very strong RT stop count at this position (Figure 3D). Intriguingly, there is also a very sharp decrease in reads at this site (Figure 3B, red box). Specifically, the read depth is ∼8,000 before A648 and ∼300 at and after it. For the reads that do extend through A648, the mutation rate at this site is elevated to ∼19% as compared to an overall mutation rate of just 0.89% on each nucleotide (Supplementary Table S1). Importantly, read depth adjacent to this site is improved in the high denaturation condition (Figure 3A and B, red arrows). Our advances in identification of modifications parallel those reported by Hauenschild et al. (19,20), although those investigators fragment the RNA while Structure-seq2 does not which may make identification of modified bases simpler. Structure-seq2 is thus able to identify positions of natural methylation.
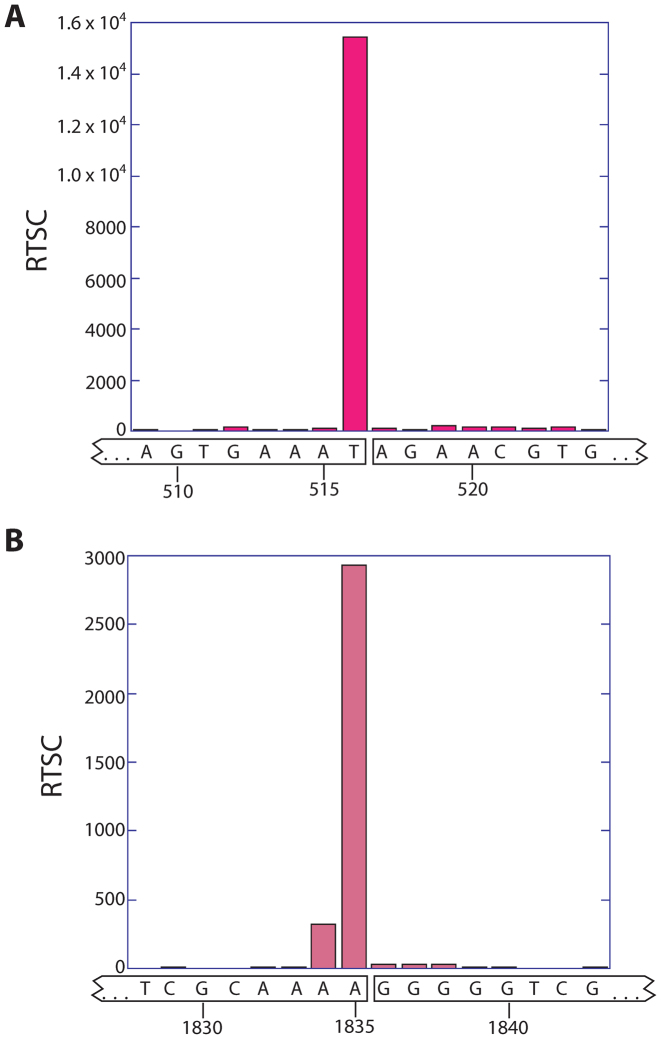
Photosynthetic plant cells are unique in that they harbor chloroplasts, which have their own ribosomes. An unusual feature of chloroplast 23S rRNA is that it has two hidden breaks, which are specific nuclease-mediated covalent breaks in the backbone of a hairpin that are necessary for efficient translation (21,22). Our Structure-seq2 data correctly identify the location of these breaks by a strong signal in the –DMS RT stop data (Figure 4A and B). Notably, these breaks would not be detectable by RNA-seq, in which the RNA is fragmented before analysis.
Figure 4.
Structure-seq2 demonstrates the presence of two hidden breaks in chloroplast rRNA. At the two locations known to harbor hidden breaks in chloroplast rRNA, the –DMS RT stop count data spike. The spike at the first hidden break (A) differs by one nucleotide from the published break site in spinach and Arabidopsis (21,28), which could be due to the slight sequence variation between species (Arabidopsis: 5′-GGGAGUGAAA*UAGAACA-3′, Rice: 5′-GGGUAGUGAAAU*AGAACG-3′, where * indicates the proposed break site). The spike at the second hidden break (B) occurs precisely at the published cleavage site for spinach and Arabidopsis (21,28).
DISCUSSION
Structure-seq2 provides a sensitive and accurate method for profiling RNA structure in vivo. While Structure-seq is a powerful tool for determining genome-wide structural information, Structure-seq2 improves the original Structure-seq protocol in several respects (5). First, a deleterious by-product was found to form in Structure-seq between excess RT primer and the ligation adaptor. Removing this by-product in Structure-seq2 significantly increases the quality of the sequenced libraries. Structure-seq2 provides two orthogonal methods to remove this by-product and thus can be tuned to the user's preferences. One of these methods purifies the desired product from the by-product by a total of three PAGE purifications, while the other saves time and material by purifying biotin-containing extension products via a streptavidin purification protocol, thus circumventing two of the three PAGE gels.
In addition to increasing library quality through by-product removal, Structure-seq2 implements optimizations that reduce ligation bias, improve read depth coverage, lower the overall mutation rate, and increase mapping rate. Using T4 DNA ligase with a hairpin ligation adaptor reduces ligation bias. Performing the RT denaturation and annealing steps with conditions that disfavor RNA self-structure (higher heat) and favor RNA-DNA hybridization (50 mM KCl) leads to an improved read depth coverage. Increasing the RT reaction temperature and using a higher fidelity PCR polymerase lowers the overall mutation rate. Using a custom sequencing primer to minimize low-diversity sequencing reads dramatically increases the mapping rate. Through the incorporation of these improvements, we are able to lower the starting material needed for adequate read counts by over 4-fold while also reducing the number of PCR cycles. These improvements are important for cases where RNA samples are limited, significantly reducing the cost of preparing the input poly(A) mRNA, and minimizing mutations arising from DNA amplification.
The high-resolution data obtained from Structure-seq2 applied to rice suggest that a previously unreported m1A is present in 25S rRNA of rice. Additionally, Structure-seq2 data contain reads closer to this natural modification than data obtained using the RT denaturation conditions found in the original version of Structure-seq. We also show that hidden breaks are detectable in chloroplast 23S rRNA using Structure-seq2. While our improvements are applied here to Structure-seq, they can be extended to other genome-wide RNA structure methods including SHAPES, CIRS-seq, HRF-seq, MAP-seq, ChemModSeq, and SHAPE-seq (14,23–27).
AVAILABILITY
RNA was folded via the StructureFold pipeline on Galaxy https://usegalaxy.org/ except as otherwise noted. Secondary structure for rRNA was obtained from the Comparative RNA website http://www.rna.icmb.utexas.edu/. Ribosomal RNA modifications were found at the 3D Ribosomal Modification Maps database https://people.biochem.umass.edu/fournierlab/3dmodmap/main.php.
ACCESSION NUMBERS
Raw sequencing reads are available on Gene Expression Omnibus (GEO) at National Center for Biotechnology Information (NCBI) with the series entry GSE95514.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Craig Praul, Director of the Huck Institute of Life Sciences Genomics Core Facility, for valuable insights on Illumina sequencing. We also thank our lab members for discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the National Science Foundation Plant Genome Research Program [NSF-IOS-1339282], with additional support from The Kaufman Foundation of The Pittsburgh Foundation [KA2016-85222] and an Innovation Award from Penn State University. Funding for open access charge: National Science Foundation Plant Genome Research Program [NSF-IOS-1339282].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bevilacqua P.C., Ritchey L.E., Su Z., Assmann S.M.. Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 2016; 50:235–266. [DOI] [PubMed] [Google Scholar]
- 2. Kwok C.K., Tang Y., Assmann S.M., Bevilacqua P.C.. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 2015; 40:221–232. [DOI] [PubMed] [Google Scholar]
- 3. Strobel E.J., Watters K.E., Loughrey D., Lucks J.B.. RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr. Opin. Biotechnol. 2016; 39:182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kubota M., Tran C., Spitale R.C.. Progress and challenges for chemical probing of RNA structure inside living cells. Nat. Chem. Biol. 2015; 11:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding Y., Kwok C.K., Tang Y., Bevilacqua P.C., Assmann S.M.. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat. Protoc. 2015; 10:1050–1066. [DOI] [PubMed] [Google Scholar]
- 6. Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M.. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014; 505:696–700. [DOI] [PubMed] [Google Scholar]
- 7. Leamy K.A., Assmann S.M., Mathews D.H., Bevilacqua P.C.. Bridging the gap between in vitro and in vivo RNA folding. Q. Rev. Biophys. 2016; 49:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang Y., Bouvier E., Kwok C.K., Ding Y., Nekrutenko A., Bevilacqua P.C., Assmann S.M.. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics. 2015; 31:2668–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwok C.K., Ding Y., Sherlock M.E., Assmann S.M., Bevilacqua P.C.. A hybridization-based approach for quantitative and low-bias single-stranded DNA ligation. Anal. Biochem. 2013; 435:181–186. [DOI] [PubMed] [Google Scholar]
- 11. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011; 17:10–12. [Google Scholar]
- 12. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sterling C.H., Veksler-Lublinsky I., Ambros V.. An efficient and sensitive method for preparing cDNA libraries from scarce biological samples. Nucleic Acids Res. 2015; 43:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poulsen L.D., Kielpinski L.J., Salama S.R., Krogh A., Vinther J.. SHAPE Selection (SHAPES) enrich for RNA structure signal in SHAPE sequencing-based probing data. RNA. 2015; 21:1042–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krueger F., Andrews S.R., Osborne C.S.. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One. 2011; 6:e16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. . The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inze D., Mueller-Roeber B., Vandepoele K.. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015; 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piekna-Przybylska D., Decatur W.A., Fournier M.J.. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008; 36:D178–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hauenschild R., Werner S., Tserovski L., Hildebrandt A., Motorin Y., Helm M.. CoverageAnalyzer (CAn): a tool for inspection of modification signatures in RNA sequencing profiles. Biomolecules. 2016; 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hauenschild R., Tserovski L., Schmid K., Thuring K., Winz M.L., Sharma S., Entian K.D., Wacheul L., Lafontaine D.L., Anderson J. et al. . The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 2015; 43:9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N.. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017; 36:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leaver C.J. Molecular integrity of chloroplast ribosomal ribonucleic acid. Biochem J. 1973; 135:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Incarnato D., Neri F., Anselmi F., Oliviero S.. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014; 15:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kielpinski L.J., Vinther J.. Massive parallel-sequencing-based hydroxyl radical probing of RNA accessibility. Nucleic Acids Res. 2014; 42:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seetin M.G., Kladwang W., Bida J.P., Das R.. Massively parallel RNA chemical mapping with a reduced bias MAP-seq protocol. Methods Mol. Biol. 2014; 1086:95–117. [DOI] [PubMed] [Google Scholar]
- 26. Hector R.D., Burlacu E., Aitken S., Le Bihan T., Tuijtel M., Zaplatina A., Cook A.G., Granneman S.. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014; 42:12138–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loughrey D., Watters K.E., Settle A.H., Lucks J.B.. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014; 42:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu J., Zhou W., Liu G., Yang C., Sun Y., Wu W., Cao S., Wang C., Hai G., Wang Z. et al. . The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol. 2015; 168:205–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.