Abstract
Pervasive transcription of genomes generates multiple classes of non-coding RNAs. One of these classes are stable long non-coding RNAs which overlap coding genes in antisense direction (asRNAs). The function of such asRNAs is not fully understood but several cases of antisense-dependent gene expression regulation affecting the overlapping genes have been demonstrated. Using high-throughput yeast genetics and a limited set of four growth conditions we previously reported a regulatory function for ∼25% of asRNAs, most of which repress the expression of the sense gene. To further explore the roles of asRNAs we tested more conditions and identified 15 conditionally antisense-regulated genes, 6 of which exhibited antisense-dependent enhancement of gene expression. We focused on the sporulation-specific gene SPS100, which becomes upregulated upon entry into starvation or sporulation as a function of the antisense transcript SUT169. We demonstrate that the antisense effect is mediated by its 3′ intergenic region (3′-IGR) and that this regulation can be transferred to other genes. Genetic analysis revealed that SUT169 functions by changing the relative expression of SPS100 mRNA isoforms from a short and unstable transcript to a long and stable species. These results suggest a novel mechanism of antisense-dependent gene regulation via mRNA isoform switching.
INTRODUCTION
Pervasive transcription is a common feature of eukaryotic genomes (1–4). Transcriptomics studies in the yeast Saccharomyces cerevisiae led to the annotation of hundreds of previously unknown non-coding RNAs (5–10) originating from bidirectional promoters that typically form within nucleosome-depleted regions (NDRs) up- and downstream of the open reading frames (ORFs) (6,7,11–13). As the yeast genome is very compact, intergenic regions (IGRs) typically span only one NDR. Combined with the propensity of NDRs for bidirectional transcription this frequently results in non-coding RNAs (ncRNAs) overlapping neighbouring genes in antisense direction. These particular types of ncRNAs are termed antisense RNAs (asRNAs). While the major termination and degradation pathways involved in non-coding transcription have been characterized, a comprehensive understanding of the functions and mechanisms that can be exerted by asRNAs is still lacking.
It is known that ncRNAs can function as regulators of the genes they overlap, typically interfering with the expression of the overlapping genes mostly by cis-acting mechanisms (14), often collectively referred to as transcriptional interference (15). Of note, all of the mechanisms are independent of RNA interference (RNAi) as yeast lacks the respective machinery (16,17).
To date, most studies showed that the expression of ncRNAs alters the chromatin environment. For example, the transcription of the ncRNA IRT1 across the IME1 promoter silences IME1 by Set1- and Set2-dependent methylation of H3 at lysine residues 4 and 36, which in turn results in the recruitment of the Set3C and Rpd3C(S) histone deacetylase (HDAC) complexes, respectively (18). ncRNA-mediated regulation of gene expression by HDAC-dependent mechanisms has also been reported, among others, for the GAL gene cluster (19,20), FLO11 (21,22) and PHO84 (23,24).
Other mechanisms include the modulation of nucleosome occupancy patterns by nucleosome remodellers. This can lead to enhanced transcription of the gene upon induction, for example when PHO5 is induced by a ncRNA in phosphate starvation (25,26), or to reduced recruitment of activators, for example in the SER3 gene (27–30).
However, the diversity of mechanisms of gene regulation by ncRNAs extends beyond chromatin. For example, exposure of yeast to osmotic stress leads to Hog1-dependent transcription of a ncRNA antisense to CDC28. This asRNA is thought to induce CDC28 expression through the establishment of a gene loop and relocation of chromatin-bound Hog1 from the 3′ untranslated region (UTR) to the +1 nucleosome region of CDC28 (31).
In many cases, the mechanism underlying the impact of ncRNA on gene expression is not fully understood. IME4, for example, is suppressed by its asRNA RME2 (32). This repression depends on the presence of an ∼400 bp sequence at the 5′ end of the ORF. RME2 transcription does not appear to alter the occupancy of TATA-binding protein at the sense promoter, which led the authors to speculate that repression is mediated by premature sense termination (33). More direct evidence for a truncation-related mechanism was provided for KCS1, where an antisense RNA seems to induce N-terminal truncation of the protein (34). Finally, ncRNAs were also suggested to suppress gene expression by preventing the recruitment of transcription factors, for example at the ADH1 promoter (35).
Considering this intriguing mechanistic variety and the fact that there are several hundred asRNAs reported in S. cerevisiae (6,36,37) it seems plausible that further mechanisms involved in asRNA-dependent gene regulation remain to be discovered, e.g. by probing more genes and/or conditions.
Recently, we employed a strategy based on an unidirectional terminator (38) and seamless gene tagging (39) to investigate the impact of the selective inhibition of >150 asRNAs on expression of the corresponding sense genes (40). Using super-folder GFP (sfGFP) tagging and quantitative microscopy under four growth conditions (YPAD/YPGal/YPE/SC) we found that roughly a quarter of asRNAs had (mostly weak) repressive effects on the protein levels of the overlapping sense gene.
Here, we extended the analysis of asRNA-dependent regulation by testing the same set of genes under a different set of growth conditions using growth on agar plates. We discovered novel regulatory targets of asRNAs and identified several genes that were positively regulated by antisense transcription. We focused on the SPS100 gene as it was strongly upregulated by antisense transcription. We investigated the gene expression dynamics and the underlying regulatory mechanism triggered by the asRNA. Our results demonstrated a novel role for the asRNA on the regulation of the abundance of different SPS100 mRNA isoforms, which extends the repertoire of mechanisms by which asRNAs can regulate sense expression.
MATERIALS AND METHODS
Yeast strains, plasmids and culturing conditions
Unless otherwise stated, yeast cultures were grown using standard procedures (41) for 3 days at 30°C in the indicated media. Colonies on agar plates were pinned using a RoToR handling robot (Singer Instruments). For growth into stationary phase (starvation), synthetic complete medium (SC) with 0.1% glucose and an inoculation density of OD600 = 0.05 was used as starting condition. For exponential growth, cells were grown in SC with 2% glucose to an OD600 of 0.5–1.0 for at least 8 h.
For thiolutin treatment the cells were grown in stationary phase for 24 h in liquid SC medium with 0.1% glucose at 30°C. Thiolutin (Abcam) was added to final concentration of 50 μM (from 10 mM stock in dimethyl sulfoxide) and aliquots of cells (10 ml) were taken with 12 min intervals for RNA extraction.
Yeast strain construction was performed using standard protocols for yeast transformation and polymerase chain reaction (PCR) targeting (42,43) and strain validation using colony PCRs, in some cases followed by sequencing. Strains are listed in Supplementary Table S1. Antisense library strains were published previously (40). Plasmids were constructed using standard procedures (44) and verified by sequencing. All plasmids are listed in Supplementary Table S2.
Whole colony fluorescence measurements and data analysis
For whole colony fluorescence measurements yeast were first grown using 96-well cell culture plates in YPD medium containing ClonNat (Jena Bioscience) (100 mg/l) at 30°C to saturation, followed by pinning onto agar plates with the appropriate growth medium using a pinning robot RoToR (Singer Instruments) and by merging four 96-well plates on one 384 colony format plate. Following incubation for 1 day the cells were pinned to fresh plates in four replicates, resulting in a final 1536 colony format plate. The agar plates were then incubated for 1–3 days depending on the experiment.
The fluorescence of individual colonies on plates was measured using a fluorescence plate reader (Infinite M1000Pro, Tecan) using the following excitation/emission wave length settings for sfGFP: 488/510 ± 10 nm; for mCherry: 587/610 ± 10 nm. For analysis, raw sfGFP intensity values were normalized to a non-fluorescent control strain distributed evenly across the plate to correct for possible position effects. Mean sfGFP values of each protein fusion were calculated between the replicates. The impact of antisense abrogation (antisense effect) was calculated as the ratio of sfGFP values of strains with the PHO5T:scr and the PHO5T insertions (40). Genes were scored as hits using the following criteria: fluorescence levels of sfGFP fusions was 2-fold above background; antisense effect value was lower than 0.8 or higher than 1.35; protein expression of sfGFP fusions with PHO5T:scr and WT constructs differed by not more than 50%; protein expression differences between the fusions with PHO5T:scr and PHO5T were significant (FDR < 10%). Statistical analyses were performed using R (R Development Core Team 2008): P-values were determined with the Student's t-test and corrected for multiple testing using the Benjamini–Hochberg approach (45).
RNA methods
RNA extraction
Total RNA was extracted from 5–10 ml of cell culture following a hot phenol protocol (46) and genomic DNA was subsequently removed with the TURBO DNA-free Kit (Life Technologies) according to the manufacturer’s instructions.
Strand-specific RT-qPCRs
DNase-treated RNA was reverse transcribed using 2 pmol gene-specific primer and 1 μg RNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Actinomycin D (20 μg/ml) was added to each reaction to ensure strand-specificity of the reverse transcription (47). Obtained cDNA samples and controls (without reverse transcriptase) were diluted 1:20 and 2.5 μl were used for amplification using a LightCycler 480 SYBR Green I Master Mix (Roche) and a LightCycler 480 II instrument (Roche, software release 1.5.0593) with an annealing temperature of 55°C. ALG9 was used as a reference gene. For the list of used primers, see Supplementary Table S3.
3′ RACE
3′ RACE was performed as previously described (48) using 5 μg of total RNA as input. PCR products were gel extracted using the QIAquick Gel Extraction Kit (Qiagen) and sent for sequencing. See Supplementary Table S3 for a list of the primers used.
Northern blotting
Northern blots were performed as previously described (49). A total of 20 μg of total RNA was used per lane and separated by electrophoresis on a formaldehyde 1.2% agarose gel. The RNA was subsequently transferred to a positively charged nylon membrane (Amershan Hybond N+, GE Healthcare) using electroblotting. Prior to hybridization the membranes were blocked using UltraHyb hybridization buffer (Ambion) for 1 h at 42°C. Hybridization was performed for 14–16 h using DIG-labelled probes (DIG Oligonucleotide 3′-End Labeling Kit, Roche). See Supplementary Table S3 for sequences of the used probes. Probes were detected using an AP-coupled Anti-DIG Fab fragments (Roche) followed by incubation with CDP-star substrate (Roche) and measurement with an LAS 4000 imaging system (Fujifilm, GE Healthcare).
Time lapse fluorescence microscopy for SPS100 mRNA expression
Imaging was performed on a DeltaVision RT microscope (Applied Precision) equipped with a 60×/1.40 NA Plan Apo oil objective (Olympus), a CoolSNAP HQ camera (Photometrics) and appropriate filters. Full fields of view were scanned through the axial axis (11 planes per time point, spaced by 500 nm, exposure time per plane 50 ms) every 10 s over the course of 30 min. Maximum projected images were used for detection of individual transcription events using u-track software (50) and detected transcriptional intensity traces were analysed using a Hidden Markov model in Matlab (MathWorks).
Protein methods
Western blotting
Proteins were extracted using the trichloroacetic acid method (51). Proteins were then separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (52) and transferred to nitrocellulose membranes using wet blotting (XCell II Blot Module, Invitrogen). Membranes were incubated overnight with primary anti-sfGFP antibodies (Abcam, ab6556) or anti-Pgk1 antibodies as loading control (ThermoFisher Scientific, Monoclonal antibody 22C5D8). Primary antibodies were detected using secondary antibodies labelled with Alexa 680 (Invitrogen, Germany) or IRDye800 (Rockland Immunochemicals Inc., USA). Detection was performed using an Odyssey Infrared Imaging Systems (Li-Cor Biosciences).
Flow cytometry
For flow cytometry of yeast cells expressing fluorescent proteins a flow cytometer (BD FACSCanto RUO HTS) equipped with the necessary laser and filters was used. 20000 cells were recorded per sample. Data were processed in Matlab (Mathworks).
Time lapse fluorescence microscopy for Sps100-sfGFP protein expression
Cells were inoculated in SC, 0.1% glucose and grown to OD600 = 0.5 before transferring them to a CellASIC microfluidic perfusion chamber (Millipore). The growth medium was then replaced by preconditioned starvation medium obtained by filtering the medium of cells grown for 3 days in SC, 0.1% glucose. Fluorescence imaging was started 6 h following this medium change. For imaging a Nikon Ti-E epifluorescence microscope equipped with a 60× ApoTIRF oil-immersed objective (1.49 NA, Nikon), a 2048 × 2048 pixel (6.5 μm) sCMOS camera (Flash4, Hamamatsu) and an autofocus system (Perfect Focus System, Nikon) with either bright field or 470/40 excitation and 525/50 emission filters (Chroma). Images from one plane in the middle of the cell were taken every 15 min over the course of 24 h. Tracking and segmentation of cells was performed automatically using a custom script based on the CellX software (53) and quantified in R (R Development Core Team 2008) and Matlab (MathWorks).
Yeast sporulation
Sporulation experiments were conducted using hybrid strains constructed with the well-sporulating strain SK1. The poorly sporulating strains containing the different modified SPS100 loci (wt/PHO5T/PHO5T:scr) in the BY4741 background were mated to an SK1 strain of the opposite mating type containing a deletion of SPS100. For sporulation, diploid strains were grown for 24 h in YPD media (30°C, 230 rpm) to saturation, diluted 1:50 with YPA media (YP with 1% potassium acetate) and grown for another 13.5 h (30°C, 230 rpm). Cells were harvested (3 min, 2000 rpm, room temperature), washed with water (room temperature) and suspended in sporulation media (1% potassium acetate). Transfer to sporulation media was considered as starting point for the sporulation time course measurements.
RESULTS
Whole colony fluorescence assays identify antisense-regulated genes across many conditions
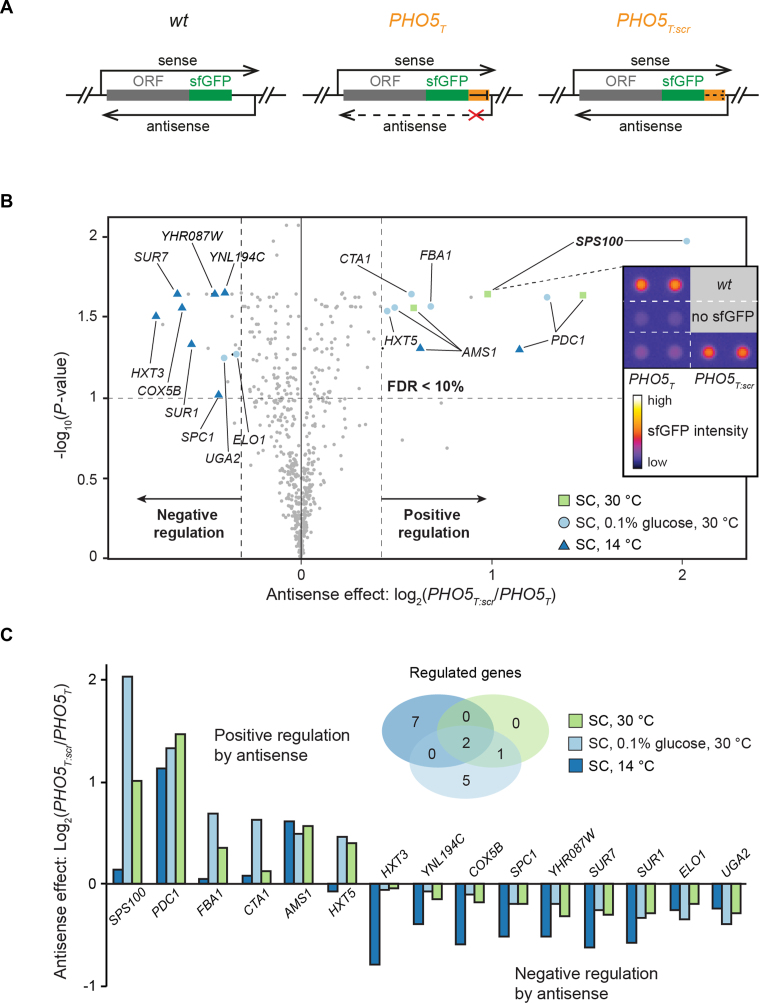
Regulation of gene expression by antisense transcription has been reported to be influenced by the growth condition. In order to identify novel targets with condition-specific regulation by antisense transcription, we used a yeast strain collection that was previously established in our laboratory (40) termed the ‘antisense library’. The library consists of strains where 188 different genes have been tagged with seamless insertions at their 3′-end, either with sfGFP alone (wt strain) or with sfGFP followed by a unidirectional transcriptional terminator element of 93 bp (PHO5T (38)) or a scrambled version of PHO5T as control (PHO5T:scr) (Figure 1A). The PHO5T efficiently terminates asRNAs originating in the 3′-IGR. The sfGFP fluorescence ratio between PHO5T:scr (AS+) and PHO5T (AS−) was used to quantify the impact of asRNA transcription, referred to as the ‘antisense effect’. The strain with sfGFP (wt strain) served as an additional control. We previously reported that ∼25% of these genes are regulated by antisense using exponentially growing yeast cells in four different growth media (YPAD, YPGal, YPE and SC) and we found that in almost all cases the asRNA had an inhibitory effect on the expression of the sense gene (40).
Figure 1.
Identification of antisense-regulated genes using whole colony fluorescence measurements. (A) The antisense library contains strains where 188 genes were seamlessly tagged with either sfGFP alone (wt), sfGFP with a unidirectional terminator (PHO5T), or a scrambled version of PHO5T (PHO5T:scr). See Huber et al. (40) for additional details. (B) Whole colony fluorescence intensity ratios of PHO5T:scr/PHO5T strains were determined for all strains of the antisense library grown under the three growth conditions indicated. Significantly regulated genes (FDR < 10%) are highlighted along with the respective growth condition (enlarged symbols). Grey dots indicate genes that did not meet some selection criteria. The inset shows the data for the Sps100-sfGFP. (C) Bar plots of the antisense effects for all significantly regulated genes found in (B) for all three conditions tested. The Venn diagram shows the overlap of regulated genes between the three growth conditions.
We reasoned that testing more conditions might reveal additional genes regulated by antisense. We opted for whole colony sfGFP fluorescence measurements because this method permits fast and efficient testing of different growth conditions. Moreover, yeast colonies represent a growth environment that is very different from their exponential growth in liquid culture (54,55).
First, we performed a coarse-grained assessment of 12 growth conditions differing in their carbon sources, osmolarity and temperature (Supplementary Table S4). We noticed that three of the conditions captured most of the differences in antisense-dependent protein expression observed in the whole dataset. These conditions were SC media with 2% glucose at 30°C and 14°C and SC with 0.1% glucose at 30°C (Figure 1B). Using plate reader fluorescence measurements, the expression of sfGFP was detected in 50 out of the 188 genes represented in the library under at least one growth condition (Supplementary Table S5). Among the genes detected, 15 showed a significant antisense effect (Figure 1B and C) and 8 of those were not previously reported to be regulated by antisense (see Supplementary Table S6 for details) (40). This suggests that the antisense-dependent regulation of these genes is strongly affected by the growth condition. Interestingly, six of the regulated genes had reduced protein levels in the PHO5T background, indicating that antisense transcription leads to an enhanced expression of the overlapping protein. This contrasts our previous findings where we found almost exclusively repressive effects imposed by the asRNAs (Supplementary Table S6 and (40)).
SPS100 expression strength is modulated by antisense in starving and sporulating cells
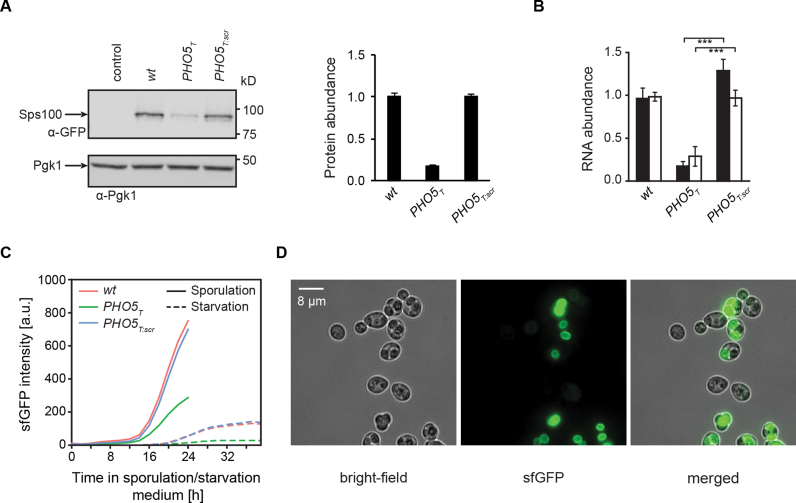
Of all the antisense regulated genes identified in this colony assay, the SPS100 gene exhibited the strongest antisense effect (Figure 1 and Supplementary Figure S1). Transcription of the asRNA of SPS100 initiates around 220 bp downstream of its STOP codon and this asRNA is called SUT169 (7). We first performed a functional analysis of the regulation of the SPS100 gene and explored growth conditions suitable for bulk analyses of SPS100-sfGFP. We found that culturing the strains for 3 days in liquid SC medium (0.1% glucose, 30°C) recapitulated the results obtained with colonies on plates, where the presence of the asRNA leads to increased SPS100-sfGFP expression, resulting in a robust antisense-dependent expression of Sps100 (Figure 2A). Strand-specific RT-qPCR confirmed that both sense and antisense RNA levels decreased in the PHO5T background (Figure 2B).
Figure 2.
Protein, sense and antisense RNA levels of SPS100 are correlated and the antisense effect is preserved in sporulation. (A) Representative immunoblot and quantification of three biological replicates of Sps100-sfGFP protein levels in Sps100-sfGFP wt, PHO5T and PHO5T:scr strains after growth under starvation conditions (SC, 0.1% glucose, 30°C). (B) Plots showing RT-qPCR of sense and antisense transcripts of SPS100-GFP in the cells from (A). Error bars indicate standard deviation of three biological replicates. (C) Sporulation and starvation time course measurements using a diploid sps100Δ/SPS100-sfGFP hybrid with wt, PHO5T and PHO5T:scr constructed by mating of the haploid SPS100-sfGFP strains (in the BY4741 background) with a sps100Δ strain (in the well-sporulating SK1 background). Fluorescence of sfGFP was quantified by flow cytometry. (D) Representative images showing the spore-autonomous localization of Sps100-sfGFP after 24 h into sporulation.
Sps100 has been previously identified as a sporulation-specific protein expressed late in sporulation with a potential role in spore wall maturation (56,57). To determine whether the gene expression regulation of SPS100 during sporulation also depends on the presence of SUT169, we measured Sps100-sfGFP expression using flow cytometry in wt, PHO5T and PHO5T:scr strains as a function of time after the induction of sporulation (Figure 2C). This revealed strong expression of Sps100-sfGFP late in meiosis and the same dependency on antisense transcription of Sps100-sfGFP as observed for starvation conditions (Figure 2C). Fluorescence microscopy revealed that most Sps100-sfGFP was produced in a spore-autonomous manner, i.e. after exiting meiosis II and the closure of the spores (Figure 2D). This suggests that the mechanism of SPS100 gene regulation by SUT169 is similar in sporulation and starvation.
Dynamics of Sps100 sense and antisense expression
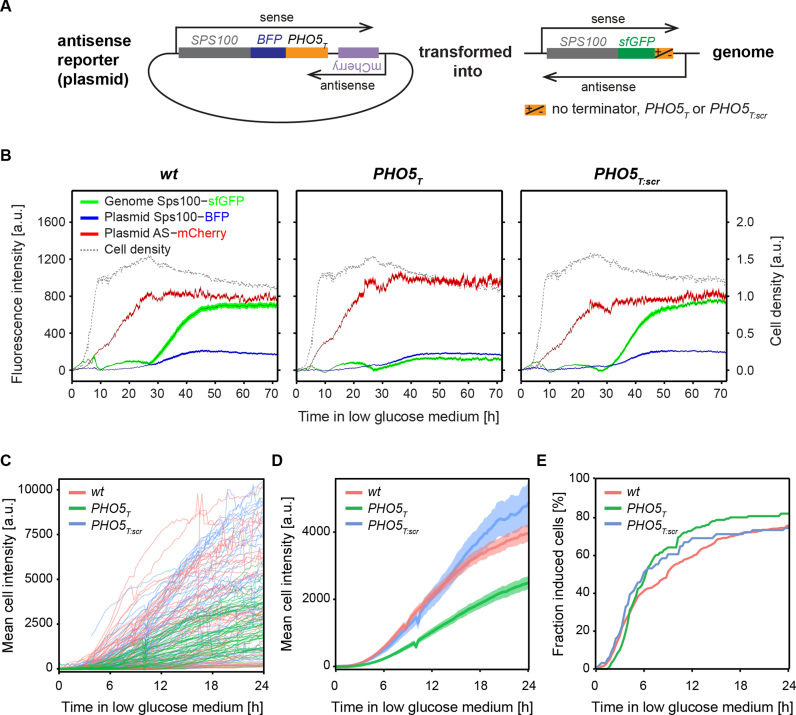
The expression of the sense SPS100 mRNA may be stimulated by concomitant expression of the antisense SUT169, or expression of SUT169 may precede expression of the mRNA. To distinguish between these possibilities, we constructed a reporter plasmid to monitor the activity of the SUT169 promoter. We cloned the SPS100 gene tagged with a blue fluorescent protein (BFP) with its up- and downstream IGRs into a centromeric plasmid (wt, PHO5T and PHO5T:scr) and inserted mCherry at the antisense transcript initiation site in antisense direction (Supplementary Figure S2). The plasmid with PHO5T was then transformed into three strains: SPS100-sfGFP wt, PHO5T and PHO5T:scr (Figure 3A). We measured the expression levels of BFP, sfGFP and mCherry for three days following transfer to liquid starvation medium (Figure 3B) using a plate reader and liquid cultures grown in 96 well culture plates. Sps100-sfGFP and Sps100-BFP-PHO5T levels were low at the beginning but increased between 25 and 40 h after transfer to starvation medium. As expected, sfGFP levels were higher in the presence of SUT169, e.g. in the wt and PHO5T:scr strains. Interestingly, antisense expression (mCherry) levels increased much earlier, between 5 and 20 h after transfer to the starvation medium. We conclude that the expression of the SPS100 asRNA precedes the expression of the SPS100 sense mRNA by 20 h under starvation growth conditions.
Figure 3.
Dynamics of SPS100 sense and antisense expression upon entry into starvation. (A) An antisense reporter plasmid with mCherry in the initiation site of the SPS100 asRNA was transformed into wt, PHO5T and PHO5T:scr SPS100 strains. (B) A fluorescence plate reader was used to monitor expression levels of mCherry, Sps100-BFP and Sps100-sfGFP in liquid cultures growing in SC, 0.1% glucose, at 30°C for 3 days. The black line represents the mean and the areas correspond to the 95% confidence interval. Cell density measured at 600 nm, non-linear scale in arbitrary units. (C) Florescence microscopy of Sps100-sfGFP expression level in single cells quantified using time-lapse imaging of cells during the transition from exponential growth into starvation. (D) Same data as in (C) but showing the mean fluorescence intensity for all cells with signals above background. Shaded areas indicate the 95% confidence interval. (E) Cumulative analysis of the fraction of cells with induced Sps100-GFP expression from (C), showing the fraction of cells that exhibit fluorescence above background as a function of time.
To obtain single cell SPS100 induction kinetics, we quantified the expression of the Sps100-sfGFP protein in wt, PHO5T and PHO5T:scr cells using time lapse fluorescence microscopy. In order to recapitulate liquid growth conditions for fluorescence measurements we used exponentially growing cells and transferred them for imaging to microfluidics chambers containing preconditioned starvation medium (‘Materials and Methods’ section). sfGFP and bright field images (for segmentation) were recorded every 15 min for 24 h in >100 cells per strain (Figure 3C). Quantitative analysis of individual cell traces revealed that the timing of induction of the Sps100-sfGFP expression (Figure 3D) and the total number of induced cells (Figure 3E) were similar for strains with and without asRNA. However, the slope of induction of protein expression, corresponding to the induction strength, was higher in the cells with intact antisense transcription (wt and PHO5T:scr) compared to the cells with prematurely terminated antisense transcription (PHO5T) (Figure 3C and D).
Together, these results demonstrate that SUT169 is expressed much earlier than the SPS100 mRNA and that the expression of SUT169 influences the total amount of expressed Sps100 protein but not the timing of induction. This argues for a role of the SUT169 in altering the transcriptional activity, the stability, or the translation efficiency of the SPS100 transcripts, rather than a function in timing or duration of SPS100 transcription.
Regulation of SPS100 expression by antisense transcription occurs in cis and can be transferred to other genes
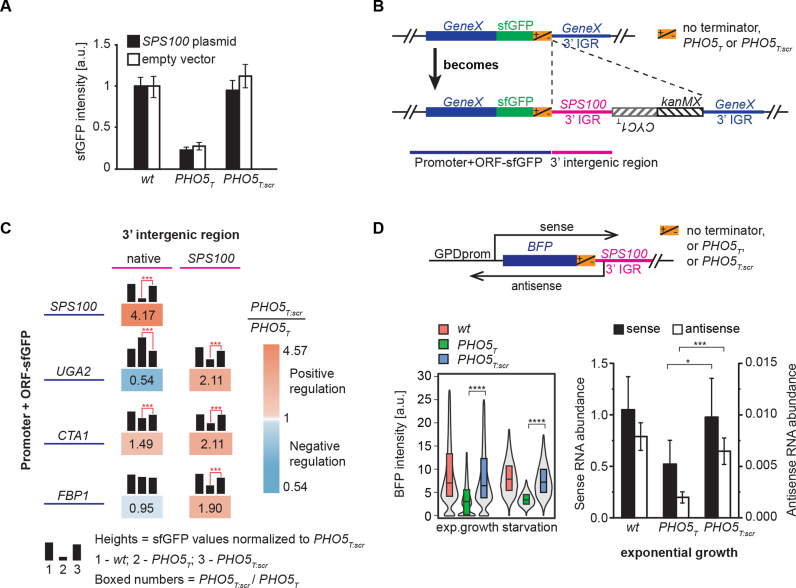
So far our data demonstrate that the transcription of SUT169 influences the expression strength of the Sps100 protein, leading to faster accumulation of the protein inside the cells. To further dissect the mechanism, we first determined whether this regulation occurs in cis or in trans. We transformed a plasmid containing the SPS100 ORF and its up- and downstream IGRs (including the asRNA) into the SPS100 strains (wt/PHO5T/PHO5T:scr). The ectopic expression of SUT169 did not rescue expression of SPS100 in the PHO5T strain (Figure 4A), suggesting that the regulation of SPS100 expression by SUT169 occurs in cis.
Figure 4.
SPS100 regulation by the 3′-IGR occurs in cis and can be transferred to other genes. (A) Strains expressing SPS100-sfGFP in wt, PHO5T or PHO5T:scr background were transformed with an empty plasmid (white boxes) or a plasmid containing SPS100-BFP including its 5′- and 3′-IGRs. sfGFP fluorescence intensities were measured using colony fluorescence measurements. Error bars denote standard deviations of 3 biological replicates. (B) Scheme of the strategy used to transfer the SPS100 3′-IGR to other genes. Using PCR targeting the SPS100 3′-IGR with either wt, PHO5T or PHO5T:scr along with a selection marker was integrated after the STOP codon of sfGFP tagged target genes (GeneX), as indicated by dashed lines. (C) Regulation imposed by the SPS100 3′-IGR on three different genes, as shown in (B), was investigated by measuring colony fluorescence. The different genes are indicated in the rows. Black bar heights denote sfGFP expression levels in the wt, PHO5T or PHO5T:scr background. Blue color: negative regulation by antisense; orange color: positive regulation. (D) A GPD promoter controlling BFP followed by the SPS100 3′ IGR was genomically integrated into the URA3 locus. Expression levels were measured by flow cytometry for exponentially growing cells and cells under starvation conditions (bottom left panel). The boxes show the first and third quartiles and the median. The grey violin plots show the distribution densities ranging from the first quartile minus 1.5 * interquartile range (IQR) to the third quartile + 1.5 * IQR. RNA levels were measured by RT-qPCR in the exponentially growing cells (bottom right panel). Error bars denote standard deviations of three biological replicates.
Next we asked whether the antisense effect on SPS100 expression is mediated solely by the 3′-IGR of SPS100. If this is the case, SPS100 3′-IGR should be able to regulate other genes in a similar manner. We employed a PCR-targeting-based approach to replace the 3’-IGR of a gene of interest by the SPS100 3′-IGR (Figure 4B). We applied this strategy to three genes: CTA1, another gene upregulated by antisense transcription in our colony fluorescence assay; UGA2, a gene that is repressed by the antisense transcription, and FBP1, a gene that does not show expression changes as a function of the PHO5T element (Figure 1B and C; Supplementary Tables S5 and 6). In all cases the SPS100 3′-IGR led to an antisense-dependent upregulation of the corresponding genes, albeit with a smaller antisense effect than observed for the SPS100 gene (Figure 4C). This indicates that the 3′-IGR is probably the most important but not the only element responsible for the SPS100 antisense effect.
Next, we generated an artificial construct containing BFP under the control of the strong GPD promoter followed by the SPS100 3′-IGR with the wt/PHO5T/PHO5T:scr sequences (Figure 4D). Using BFP fluorescence as a readout we observed that ∼50% of the antisense effect was still present and gene expression was not restricted to low glucose growth conditions (Figure 4D). Transcript levels during exponential growth were also quantified by RT-qPCR (Figure 4D). These results indicate that the SPS100 3′-IGR contains a dominant signal which modulates the expression level of the gene whereas the starvation condition is required to induce the promoter of the SPS100 gene.
Gene loops have been previously implicated in gene regulation by asRNA (31). To determine whether antisense-dependent upregulation of SPS100 expression occurs via formation of a chromatin loop, we measured chromatin contacts in the SPS100 locus by chromosome conformation capture followed by PCR (Supplementary Figure S3). We were able to detect gene looping between the promoter and terminator regions of the SPS100 gene, but the formation of a gene loop did not depend on the presence of the PHO5T element (Supplementary Figure S3), likely excluding a role of gene loops in antisense regulation of SPS100 expression.
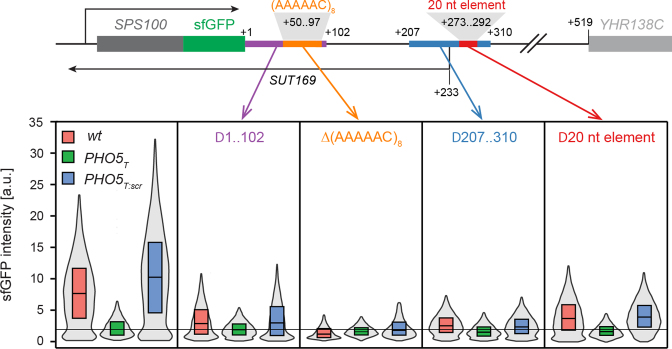
SPS100 regulation is mediated by two short nucleotide motifs in its 3′-IGR
Next, we searched for sequence determinants in the 3′-IGR that might mediate the antisense effect. We first deleted pieces of ∼100 nts in the SPS100 3′-IGR (total length: 518 nt) in a plasmid containing SPS100-BFP followed by the SPS100 3′-IGR (Supplementary Figure S4). Deletion of a centrally located ∼100 nt element (nucleotides 207–310 as counted from the SPS100 STOP codon) as well as of a 20 nt element within this sequence strongly reduced the antisense effect on SPS100 expression (Supplementary Figure S4, red boxes). We validated this finding by introducing the same deletions into the genomic loci in SPS100-sfGFP wt, PHO5T or PHO5T:scr strains using the strategy shown in Figure 4B and by measuring Sps100-sfGFP protein levels using flow cytometry. We observed a strong reduction of the antisense effect for both the 100 and the 20 nt deletions (Figure 5), with the 20 nt deletion having a slightly milder effect.
Figure 5.
The antisense effect is mediated by two different sequence elements of the SPS100 3′-IGR. Selected parts of the SPS100 3′-IGR in SPS100-sfGFP wt, PHO5T and PHO5T:scr strains were deleted (scheme at the top) using the strategy outlined in Figure 4B. sfGFP intensities were measured by flow cytometry (bottom panel). The leftmost box in the bottom panel shows the strains without any 3′-IGR manipulations. The boxes show the first and third quartiles and the median. The grey violin plots show the distribution densities ranging from the first quartile minus 1.5 * interquartile range (IQR) to the third quartile + 1.5 * IQR.
We noticed that positions 50–97 in the SPS100 3′-IGR consisted of a conspicuous (AAAAAC)8 tandem repeat. To determine if the presence of this tandem repeat is required for the antisense effect on SPS100 expression, we deleted this repeat as well as the first 102 nt of the SPS100 3′-IGR, even though the latter deletion had only a mild effect in the plasmid-based approach (Supplementary Figure S4). Strikingly, the deletion of the (AAAAAC)8 repeat completely abolished the antisense effect (Figure 5). Deletion of the first 102 nt also strongly reduced the antisense effect but to a smaller extent (Figure 5). In summary, we identified two regions that play an important role in the antisense-dependent upregulation of SPS100 expression.
SPS100 regulation by antisense correlates with a change in isoform abundances
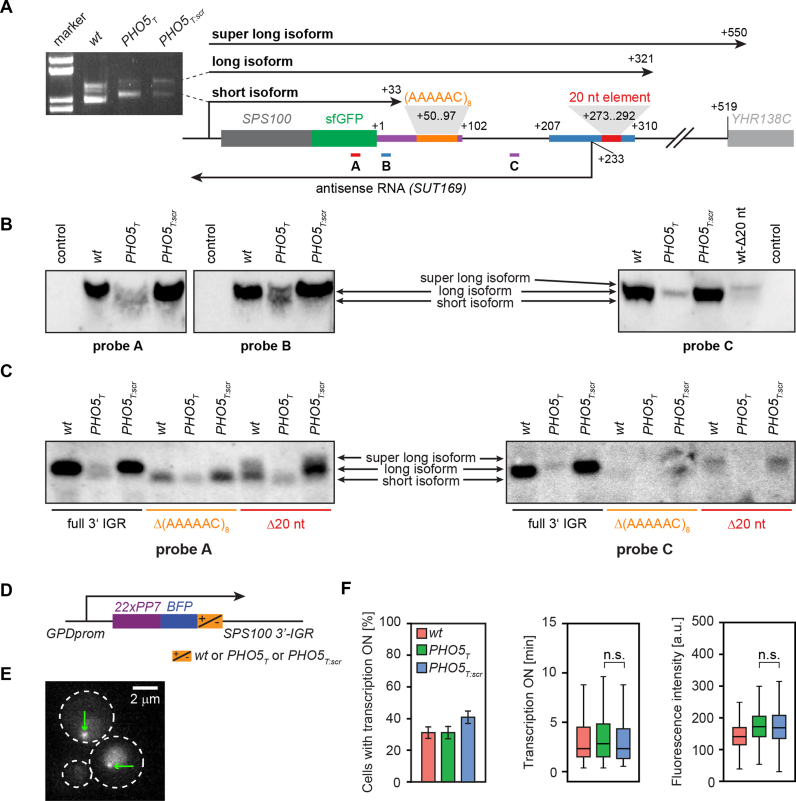
Previously, several mRNA isoforms differing in the position of their 3′-ends were reported for the SPS100 gene (56). We therefore asked whether antisense-dependent upregulation of SPS100 occurs in an isoform-specific manner. First, we mapped the SPS100 mRNA 3′ boundaries by rapid amplification of cDNA ends (3′-RACE (48)) followed by Sanger sequencing. We detected a short and a long transcript isoform with the same 3′-mRNA ends in all three strains (wt, PHO5T and PHO5T:scr) (Figure 6A), indicating that the PHO5T and PHO5T:scr elements do not influence the 3′-end formation of the SPS100 mRNA isoforms. We also mapped the SUT169 3′ end and, in agreement with existing tiling array data (7), it ends −744 bp upstream of the SPS100 start codon (data not shown).
Figure 6.
Antisense transcription promotes expression of the long isoform of SPS100. (A) Scheme of the locus of SPS100-sfGFP (wt) with the sequence motifs of Figure 5 and the three isoforms detected by 3′ RACE indicated. An example gel for PCR products corresponding to the short and the long isoform is shown to the left. The super long isoform was detected in a separate 3′ RACE experiment (data not shown). (B) Northern blots on SPS100 mRNAs with three different probes as indicated in (A). (C) Northern blots for SPS100-sfGFP wt, PHO5T or PHO5T:scr strains with either an unmodified 3′-IGR or a 3′-IGR carrying a deletion of the 20 nt element or the (AAAAAC)8 repeat shown in (A). Loading controls for (B) and (C) are shown in Supplementary Figure S5. (D) A GPD-22PP7-BFP-SPS100 3′ IGR construct was used for monitoring mRNA transcription in vivo. (E) Selected microscopy image, greenarrows indicate the transcription site. White dashed lines border yeast cell outer membranes. (F) Quantification of the cells with active transcription (bar plot, error bars denote standard deviations), the duration of active transcription (left boxplot) and the intensity of transcription site (right boxplot). Boxes show the first and third quartiles and the median. Whiskers indicate the first quartile minus 1.5 * interquartile range (IQR) and the third quartile + 1.5 * IQR.
To examine the relative amounts of the SPS100 mRNA isoforms, we performed Northern blotting using three probes that targeted different regions of the sense mRNA transcripts, thus detecting either both mRNA isoforms or only the long isoform (Figure 6A, probes A–C). Interestingly, only the long isoform was detected in the SPS100-sfGFP wt and PHO5T:scr strains whereas both short and long isoforms were present in the PHO5T background (Figure 6B, see Supplementary Figure S5a for loading controls and lower contrast). This suggests that the expression of the long isoform is stimulated by SUT169 and that Sps100-GFP upregulation in starvation results from the long isoform. This interpretation was further supported by RT-qPCR (Supplementary Figure S6). SUT169, in contrast, seems to be expressed as one isoform (Supplementary Figure S5b).
Next, we investigated the functions of the 20 nt element and the (AAAAAC)8 repeat. Northern blotting in the SPS100-sfGFP wt, PHO5T and PHO5T:scr strains revealed that deleting the 20 nt element not only reduced sense mRNA levels in the wt and PHO5T:scr backgrounds but also led to the appearance of a third isoform which had also been detected in the 3′ RACE experiments (‘super long isoform’, Figure 6A), even though the long isoform still predominated (Figure 6B, rightmost blot and Figure 6C and Supplementary Figure S5c). Deletion of the 20 nt element did not prevent the formation of normal amounts of SUT169 as determined by strand-specific RT-qPCR (Supplementary Figure S7). Therefore, we conclude that the 20 nt element is not required for regulation of SPS100 expression by antisense transcription. Rather, it seems to be required for proper transcriptional termination of the long SPS100 mRNA isoform and hence results in the formation of an unstable super long isoform. In support of this, the nucleotide sequence TACAGTA within the 20 nt element closely resembles the consensus sequence for the efficiency element of yeast termination signals (TAYRTA, Y = pyrimidine, R = purine (58)).
In contrast, northern blotting experiments in the Δ(AAAAAC)8 strains, which completely lack the antisense effect on SPS100 expression (Figure 5), indicated that the long isoform is not expressed in those strains (Figure 6C and Supplementary Figure S5c). Strand-specific RT-qPCR showed that deletion of this repeat leads to a decrease in the antisense transcript levels in the wt and PHO5T:scr strains to the levels observed in the PHO5T strain (Supplementary Figure S7). This supports a model where antisense transcription stimulates the expression of the long sense mRNA isoform and suggests a function of the repeat either in the stability of the SUT169 or in the regulation of the mRNA isoform switch, or both. To provide additional evidence that this mechanism depends on proper antisense transcript formation and not on the presence of the PHO5T element, we constructed strains where a portion of the 3′ IGR was replaced by a heterologous, bidirectional terminator. In those strains, antisense transcript expression was abolished and, accordingly, no antisense effect on sense gene expression was observed, suggesting a direct involvement of SUT169 in the regulation of SPS100-sfGFP expression (Supplementary Figure S8).
These results indicate that the antisense effect on SPS100 gene expression is mediated by processes in the 3′-IGR region of the gene and that interfering with these processes will not affect the transcriptional activity of the gene in its 5′-region. To test whether SUT169 interferes with SPS100 transcriptional activity we monitored mRNA transcriptional activity in the 5′-region of the gene by inserting PP7 elements and a 3mCherry reporter fused to the PP7 binding protein. We took advantage of the transferability of the antisense effect and used artificial BFP constructs controlled by the GPD promoter (Figure 4D) (59,60) to obtain sufficient levels of transcriptional activity for live cell microscopy quantification using time lapse movies (Figure 6E and Supplementary Movie S1). We quantified the number of cells with active transcription, the time during which transcription was active and the fluorescence intensity of the transcription spot. This showed that none of those parameters did significantly differ between the PHO5T and PHO5T:scr strains (Figure 6F), suggesting that the abrogation of SUT169 does not influence the transcriptional activity of the SPS100 promoter.
Antisense expression stimulates the formation of a stable long mRNA isoform
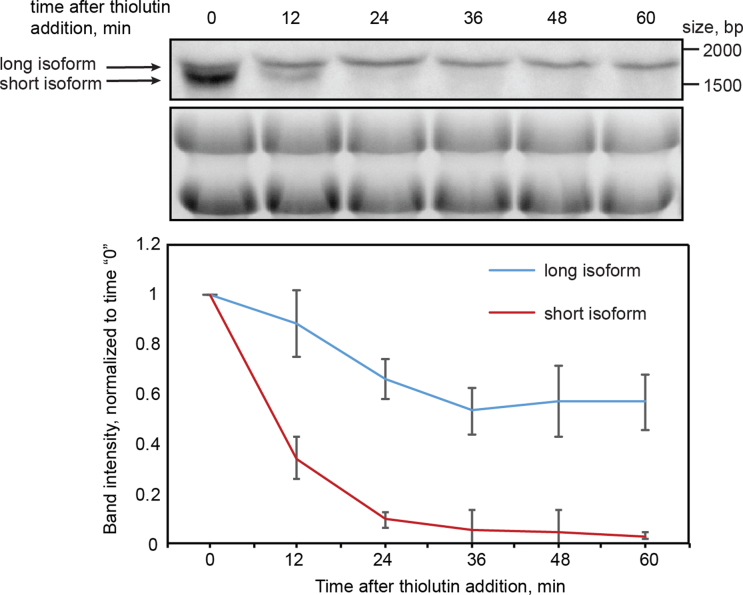
Our experiments show that both absolute and relative abundances of the mRNA isoforms are affected by antisense transcription (Figure 6B) whereas the transcriptional activity of the mRNA promoter appeared unchanged (Figure 6F). Therefore, a possible explanation for the antisense effect seen at the protein level is that the longer SPS100 isoform—which is more abundant in an antisense expressing background (wt and PHO5T:scr)—is more stable than the short isoform. This would give rise to higher levels of the long transcript and consequently more Sps100 protein. To test this possibility, we performed a thiolutin chase experiment. Thiolutin has been used to quantify mRNA turnover (61), because it causes a rapid stop of Pol II transcription that is mediated indirectly via inhibition of the proteasome (62). First of all, we needed to achieve a better separation of the mRNA isoforms by northern blotting to be able to follow their stabilities in a time course experiment. We therefore made new strains in which SPS100 was tagged with PHO5T and PHO5T:scr but without sfGFP. The resulting shortening of the mRNAs allowed a better resolution of the RNA isoforms by gel electrophoresis. Northern blotting confirmed that the antisense effect is still present in the absence of the sfGFP tag and better separation of the short and long isoform bands was observed (compare Figure 6B with Supplementary Figure S9). For the thiolutin chase experiment we used the PHO5T strain, in which both isoforms can be detected (Figure 6B), thus permitting direct comparison of their degradation as a function of time after the addition of thiolutin. We observed a rapid disappearance of the short SPS100 mRNA isoform within 24 min after addition of thiolutin, whereas the long isoform was essentially stable (Figure 7). This shows that the short mRNA isoform that is produced in the absence of antisense is indeed less stable than the long isoform. Thus, antisense stimulates expression of the longer and more stable SPS100 mRNA isoform, resulting in enhanced Sps100 protein expression as a function of the cognate SUT169 transcript.
Figure 7.
Long mRNA isoform of SPS100 is more stable than the short one. Northern blots of a thiolutin chase experiment in the SPS100-PHO5T strain. Samples were taken for RNA extraction in indicated time intervals after thiolutin addition to the media. The upper panel is the blot probed with probe A (see Figure 6), the long and short isoforms are labelled with arrows. The lower panel is the loading control (RNA gel stained with ethidium bromide). Quantification of lower and upper bands from the Northern blots and from another independent replicate is shown on the plot below. Intensity values were normalized to the time point ‘0’, errors bars indicate standard deviations.
DISCUSSION
A growing number of studies have documented the diversity and complexity of mechanisms underlying regulation of gene expression by non-coding transcription (reviewed in (3,4,14,63)). Within this world, ncRNAs that overlap coding regions in antisense direction are particularly intriguing. The lack of the RNA interference machinery in yeast provides a good opportunity to characterize these cases further and to explore the fascinating variety of antisense-dependent gene regulation mechanisms.
Genes that are positively regulated by antisense are rare according to the literature, with only a few documented cases, which require specific growth or stress conditions (25,31). Our previous systematic analysis of the regulatory function of a large number of asRNAs revealed that ∼25% of asRNAs have an inhibitory function (40). In this study, regulation was detected under exponential growth conditions and the inhibitory effect was often not fully abrogating the expression of the sense gene. Moreover, for the majority of these cases the likelihood of regulation was found to be correlated with antisense transcription across the sense transcript start site. This is in line with other studies and suggests a generally inhibitory and rather mild regulatory function in most observed cases of antisense RNA regulation. However, there is also a number of genes with more pronounced and often condition specific antisense-dependent regulation (18,25,31,32).
In contrast to those previous studies, our colonies were grown on agar plates for 3 days, an environment that has characteristics of starvation conditions (54,55). For some of the genes these conditions established a regulatory environment that subjected them to antisense-dependent regulation (Supplementary Table S6). In other cases, a gene may be capable of regulation during exponential growth but is not expressed at the right levels for this regulation to occur. SPS100 seems to belong to this group as the antisense effect was preserved in exponentially growing cells when mRNA expression was driven by a GPD promoter (Figure 4D).
The mechanism regulating SPS100 expression appears to be different from other established antisense-dependent mechanisms. Instead of a process involving the promoter region of the gene and influencing promoter and transcriptional activity, our experiments indicate that regulation is restricted to the 3′-IGR via the formation of different mRNA isoforms with different stabilities. This is in concordance with the notion that different mRNA isoforms can behave differently, ranging from RNA stability to localization and translational activity, all of which could influence how much mRNA is present and how much protein is produced per molecule of mRNA. For example, degradation of mRNAs is enhanced by 3′ untranslated region (3′-UTR) sequence element binding proteins such as the PUF protein family members (64). However, binding to specific proteins may also stabilize mRNAs or change their localization. In yeast no clear correlation between length of 3′-UTR and stability of mRNAs is seen, arguing against a model where sheer length of 3′-UTRs determines the likelihood to harbor destabilizing signals, as proposed for other organisms (65).
Our deletion mapping and 3′-IGR transplantation experiments are consistent with SPS100 regulation being restricted to processes in the 3′-IGR. However, the PHO5T element is located further upstream, at the boundary between the 3′-IGR and the coding region. Consequently, we originally feared that the PHO5T element—in spite of what had been reported (38)—does exhibit unexpected termination activity also in sense direction that was not seen when fused to other loci. Several arguments speak against this possibility. First, autonomous termination activity of the element was ruled out by Northern blots using probes binding both up- and downstream of PHO5T as well as 3′ RACE results (Figure 6A and B). Second, SPS100 3′-IGR specific regulation could be observed under other growth conditions, provided that the sense gene is actively transcribed (Figure 4D). Third, only a handful of the genes in the antisense library exhibited a 3′-IGR-dependent upregulation that could be abrogated with the PHO5T. If the PHO5T element did exhibit a starvation-specific activity not present in the PHO5T:scr, one would expect to see a stimulating PHO5T -specific effect for many other genes under starvation conditions. This is clearly not the case. Fourth, abrogation of the antisense transcription start site with transcriptionally silent bidirectional terminators (Supplementary Figure S8) also abrogated most of the PHO5T dependent effect. At the same time, deletion of the (AAAAAC)8 repeat located 50 nt downstream of the STOP codon led to a reduction of SUT169 levels to the same amounts as in the PHO5T background, a complete abrogation of antisense-dependent regulation, and, importantly, the concomitant loss of detectable amounts of the long isoform (Figures 5 and 6C). While these points do not constitute a complete formal proof, they strongly support our interpretation that the down regulation in the PHO5T background is antisense-dependent and caused by a switch in isoform preferences. The effect of these isoforms must be dominant, as demonstrated by the transferability of the antisense effect.
Like the majority of antisense-regulated yeast genes (14) we found that SPS100 is regulated by its antisense RNA in cis. Due to the transferability of the effect we conclude that isoform switching is the responsible mechanism also for other genes when tagged with the SPS100 3′-IGR, and that differential stability causes the increase of mRNA and protein detected for SPS100 upon starvation.
Given the novelty of the regulatory process we currently can only speculate about the underlying mechanism that causes the polymerase to form the long isoform. Our results show that the (AAAAAC)8 repeat is crucial for the antisense effect and that it is required for efficient antisense expression, although we cannot exclude that the deletion of the repeat could have affected the processing or stability of the mRNA. A BLAST search for similar motifs in the yeast genome did not yield any significant results (data not shown). Moreover, it is formally possible that the SUT169 or the 3′UTR of the long SPS100 isoform could encode short peptides that play a role in regulation. Given that antisense RNA levels were always lower than sense levels in our RT-qPCR experiments we do not think that the transcript itself plays an important role in the mechanism. Rather, we speculate that transcription of SUT169 results in an epigenetic event, such as certain chromatin marks, an alteration of the nucleosome architecture, or on post-translational RNA Pol II modifications, all of which have been shown to potentially influence the choice of the polyadenylation site (reviewed in (58)). In a genetic screen that we performed to identify regulators of SPS100 regulation by antisense RNA did not identify known regulators such as members of the HDA and COMPASS complexes. Rather, we found an enrichment for components of the mediator complex as well as mRNA regulation associated genes (e.g. CSE2, LEO1, MED1, NUT1, CAF40, PAN2 or RTT103). None of those genes completely abrogated the antisense effect (data not shown). Together with the numerous functions exerted by mediator, this suggests that SPS100 is regulated by novel, potentially intricate, mechanism(s). We hope that this work provides a starting point for the dissection of the underlying molecular mechanisms.
From a more general perspective, our findings have interesting implications for the role of antisense transcription and how this could relate to the observed plasticity of the yeast transcriptome. The set of all yeast transcript isoforms outnumbers the number of yeast genes by more than an order of magnitude (63) and non-coding RNAs may contribute to this diversity by regulation of isoform variability of coding but also non-coding RNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Monika Langlotz and the ZMBH FACS facility for support. We also like to thank Michael Brunner and Axel Diernfellner for advice and supply of thiolutin.
Footnotes
Present addresses:
Daria Bunina, Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Meyerhofstraße 1, 69117 Heidelberg, Germany.
Florian Huber, Genome Biology Unit, European Molecular Biology Laboratory (EMBL), Meyerhofstraße 1, 69117 Heidelberg, Germany.
Martin Štefl, Department of Biophysical Chemistry, J. Heyrovsky Institute of Physical Chemistry of the CAS, Dolejškova 2155/3, 182 23 Prague 8, Czech Republic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Deutsche Forschungsgemeinschaft [DFG-KN498/8-1, DFG-KN489/11-1 to M.K.]; CellNetworks/EcTop 2 and the Alexander von Humboldt Foundation (to M.S.); Boehringer Ingelheim Fonds PhD Fellowship (to D.B.); HBIGS Graduate School PhD Fellowship (to F.H.). Funding for open access charge: Deutsche Forschungsgemeinschaft [DFG-KN498/11-1].
Conflict of interest statement. None declared.
REFERENCES
- 1. David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M.. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S. et al. . Global identification of human transcribed sequences with genome tiling arrays. Science. 2004; 306:2242–2246. [DOI] [PubMed] [Google Scholar]
- 3. Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009; 10:833–844. [DOI] [PubMed] [Google Scholar]
- 4. Jensen T., Jacquier A., Libri D.. Dealing with pervasive transcription. Mol. Cell. 2013; 52:473–484. [DOI] [PubMed] [Google Scholar]
- 5. Davis C.A., Ares M.J.. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neil H., Malabat C., d, Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A.. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009; 457:1038–1042. [DOI] [PubMed] [Google Scholar]
- 7. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M.. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B. et al. . Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005; 121:725–737. [DOI] [PubMed] [Google Scholar]
- 9. Colin J., Candelli T., Porrua O., Boulay J., Zhu C., Lacroute F., Steinmetz L.M., Libri D.. Roadblock termination by reb1p restricts cryptic and readthrough transcription. Mol. Cell. 2014; 56:667–680. [DOI] [PubMed] [Google Scholar]
- 10. van Dijk E.L., Chen C.L., d'Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., Bertrand C., Silvain M., Legoix-Ne P., Loeillet S. et al. . XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011; 475:114–117. [DOI] [PubMed] [Google Scholar]
- 11. Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes H., Nislow C.. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007; 39:1235–1244. [DOI] [PubMed] [Google Scholar]
- 12. Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J. et al. . The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009; 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mavrich T.N., Ioshikhes I.P., Venters B.J., Jiang C., Tomsho L.P., Qi J., Schuster S.C., Albert I., Pugh B.F.. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008; 18:1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Castelnuovo M., Stutz F.. Role of chromatin, environmental changes and single cell heterogeneity in non-coding transcription and gene regulation. Curr. Opin. Cell Biol. 2015; 34:16–22. [DOI] [PubMed] [Google Scholar]
- 15. Shearwin K.E., Callen B.P., Egan J.B.. Transcriptional interference–a crash course. Trends Genet. 2005; 21:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drinnenberg I.a., Fink G.R., Bartel D.P.. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011; 333:1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R., Bartel D.P.. RNAi in budding yeast. Science. 2009; 326:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Werven F.J., Neuert G., Hendrick N., Lardenois A., Buratowski S., Van Oudenaarden A., Primig M., Amon A.. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012; 150:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M.. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008; 32:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pinskaya M., Gourvennec S., Morillon A.. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009; 28:1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bumgarner S.L., Dowell R.D., Grisafi P., Gifford D.K., Fink G.R.. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bumgarner S.L., Neuert G., Voight B.F., Symbor-Nagrabska A., Grisafi P., Van Oudenaarden A., Fink G.R.. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell. 2012; 45:470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F.. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007; 131:706–717. [DOI] [PubMed] [Google Scholar]
- 24. Castelnuovo M., Rahman S., Guffanti E., Infantino V., Stutz F., Zenklusen D.. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat. Struct. Mol. Biol. 2013; 20:851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uhler J.P., Hertel C., Svejstrup J.Q.. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8011–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Small E., Xi L., Wang J.. Single-cell nucleosome mapping reveals the molecular basis of gene expression heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2462–E2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hainer S.J., Charsar B. a., Cohen S.B., Martens J. a.. Identification of mutant versions of the Spt16 histone chaperone that are defective for transcription-coupled nucleosome occupancy in Saccharomyces cerevisiae. G3. 2012; 2:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hainer S.J., Pruneski J.A., Mitchell R.D., Monteverde R.M., Martens J.A.. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011; 25:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martens J.A., Wu P.Y.J., Winston F.. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005; 19:2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thebault P., Boutin G., Bhat W., Rufiange A., Martens J., Nourani A.. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell. Biol. 2011; 31:1288–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nadal-Ribelles M., Solé C., Xu Z., Steinmetz L.M., deNadal E., Posas F.. Control of Cdc28 CDK1 by a stress-induced lncRNA. Mol. Cell. 2014; 53:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hongay C.F., Grisafi P.L., Galitski T., Fink G.R.. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006; 127:735–745. [DOI] [PubMed] [Google Scholar]
- 33. Gelfand B., Mead J., Bruning A., Apostolopoulos N., Tadigotla V., Nagaraj V., Sengupta A.M., Vershon A.K.. Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol. Cell. Biol. 2011; 31:1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishizawa M., Ikeya Y., Okumura T., Kimura T.. Post-transcriptional inducible gene regulation by natural antisense RNA. Front. Biosci. (Landmark Ed.). 2015; 20:1–36. [DOI] [PubMed] [Google Scholar]
- 35. Bird A.J., Gordon M., Eide D.J., Winge D.R.. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006; 25:5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M.. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z., Wei W., Gagneur J., Clauder-Münster S., Smolik M., Huber W., Steinmetz L.M.. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011; 7:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Irniger S., Egli C.M., Braus G.H.. Different classes of polyadenylation sites in the yest Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:3060–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khmelinskii A., Meurer M., Duishoev N., Delhomme N., Knop M.. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS One. 2011; 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huber F., Bunina D., Gupta I., Theer P., Steinmetz L.M., Knop M.. Protein abundance control by non-coding antisense protein abundance control by non-coding antisense transcription. Cell Rep. 2016; 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sherman F. Getting started with yeast. Methods Enzymol. 2002; 350:3–41. [DOI] [PubMed] [Google Scholar]
- 42. Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al. . A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004; 21:947–962. [DOI] [PubMed] [Google Scholar]
- 43. Huber F., Meurer M., Bunina D., Kats I., Maeder C.I., Stefl M., Mongis C., Knop M.. PCR duplication: A one-step cloning-free method to generate duplicated chromosomal loci and interference-free expression reporters in yeast. PLoS One. 2014; 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Green M.R., Sambrook J.. Molecular Cloning: a Laboratory Manual. 2012; 4th ednNY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 45. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995; 57:289–300. [Google Scholar]
- 46. Collart M.A., Oliviero S.. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. 2001; 23:doi:10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 47. Perocchi F., Xu Z., Clauder-Munster S., Steinmetz L.M.. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007; 35:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scotto-Lavino E., Du G., Frohman M.A.. 3. end cDNA amplification using classic RACE. Nat. Protoc. 2006; 1:2742–2745. [DOI] [PubMed] [Google Scholar]
- 49. Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J.. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008; 32:465–477. [DOI] [PubMed] [Google Scholar]
- 50. Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G.. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008; 5:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E.. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999; 15:963–972. [DOI] [PubMed] [Google Scholar]
- 52. Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685. [DOI] [PubMed] [Google Scholar]
- 53. Mayer C., Dimopoulos S., Rudolf F., Stelling J.. Using CellX to quantify intracellular events. Current Protocols in Molecular Biology. 2001; 101, Hoboken: John Wiley & Sons, Inc; 14.22.1–14.22.20. [DOI] [PubMed] [Google Scholar]
- 54. Palkova Z., Devaux F., Icicova M., Minarikova L., Le Crom S., Jacq C.. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol. Biol. Cell. 2002; 13:3901–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Traven A., Janicke A., Harrison P., Swaminathan A., Seemann T., Beilharz T.H.. Transcriptional profiling of a yeast colony provides new insight into the heterogeneity of multicellular fungal communities. PLoS One. 2012; 7:e46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Law D.T., Segall J.. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol. Cell. Biol. 1988; 8:912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I.. The transcriptional program of sporulation in budding yeast. Science. 1998; 282:699–705. [DOI] [PubMed] [Google Scholar]
- 58. Mischo H.E., Proudfoot N.J.. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim. Biophys. Acta. 2013; 1829:174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lim F., Peabody D.S.. RNA recognition site of PP7 coat protein. Nucleic Acids Research. 2002; 30:4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Larson D.R., Zenklusen D., Wu B., Chao J.A., Singer R.H.. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011; 332:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Trcek T., Larson D.R., Moldon A., Query C.C., Singer R.H.. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell. 2011; 147:1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lauinger L., Li J., Shostak A., Cemel I.A., Ha N., Zhang Y., Merkl P.E., Obermeyer S., Stankovic-Valentin N., Schafmeier T. et al. . Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat. Chem. Biol. 2017; 13:709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pelechano V., Steinmetz L.M.. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013; 14:880–93. [DOI] [PubMed] [Google Scholar]
- 64. Ulbricht R.J., Olivas W.M.. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA. 2008; 14:246–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gupta I., Clauder-Munster S., Klaus B., Jarvelin A.I., Aiyar R.S., Benes V., Wilkening S., Huber W., Pelechano V., Steinmetz L.M.. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol. Syst. Biol. 2014; 10:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.