Abstract
Origin of new biological functions is a complex phenomenon ranging from single-nucleotide substitutions to the gain of new genes via horizontal gene transfer or duplication. Neofunctionalization and subfunctionalization of proteins is often attributed to the emergence of paralogs that are subject to relaxed purifying selection or positive selection and thus evolve at accelerated rates. Such phenomena potentially could be detected as anomalies in the phylogenies of the respective gene families. We developed a computational pipeline to search for such anomalies in 1,834 orthologous clusters of archaeal genes, focusing on lineage-specific subfamilies that significantly deviate from the expected rate of evolution. Multiple potential cases of neofunctionalization and subfunctionalization were identified, including some ancient, house-keeping gene families, such as ribosomal protein S10, general transcription factor TFIIB and chaperone Hsp20. As expected, many cases of apparent acceleration of evolution are associated with lineage-specific gene duplication. On other occasions, long branches in phylogenetic trees correspond to horizontal gene transfer across long evolutionary distances. Significant deceleration of evolution is less common than acceleration, and the underlying causes are not well understood; functional shifts accompanied by increased constraints could be involved. Many gene families appear to be “highly evolvable,” that is, include both long and short branches. Even in the absence of precise functional predictions, this approach allows one to select targets for experimentation in search of new biology.
Keywords: evolutionary rate, neofunctionalization, subfunctionalization, archaea, clade-specific acceleration of evolution, tree anomalies
Introduction
The molecular clock concept holds that, in the absence of functional change, orthologous gene families evolve at rates that are constant up to the magnitude of random fluctuations (Kimura 1983; Bromham and Penny 2003). Accordingly, substantial deviations from the family-specific evolutionary rates are thought to be caused by changes in gene functions. Often, such changes are linked to gene duplication resulting in the emergence of paralogs. Elaborate theoretical models have been developed to account for the evolution of paralogs and tested by comparative sequence analysis. Paralogs are thought to evolve via the neofunctionalization or subfunctionalization routes (Ohno 1970; Force etal. 1999; Lynch and Conery 2000; Lynch and Katju 2004; Innan and Kondrashov 2010). Neofunctionalization implies a pronounced asymmetry between the paralogs whereby one paralog retains the original function whereas the other one evolves a new function. This scenario is thought to be associated with an acceleration of evolution of the innovating paralog caused by the removal of purifying selection pressure associated with the retention of the original function and possibly positive selection driving the evolution of the new function (Ohno 1970). The subfunctionalization scenario that appears to be more common than neofunctionalization involves more symmetrical evolution of the two paralogs whereby each loses complementary subfunctions of the common, multifunctional ancestor. In this case, the evolution of both paralogs appears to accelerate after duplication albeit not to the degree of the innovating paralog in the neofunctionalization scenario. Acceleration of evolution predicted by these models indeed has been observed in genome-wide analyses of evolutionary rates (Lynch and Conery 2000; Kondrashov etal. 2002).
In archaea and bacteria, homologous genes in the same genome are, in some cases, true paralogs that arise from intragenomic duplication but more often appear to be “pseudoparalogs” acquired from different organisms via horizontal gene transfer (HGT) (Makarova etal. 2005; Treangen and Rocha 2011). In contrast to the detailed studies on paralogy, evolution of pseudoparalogs has not been addressed in detail in the theoretical evolutionary biology context. Nevertheless, it can be expected from general considerations and on the basis of several observations that, similar to genuine paralogs, pseudoparalogs experience a period of accelerated evolution after HGT (Wiedenbeck and Cohan 2011; Mozhayskiy and Tagkopoulos 2012). Additionally and perhaps more commonly, pseudoparalogs can create the appearance of accelerated evolution simply due to their acquisition from donor lineages that are evolutionarily distant from the recipient.
Assuming that a substantial change of the evolutionary rate in a particular gene family is likely to be linked to a functional change, whether or not it involves paralogs or pseudoparalogs, the reverse is expected to hold as well: significant changes in the evolutionary rate (typically, acceleration) can be construed as indications of functional innovations. Indeed, some examples of extremely fast evolution of genes responsible for key cellular function has been described for archaea and bacteria (Makarova and Koonin 2010, 2013; Makarova etal. 2012, 2015; Wolf etal. 2013). To our knowledge, however, a comprehensive, genome-wide analysis of such events has not been reported so far. Furthermore, gene-specific changes in evolutionary rates are confounded by lineage-specific rates that are common in diverse groups of organisms, in particular in bacteria and archaea (Groussin and Gouy 2011; Denef and Banfield 2012; Snir etal. 2012; Brown and Wernegreen 2016). Thus, to identify gene-specific accelerations or decelerations that could have functional implications, it is essential to calibrate the measurements against the expectations based on the lineage-specific rates.
Here, we aimed to analyze systematically evolutionary rate anomalies in phylogenetic trees for the archaeal gene families that predate the divergence of major archaeal groups, using the comparative genomic framework provided by the archaeal clusters of orthologous genes (arCOGs) (Makarova etal. 2015). To identify significant evolutionary rate changes, we implemented a computational pipeline that explores the topology and branch lengths of phylogenetic trees. We present an overview of the evolutionary rate variation across archaeal lineages, arCOGs, and functional groups of genes, as well as detailed examination of several cases of potential biological interest.
Materials and Methods
Orthologous Gene Families and Phylogenetic Trees
The list of the 168 analyzed archaeal genomes and taxonomic information are provided in supplementary table 1, Supplementary Material online. The 13,443 archaeal orthologous families (arCOGs), the phylogenetic tree of archaea, and the derivation of genome weights have been described previously (Makarova etal. 2015). Briefly, the archaeal phylogeny was reconstructed from a concatenated alignment of ribosomal proteins; clade weights (down to the individual genomes) were iteratively derived from this tree proportionally to the sum of branch lengths in the clades (i.e., to the amount of the evolutionary change in the respective clade). The tree was used as the reference to define archaeal taxa for further analysis. Multiple protein sequence alignments for selected arCOGs were constructed using the MUSCLE program with default parameters (Edgar 2004); poorly aligned sequences and unreliable alignment positions were removed as previously described (Yutin etal. 2008). Phylogenetic trees were constructed using the FastTree program (Price etal. 2010), with gamma-distributed site rates and WAG evolutionary model, and rooted using a modified midpoint procedure.
Quantification of Taxa Representation
All archaeal genomes in the data set were classified into (presumably monophyletic) taxa. To quantify the representation of a taxon in an arCOG, a representation index was introduced. Informally, the representation index shows the fraction of genomes from the given taxon that is present in the given arCOG on the scale from 0 to 1, taking into account that genomes of different taxa in general are unevenly sampled. For example, if an arCOG is represented in Sulfolobus tokodaii but not in any of the 10 S. islandicus genomes, whereas another arCOG is represented in all S. islandicus genomes but not in the S. tokodaii genome, their representation should be roughly equivalent (present in one of the two species) despite the fact that the sampling of S. islandicus genomes is 10 times that of the S. tokodaii. Formally, the total weight for taxon T in the complete set of available genomes is defined as , where wi is the weight of the i-th genome. Each arCOG contains genes from a set of genomes; the representation index for taxon T in arCOG C is defined as , where WT,C is the sum of weights of genomes from taxon T present in the tree of the arCOG C (ignoring paralogs within, that is, counting each genome only once). A taxon with RT,C ≥ 0.75 was considered “well-represented” in the given arCOG.
Detection of Monophyletic Groups (Clades)
Genomes that belong to a particular taxon can be distributed on a tree in a variety of ways. For the purposes of this analysis, we were particularly interested in cases where the genes from a monophyletic group of archaea (taxon) codiverged from their homologs from other taxa concomitantly with the taxon divergence and then evolved largely within the respective genomes (i.e., were not involved in extensive cross-taxa HGT). In a phylogenetic tree, such cases present as clades that 1) contain genes from all or most of the genomes from a particular taxon (indicating an origin in the taxon ancestor) and 2) contain none or few genes from other taxa (suggesting low level of intertaxa HGT). For the purpose of automatic analysis of the trees, we formalized these properties in the taxon coverage and purity indices which are associated with individual clades.
For any clade b in the tree of arCOG C, a coverage index of a taxon T can be calculated as . The extent to which clade b in the tree of arCOG C predominantly belongs to a taxon T can be quantified as a purity index . A combined quality index for clade b as a representative of taxon T in the tree of arCOG C can be calculated as the product of its coverage and purity, QT,b,C = CT,b,C PT,b,C (analogous to the G-measure) (Powers 2011). An QT,b,C value of 1 implies that the given clade contains all species from the taxon T that are present in the given arCOG and no species from other taxa. For each taxon T that was found to be well-represented in a tree of an arCOG C (RT,C ≥ 0.75), the deepest clade b satisfying QT,b,C ≥ 0.75 was considered to be a representative clade. If paralogs in an arCOG form taxon-specific subtrees, this taxon can be represented by more than one clade; conversely, in some arCOGs, genomes from well-represented taxa can be scattered across the tree and never form a clade. Trees with at least two well-represented taxa that formed clades were included in further analysis.
Decomposition of Observed Clade Heights
A good first approximation of the evolution of a gene with a conserved function and in the absence of widespread HGT is provided by a simple model (Wolf etal. 2013) in which each gene (arCOG) evolves at its own characteristic, nearly constant rate, so that the expectation of a branch length corresponding to a particular amount of evolutionary time elapsed between the ancestor and the descendant can be estimated as the product of the rate and the time. A variety of deviations that include both rate variation and the uncertainty of distance measurements can be rolled into a multiplicative, log-normally distributed error factor with the expected value of 1 (Wolf etal. 2013). Neither the evolutionary rate nor the evolutionary time can be estimated directly except at very short time scales (usually years to decades); however, observing the evolution of many genes in many taxa makes them amenable to the following analytic procedure. The height of a taxon clade in the tree (i.e., the average distance from the base of the branch leading to the common ancestor to the tips of the tree) combines two unknown factors (the evolutionary rate and the time since this lineage diverged from the rest of the archaea) and some additional, presumably random but also unknown, deviation. However, it is generally reasonable to assume that among the orthologs from different taxa (i.e., across an arCOG tree), the evolutionary rate is uniform. Likewise, the elapsed time since divergence for sequences from a given taxon is the same across the genes (arCOGs), provided that the evolution did not involve much HGT. An overabundance of observations (number of arCOGs times the average number of taxa per arCOGs tree) relative to the number of unknown parameters (number of arCOGs plus the number of taxa) allows for a statistical estimate of both the arCOG-specific relative evolution rates and the taxon-specific effective divergence times.
Formally, the observed height of a clade of taxon T in arCOG C (HT,C) was decomposed into the product of the arCOG-specific relative evolution rate (rC), taxon-specific effective divergence time (tT), and clade-specific deviation factor (eT,C). In log scale, this relationship can be represented as . Minimizing the deviations across the set of clades gives the estimates of relative evolution rates and effective divergence times (vectors r and t, respectively) that determine the expected clade height rC tT. The solution for r, t, and e was obtained by solving the system of linear equations that results from differentiating the sum of square deviations with respect to rC and tT.
Comparison with Normal Distribution
A single-peak distribution with exponentially declining tails can be characterized by its mean and standard deviation. The mean value is relatively robust to outliers but the standard deviation estimates are strongly affected because they involve squares of the deviations from the mean. Finding a normal equivalent of a distribution requires a robust measure of variation that is less sensitive to outliers compared with the standard deviation.
Normal distribution implies a specific relationship between the interquartile distances and the standard deviation [, , where is a cumulative distribution function for a normal distribution with the mean and standard deviation ]. This relationship allows one to estimate a normal-equivalent standard deviation (where and are the first and the third quartiles, respectively) for the central part of the distribution such that the shape of the distribution is insensitive to outliers in the tails. For observations with or, where is determined by the relation , where is the number of observations, the expectation is <1 in a sample of size taken from a normal distribution with the mean and standard deviation .
Detailed Analysis of arCOGs
The arCOGs containing clades with the largest deviations from the expectation were selected for detailed analysis. Gene neighborhoods, including three genes upstream and downstream of the selected arCOGs were extracted from RefSeq genomes and annotated using arCOGs (Makarova etal. 2015) and the NCBI CD database (Marchler-Bauer etal. 2015). To construct expanded phylogenetic trees, more distant homologs belonging to the same COG (Galperin etal. 2015) were extracted and clustered using UCLUST (Edgar 2010), with the similarity threshold of 0.5. Sequences within the clusters were aligned using MUSCLE (Edgar 2004), and phylogenetic trees were constructed using FastTree (Price etal. 2010). Additional phylogenetic analysis was performed using the PhyML program (LG substitution model, gamma-distributed site rates, empirical frequencies) (Guindon etal. 2003). Multiple alignments of the sequences from different clusters were compared with each other using hhsearch (Soding 2005); pairwise profile-profile similarity scores were used to construct UPGMA similarity dendrograms.
Results
Taxa Representation in Archaeal Orthologous Gene Clusters
For the purpose of this analysis, all archaeal genomes were classified into 16 operationally defined groups (supplementary table 1, Supplementary Material online) that might or might not have formal taxonomic standing, but form largely accepted clades in the archaeal species tree (Guy and Ettema 2011; Yutin etal. 2012; Makarova etal. 2015; Petitjean etal. 2015; Raymann etal. 2015). Among these, 13 (except for Korarchaeota, Nanoarchaeota, and Methanopyrales) consist of three or more genomes; the further analysis described below involved only these groups. Of the 13, 443 arCOGs, in 1, 834 two or more taxa are well-represented (supplementary table 2, Supplementary Material online); all 13 groups are well-represented in 217 arCOGs, which coincides with the size of archaeal core gene set (Makarova etal. 2015).
Monophyletic Groups in arCOGs
We then developed formal definitions of taxa representation by clades (fig. 1; see Methods for details). In 1,587 of the 1,834 arCOGs, in which two or more taxa are well-represented, at least two taxa form clades. The number of arCOGs, in which at least one taxon is well-represented, ranges from 532 in Thermoplasmata to 1,120 in Methanocellales, a factor of ∼2. In contrast, the number of arCOGs, in which at least one taxon includes a clade, ranges from 159 in Desulfurococci to 1,034 in Methanocellales, a factor of ∼6.5. This drastic difference between the major archaeal taxa most likely reflects a broad range of taxon-specific levels of HGT. The fraction of arCOGs, in which a taxon forms a clade(s), among the arCOGs that include a well-represented taxon ranges from 25% for Desulfurococci to 95% for Halobacteria, and strongly, negatively correlates with the diversity of the genomes in the clade (rPearson = −0.73 for the sum of branch lengths in the ribosomal protein tree; supplementary tables 3 and 4, Supplementary Material online).
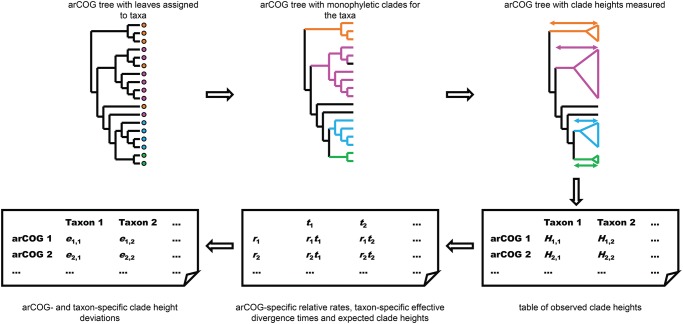
Fig. 1.
—Computational pipeline for identification of evolutionary rate deviations from phylogenetic trees. The main steps of the computational pipeline developed and applied here for the detection of evolutionary rate deviations in phylogenetic trees of arCOGs are shown. (A) Detection of monophyletic groups (clades) in arCOG trees. The deepest clades satisfying the purity and coverage criteria are identified for all well-represented taxa in all arCOG trees. Empty circles indicate the tree clades that are analyzed; filled circles indicate the clades that were identified for the given taxa in the given tree. (B) Calculation of the distance deviation for selected clades. The table of observed clade heights is decomposed into the expected distances rC tT and deviations eT, C; distance deviations (in the log scale) for all clades are recorded.
Altogether, the analyzed data set includes 9,209 clades from 1,587 arCOGs. A taxon can be represented by up to seven paralogous clades in a single arCOG (arCOG00194, ATPase component of an ABC-type multidrug transport system in Methanocellales). However, the majority of the clades (7,805, or 85%) represent the only monophyletic group for the corresponding taxon in an arCOG (supplementary table 3, Supplementary Material online).
Evolutionary Rates, Effective Divergence Times, and Clade-Specific Deviations
For each clade in all arCOGs, the clade height was calculated as the weighted average distance from the ancestral node of the clade to the tips (using the weights of genomes) plus the length of the branch, incoming to the ancestral node. The observed clade heights were decomposed into relative arCOG evolution rates, taxon divergence times, and deviation factors. More formally, , where HT,C is the observed height of a clade of taxon T in arCOG C, rC is the relative evolution rate of arCOG C, tT is the effective time of divergence of the taxon T from other archaea (in arbitrary units, combining the astronomical time since divergence with the taxon-specific variations of evolution rates), and eT,C is the deviation of the observed clade height (HT,C) from the expected clade height (rC tT). The values of vectors r and t that minimize were obtained numerically as described under Methods.
Decomposition of the observed clade heights yields taxon-specific effective divergence times (tT, supplementary table 4, Supplementary Material online), arCOG-specific relative evolution rates (rC, supplementary table 5, Supplementary Material online), and clade-specific deviations of the observed distance from the one predicted for a given arCOG and taxon (eT,C, supplementary table 3, Supplementary Material online). Effective divergence times are similar between taxa within a factor of 2, suggesting that the selected taxa represent distinct groups of archaea with similar degrees of separation from their respective closest relatives. In contrast, the relative evolution rates differ by a factor of 9 between arCOGs, indicating that the intrinsic differences are considerable even among the relatively highly conserved genes (those that are well-represented in two distinct taxa). In agreement with previous observations (Novichkov etal. 2004; Wolf etal. 2009; Snir etal. 2012, 2014), the relative evolution rate distributions significantly differ, albeit overlap, between genes that belong to different functional categories. Predictably, on an average, the translation-related genes are the slowest-evolving, whereas genes with unknown functions are the fastest-evolving group.
To test the results for robustness, we constructed phylogenetic trees using PhyML for the six arCOGs that are discussed in detail below (arCOG01758, arCOG01833, arCOG01981, arCOG04045, arCOG04406, and arCOG04407) starting from the same alignments that were used for the construction of the FastTree trees. The same protocol was the applied for the analysis of the resulting trees. Of the 55 clades identified in the FastTree trees, 53 (96%) were found to be monophyletic in the PhyML trees as well; one extra clade was found in the PhyML trees that was absent in the FastTree trees (supplementary table 6, Supplementary Material online). In the shared set of 53 clades, the observed clade heights are highly correlated between the PhyML trees and the FastTree trees (supplementary table 6, Supplementary Material online), with the Pearson correlation coefficient of 0.96 between the clade heights in the log scale. Although, on an average, clade heights estimated using PhyML were greater than those estimated using FastTree by a factor of 1.28, all analysis reported here relies on the relative magnitudes, which turned out to be remarkably robust.
Distribution of Clade-Specific Deviations of Evolutionary Rates
The observed clade heights (supplementary table 3, Supplementary Material online) range from being shorter than expected by a factor of 4 to being longer than expected by a factor of 5 (−1.6 to 1.4 in natural log scale). Overall, the distribution of the deviations is approximately lognormal, with the standard deviation of 0.26 log units. Despite the substantial ecological and physiological differences between the taxa and the molecular and functional differences between the genes, the heights of 90% of the clades are within a factor of 1.5 of the expected value (fig. 2). Nevertheless, this distribution has considerably fatter tails compared with a normal distribution, with an excess kurtosis of 1.6. This deviation from the normal distribution implies that the largest among the observed deviations do not result from accumulation of multiple small, random, independent fluctuations but rather reflect biological factors that cause acceleration or deceleration of evolution of particular genes in particular taxa. For the magnitudes of deviation corresponding to the expectation of <1 for the normal distribution, we observed 23 short clades and 43 long clades (see Methods for details).
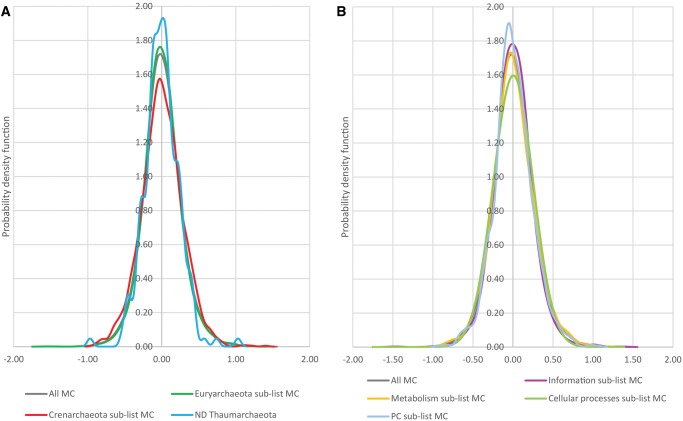
Fig. 2.
—Distribution of distance deviations across archaeal lineages and functional categories of genes. (A) Probability density functions of the distance deviations (log scale) for the clades belonging to the three major archaeal phyla (Euryarchaeota, Crenarchaeota, and Thaumarchaeota). (B) Probability density functions of the distance deviation (log scale) for the clades belonging to the four functional classes of arCOGs (Information Storage and Processing, Cellular Processes and Signaling, Metabolism, Poorly Characterized [PC]).
Among the 7,805 clades that have no paralogs in the corresponding arCOGs, the deviation rate is considerably less (90% of the deviations are between −0.38 and 0.38 log units) than in the remaining 1,404 clades that come from within-arCOG paralogs (90% of the deviations between −0.53 and 0.65). Among the 1% of the clades with the largest acceleration and 1% of the clades with the largest deceleration of evolution (thereafter, the extreme deviation subset), 47% have (pseudo)paralogs (compared with 15% in the entire set). These findings are compatible with the theoretical models in which, in the presence of paralogs, the constraints on the tempo, and mode of gene evolution are substantially relaxed (Ohno 1970; Lynch and Force 2000; Innan and Kondrashov 2010).
The taxon-specific distributions of deviations center ∼0 on the log scale (factor of 1) by construction (the taxon-specific effective divergence time is chosen to minimize the deviations). The standard deviations of these distributions (supplementary table 7, Supplementary Material online), however, differ considerably, ranging from 0.17 log units in Thermoplasmata to 0.30 in Thermococci and Sulfolobi. The taxa with the widest distribution of deviations (Thermococci, Sulfolobi, and Methanocellales) are also overrepresented in the extreme deviation subset, accounting for 61% of the clades versus the expected 33%. This overrepresentation correlates with the extent of paralogy in this data set, whereby all three taxa with the widest ranges of deviations encompass over 20% of paralogous clades (compared with 15% in the complete set). The Pearson correlation between the standard deviation of the distribution of deviations and the fraction of paralogous clades in taxa was 0.88, thus explaining ∼77% of the variance among the taxa.
Likewise, distributions of deviations for arCOGs that belong to different functional categories all centered ∼0 (in log scale) but differed in width. The widest distributions were observed for the categories D (Cell cycle control, cell division, chromosome partitioning), T (Signal transduction mechanisms), and V (Defense mechanisms), and not surprisingly, arCOGs from these categories contain the largest fractions of paralogous clades. There was a highly significant positive correlation between the standard deviation of the distribution of evolutionary rate deviations and the fraction of paralogous clades in functional categories of genes (Pearson correlation 0.72, explaining ∼52% of the variance between COG categories).
Clades with the Largest Deviations from the Expectation
In addition to the general characteristics described earlier, we were interested in detailed analysis of the individual gene families that most strongly deviate from the expected evolutionary rate, in an attempt to decipher potential new or modified functions behind these dramatic accelerations or decelerations of evolution (tables 1 and 2). This analysis involves construction of additional phylogenetic trees that include bacterial orthologs of the analyzed archaeal genes and in depth comparative analysis of gene context and domain architectures of the respective proteins. The functional and evolutionary inferences for 20 arCOGs characterized by the greatest relative positive deviations (acceleration) from the expected evolutionary rates (hereinafter “fast evolving,” when we are confident that HGT is not involved, or “long branches” for other cases) are summarized in table 1. Ten arCOGs with the greatest relative negative deviation (deceleration; “slow evolving” and “short branches,” respectively) are described in table 2. Notably, 13 of these 33 arCOGs contain more than one highly deviant subtree, suggesting that these families are “highly evolvable,” that is, prone to large-scale (top or bottom 1% of the deviation range) evolutionary rate changes that occur independently and repeatedly in different lineages.
Table 1.
Top 20 arCOGs with Long Branches
| arCOG, Protein, Annotation | Clade with Long and/or Short Branches (rank)a | Commentsb | |
|---|---|---|---|
| 1 | arCOG01981, Tfb, Transcription initiation factor TFIIB | ↑(1) Thermoprotei; | Family specific to archaea and eukaryotes. Multiple duplications in different archaeal lineages. |
| ↓(2) Methanocellales; | Sub- or neofunctionalization in the duplicated copies. See text for details. | ||
| ↓(3) Methanomicrobiales; | |||
| ↑(6) Sulfolobi; | |||
| ↓(6) Methanosarcinales; | |||
| ↑(9) Halobacteria; | |||
| ↑(11) Thaumarchaeota; | |||
| ↑(16) Thermococci | |||
| 2 | arCOG01758, RpsJ, Ribosomal protein S10 | ↑(2) Halobacteria; | No evidence of HGT. The only duplication in archaea most likely occurred in the common ancestor of Halobacteria. Likely neofunctionalization of the fast-evolving paralog that is implicated in a house-cleaning pathway. See text for details. |
| ↓(75) Methanocellales | |||
| 3 | arCOG01331, HtpX, Zn-dependent protease with chaperone function | ↑(3) Thermococci; | Thermococcal sequences from the fast-evolving clade lack the N-terminal membrane-associated domain but are predicted to be active proteases. Probably regulated by a dedicated transcriptional regulator (arCOG05764), which is encoded in the same predicted operon. Likely sub- or neofunctionalization of this paralog. |
| ↓(14) Halobacteria | |||
| 4 | arCOG02062, TusA, TusA-related sulfurtransferase | ↑(4) Sulfolobi; | The slow-evolving and the “regular-evolving” paralogs in Sulfolobi are both encoded in heterodisulfide reductase locus and are apparently involved in Fe-S cluster assembly. The fast-evolving paralog is probably involved in a different pathway with DsrE/DsrF/DrsH-like protein; this genomic and functional association has been described in Sulfolobi (Liu etal. 2014). This paralog might have been horizontally transferred from bacteria to Sulfolobi, but the tree is inconclusive due to small number of informative sites. |
| ↓(35) Sulfolobi | |||
| 5 | arCOG01075, NUDIX family hydrolase | ↑(5) Methanocellales; | Probable HGT from bacteria to Methanocellales followed by gene duplication. Methanocellales species encode one or two additional pseudo-paralogs that are not monophyletic with the fast-evolving paralog and might have been acquired from different sources. All proteins in the family predicted to be active enzymes. Probable subfunctionalization |
| ↓(66) Archaeoglobi | |||
| 6 | arCOG02579, Fe-S-cluster containing protein | ↑(7) Methanocellales | Multiple lineage-specific paralogs in Methanocellales specifically, including two highly diverged paralogs in the fast-evolving clade. HGT from any group of archaea or bacteria could not be demonstrated due to unreliable trees. Probable neofunctionalization. |
| 7 | arCOG00415, RadA, RadA/RecA recombinase, contains N-terminal HHH domain | ↑(8) Thermoprotei; | Duplication in the ancestor of Thermoprotei, additional duplications in several Pyrobaculum species. Active ATPase, possible neofunctionalization by loss of RadB paralog in Thermoprotei (arCOG00417). |
| ↓(55) Methanocellales | |||
| 8 | arCOG04407, TtdA, Tartrate dehydratase alpha subunit/Fumarate hydratase class I, N-terminal domain | ↑(10) Methanobacteria | Possible HGT from bacteria. Same pattern as in arCOG04406 [↑(96) Methanobacteria], involved in the same pathway and located next to arCOG04407 in several archaeal genomes. Probable coevolution of these two genes. See text for details. |
| 9 | arCOG00476, UbiA, 4-hydroxybenzoate polyprenyltransferase or related prenyltransferase | ↑(12) Archaeoglobi | Probable duplication in the ancestor of Archaeoglobi resulting in sub- or neofunctionalization. Due to high sequence divergence, HGT cannot be confidently established. |
| 10 | arCOG00670, PgsA, Phosphatidylglycerophosphate synthase | ↑(13) Methanocellales | Methanocellales possess a third paralog, in addition to the two ancestral ones. HGT from bacteria that also have multiple paralogs. The Methanocellales-specific paralog likely has a distinct function (neofunctionalization) linked to that of HemG, (protoprophyrinogen IX oxidase) and regulated by AcrR with it forms a predicted operon conserved in Methanocellales. |
| 11 | arCOG00945, AslB, Radical SAM superfamily enzyme | ↑(14) Archaeoglobi | Patchy distribution in archaea, with many cases of HGT from bacteria including the fast-evolving Archaeoglobi lineage. The cellular function of this protein is unknown. |
| 12 | arCOG01580, DNA-binding transcriptional regulator, Lrp family | ↑(15) Methanocellales; | Multiple paralogs. No evidence of HGT from bacteria. Likely neo- or subfunctionalization in both fast-evolving lineages. |
| ↑(23) Thermococci | |||
| 13 | arCOG04143, DNA topoisomerase VI, subunit A | ↑(17) Methanocellales; | Methanocellales is the only major archaeal lineage with two paralogs. Both branches from Methanocellales group with bacteria but HGT from Methanocellales to bacteria appears more likely given the archaeal provenance of the gene. Probable subfunctionalization. |
| ↓(27) Sulfolobi; | |||
| ↑(81) Methanocellales | |||
| 14 | arCOG03072, MnhC, Multisubunit Na+/H+ antiporter, MnhC subunit | ↑(18) Thermococci | HGT of the entire operon from either Halobacteria or bacteria. Probable subfunctionalization. |
| 15 | arCOG01700, SpeB, Arginase family enzyme | ↑(19) Thermococci | A second, highly diverged copy, in addition to the typical archaeal version. Predicted to be an active enzyme. Lacks ∼80 aa N-terminal region responsible for dimerization (Dowling etal. 2008). Possible HGT from bacteria and neofunctionalization. |
| 16 | arCOG04045, IlvD, Dihydroxyacid dehydratase/phosphogluconate dehydratase | ↑(20) Methanocellales | HGT from bacteria to Methanocellales.Possible neofunctionalization.See text for details. |
| 17 | arCOG01452, Cas1, CRISPR-Cas system associated protein Cas1, integrase | ↑(21) Methanomicrobiales | Casposon-associated Cas1. Clear case of neofunctionalization. Most likely, Cas1 exapted by the CRISPR-Cas system from a casposon (Krupovic etal. 2014). |
| 18 | arCOG01165, DNA topoisomerase VI, subunit B | ↑(22) Methanocellales | See arCOG04143 above. |
| 19 | arCOG00354, GTPase SAR1 or related small GTPase | ↑(24) Methanococci | Likely a single HGT from bacteria to the three taxa Methanococci, Thermococci, and Methanopyrales, followed by acceleration of evolution and replacement of the ancestral gene in the three taxa mentioned earlier. Possible neofunctionalization. Sar1 might function as analog of the actin-like ATPase MreB given that MreB is missing in most Methanococci (Makarova etal. 2010). |
| 20 | arCOG00419, Hit, HIT family hydrolase | ↑(25) Methanocellales; | Multiple paralogs. HGT from bacteria to Methanocellales which only retain the fast-evolving variant. Located in a conserved neighborhood with two ABC-type multidrug transport system component (arCOG00194 and arCOG04450). Likely neofunctionalization. |
| ↓(23) Thermococci |
Upward arrow shows an exceptionally long branch and downward arrow shows an exceptionally short branch.
For additional information, see ftp://ftp.ncbi.nlm.nih.gov/pub/wolf/_suppl/archdev.
Table 2.
Top 10 arCOGs with Short Branches
| Top 10 arCOGs with Short Branches | |||
|---|---|---|---|
| arCOG, Protein, Annotation | Clade with Short and/or Long Branches (rank)a | Commentsb | |
| 1 | arCOG01171, KaiC, KaiC-superfamily ATPase implicated in signal transduction | ↓(1) Thermococci; | Large family with multiple paralogs (McRobbie etal. 2009). Likely diversification of the family through sub- and neofunctionalization, with acceleration and deceleration of the evolution of the different copies. |
| ↑(35) Methanocellales; | |||
| ↓(57) Archeaoglobi | |||
| 2 | arCOG02391, CheY, Rec domain | ↓(4) Methanomicrobiales; | Large family with multiple paralogs. The short branches in Methanomicrobiales clade correspond to an ancestral variant, located in a conserved neighborhood of genes involved in chemotaxis. |
| ↑(34) Methanocellales | |||
| 3 | arCOG04272, CobT, NaMN: DMB | ↓(5) Thermococci | Likely xenologous gene displacement via HGT from Crenarchaea to Thermococci. Possible neofunctionalization. |
| 4 | arCOG01568, MscS, Small-conductance mechanosensitive channel | ↓(7) Archaeoglobi | Likely multiple HGT events between archaea and bacteria. Potential coevolution with uncharacterized protein of arCOG01766 family. Possible subfunctionalization. |
| 5 | arCOG01057, HxlR, DNA-binding transcriptional regulator, HxlR family | ↓(8) Methanomicrobiales | Probable HGT from bacteria to Methanomicrobiales+ Methanosarcinales. Encoded in a predicted operon with hydroxylamine reductase (arCOG02430).Likely subfunctionalization by recruitment as a transcriptional regulator for hydroxylamine reductase. |
| 6 | arCOG01715, SufB, Cysteine desulfurase activator SufB | ↓(9) Thaumarchaeota; | Duplication in the common ancestor of Thaumarchaeota and Crenarchaeota. Subfunctionalization of the two paralogs is likely given considerable sequence divergence (Iwasaki 2010). |
| ↑(44) Sulfolobi | |||
| 7 | arCOG01107, ArgE, Acetylornithine deacetylase/Succinyl-diaminopimelate desuccinylase or related deacylase | ↓(10) Thermococcales | Large family with multiple paralogs many apparent cases of HGT. Metal-binding site conserved in all enzymes (Bienvenue etal. 2003) with minor divergences. Subfunctionalization could be related to a tight functional link with SAM-dependent methyltransferase protein (arCOG00111) present in the predicted operon. |
| 8 | arCOG02267, PitA, Phosphate/sulphate permease | ↓(11) Archaeoglobi | Two paralogs in most of Archaeoglobi. Likely coevolves with PhoU-like protein (arCOG02640) involved in regulation of phosphate transport. Possible subfunctionalization accompanied by stronger purifying selection. |
| 9 | arCOG04308, -, Uncharacterized protein | ↓(12) Sulfolobi; | No paralogs, archaea-specific. In a conserved neighborhood with an uncharacterized Zn-finger containing protein (arCOG00578). No new function predicted, the cause behind strong purifying selection is not clear. |
| ↓(60) Methanocellales | |||
| 10 | arCOG04243, RplS, Ribosomal protein S9 | ↓(13) Thermococci | No paralogs, archaea-specific. No new function predicted, the cause behind strong purifying selection is not clear. |
Upward arrow shows an exceptionally long branch and downward arrow shows an exceptionally short branch.
For additional information, see ftp://ftp.ncbi.nlm.nih.gov/pub/wolf/_suppl/archdev.
All 20 arCOGs with long branches include at least one additional instance of the same arCOG, that is, a (pseudo)paralog, in the respective archaeal lineage, and thus have the prerequisite for sub- or neofunctionalization (Ohno 1970; Lynch and Force 2000). About 2/3 of the cases, for which the additional phylogenetic analysis was conclusive, involve apparent acquisition of a pseudoparalog via HGT; the remaining 1/3 are likely to result from actual fast evolution (table 1, see Supplementary Material online).
With a few exceptions, we were unable to predict the presumed new function for the long-branch variant. Perhaps the most striking case, for which such prediction succeeded, has been already described (Krupovic etal. 2014), albeit discovered without reliance on the predictive framework presented here. This is the second copy of the cas1 gene in Methanomicrobiales. The Cas1 protein is the endonuclease involved in spacer integration (adaptation) in CRISPR–Cas systems. However, the longer Cas1 branch in Methanomicrobiales is associated with transposable elements that have been dubbed Casposons. This prediction was made in silico by in depth analysis of this particular Cas1 subfamily in Methanomicrobiales, Methanosarcinales, and in some bacteria (Krupovic etal. 2014). Subsequently, the horizontal mobility of the Casposons in Methanosarcinales has been demonstrated by comparative genomic analysis (Krupovic etal. 2016), and the endonuclease and integrase activities of the Casposon-encoded Cas1 have been experimentally validated (Hickman and Dyda 2015; Beguin etal. 2016). Given the extreme functional complexity of the CRISPR-Cas, these are likely to be derived forms, so in the proposed evolutionary scenario, Casposons are posited to be the donors of the integrase. However, even regardless of the direction of evolution, the diversification of Cas1 presents a clear case of neofunctionalization.
In most of the analyzed cases of anomalously short branches, at least one other paralog is present in the same lineage, which usually evolves as expected or faster. Furthermore, in most of these cases, there were other archaeal lineages in the same arCOG that encompassed fast-evolving or slow-evolving paralogs, suggesting parallel evolution (table 2, see Supplementary Material online). In a few cases, no duplication in the respective clade was identified, suggestive of clade-specific increased purifying selection. Examples include two conserved gene families, ribosomal protein S9 (arCOG04243), and an uncharacterized protein (arCOG04308) (table 2).
Below we describe five more detailed case studies that represent different evolutionary scenarios for the emergence of clade-specific protein subfamilies with distinctly different branch lengths in the respective phylogenetic trees.
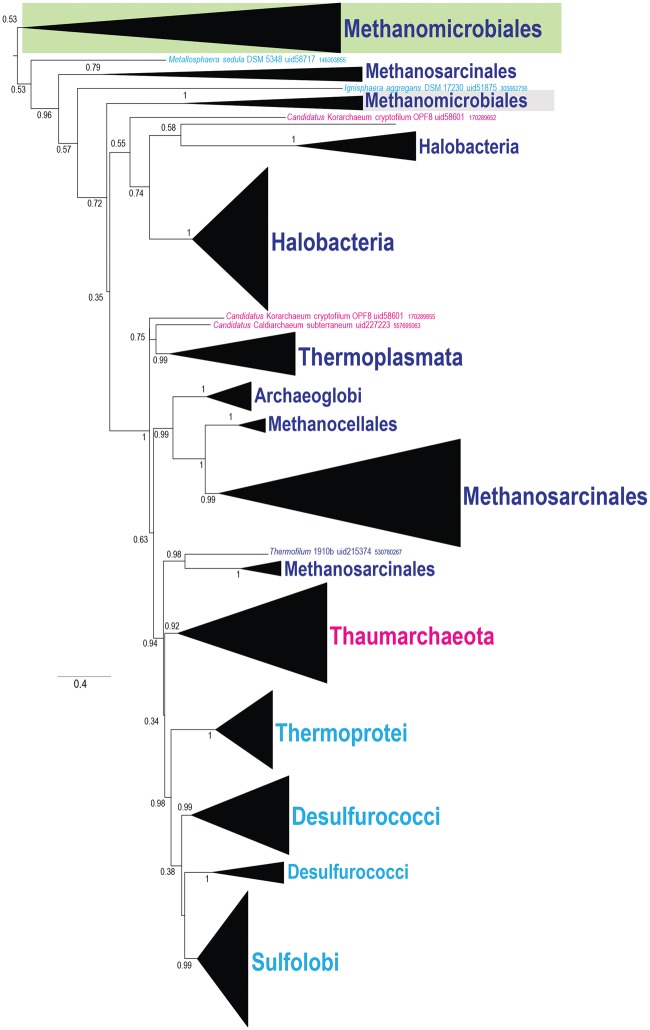
Putative Neofunctionalization of Ribosomal Protein S10
Ribosomal protein S10 (arCOG01758) is represented by a single copy in most archaea but Halobacteria encode a second, fast-evolving paralog. In this case, there is no evidence of HGT because both bacteria and archaea are monophyletic in the S10 tree and so are the major archaeal taxa (fig. 3A). The fast-evolving S10 paralog gene is located in a conserved neighborhood that also includes gene coding for a NUDIX family hydrolase (arCOG01078) and a metal-dependent amidase (arCOG01108). In contrast, the paralog evolving at the expected rate is located in the conserved context of other ribosomal protein genes in a locus syntenic with the orthologous loci in other archaeal lineages (fig. 3B). Thus, neofunctionalization of the faster evolving paralog is most likely. Considering the conservation of the gene neighborhood, the halobacterial S10 paralog, together with the NUDIX hydrolase and amidase, might be involved in “house-cleaning” pathways, that is, elimination of toxic compounds (Galperin etal. 2006), which is a characteristic cellular role of enzymes in these families (Carter etal. 2007; Srouji etal. 2016). Alternatively, given the RNA-binding capacity of S10, this set of proteins could be involved in an uncharacterized RNA modification pathway.
Fig. 3.
—Phylogeny and conserved genomic neighborhoods of ribosomal protein S10 and its paralog. (A) Schematic phylogenetic tree of the S10 ribosomal protein family in Bacteria and Archaea. Collapsed branches are shown by triangles and denoted by the taxon name. Color code: Archaea, black; Bacteria, light blue. (B) Schematic phylogenetic tree of the S10 ribosomal protein family in Archaea. Collapsed branches are shown by triangles and denoted by the taxon name. The branch identified as fast evolving is shown by a green rectangle and the regular branch—by gray rectangle. Color code: Euryarchaeota, dark blue; Crenarchaeota, light blue; Thaumarchaeota and Caldiarchaeum subterraneum, magenta; Korarchaeota, brown; Nanoarchaeota, pink. The conserved gene neighborhoods are shown next to the respective branches. Genes in these neighborhoods are shown schematically by arrows (not to scale). Abbreviations: RpsJ, ribosomal protein S10; TEF1, translation elongation factor EF-1 alpha, GTPase; FusA, translation elongation factor G, EF-G (GTPase); RpsG, ribosomal protein S7; AbgB, metal-dependent amidase/aminoacylase/carboxypeptidase; 1078, arCOG01078 NUDIX family hydrolase.
Functional Diversification through Multiple Duplications: The Transcription Initiation Factor TFIIB
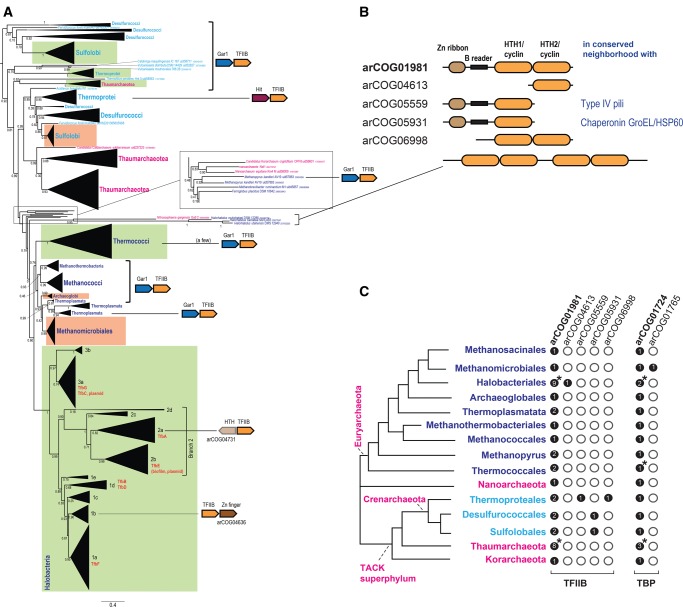
The transcription initiation factor TFIIB family (arCOG01981) is involved in the largest number of evolutionary anomalies among the key housekeeping genes in Archaea (table 1 and fig. 4A;supplementary table 3, Supplementary Material online). TFIIB (TFB) is an essential component of the initiation complexes of archaeal and eukaryotic RNA polymerases (RNAP) which share a common core architecture (Lane and Darst 2010; Werner and Grohmann 2011). No TFIIB or TBP (TATA-binding protein, another key component of the complex) orthologs are found in bacteria, so horizontal transfer from bacteria is unlikely to be a factor in the evolution of this protein family in archaea (Werner and Grohmann 2011). There is no evidence of HGT between major archaeal lineages either as the topology of the TFIIB family tree closely follows the archaeal taxonomy (fig. 4A).
Fig. 4.
—Phylogeny, conserved genomic neighborhoods, domain organization, and phyletic patterns of the TFIIB family. (A) Schematic phylogenetic tree of the TFIIB family. The maximum likelihood tree is from previous work (Makarova etal. 2015). Collapsed branches are shown by triangles and denoted by the corresponding taxon name. Branches for multiple Halobacterial paralogs are labelled by numbers (1–3) and letters (a–e). For functionally characterized genes of Halobacterium salinarum NRC‐1 (Turkarslan etal. 2014), the respective gene name is indicated in orange to the right of the corresponding branch. Branches identified in this work as fast evolving are shown by green rectangles, and slow evolving branches are shown by orange rectangles. Color code is the same as in figure 3. The scale bar indicates a “number of substitutions per site.” The “//” symbol indicates a long branch that is shown not to scale. Conserved gene neighborhoods are shown next to the respective branches (not included for branches in which the neighborhood was not conserved). Genes in these neighborhoods are depicted schematically by arrows (not to scale). Homologous genes are shown by the same color. Abbreviations: HTH, helix turn-helix; GAR1, small nucleolar RNP required for pre-mRNA processing; Hit, HIT family hydrolase. (B) Domain organization of TFIIB and its paralogs . (C) Phyletic patterns of TFIIB-like and TBP-like genes in Archaea. Filled circles show presence and empty circles show absence of the given arCOG in the respective archaeal lineage. The mean number of paralogs is indicated inside each circle. Asterisks indicate that the number of paralogs significantly varies in different genomes of the respective lineage.
It has been recently demonstrated that TFIIB is homologous to bacterial σ factors with which it shares two HTH/cyclin domains (fig. 4B). The HTH2/cyclin domain binds to the so-called BRE (B recognition element) DNA site, whereas the HTH1/cyclin domain interacts with the RNAP (Werner and Grohmann 2011). Both cyclin domains interact with the TBP bound to the TATA box (Goede etal. 2006). In addition, TFIIB contains an N-terminal Zn-ribbon which interacts with the dock domain of the RNAP and the so-called B-reader region interacting with the RNAP II template tunnel (Kostrewa etal. 2009; Sainsbury etal. 2013).
Many archaea possess lineage-specific TFIIB paralogs, most of which retain the same domain organization; in addition, four arCOGs consist of distant homologs of TFIIB, with different domain organizations (fig. 4C). In each of the five fast-evolving archaeal lineages, namely Halobacteria, Thermococci, Sulfolobi, Thermoprotei, and Thaumarchaeota, there are at least two TFIIB paralogs in arCOG01981 (see Supplementary Material online). In Sulfolobi, Thermoprotei, and Thaumarchaeota one paralog (or a group of paralogs) evolves substantially faster than the other, whereas in other archaeal lineages that encode a single TFIIB, this gene typically evolves slower (fig. 4A). In Halobacteria, all paralogs generally evolve faster than expected and thus have been detected by our pipeline as a single fast-evolving branch, but within the halobacterial branch, additional differences in the rates of evolution seem to exist (e.g., the TfbE/TfbA branch is longer than TfbC/TfbG branch) (fig. 4C). We found evidence of acceleration of evolution both in paralogs retained in the probable ancestral context (next to the gar1 gene encoding a small nucleolar RNP that is required for pre-mRNA processing; Baker etal. 2005) and in paralogs lodged in other genomic locations, for which functional change could be readily predicted. Thus, in situ acceleration of evolution is a common phenomenon for the TFIIB genes. Finally, the paralogization of TFIIB appears to have been accompanied by a parallel lineage-specific paralogization of TBP genes in Halobacteria (Facciotti etal. 2007) and Thaumarchaeota although not to the extent of TFIIB. Given that the two factors physically interact, this could be a consequence of their coevolution in these two lineages (fig. 4C).
In most cases, the nature of putative functional differences between the lineage-specific TFIIB paralogs that are expected to be linked to lineage-specific accelerations of evolution remains unknown. However, on several occasions, a conserved context of the fast-evolving TFIIB paralogs prompts prediction of their functions (fig. 4A and C). For example, in halobacterial branches 1 b and 2a, TFIIB is encoded in a predicted operon with other DNA-binding proteins that could be involved in the regulation of these TFIIB genes or modulate their activity as shown for several proteins affecting TFIIB activity in other archaea (Ouhammouch etal. 2003; Ochs etal. 2012). In Thermoprotei, a distant paralog of TFIIB (arCOG05559) is strongly linked to a type IV pili system (Makarova etal. 2016) and can be predicted to specifically regulate the expression of this system, in accord with several experimental observations on involvement of TFIIB in the regulation of genes essential for motility/adhesion (Hidese etal. 2014). Experimental data on TFIIB paralogs in archaea are scarce. In Thermococcus kodakarensis, the two paralogous TFIIB are essential for growth at high temperatures and differentially regulate expression of other genes (Hidese etal. 2014). These findings imply functional diversification of the paralogs although both of them appear to retain the same general function, which appear to be better compatible with the subfunctionalization scenario.
Halobacteria possess the largest number of TFIIB paralogs. Although the entire group shows acceleration of TFIIB evolution, branch 2 (the monophyletic group containing clades 2a, b, c, and d) is particularly long. Many of the TFIIB genes that comprise this branch are encoded by genes located on plasmids that generally tend to evolve faster than genes on the main chromosome. A comprehensive functional analysis of seven TFIIB genes in Halobacterium salinarum NRC‐1 has shown that they form a complex gene regulatory network, in which each factor is adapted to specific environmental conditions (Turkarslan etal. 2014). Furthermore, promoter regions of these genes appear to evolve independently and can therefore increase the flexibility and adaptation potential of the regulatory network (Turkarslan etal. 2014). Thus, in this case, once again, our observation on accelerated evolution of paralogs and experimental evidence both suggest a subfunctionalization scenario.
The present analysis of evolutionary rates points to several new aspects in the evolution of the TFIIB family. Independent emergence of TFIIB paralogs in multiple archaeal lineages suggests that their subfunctionalization is a widespread evolutionary mechanism to fine-tune gene expression under different conditions. We show that the rate changes in TFIIB evolution are not necessarily associated with relocation to a new gene context, and some TFIIB genes start evolving faster in situ, that is, within the ancestral context. Additionally, we describe several diverged and potentially neofunctionalized paralogs of TFIIB with altered domain organizations. These events parallel the burst of diversification in cyclin domain-containing proteins that is observed in eukaryotes (Gunbin etal. 2011; Harashima etal. 2013).
Acceleration of Evolution in the Hsp20 Family of Molecular Chaperones in Methanomicrobiales
Both long and short branches belonging to the top 1% of the extremely deviated clades were detected in the phylogeny of the Hsp20 family (IbpA, arCOG01833) (fig. 5 and supplementary table 1, Supplementary Material online). The Hsp20 proteins belong to the small Heat Shock Proteins (sHSP) family of chaperones that are involved in stress response in all three domains of life (de Jong etal. 1998; MacRae 2000; Rohlin etal. 2005; Ribeiro etal. 2011; Li etal. 2012; Jaspard and Hunault 2016). The sHSPs form large homo-oligomers that are implicated in prevention of aggregation of misfolded proteins (Jaspard and Hunault 2016). Based on domain organization and sequence similarity, sHSPs have been partitioned into 21 classes; all archaeal members of this family belong to class 11 (Jaspard and Hunault 2016).
Fig. 5.
—Schematic phylogenetic tree of the Hsp20 family. The maximum likelihood tree has been built in the course of the previous work (Makarova etal. 2015). Collapsed branches are shown by triangles and denoted by the taxon name and a number. Branch that was identified in this work as fast evolving is shown by a green rectangles and the regular branch—by gray rectangle. Color code is the same as in figure 3.
The phylogeny of COG0071, in which a representative sample of bacterial and archaeal sequences was included, supports the monophyly of most of the archaeal Hsp20s (see Supplementary Material online). In this tree, Methanomicrobiales are represented by two clade-specific branches. One of these branches groups with bacterial sequences suggesting acquisition via HGT. The second branch that was identified by our method as the longest in the arCOG01833 phylogeny groups with most of the other archaea and probably is the ancestral form that experienced a substantial acceleration of evolution in Methanomicrobiales, conceivably, following the acquisition of the bacterial pseudo-paralog.
The Hsp20 protein of Sulfolobus solfataricus (SSO2427) has been shown to possess chaperone activity invitro and protects Escherichia coli from both cold and heat stress (Li etal. 2012). The functional connotations of the Hsp20 family expansion in some archaeal lineages and the dramatic acceleration of evolution in Methanomicrobiales remain unclear.
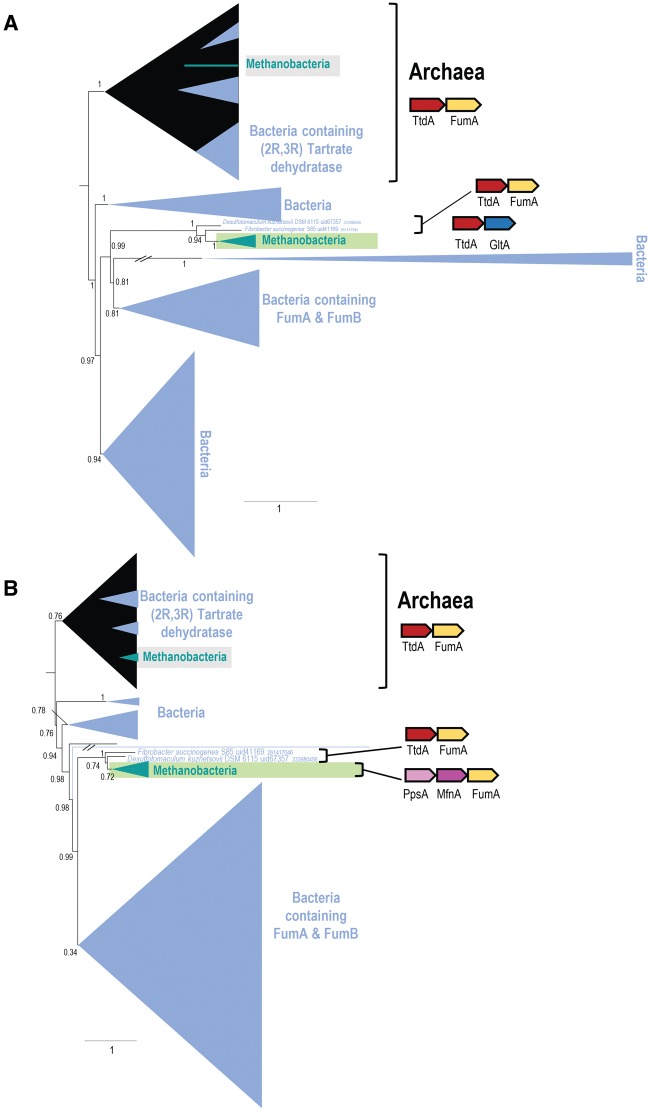
Coevolution of Two Genes: paralogous Tartrate Dehydratases/Fumarate Hydratases in Methanobacteria
In the trees of the TtdA family that consists of α-subunits of tartrate dehydratase (arCOG04407) and N-terminal domains of class I fumarate hydratase, and FumA family, which includes β-subunits of tartrate dehydratase and C-terminal domains of class I fumarate hydratase (arCOG04406), the Methanobacteria clades were detected as long branches (see supplementary table 3 and Supplementary Material online). Both these families are represented by two paralogs in all methanobacterial genomes (with the only exception of Methanosphaera stadtmanae DSM 3091), hereinafter RB (Regular Branch) and LB (Long Branch), whereas all other archaea encode only one enzyme of each family (fig. 6A and B and supplementary fig. 1A and B, Supplementary Material online). In most of the bacteria, these two proteins are fused and comprise class I fumarase (fumD gene product) (Kronen and Berg 2015), a key enzyme of the citric acid cycle that catalyzes the reversible hydration of fumarate to malate but also can convert mesaconate to (S)-citramalate (Tseng etal. 2001). Several closely related bacterial homologs belong to the same family but have been shown to possess (2 S, 3 S)-tartrate dehydratase activity (Kronen and Berg 2015; Kronen etal. 2015). In archaea, these proteins are assumed to possess fumarate hydratase activity and have been implicated in the dicarboxylate/4-hydroxybutyrate cycle pathway in Ignicoccus hospitalis (Huber etal. 2008). Recent phylogenetic analysis of the FumA family resulted in the identification of a distinct branch that consists of fumarase/mesaconases and (2 R, 3 R)-tartrate dehydratases (Kronen and Berg 2015). All archaeal representatives in this clade are monophyletic and are paraphyletic to both fumarase/mesaconase and (2 R, 3 R)-tartrate dehydratase branches (Kronen and Berg 2015). We built trees of the FumA and TtdA families (COGs 1838 and 1951, respectively; fig. 6A and B) for all corresponding archaeal and bacterial sequences, a much more inclusive set than the one analyzed previously (see Supplementary Material online). The LB paralogs of FumA in Methanobacteria do not group with neither archaeal homologs nor functionally characterized bacterial fumarase/mesaconase and (2 R, 3 R)-tartrate dehydratases, but rather fall within an uncharacterized bacterial clade (fig. 6A). A similar topology is observed for the LB of the TtdA family (see Supplementary Material online). These congruent topologies of the two trees suggest that the LB of both families were transferred to Methanobacteria from bacteria, most likely, as an operon. Indeed, in two bacteria (Fibrobacter succinogenes S85 and Desulfotomaculum kuznetsovii DSM6115) that group with the Methanobacteria LB clade in both trees, these two genes from an operon although in Methanobacteria they do not (see Supplementary Material online). Thus, it appears that operon acquisition was followed by genomic rearrangement and accelerated evolution of both genes in the common ancestor of Methanobacteria.
Fig. 6.
—Schematic phylogeny of TtdA (A) and FumA (B) families in Archaea and bacteria. Collapsed branches are shown by triangles and denoted by the taxon name. Branches identified as long is shown by a green rectangle and the regular branch—by gray rectangle. Color code: Methanobacteria, green; rest of the Archaea, black; Bacteria, light blue. Functions for the major clades are assigned according to Kronen etal. (Kronen and Berg 2015). The “//” symbol indicates a long branch that is shown not to scale. Two extremely long branches are interrupted; their actual lengths are ∼5-fold greater than shown (see Supplementary Material online). Conserved gene neighborhoods are shown next to the respective branches. Genes in these neighborhoods are shown schematically by arrows (not to scale). Homologous genes are shown by the same color. Abbreviations: PpsA, phosphoenolpyruvate synthase/pyruvate phosphate dikinase; MfnA, L-tyrosine decarboxylase, PLP-dependent protein; GltA, Citrate synthase.
The LB paralog of TtdA is encoded in a conserved gene context, which includes citrate synthase gltA, whereas the LB paralog of fumA is located next to the phosphoenolpyruvate synthase/pyruvate phosphate dikinase gene ppsA, suggesting that both LB proteins are bona fide fumarate hydratases and are involved in the citric acid cycle (fig. 6A and B). The RB paralogs of ttdA and fumA are encoded in two different contexts that are conserved only in Methanobacteria (see supplementary fig. 1A and B, Supplementary Material online). However, the RB paralogs of FumA and TtdA clearly belong to the respective ancestral archaeal clades, so it appears most likely that these paralogs are also fumarate hydratase subunits as implied by the analysis of I. hospitalis dicarboxylate/4-hydroxybutyrate cycle (Huber etal. 2008). Alternatively, these proteins might function in the citric acid cycle because many archaea encode other enzymes for this pathway, and no alternative enzyme with fumarate hydratase activity has been reported (Makarova etal. 2015). However, the archaeal clade also includes bacterial genes, probably acquired from archaea through HGT, that encode enzymes shown to possess tartrate dehydratase activity (Kronen and Berg 2015). Thus, the functions of the two paralogs in Methanobacteria as well as in other archaea are not entirely clear but acquisition of bacterial homologs could have resulted in functional diversification of the two paralogs.
To summarize this case, the long FumA and TtdA branches in Methanobacteria are explained by a HGT event, which apparently was followed by disruption of the ancestral operon organization and acceleration of the evolution of the two functionally linked enzymes. This acceleration likely was not accompanied by a change of their principal function, but by more subtle changes in substrate preferences and/or differential regulation of expression under special conditions.
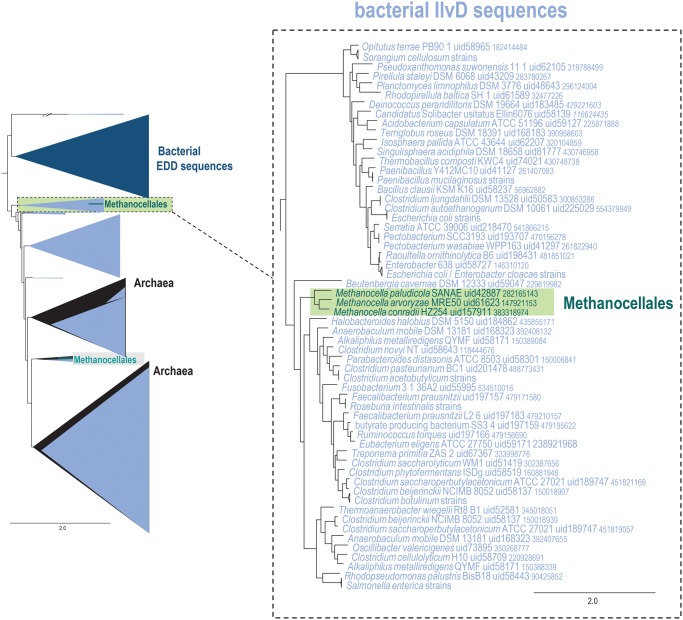
Acquisition of a Xenolog: Dihydroxyacid Dehydratase/Phosphogluconate Dehydratase IlvD
In the tree of dihydroxyacid dehydratase/phosphogluconate dehydratase IlvD (arCOG04045), Methanocellales form a long branch. IlvD catalyzes dehydratation of α,β-dihydroxy β-methylvalerate or α,β-dihydroxyisovalerate to α-keto β-methylvalerate and α,β-ketoisovalerate, the third step of isoleucine and valine biosynthesis pathways, respectively (Myers 1961). This protein is conserved in all three domains of life and has been biochemically characterized in many organisms (Myers 1961; Kanamori and Wixom 1963; Armstrong etal. 1977; Tarleton and Ely 1991; Flint etal. 1993; Oliver etal. 2012) including two archaea, Methanococcus sp. (Xing and Whitman 1991) and Sulfolobus solfataricus (Kim and Lee 2006).
IlvD is present in only one copy in most archaea, with several exceptions including all Methanocellales, which have two paralogs. In addition to IlvD, bacteria often possess a homologous gene for 6-phosphogluconate dehydratase EDD, which is involved in the Entner–Doudoroff pathway (Egan etal. 1992). Both enzymes belong to COG0129, for which we built a phylogenetic tree including representatives from bacteria and archaea (fig. 7, see Supplementary Material online). In this tree, the archaeal sequences divide into two major groups within the IlvD clade (fig. 7). A similar tree topology, without representatives from Methanocellales that were unavailable at the time, has been observed previously for this family (Kim and Lee 2006). The long branch of Methanocellales detected by our approach clearly groups with bacteria, largely Firmicutes, suggesting HGT from bacteria to the common ancestor of Methanocellales (fig. 7). Thus, the long branch of Methanocellales in the phylogeny of arCOG04045 apparently results from acquisition of an additional pseudo-paralog from bacteria by the common ancestor of Methanocellales. This horizontally transferred IlvD enzyme might possess distinct properties or substrate specificity, which remains to be studied experimentally.
Fig. 7.
—Schematic phylogeny of the IlvD family in Archaea and bacteria. Designations are the same as in figures 3–5. Collapsed branches are shown by triangles. IlvD, light blue, EDD, dark blue. Color code: Methanocellales, green; the rest of the archaea, black; bacteria, light blue. The “//” symbol indicates a long branch that is shown not to scale. Two extremely long branches are interrupted; their actual lengths are ∼2-fold greater than shown (see Supplementary Material online).
Discussion
Monophyly of the Major Archaeal Lineages in the arCOG Trees
One of the goals of this work was to quantify the tempo and mode of evolution of major archaeal lineages, under the assumption that these lineages have comparable standing within the archaeal diversity. We selected 13 archaeal taxa consisting of three or more genomes and analyzed gene families in which at least two of these taxa were well-represented (at least 75% of the effective number of genomes). Effective divergence times, inferred for these clades, differed by a factor of 1.9 which makes them, indeed, comparable in terms of the degree of separation from each other (supplementary table 4, Supplementary Material online).
However, these clades differ substantially with respect to the number of sequenced genomes (from 3 to 27) and, more importantly, the patterns of genome sampling. For example, Thermococci and Delulfurococci are represented by 16 genomes each, but the sums of the branch lengths in the ribosomal protein tree, corresponding to these clades, differ by a factor of 5.6, indicating that the Delulfurococci are almost 6-fold more diverse than the Thermococci. The taxa that cover more evolutionary history than others had more chance to experience xenologous gene displacement and therefore are expected to be underrepresented with respect to the monophyletic groups in the gene trees. Indeed, among the arCOGs in which Thermococci are well-represented, Thermococci form a clade in 90%, whereas among the arCOGs with well-represented Desulfurococci, a desulfurococcal clade exists in only 25%. Overall, the differences in the intraclade diversity explain most of the disparities in the representation of the taxa in the analysis.
Evolutionary Rate Variability
The distribution of the deviations of the observed clade heights from the expected values is smooth, and in the middle, is approximately lognormal and relatively narrow, with 90% of the clades falling within a factor of 1.5 of the expectation (fig. 2). In agreement with previous observations (Wolf etal. 2013), the combination of the arCOG-specific relative rate and the taxon-specific effective divergence times predicts the clade heights remarkably well, explaining 66% of the variance in the observations. However, both tails of the distribution are clearly wider than expected in a normal distribution, suggesting that the largest deviations result from mechanisms that are distinct from simple accumulation of random fluctuations and are likely to have biological underpinning.
In particular, the occurrence of (pseudo)paralogs seems to be among the major factors that shape the distributions of the deviations. It strongly correlates with the variance of rates between the taxa (largest variation in the Sulfolobi, the Thermococci, and the Methanocellales) and between functional groups of genes (largest variation in the cell cycle control, signal transduction and defense categories). Paralogs and pseudo-paralogs are predictably most common in the extremes of the distribution of the deviations from the expected evolutionary rate, accounting for the largest apparent accelerations and decelerations, in agreement with the key role of gene duplication in neo- and subfunctionalization (Ohno 1970; Lynch and Force 2000; Innan and Kondrashov 2010). The exact evolutionary scenario, that is, subfunctionalization versus neofunctionalization, is difficult to infer unambiguously in most of the cases where acceleration of evolution was detected. However, in general, subfunctionalization appears to be a more common route of evolution than neofunctionalization (Force etal. 1999; Lynch etal. 2001; Lynch 2007), and many examples of accelerated evolution in archaea considered here are at least compatible with the subfunctionalization scenario. It should be noted that all case studies discussed here include genes that are conserved in a group of archaeal genomes which rules out pseudogenization as a cause of the observed acceleration of evolution. However, inactivation of the ancestral function can be one of such causes as discussed for several examples.
The observable effects of paralogy and pseudo-paralogy are similar and consist in the presence of unexpectedly long branches in trees but the underlying processes are quite different. The case of paralogs involves a bona fide acceleration of evolution, whereas in the case of pseudo-paralogs, the same effect can result simply from acquisition of a gene via HGT from a distant group. The question remains open as to the frequency and magnitude of the actual acceleration of evolution following HGT. Finally, several cases of evolutionary acceleration that do not appear to be associated with detectable duplication or HGT events were identified as well. The mechanisms behind such phenomena remain unclear but might involve, for example, gain of a functionally redundant but not paralogous gene, or else, correlated changes in functionally linked genes.
Substantial deceleration of evolution is less frequent than acceleration but nevertheless, multiple cases of such apparent slow-down were detected. As in the case of acceleration in the absence of (pseudo)paralogs, the mechanisms of deceleration remain obscure. In general terms, it seems likely that evolution of a gene could slow down in the process that could be considered the reverse of subfunctionalization. A loss or inactivation of functionally analogous genes could strengthen the constraints on the remaining copy. Neofunctionalization, or perhaps more precisely, acquisition of an additional function, resulting in stronger purifying selection once the additional function is fixed, also appears to be a possibility. To test these conjectures, genome wide analysis of correlated changes between genes is required.
Notably, in many cases, the same gene family includes lineages with both accelerated and decelerated evolution (tables 1 and 2), suggestive of enhanced functional and perhaps structural plasticity of the respective proteins. It has to be noted that all analysis reported here includes acceleration or deceleration of evolution relative to the characteristic evolutionary rate of a given gene family; by design, changes of evolutionary rate that involve an entire family could not be detected by our protocol.
The measurable variation of evolutionary distances comes from many sources. Sampling fluctuations due to finite gene lengths contribute, roughly, only a half of measured variation even at relatively short evolutionary distances and in relatively highly conserved genes (Wolf etal. 2013). In addition to variations in selective pressure due to functional changes, other sources of observed rate variation include fluctuations of the effectiveness of selection due to varying population size, biased gene conversion, recombination and other intragenomic processes, HGT from lineages with different inherent evolutionary rates, tree reconstruction and distance estimation artifacts, and more. All these factors obscure the true functional signal or might even conspire to create an illusion of functionally relevant change where none occurred. The existing techniques to directly estimate the (presumably, functionally determined) selective pressure (Li etal. 1985; Zhang 2000; Hanada etal. 2007) are not practical for an analysis on the scale of the present study and at the long characteristic evolutionary distances analyzed here. Specifically, the classical measure of the selection pressure that affects the sequence of a protein-coding gene, the dN/dS ratio, is based on the assumption that, compared with the amino acid substitutions, mutations in the synonymous codon positions are effectively neutral (Li etal. 1985). The dN/dS ratio has well-understood statistical properties and a well-defined neutral expectation that can serve as a universal reference point (dN/dS = 1). The principal technical drawback of this measure is that dS estimates are extremely noisy, primarily because synonymous substitutions saturate fast and therefore the synonymous distance estimates are subject to large statistical error. Limiting the comparison to closely related sequences (dS ≪ 1) presents a different problem as the number of nonsynonymous substitutions becomes very small (again, increasing the estimate error) and the presence of nonfixed population polymorphisms in the mixture adds to the uncertainty. To mitigate these problems, the ratio between the rates of radical and conservative amino acid substitutions has been proposed to evaluate the selection pressure (Zhang 2000). As both estimates involve amino acid substitutions that occur much less frequently than (largely neutral) synonymous nucleotide substitutions, the number of mutations remains tractable at much longer evolutionary distances. However, unlike the dN/dS ratio, the neutral expectation for Kr/Kc is not well defined because it depends on the amino acid composition and structural properties of the analyzed gene product (Dagan etal. 2002). Furthermore, because these estimates, ultimately, rely on the reconstructions of ancestral sequences at each node of the phylogenetic tree, they become progressively less reliable at increasing evolutionary distances. Examples of successful applications of Kr/Kc analysis typically involve distances in the range that is characteristic of the divergence of mammals (Zhang 2000; Hanada etal. 2007; Nabholz etal. 2013; Mohlhenrich and Mueller 2016). This methodology is not applicable to the evolutionary scale of the radiation of archaeal phyla, which is relevant for the present work. Because of this limitation and considering the various uncertainties mentioned earlier, many of the functional inferences proposed here remain tentative.
Functional Changes Associated with Evolutionary Rate Deviations: Patterns and Problems with Prediction
Despite considerable effort, we were able to propose specific new functions only for a minority of the accelerated and decelerated lineages detected in this work. On several occasions, for example, ribosomal protein S10 or some TFIIB paralogs, such prediction was enabled by the change in the genomic context or substantial alteration of the domain organization of the accelerated gene. Among the genes with extreme deviation of evolutionary rates from the expectation, there is little evidence of change in the biochemical (enzymatic or binding) activity, even if the cellular function appears to change. Thus, in most cases, the observed substantial change of the evolutionary rate seems to have been caused by relatively subtle modulation of functional constraints compared with “normally” evolving paralogs. Such limited in scope but evolutionarily consequential functional effects might include change of substrate or binding specificity, the network of interaction partners or regulation. Although not numerous, the genes with major changes in evolutionary regimes identified here present interesting targets for follow-up experiments.
Concluding Remarks
Using the computational framework we developed to explore the distribution of evolutionary rates for genes families and to identify clades deviating from the expectation, we performed a comprehensive analysis of the evolutionary rates of archaeal genes. This analysis revealed many genes that evolved significantly faster or slower than expected from the normal distribution of rates for the given family, which is, probably, indicative of biological causes behind the observed difference in evolutionary rates. Changes in the rate of evolution are often, although not always, associated with gene paralogy or pseudo-paralogy, in a general agreement with the neofunctionalization and subfunctionalization regimes of evolution of paralogs. Inference of the specific functional changes for accelerated and decelerated genes proved difficult, given that most of them appear to retain the major biochemical activity characteristic of the respective family. Nevertheless, on several occasions, change of the genomic context allowed us to substantiate the neofunctionalization mode of evolution and to predict the new function, at least in general terms. The approach developed and applied here enables selection of targets for experimentation in search of new biology, even in the absence of precise functional predictions.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
C.P., K.S.M., Y.I.W., and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Literature Cited
- Armstrong FB, Muller US, Reary JB, Whitehouse D, Crout DHG.. 1977. Stereoselectivity and stereospecificity of the alpha, beta-dihydroxyacid dehydratase from Salmonella typhimurium. Biochim Biophys Acta 498(2):282–293. [DOI] [PubMed] [Google Scholar]
- Baker DL, et al. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19(10):1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguin P, Charpin N, Koonin EV, Forterre P, Krupovic M.. 2016. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 44(21):10367–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenue DL, Gilner DM, Davis RS, Bennett B, Holz RC.. 2003. Substrate specificity, metal binding properties, and spectroscopic characterization of the DapE-encoded N-succinyl-L, L-diaminopimelic acid desuccinylase from Haemophilus influenzae. Biochemistry 42:10756–10763. [DOI] [PubMed] [Google Scholar]
- Bromham L, Penny D.. 2003. The modern molecular clock. Nat Rev Genet. 4(3):216–224. [DOI] [PubMed] [Google Scholar]
- Brown BP, Wernegreen JJ.. 2016. Deep divergence and rapid evolutionary rates in gut-associated Acetobacteraceae of ants. BMC Microbiol. 16(1):140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter EL, et al. 2007. Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p-aminobenzoyl-glutamate. J Bacteriol. 189(9):3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Talmor Y, Graur D.. 2002. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol. 19(7):1022–1025. [DOI] [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JA.. 1998. Genealogy of the alpha-crystallin–small heat-shock protein superfamily. Int J Biol Macromol. 22(3-4):151–162. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Banfield JF.. 2012. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336(6080):462–466. [DOI] [PubMed] [Google Scholar]
- Dowling DP, Di Costanzo L, Gennadios HA, Christianson DW.. 2008. Evolution of the arginase fold and functional diversity. Cell Mol Life Sci. 65(13):2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Egan SE, et al. 1992. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: sequence analysis and localization of promoters for the edd-eda operon. J Bacteriol. 174(14):4638–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facciotti MT, et al. 2007. General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci U S A. 104(11):4630–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK.. 1993. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem. 268(20):14732–14742. [PubMed] [Google Scholar]
- Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV.. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43(D1):D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Moroz OV, Wilson KS, Murzin AG.. 2006. House cleaning, a part of good housekeeping. Mol Microbiol. 59(1):5–19. [DOI] [PubMed] [Google Scholar]
- Goede B, Naji S, von Kampen O, Ilg K, Thomm M.. 2006. Protein-protein interactions in the archaeal transcriptional machinery: binding studies of isolated RNA polymerase subunits and transcription factors. J Biol Chem. 281(41):30581–30592. [DOI] [PubMed] [Google Scholar]
- Groussin M, Gouy M.. 2011. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol Biol Evol. 28(9):2661–2674. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, Rannala B.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Gunbin KV, Suslov VV, Turnaev II, Afonnikov DA, Kolchanov NA.. 2011. Molecular evolution of cyclin proteins in animals and fungi. BMC Evol Biol. 11:224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Ettema TJ.. 2011. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 19(12):580–587. [DOI] [PubMed] [Google Scholar]
- Hanada K, Shiu SH, Li WH.. 2007. The nonsynonymous/synonymous substitution rate ratio versus the radical/conservative replacement rate ratio in the evolution of mammalian genes. Mol Biol Evol. 24(10):2235–2241. [DOI] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A.. 2013. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23(7):345–356. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Dyda F.. 2015. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 43(22):10576–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidese R, et al. 2014. Different roles of two transcription factor B proteins in the hyperthermophilic archaeon Thermococcus kodakarensis. Extremophiles 18(3):573–588. [DOI] [PubMed] [Google Scholar]
- Huber H, et al. 2008. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci U S A. 105(22):7851–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11(2):97–108. [DOI] [PubMed] [Google Scholar]
- Iwasaki T. 2010. Iron-sulfur world in aerobic and hyperthermoacidophilic archaea Sulfolobus. Archaea 2010:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspard E, Hunault G.. 2016. sHSPdb: a database for the analysis of small heat shock proteins. BMC Plant Biol. 16(1):135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori M, Wixom RL.. 1963. Studies in valine biosynthesis. V. Characteristics of the purified dihydroxyacid dehydratase from spinach leaves. J Biol Chem. 238:998–1005. [PubMed] [Google Scholar]
- Kim S, Lee SB.. 2006. Catalytic promiscuity in dihydroxy-acid dehydratase from the thermoacidophilic archaeon Sulfolobus solfataricus. J Biochem. 139(3):591–596. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV.. 2002. Selection in the evolution of gene duplications. Genome Biol. 3(2):RESEARCH0008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D, et al. 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462(7271):323–330. [DOI] [PubMed] [Google Scholar]
- Kronen M, Berg IA.. 2015. Mesaconase/fumarase FumD in Escherichia coli O157: H7 and promiscuity of Escherichia coli class I fumarases FumA and FumB. PLoS One 10(12):e0145098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronen M, Sasikaran J, Berg IA, Liu S-J.. 2015. Mesaconase activity of class I fumarase contributes to mesaconate utilization by Burkholderia xenovorans. Appl Environ Microbiol. 81(16):5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV.. 2014. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 12:36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV.. 2016. Recent mobility of casposons, self-synthesizing transposons at the origin of the CRISPR-Cas immunity. Genome Biol Evol. 8(2):375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, Darst SA.. 2010. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 395(4):671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D-C, Yang F, Lu B, Chen D-F, Yang W-J.. 2012. Thermotolerance and molecular chaperone function of the small heat shock protein HSP20 from hyperthermophilic archaeon, Sulfolobus solfataricus P2. Cell Stress Chaperones 17(1):103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC.. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 2:150–174. [DOI] [PubMed] [Google Scholar]
- Liu LJ, et al. 2014. Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J Biol Chem. 289(39):26949–26959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome archiecture. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A.. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154(1):459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Katju V.. 2004. The altered evolutionary trajectories of gene duplicates. Trends Genet. 20(11):544–549. [DOI] [PubMed] [Google Scholar]
- Lynch M, O’Hely M, Walsh B, Force A.. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159(4):1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae TH. 2000. Structure and function of small heat shock/alpha-crystallin proteins: established concepts and emerging ideas. Cell Mol Life Sci. 57(6):899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV.. 2010. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct. 5(1):33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV.. 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol. 5(11):a012963.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Albers SV.. 2016. Diversity and evolution of type IV pili systems in Archaea. Front Microbiol. 7:667.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV, Kelman Z.. 2012. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct. 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV.. 2015. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between thermococcales, methanococcales, and methanobacteriales. Life (Basel) 5(1):818–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV.. 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33(14):4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV.. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 8(10):731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43(D1):D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobbie AM, et al. 2009. Structural and functional characterisation of a conserved archaeal RadA paralog with antirecombinase activity. J Mol Biol. 389(4):661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlhenrich ER, Mueller RL.. 2016. Genetic drift and mutational hazard in the evolution of salamander genomic gigantism. Evolution 70(12):2865–2878. [DOI] [PubMed] [Google Scholar]
- Mozhayskiy V, Tagkopoulos I.. 2012. Horizontal gene transfer dynamics and distribution of fitness effects during microbial in silico evolution. BMC Bioinformatics 13(Suppl 10):S13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JW. 1961. Dihydroxy acid dehydrase: an enzyme involved in the biosynthesis of isoleucine and valine. J Biol Chem. 236:1414–1418. [PubMed] [Google Scholar]
- Nabholz B, Uwimana N, Lartillot N.. 2013. Reconstructing the phylogenetic history of long-term effective population size and life-history traits using patterns of amino acid replacement in mitochondrial genomes of mammals and birds. Genome Biol Evol. 5(7):1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, et al. 2004. Genome-wide molecular clock and horizontal gene transfer in bacterial evolution. J Bacteriol. 186(19):6575–6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs SM, et al. 2012. Activation of archaeal transcription mediated by recruitment of transcription factor B. J Biol Chem. 287(22):18863–18871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin-Heidelberg-New York: Springer-Verlag. [Google Scholar]
- Oliver JD, et al. 2012. The Aspergillus fumigatus dihydroxyacid dehydratase Ilv3A/IlvC is required for full virulence. PLoS One 7(9):e43559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP.. 2003. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci U S A. 100(9):5097–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D, Brochier-Armanet C.. 2015. Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol Biol Evol. 32(5):1242–1254. [DOI] [PubMed] [Google Scholar]
- Powers DMW. 2011. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness & correlation. J Mach Learn Technol. 2:37–63. [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymann K, Brochier-Armanet C, Gribaldo S.. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci U S A. 112(21):6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro DA, et al. 2011. The small heat shock proteins from Acidithiobacillus ferrooxidans: gene expression, phylogenetic analysis, and structural modeling. BMC Microbiol. 11(1):259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlin L, et al. 2005. Heat shock response of Archaeoglobus fulgidus. J Bacteriol. 187(17):6046–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P.. 2013. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493(7432):437–440. [DOI] [PubMed] [Google Scholar]
- Snir S, Wolf YI, Koonin EV.. 2012. Universal pacemaker of genome evolution. PLoS Comput Biol. 8(11):e1002785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snir S, Wolf YI, Koonin EV.. 2014. Universal pacemaker of genome evolution in animals and fungi and variation of evolutionary rates in diverse organisms. Genome Biol Evol. 6(6):1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21(7):951–960. [DOI] [PubMed] [Google Scholar]
- Srouji JR, Xu A, Park A, Kirsch JF, Brenner SE.. 2016. The evolution of function within the Nudix homology clan. Proteins 85(5):775–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton JC, Ely B.. 1991. Isolation and characterization of ilvA, ilvBN, and ilvD mutants of Caulobacter crescentus. J Bacteriol. 173(3):1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Rocha EP.. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7(1):e1001284.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CP, Yu CC, Lin HH, Chang CY, Kuo JT.. 2001. Oxygen- and growth rate-dependent regulation of Escherichia coli fumarase (FumA, FumB, and FumC) activity. J Bacteriol. 183(2):461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkarslan S, et al. 2014. Niche adaptation by expansion and reprogramming of general transcription factors. Mol Syst Biol. 7(1):554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F, Grohmann D.. 2011. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 9(2):85–98. [DOI] [PubMed] [Google Scholar]
- Wiedenbeck J, Cohan FM.. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 35(5):957–976. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ.. 2009. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 106(18):7273–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Snir S, Koonin EV.. 2013. Stability along with extreme variability in core genome evolution. Genome Biol Evol. 5(7):1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing RY, Whitman WB.. 1991. Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J Bacteriol. 173(6):2086–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV.. 2008. The deep archaeal roots of eukaryotes. Mol Biol Evol. 25(8):1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI.. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7(5):e36972.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2000. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol. 50(1):56–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.