Abstract
Animal mitogenomes are generally thought of as being economic and optimized for rapid replication and transcription. We use long-read sequencing technology to assemble the remarkable mitogenomes of four species of seed beetles. These are the largest circular mitogenomes ever assembled in insects, ranging from 24,496 to 26,613 bp in total length, and are exceptional in that some 40% consists of non-coding DNA. The size expansion is due to two very long intergenic spacers (LIGSs), rich in tandem repeats. The two LIGSs are present in all species but vary greatly in length (114–10,408 bp), show very low sequence similarity, divergent tandem repeat motifs, a very high AT content and concerted length evolution. The LIGSs have been retained for at least some 45 my but must have undergone repeated reductions and expansions, despite strong purifying selection on protein coding mtDNA genes. The LIGSs are located in two intergenic sites where a few recent studies of insects have also reported shorter LIGSs (>200 bp). These sites may represent spaces that tolerate neutral repeat array expansions or, alternatively, the LIGSs may function to allow a more economic translational machinery. Mitochondrial respiration in adult seed beetles is based almost exclusively on fatty acids, which reduces the need for building complex I of the oxidative phosphorylation pathway (NADH dehydrogenase). One possibility is thus that the LIGSs may allow depressed transcription of NAD genes. RNA sequencing showed that LIGSs are partly transcribed and transcriptional profiling suggested that all seven mtDNA NAD genes indeed show low levels of transcription and co-regulation of transcription across sexes and tissues.
Keywords: mitochondria, junk DNA, palindromes, Callosobruchus, Acanthoscelides, Bruchinae, intergenic spacers, metabolism, Coleoptera
Introduction
An outstanding question in evolutionary genetics concerns the function, if any, and evolution of the various classes of noncoding DNA that often form a major component of nuclear genomes across diverse taxa (ENCODE 2012; Graur etal. 2013). Mitochondrial genomes of animals are generally very different from nuclear genomes in this regard (Kolesnikov and Gerasimov 2012). They are typically compact and streamlined and are considered devoid of noncoding DNA, such as introns and intergenic tandem repeat elements, apart from the AT-rich control region (Zhang and Hewitt 1997). Insect mitogenomes are relatively invariant in terms of gene content and gene organization and include few intergenic regions or gene overlaps. They are approximately 15–17 kbp in size and size variation is largely due to different degrees of expansion of the control region (Zhang and Hewitt 1997; Boore 1999; Cameron 2014).
Here, we describe and analyze the remarkable mitogenome of seed beetles (Chrysomelidae; Bruchinae). We used long-read PacBio sequencing to de novo assemble the mitogenome of four species, short-read DNA sequencing to study within-species variation and RNA sequencing to analyze variation in transcript abundance in one of these species. We demonstrate that the mitogenomes of these beetles are exceptionally large and show that this is due to the presence of two long stretches of intergenic noncoding DNA, consisting of a complex pattern of multiple tandem repeats. The presence of noncoding elements in the mitogenome may offer novel insights into the evolution of noncoding DNA, as the mitogenome is haploid, autonomous, relatively well understood, and restricted in size and gene content.
Materials and Methods
We used a Pacific Biosciences RSII sequencer to sequence the mitogenomes of the three Callosobruchus species, employing the SMRT-analysis HGAP3 pipeline for assembly, and a Pacific Biosciences Sequel sequencer to sequence the mitogenome of A. obtectus, using the SMRT-analysis HGAP4 pipeline for assembly. The mitogenomes were annotated using DOGMA (Wyman etal. 2004) and MITOS (Bernt etal. 2013). An Illumina HiSeq2500 platform (v4 sequencing chemistry) was used for the resequencing of the three C. maculatus populations which were assembled using MITObim V 1.8 (Hahn etal. 2013) and MIRA V 4.0.2 (Chevreux etal. 1999). The RNA sequencing involved standard RNA extraction and library preparation protocols followed by Illumina HiSeq2500 sequencing, described in detail in Sayadi etal. (2016) and Immonen etal. (2017) (Supplementary Material online).
Results
Mitogenome Organization
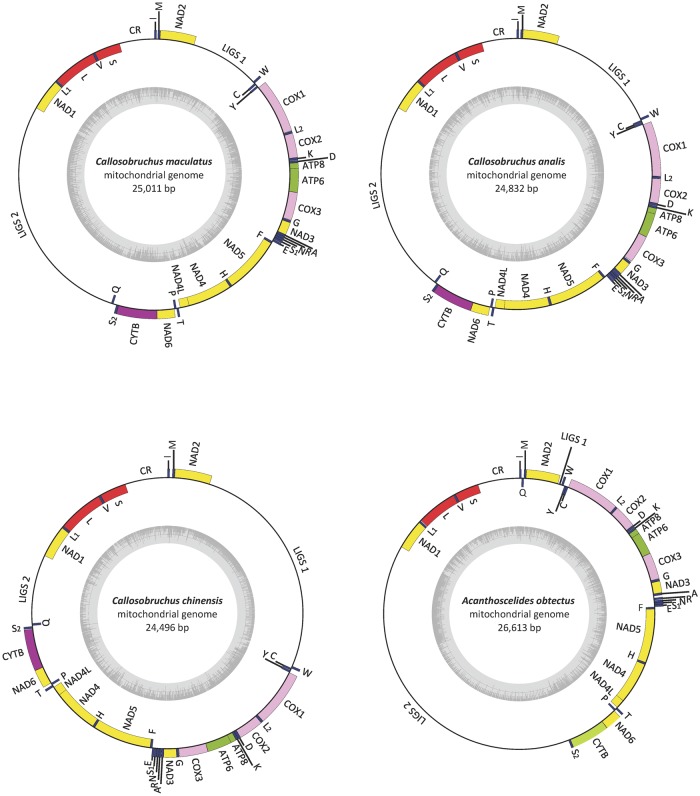
The annotated mitogenomes of the seed beetles Callosobruchus maculatus, C. analis, C. chinensis, and Acanthoscelides obtectus are presented in figure 1A. The four beetles have the largest single chromosome mitogenomes fully assembled in insects and fall within the top 0.5 percentile among all animals, with a size of 25,011 bp for C. maculatus, 24,832 bp for C. analis, 24,496 bp for C. chinensis and 26,613 bp for A. obtectus. A few other insects have mitogenomes even larger than this, but these are then either fragmented into multiple minichromosomes (in lice; e.g., Shao etal. 2017) or have not yet been assembled (weevils; Boyce etal. 1989). The mitogenomes all showed a gene order canonical for insects, except for tRNAq which was displaced in the genus Callosobruchus. The mitogenomes contain 22 tRNA genes, 13 protein-coding genes (PCGs), 16S and 12S ribosomal RNA, a control region (CR) (C. maculatus: 1,031 bp, C. analis: 1,024 bp, C. chinensis: 1,230 bp, A. obtectus: 1,306 bp) and two very long intergenic spacers (LIGS1, LIGS2) composed of tandem repeat arrays. The first LIGS is located between NAD2 and tRNAw, and the second LIGS between tRNAq/s and NAD1. All PCGs started with a standard initiation codon (ATN), except for COX1 which was initiated by an (AAT) codon. COX2, NAD4, and NAD5 were terminated by an incomplete termination codon (T), except for NAD4 in C. analis which was terminated by a (TAG) codon. The degree of overlap between adjacent PCGs was low and the mitogenomes also contained a number of short intergenic spacers, of which most were only a few bp while a few ranged up to 93 bp in length (Supplementary Material online). The CR of C. maculatus share 67.23% sequence identity with C. analis, and 61.96% sequence identity with C. chinensis. The CR of C. analis share 63.52% sequence identity with C. chinensis. The CR of A. obtectus showed even lower sequence identity with the three Callosobruchus species (54.85–56.25%).
Fig. 1.
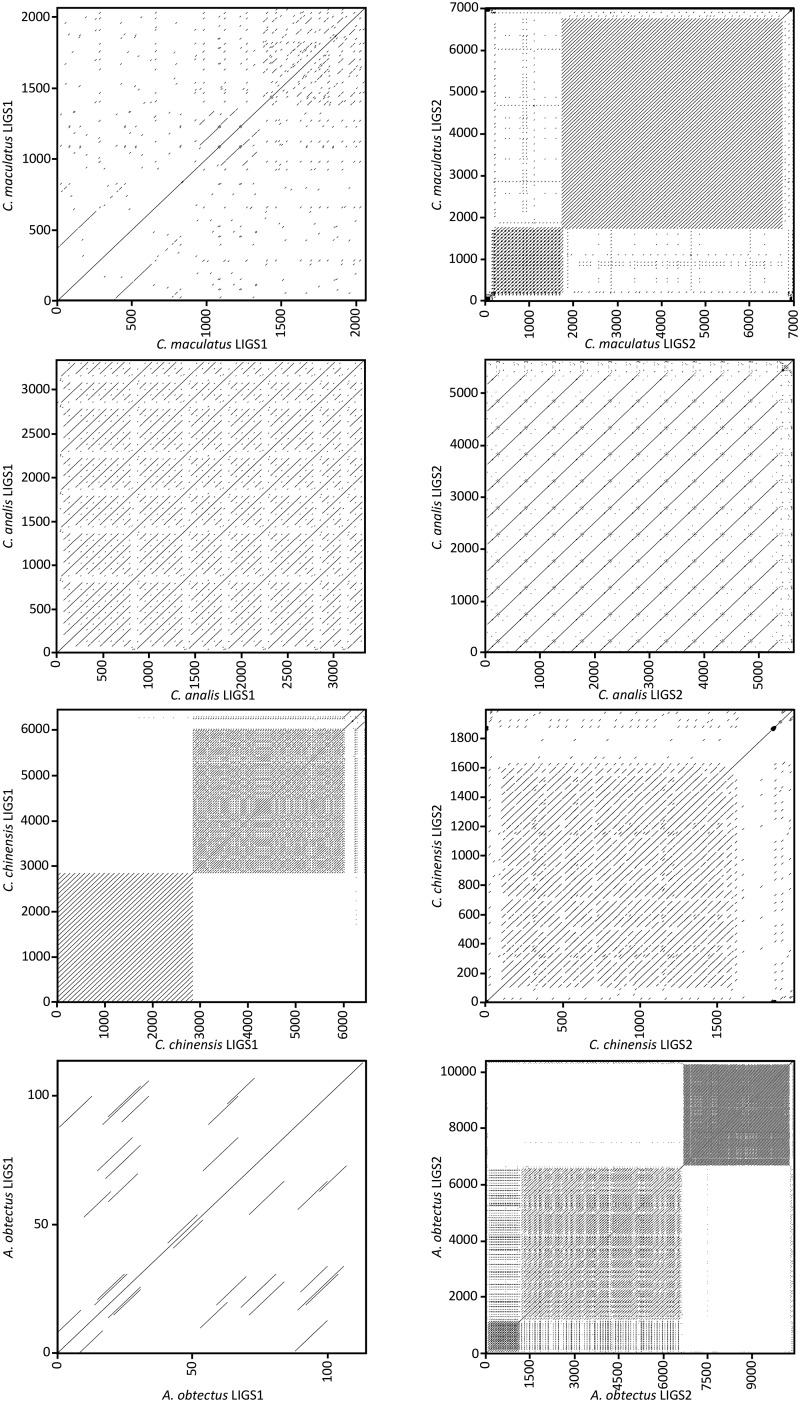
—(A) Annotated mitogenomes of Callosobruchus maculatus, C. analis, C. chinensis and A. obtectus. Yellow, green, purple, and pink regions represent protein coding genes, red rRNAs, and dark blue tRNAs. Unannotated parts represent the control region (CR) and the two long intergenic repeat regions (LIGSs). Inner circle (grey histogram) illustrates relative GC content. (B) The tandem repeat structure of LIGS1 (left) and LIGS2 (right) in the four species is here visualized by dot-plots of their self-aligned sequences. Here, parallel diagonal lines indicate arrays of tandem repeat motifs where the distance between lines equals the length of the motif.
The PacBio sequenced population of C. maculatus was South India. We also sequenced and assembled the mitogenome of two isofemale lines from each of three additional populations of C. maculatus, with different geographic origin (Brazil, California, and Yemen). The four populations differed somewhat in mitogenome size: 25,011 bp for South India; 24,947 bp for Brazil; 25,026 bp for California; and 25,069 bp for Yemen. However, all four exhibited the same genome organization and start and stop codon usage (Supplementary Material online).
The seven mitogenomes assembled and analyzed here are deposited at GenBank under accession numbers KY856743, KY856744, KY856745, KY942060, KY942061, KY942062, and MF925724.
Intergenic Repeat Regions
The two LIGSs were characterized by their highly variable size (114–10,408 bp), their low sequence identity, their tandem repeats sequence composition and an AT bias (75.2% to 87.5%) that is even higher than the control region. Pairwise comparison of these regions between the four mitogenomes did not show any significant sequence similarities and we found no evidence for conserved sequence blocks. All attempts to annotate the LIGSs failed: we found no blast hits using default parameters, no open reading frames and no tRNAs or rRNAs were predicted in these regions. When including also regions of the LIGS showing low compositional complexity in blast searches, a few short LIGS sequence blocks from all species mapped significantly to the control region of the mitogenome of some other insects. A tandem repeat (TR) search revealed that the LIGSs were chiefly composed by a large number of TRs (fig. 1B), which showed no significant sequence similarities among them. LIGS1 in C. maculatus contained predicted TRs ranging from 12 to 372 bp in length that were repeated between 1.9 and 5.4 times, C. analis from 131 to 430 bp repeated between 2.4 and 9.8 times, and C. chinensis from 2 to 103 bp repeated between 16 and 60.8 times. LIGS2 in C. maculatus contained TRs ranging from 2 to 164 bp in length being repeated between 18.5 and 51 times, C. analis from 12 to 35 bp repeated between 2.1 and 4.8 times, C. chinensis from 2 to 209 bp repeated between 3.3 and 28.7 times and A. obtectus from 13 to 155 bp repeated between 1.9 and 70.4 times. In C. analis, for example, LIGS1 contains nine full copies and a partial copy of a TR of 262 bp, which alone forms 81% of LIGS1. In C. chinensis, three TRs collectively make up 93% of the sequence of LIGS1. The first (103 bp) is repeated 27.6 times, the second (52 bp) 60.8 times and the third (2 bp) 16 times. LIGS2 showed a similar composition in C. maculatus and C. chinensis, in being almost completely formed by predicted TRs. In C. maculatus, four distinct TRs made up 95% of LIGS2 with, for example, a single 164 bp TR being repeated 30.4 times. In C. analis, however, LIGS2 contained a much lower number of predicted TRs. The very long LIGS2 of A. obtectus is dominated by two blocks of TR, a 90 bp motif repeated 60.8 times and 51 bp motif repeated 70.4 times, both clearly visible in the dot plot (fig. 1B). Finally, a search for inverted repeats within the LIGSs identified a very large number of cases of reverse complimentary repeat motifs (palindromes), and the predicted DNA folding of the LIGSs showed multiple markedly extended hairpin structures (Supplementary Material online).
In addition to the striking interspecific variation described above, the LIGSs also showed some variation across the four populations of C. maculatus, both in size and TR themes. Sequence similarities between the LIGSs from different populations were high (>94%) and the populations showed the same four TRs in LIGS1. LIGS2 showed more variation. For example, the first TR in South India (2 × 51 bp) was replaced by longer TRs in Brazil (2 × 93 bp) and California (2.2 × 91 bp), while Yemen showed an even longer TR in this site (2 × 127 bp). A unique TR (1.9 × 34 bp) adjacent to the first TR, was present only in California and Yemen. Overall, both LIGSs were somewhat longer in the Yemen population (2,073 and 7,060 bp, respectively) (Supplementary Material online).
We stress that the LIGSs are not artifacts or the result of misassemblies. Assemblers generated closed circular genomes and mapping back PacBio reads showed that sequencing depth was uniformly high along the entire mitogenome, including the LIGSs. Moreover, a very large number of reads spanned much of the entire mitogenomes, including entire LIGSs and the regions flanking the LIGSs (Supplementary Material online). The longest reads that mapped to the mitogenomes of C. maculatus, C. analis, C. chinensis, and A. obtectus were 24,673, 19,830, 23,504, and 24,994 bp, respectively. No reads that mapped to the mitogenomes contained nonmitogenomic sequences.
Selection on mtDNA Genes
All 13 PCGs showed substantial sequence variation, both within and across species. Within C. maculatus, nucleotide diversity (π) was 0.014–0.071 for synonymous sites and 0–0.003 for nonsynonymous sites. Notably, all but four PCGs showed nonsynonymous substitutions (1–9 substitutions) across the four C. maculatus populations, which is interesting considering that previous experimental work has demonstrated that these haplotypes are indeed under selection (Kazancıoğlu and Arnqvist 2014) and are functionally distinct in terms of their effect on growth rate (Dowling etal. 2007), metabolic rate (Arnqvist etal. 2010; Immonen etal. 2016a), behavior (Løvlie etal. 2014) and senescence (Immonen etal. 2016b).
Most values of global ω (i.e., dN/dS), assessed across species, were considerably lower than ω < 0.1 for all 13 PCGs and we found no estimate of ω > 0.4. Models allowing ω to vary across sites showed a significant improvement in fit over those assuming a single value of ω for most PCGs. However, in no case did our analyses indicate that any site of any gene was under significant positive selection, using Naive Empirical Bayes analysis of ω > 1. Similarly, the ratio of nonsynonymous to synonymous nucleotide diversity (πN/πS) across the four populations of C. maculatus was very low for all genes (0.014–0.118). These analyses thus provided evidence for strong purifying selection on PCGs, although the strength of this purifying selection varied across sites within PCGs (Supplementary Material online).
Transcription of the Mitogenome
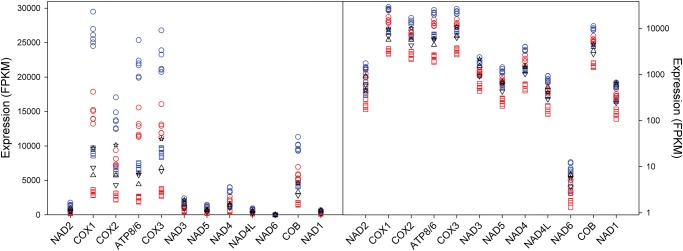
Transcription of the PCGs was assessed by mapping mRNA sequencing reads to the assembled mtDNA genes in C. maculatus. Transcript abundance varied substantially across PCGs, with the 7 NAD genes being present at much lower levels than the other mtDNA genes (fig. 2). This is not likely simply the results of a lack of polyadenylation, as transcripts of several NAD genes carry poly(A) tails in C. maculatus (Supplementary Material online) as well as in other insects (Stewart and Beckenbach 2009; Torres et al 2009; Gao etal. 2016). Further, relative gene expression of mtDNA PCGs was higher in males than in females, higher in the head and thorax than the abdomen and this effect of tissue was stronger in males than in females (table 1 and fig. 2). The NAD genes were the least differentially expressed of all PCGs (supplementary table 7, Supplementary Material online). Cluster analyses of covariation in expression of genes across samples showed that the 7 NAD genes were coexpressed relative to the other mtDNA genes (Supplementary Material online). We found very little evidence for polycistronic transcription of PCGs, as adjacent genes 1) were assembled as distinct transcripts (Sayadi etal. 2016), 2) showed distinct expression, and 3) did not tightly covary in their level of expression. An exception to this was ATP6 and ATP8, which were clearly transcribed as a joint bicistronic transcript (Stewart and Beckenbach 2009).
Fig. 2.
—Relative transcript abundance of mtDNA genes (expressed as FPKM values) across sexes, tissues, and life stages, based on mapping RNA reads back to the assembled genes on a linear (left) and a log (right) scale. Red symbols denote females, blue males, squares are samples from the abdomen, circles are samples from the head and thorax, stars represent larvae, triangles down pupae, and triangles up a mixed adult sample. Transcript abundance differed dramatically across genes (Kruskal–Wallis ANOVA, P < 0.001), with all NAD genes showing very low abundance.
Table 1.
Multivariate Analysis of Variance (MANOVA) of Variation in Transcript Abundance (FPKM) of the 13 mtDNA PCGs Across Sexes, Tissue (Head/Thorax vs. Abdomen) and Mating Status (i.e., Virgin vs. Mated)
| Source | Wilk‘s λ | F | Df | P |
|---|---|---|---|---|
| Sex | 0.0117 | 42.1 | 12, 6 | <0.001 |
| Tissue | 0.0013 | 376.7 | 12, 6 | <0.001 |
| Mating | 0.0733 | 6.3 | 12, 6 | 0.017 |
| Mating × Sex | 0.0751 | 6.1 | 12, 6 | 0.018 |
| Tissue × Sex | 0.0383 | 12.5 | 12, 6 | 0.003 |
| Mating × Tissue | 0.1081 | 4.1 | 12, 6 | 0.047 |
We assessed the possibility that parts of the LIGSs are transcribed by blasting all de novo assembled transcripts (Sayadi etal. 2016; Immonen etal. 2017) against the LIGSs in C. maculatus (South India). We found strong evidence for transcription of some regions of both LIGSs. A 643 bp transcript (TR34743|c0_g1_i1) mapped to part of the 2.9 × 372 bp tandem repeat theme that initiates LIGS1. The first 155 bp of this transcript shows 89% sequence identity with the terminal part of NAD2. Transcript abundance was low, but this transcript was present in all 27 samples (FPKM range = 33–454). Three different transcripts mapped to LIGS2, ranging from 276 (TR2501|c4_g1_i1) over 342 (TR45502|c0_g1_i1) to 364 (TR71824|c2_g2_i2) bp in size. Again, the three transcripts were present in all 27 samples, but transcript abundances were low (FPKM range = 6–161). All of the four transcripts that mapped to the LIGSs were fully covered by the LIGSs and all hits showed e-values < 10−138. None contained any candidate open reading frames (ORFs).
Discussion
The mitochondrion is the powerhouse of the eukaryotic cell. Mitogenomes show very high rates of replication and mtDNA transcription limits metabolic processes in tissues with high energy demands. For these reasons, mitogenomes of animals are under selection for small size and are generally devoid of noncoding sequences (Rand 1993; Zhang and Hewitt 1997). The mitogenomes of the seed beetle species studied here are extraordinary (Nardi etal. 2012) in that two very long intergenic noncoding and TR-rich elements form a very large part of their complete sequence. The CR of insect mitogenomes typically contains TRs (Zhang and Hewitt 1997). The fact that the CRs of the seed beetle mitogenomes are devoid of TRs while the LIGSs are largely composed of TRs suggests that LIGSs may once have originated through a translocation of TRs from the CR, a suggestion also supported by the significant sequence similarity between a few short sequence blocks of LIGSs and the CR of some other insects. Irrespective of their origin, however, four observations are difficult to reconcile with the hypothesis that the LIGSs are the result of purely neutral evolution. Below, we first discuss these four facets and then suggest a potential mechanism by which these LIGSs might offer a selective advantage in this group of insects.
First, the LIGSs have been retained for a very long time. Callosobruchus maculatus and C. analis are closely related species that diverged approximately 5 Ma (Tuda etal. 2006) while C. chinensis diverged from these approximately 22 Ma and A. obtectus diverged from the three Callosobruchus species some 45 Ma (Kergoat etal. 2005). Long-lasting retention of shorter intergenic mitogenomic spacers has previously been observed in vertebrates (Kumazawa etal. 1996; McKnight and Shaffer 1997; Jørgensen etal. 2014). The LIGSs in seed beetles have persisted despite what must have been repeated contractions and expansions of the LIGSs, because their size varies, their repeat themes are distinct (fig. 1B) and their sequence similarity is very low. A dynamic evolution of the LIGSs is also suggested by the fact that they showed some variation across populations within one of our species and is consistent with rapid turnover of several types of repeat sequences in general, including the mitogenomic CR (Solignac et al 1986; McKnight and Shaffer 1997). We note that other members of the family Chrysomelidae, as far as we know, show conventional mitogenomes (15.7–16.8 kbp) and apparently lack LIGSs. Second, in light of the above, the fact that the mitogenomes of the four species are so similar in size is noteworthy. Although the LIGSs vary greatly in size, the sum of the two LIGSs varies little across species (fig. 1A). Remarkably, the size of LIGS1 is perfectly and negatively related to the size of LIGS2 across the four species and a phylogenetic least-squares regression provided strong evidence for negative correlated evolution between the size of LIGS1 and LIGS2 (r = −0.99, P = 0.006; Supplementary Material online). This concerted evolution implies that some form of size-related functional constraint, such as increased costs of replication beyond a certain total mitogenome size, must affect the evolution of the LIGSs.
Third, our analyses of the molecular hallmarks of selection showed that the PCGs of the mitogenome of these seed beetles has experienced strong purifying selection in the past, as is typical for insect mitogenomes (Bazin etal. 2006; Meiklejohn etal. 2007; James etal. 2016). This nonrecombining genome encodes some of the key building blocks of the main energy producing pathway, the ATP producing OXPHOS pathway, and it is therefore unsurprising that a well-functioning mitogenome has been imperative. This suggests that LIGSs should have been purged by selection if conferring even a marginal a net cost.
Fourth, although less remarkable in size, a few cases of LIGSs have previously been reported in insect mitogenomes. What is remarkable, however, is that the intergenic locations of several of these LIGSs coincide with that found in seed beetles. Between Nad2 and Cox1, Wan etal. (2012) described a LIGS of 2.8 kbp in an earwig (Dermaptera), Linard etal. (2016) a LIGS of 2.7 kbp in a beetle (Coleoptera), Bae etal. (2004) a LIGS of 1.7 kbp in a beetle (Coleoptera) and Cameron etal. (2008) a LIGS of 0.3 kbp in a wasp (Hymenoptera). Similarly, between Cob and Nad1, Linard etal. (2017) described a 1.4 kbp LIGS in a caddis fly (Trichoptera), Cameron etal. (2008) a 0.8 kbp LIGS in a wasp (Hymenoptera) and Dotson and Beard (2001) a 0.3 kbp LIGS in a true bug (Hemiptera). These species belong to five different insect orders and LIGSs in these precise intergenic locations must therefore have evolved independently multiple times in insects.
Long intergenic repeat regions in these sites in the mitogenome have clearly evolved independently several times in insects and have been retained in seed beetles for >45 my in the face of purifying selection, despite the fact that the evolution of LIGSs is clearly not conserved at the sequence level. We see two potential explanations for our findings. One possibility is that selectively neutral repeat expansions are somehow tolerated in these particular sites in the mitogenome, thus constituting intergenic “hot-spots” for neutral expansion of repeat arrays in insects. However, several circumstances challenge a view of the LIGSs as neutral “junk” DNA. Most importantly, given the very high levels of replication of the mitogenome, purifying selection on this nonrecombining and haploid genome should purge nonfunctional mtDNA at the population level even if the additional costs of replication are minor (Bergstrom and Pritchard 1998). Indeed, experimental work has demonstrated that purifying selection efficiently removes weakly deleterious mtDNA mutations (Stewart etal. 2008). This, in essence, forms the basis for our understanding of why the archetypal mitogenome is compact and deprived of noncoding DNA (Rand 1993; Zhang and Hewitt 1997) and why it has resisted mutational meltdown via Muller’s ratchet. The observed pattern of concerted size evolution of the LIGSs is also incompatible with this possibility. We thus suggest that it is very unlikely that the LIGSs are purely a result of genetic hitchhiking.
The other possibility is that the LIGSs represent functional DNA, although the selected effect would be more related to the presence or absence of LIGSs rather than the precise sequence of nucleotides that form the LIGSs (i.e., “indifferent DNA”, sensu Graur etal. [2015]) given the absence of significant sequence conservation. We suggest that the unusual metabolic biology of seed beetles, as well as that of several of the other insects with LIGSs in these sites, points to the possibility that LIGSs may help regulate the transcriptional and/or translational machinery of the mitogenome. Our reasoning is as follows. The mitochondrion uses oxidative phosphorylation (OXPHOS) to feed the reformation of ATP by ATP synthase. Here, enzyme complex I (NADH dehydrogenase) oxidizes NADH to generate ubiquinol which is fed to complex III (cytochrome c reductase). Alternatively, complex III can be fed by complex II (succinate dehydrogenase—also part of the Krebs cycle), which oxidizes FADH2 to generate ubiquinol. Importantly, the relative amounts of FADH2 and NADH generated during the breakdown of glucose differ from that during fatty acid oxidation and amino acid catabolism, such that relatively less NADH is formed when fat or protein is utilized as metabolic substrates compared to carbohydrates (Speijer etal. 2014). Seed beetles live their entire juvenile life inside a single legume seed, where they consume the protein-rich cotyledon. Adults are facultatively aphagous and instead utilize large deposits of stored lipids, accumulated during the larval stage, as their main source of energy. Studies of seed beetle metabolism have confirmed that larvae use a mixture of proteins and lipids, and only to smaller extent carbohydrates, for their metabolic demands and that of adults is based almost entirely on lipids (Wightman 1978a, 1978b; Yates etal. 1989; Immonen etal. 2016a). Thus, NADH dehydrogenase plays a relatively minor role in OXPHOS in seed beetle life histories, compared to taxa with a more carbohydrate based metabolism.
Imbalanced transcription and translation of necessary OXPHOS components can affect reactive oxygen species production and cause a fitness decline (Bonawitz etal. 2006). The LIGSs may thus serve to economize the translational machinery of the mitogenome by reducing polycistronic transcription and/or posttranscriptional modification (Stewart and Beckenbach 2009), thereby allowing more flexible adjustment of the composition of the electron transport chain complex. In particular, the LIGSs may allow a decreased transcription of the NAD genes given a lipid- and protein-based metabolism. Four aspects of our transcript abundance analyses are at least consistent with such a scenario. First, polycistronic transcription was very limited. Second, transcripts from NAD genes all showed a very low abundance. Third, variation in transcript abundance of NAD genes suggested coregulation across sexes and tissues. Fourth, males showed a higher transcript abundance of non-NAD PCGs than females (fig. 2), suggesting transcriptional decoupling between the NAD genes and other mtDNA PCGs consistent with the fact that male seed beetles show a higher metabolic rate (Berger etal. 2014; Arnqvist etal. 2017) and consume their lipid metabolite stores more rapidly than females (Lazarević etal. 2012).
The mode by which LIGSs might affect transcription of PCGs is unknown. The fact that they are very AT rich suggests that they may provide secondary structure that, directly or indirectly, guide DNA-binding proteins that compartmentalize or otherwise affect transcription (Nardi etal. 2012; Liu etal. 2013). Intergenic mitogenomic spacers have been suggested to function as alternative sites of initiation of replication and/or transcription in vertebrates (Kumazawa etal. 1996; McKnight and Shaffer 1997) and palindromic mtDNA sequences occur in a range of taxa (Nardi etal. 2012). Models of the secondary structures of the LIGSs did indeed unveil multiple and sometimes dramatically extended hairpin loops (supplemental material). This possibility is also supported by the sequence similarity between a few blocks of the LIGSs and that of the CR of other insects. Yet, the LIGSs are demonstrably at least partly transcribed, albeit at seemingly low levels. We do note, however, that our RNA sequencing involved poly(A) enrichment, and although transcripts of mtDNA PCGs showed poly(A) tails (Supplementary Material online) transcription of LIGSs are less likely to involve polyadenylation. Our data therefore does not allow firm quantitative conclusions regarding transcription of the LIGSs. Our observation could represent transcriptional noise resulting from low RNA polymerase fidelity, but this is less likely given that the four assembled transcripts that mapped to the LIGSs were present in all samples. One possibility is that the LIGSs encode long noncoding RNAs, which might affect transcription of PCGs in the mitogenome (Jørgensen etal. 2014; Gao etal. 2016; Liu etal. 2017). The LIGSs could also contain small noncoding RNAs, which may affect transcription or translation, but reliable detection of such motifs is complicated by the very high AT content of the LIGSs. Nevertheless, the fact that the LIGSs are at least partly transcribed makes them interesting potential sources of mitochondrial noncoding RNAs.
Most mitogenomes to date have been assembled using either PCR and “primer walking” or short-read sequencing techniques, often using reference genomes. These methods perform well when assembling simple mitogenomes but do not yield complete de novo assemblies of more complex mitogenomes, especially when containing tandem repeat units (Bernt etal. 2013; Hahn etal. 2013). This is well illustrated by two recently presented mitogenome assemblies of C. chinensis and A. obtectus, based on PCR and “primer walking” but also paired-end short-read sequencing (Li etal. 2016; Yao etal. 2017). Yao etal. (2017) reports an apparent 13 bp overlap between tRNAs and NAD1, at the site of LIGS2 in A. obtectus. The assembly of C. chinensis by Li etal. (2016) has a major gap which includes all of LIGS1, as well as the flanking regions. Interestingly, their assembly does contain an intergenic spacer of >600 bp which is identical to the initial 505 bp and the terminal 126 bp of LIGS2, but does not include the repeat-rich 1,364 bp central part of LIGS2. Paired-end short-read sequencing reads of both LIGSs were present but did not allow the assembly of these TR-rich regions. For the parts of these two assemblies that are shared with ours (16,148 bp for C. chinensis and 16,130 bp for A. obtectus), sequence identities are very high indeed (99.8% and 99.5%, respectively). Thus, this illustrates the superior ability of long-read sequencing to capture and handle long tandem repeat arrays. We predict that the increased use of long-read sequencing techniques will uncover many more cases of unconventional mitogenomes in the near future.
In conclusion, the remarkable mitogenomes of seed beetles illustrate that the canonical view of insect mitogenomes as being under selection for small size and being devoid of noncoding sequences is incomplete. Our results further suggest that natural selection have acted to retain large amounts of repeat DNA in these mitogenomes and we suggest that these tandem repeat arrays may act to regulate transcription or translation of PCGs. We predict that the future will reveal many more instances of repeat-rich insect mitogenomes and hope that these can help uncover the functional significance of noncoding mtDNA.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
Laboratory assistance was provided by J. Rönn. N. Alvarez, S. Glemin, and A. Suh provided thoughtful comments on our manuscript. We thank all members of the GENCON lab, for helpful discussion. X.-J. Li and Z.-M. Wei kindly shared the results of their assembly efforts with C. chinensis with us. This study was supported by a European Research Council Advanced Investigator Grant (GENCON AdG-294333) to G.A. and a grant from the Swedish Research Council (621-2014-4523) to G.A. We thank the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) at Uppsala University for providing computational resources. Sequencing was performed by the National Genomics Infrastructure (NGI)/Uppsala Genome Center and the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University. This study was supported by the Wallenberg Advanced Bioinformatics Infrastructure (WABI). These national infrastructures are supported by the Swedish Research Council (VR RFI) and the Knut and Alice Wallenberg Foundation.
Literature Cited
- Arnqvist G, et al. 2010. The genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution 64(12): 3354–3363. [DOI] [PubMed] [Google Scholar]
- Arnqvist G, Stojković B, Rönn J, Immonen E.. 2017. The pace-of-life: a sex-specific link between metabolic rate and life history in bean beetles. Func Ecol. doi: 10.1111/1365-2435.12927. [Google Scholar]
- Bae JS, Kim I, Sohn HD, Jin BR.. 2004. The mitochondrial genome of the firefly, Pyrocoelia rufa: complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol Phylogenet Evol. 32(3): 978–985. [DOI] [PubMed] [Google Scholar]
- Bazin E, Glémin S, Galtier N.. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312(5773): 570–572. [DOI] [PubMed] [Google Scholar]
- Berger D, Berg EC, Widegren W, Arnqvist G, Maklakov AA.. 2014. Multivariate intralocus sexual conflict in seed beetles. Evolution 68(12): 3457–3469. [DOI] [PubMed] [Google Scholar]
- Bergstrom CT, Pritchard J.. 1998. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics 149(4): 2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, et al. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2): 313–319. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Rodeheffer MS, Shadel GS.. 2006. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol Cell Biol. 26(13): 4818–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8): 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce TM, Zwick ME, Aquadro CF.. 1989. Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics 123(4): 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SL, et al. 2008. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 51(10): 800–808. [DOI] [PubMed] [Google Scholar]
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Ann Rev Entomol. 59: 95–117. [DOI] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S.. 1999. Genome sequence assembly using trace signals and additional sequence information. German Conf Bioinf 99: 45–56. [Google Scholar]
- Dotson EM, Beard C.. 2001. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol Biol. 10(3): 205–215. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Abiega KC, Arnqvist G.. 2007. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution 61(1): 194–201. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, et al. 2016. PacBio full-length transcriptome profiling of insect mitochondrial gene expression. RNA Biol. 13(9): 820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Zheng Y, Azevedo RB.. 2015. An evolutionary classification of genomic function. Genome Biol Evol. 7(3): 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, et al. 2013. On the immortality of television sets: "function" in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 5(3): 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B.. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads: a baiting and iterative mapping approach. Nucleic Acids Res. 41(13): e129.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Collet M, Goenaga J, Arnqvist G.. 2016b. Direct and indirect genetic effects of sex-specific mitonuclear epistasis on reproductive ageing. Heredity 116: 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Rönn J, Watson C, Berger D, Arnqvist G.. 2016a. Complex mitonuclear interactions and metabolic costs of mating in male seed beetles. J Evol Biol. 29(2): 360–370. [DOI] [PubMed] [Google Scholar]
- Immonen E, Sayadi A, Bayram H, Arnqvist G.. 2017. Mating changes sexually dimorphic gene expression in the seed beetle Callosobruchus maculatus. Genome Biol Evol. 9(3):677–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JE, Piganeau G, Eyre-Walker A.. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25(1): 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen TE, et al. 2014. An evolutionary preserved intergenic spacer in gadiform mitogenomes generates a long noncoding RNA. BMC Evol Biol. 14: 182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazancıoğlu E, Arnqvist G.. 2014. The maintenance of mitochondrial genetic variation by negative frequency-dependent selection. Ecol Lett. 17(1): 22–27. [DOI] [PubMed] [Google Scholar]
- Kergoat GJ, Alvarez N, Hossaert-McKey M, Faure N, Silvain JF.. 2005. Parallels in the evolution of the two largest New and Old World seed‐beetle genera (Coleoptera, Bruchidae). Mol Ecol. 14(13): 4003–4021. [DOI] [PubMed] [Google Scholar]
- Kolesnikov AA, Gerasimov ES.. 2012. Diversity of mitochondrial genome organization. Biochem Mosc. 77(13): 1424–1435. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T.. 1996. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 13(9): 1242–1254. [DOI] [PubMed] [Google Scholar]
- Lazarević J, Tucić N, Šešlija Jovanović D, Večeřa J, Kodrík D.. 2012. The effects of selection for early and late reproduction on metabolite pools in Acanthoscelides obtectus Say. Insect Sci. 19(3): 303–314. [Google Scholar]
- Li X, Ou J, Wei Z, Li Y, Tian Y.. 2016. The mitogenomes of three beetles (Coleoptera: Polyphaga: Cucujiformia): new gene rearrangement and phylogeny. Biochem Syst Ecol. 69: 101–107. [Google Scholar]
- Linard B, Arribas P, Andújar C, Crampton‐Platt A, Vogler AP.. 2016. Lessons from genome skimming of arthropod‐preserving ethanol. Mol Ecol Res. 16(6): 1365–1377. [DOI] [PubMed] [Google Scholar]
- Linard B, Arribas P, Andújar C, Crampton-Platt A, Vogler AP.. 2017. The mitogenome of Hydropsyche pellucidula (Hydropsychidae): first gene arrangement in the insect order Trichoptera. Mitochondrial DNA 28(1): 71–72. [DOI] [PubMed] [Google Scholar]
- Liu C, Chang J, Ma C, Li L, Zhou S.. 2013. Mitochondrial genomes of two Sinochlora species (Orthoptera): novel genome rearrangements and recognition sequence of replication origin. BMC Genomics 14: 114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, et al. 2017. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355(6320): aah7111.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løvlie H, Immonen E, Gustavsson E, Kazancioğlu E, Arnqvist G.. 2014. The influence of mitonuclear genetic variation on personality in seed beetles. Proc R Soc Lond B. 281. doi: 10.1098/rspb.2014.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight ML, Shaffer HB.. 1997. Large, rapidly evolving intergenic spacers in the mitochondrial DNA of the salamander family Ambystomatidae (Amphibia: Caudata). Mol Biol Evol. 14(11): 1167–1176. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM.. 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23(6): 259–263. [DOI] [PubMed] [Google Scholar]
- Nardi F, Carapelli A, Frati F.. 2012. Repeated regions in mitochondrial genomes: distribution, origin and evolutionary significance. Mitochondrion 12(5): 483–491. [DOI] [PubMed] [Google Scholar]
- Rand DM. 1993. Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol. 37(3): 281–295. [DOI] [PubMed] [Google Scholar]
- Sayadi A, Immonen E, Bayram H, Arnqvist G.. 2016. The de novo transcriptome and its functional annotation in the seed beetle Callosobruchus maculatus. PLoS One 11(7): e0158565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R, Li H, Barker SC, Song S.. 2017. The mitochondrial genome of the guanaco louse, Microthoracius praelongiceps: insights into the ancestral mitochondrial karyotype of sucking lice (Anoplura, Insecta). Genome Biol Evol. 9(2): 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M, Monerot M, Mounolou JC.. 1986. Concerted evolution of sequence repeats in Drosophila mitochondrial DNA. J Mol Evol. 24(1–2): 53–60. [DOI] [PubMed] [Google Scholar]
- Speijer D, Manjeri GR, Szklarczyk R.. 2014. How to deal with oxygen radicals stemming from mitochondrial fatty acid oxidation. Philos Trans R Soc Lond B. 369(1646): 20130446.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Beckenbach AT.. 2009. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene 445(1–2): 49–57. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Larsson NG.. 2008. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 9(9): 657–662. [DOI] [PubMed] [Google Scholar]
- Torres TT, Dolezal M, Schlötterer C, Ottenwälder B.. 2009. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucleic Acids Res. 37(22): 7509–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuda M, Rönn J, Buranapanichpan S, Wasano N, Arnqvist G.. 2006. Evolutionary diversification of the bean beetle genus Callosobruchus (Coleoptera: Bruchidae): traits associated with stored-product pest status. Mol Ecol. 15(12): 3541–3551. [DOI] [PubMed] [Google Scholar]
- Wan X, Kim MI, Kim MJ, Kim I.. 2012. Complete mitochondrial genome of the free-living earwig, Challia fletcheri (Dermaptera: Pygidicranidae) and phylogeny of Polyneoptera. PLoS One 7(8): e42056.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman JA. 1978a. The ecology of Callosobruchus analis (Coleoptera: Bruchidae): morphometrics and energetics of the immature stages. J Anim Ecol. 47: 117–129. [Google Scholar]
- Wightman JA. 1978b. The ecology of Callosobruchus analis (Coleoptera: Bruchidae): energetics and energy reserves of the adults. J Anim Ecol. 47: 131–142. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20(17): 3252–3255. [DOI] [PubMed] [Google Scholar]
- Yao J, Yang H, Dai R.. 2017. Characterization of the complete mitochondrial genome of Acanthoscelides obtectus (Coleoptera: Chrysomelidae: Bruchinae) with phylogenetic analysis. Genetica doi: 10.1007/s10709-017-9975-9. [DOI] [PubMed] [Google Scholar]
- Yates LR, Daza M, Saiz F.. 1989. The energy budget of adult Pseudopachymerina spinipes (Er.)(Coleoptera: Bruchidae). Can J Zool. 67(3): 721–726. [Google Scholar]
- Zhang DX, Hewitt GM.. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25(2): 99–120. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.