Abstract
Background and Aims Understanding the direct consequences of polyploidization is necessary for assessing the evolutionary significance of this mode of speciation. Previous studies have not studied the degree of between-population variation that occurs due to these effects. Although it is assumed that the effects of the substances that create synthetic polyploids disappear in second-generation synthetic polyploids, this has not been tested.
Methods The direct consequences of polyploidization were assessed and separated from the effects of subsequent evolution in Vicia cracca, a naturally occurring species with diploid and autotetraploid cytotypes. Synthetic tetraploids were created from diploids of four mixed-ploidy populations. Performance of natural diploids and tetraploids was compared with that of synthetic tetraploids. Diploid offspring of the synthetic tetraploid mothers were also included in the comparison. In this way, the effects of colchicine application in the maternal generation on offspring performance could be compared independently of the effects of polyploidization.
Key Results The sizes of seeds and stomata were primarily affected by cytotype, while plant performance differed between natural and synthetic polyploids. Most performance traits were also determined by colchicine application to the mothers, and most of these results were largely population specific.
Conclusions Because the consequences of colchicine application are still apparent in the second generation of the plants, at least the third-generation polyploids should be considered in future comparisons. The specificities of the colchicine-treated plants may also be caused by strong selection pressures during the creation of synthetic polyploids. This could be tested by comparing the initial sizes of plants that survived the colchicine treatments with those of plants that did not. High variation between populations also suggests that different polyploids follow different evolutionary trajectories, and this should be considered when studying the effects of polyploidization.
Keywords: Anti-mitotic agent, common garden experiment, Fabaceae, flow cytometry, individual growth rate, neopolyploid, reproductive fitness, trait evolution
INTRODUCTION
Understanding the consequences of polyploidization is necessary because it is an important determinant of biodiversity (Ramsey and Ramsey, 2014). Despite the abundant literature that exists on this topic (reviewed in Ramsey and Ramsey, 2014), the validity of most existing knowledge is limited because it is difficult to distinguish the consequences of polyploidization and the effects of independent evolutionary history on polyploid lineages (but see Husband et al., 2008, 2016; Maherali et al., 2009; Oswald and Nuismer, 2011).
A useful approach to understand the consequences of polyploidization is using synthetic polyploids (Husband et al., 2008). By including synthetic polyploids in ecological studies, we can distinguish traits that are due to polyploidization (i.e. traits that are the same in natural and synthetic polyploids but different in natural diploids) from traits originating from subsequent evolution of the polyploid lineage (i.e. traits that are the same in natural diploids and synthetic tetraploids but different in natural tetraploids). While synthetic polyploids have long been used for plant breeding (e.g. Semeniuk and Arisumi, 1968; Lignowski and Scott, 1972; Levin, 1983; Lumaret, 1988; Levin, 2002), ecological studies using synthetic polyploids have just appeared in the last decade and in only a few model systems (e.g. Husband et al., 2008, 2016; Tate et al., 2009; Maherali et al., 2009; Oswald and Nuismer, 2011; Cohen et al., 2013).
Synthetic polyploids are induced by using colchicine, trifluralin or oryzalin, i.e. anti-mitotic substances that block the cell cycle in young plants (Semeniuk and Arisumi, 1968; Lignowski and Scott, 1972; Jaskani et al., 2005; Madon et al., 2005; Amiri et al., 2010). Application of the anti-mitotic substances to plants has many direct consequences for plant phenotypes that cannot be regarded as a consequence of polyploidization (Ramsey and Schemske, 2002). To solve this, Husband et al. (2008) suggested that the effects of the anti-mitotic substances may be eliminated by crossing two synthetic polyploids from the same population and following the performance of the offspring of these parents. The properties of these plants are assumed to be a consequence of polyploidization and not of an anti-mitotic substance (Husband et al., 2008). While this assumption seems reasonable, it has rarely been tested. Therefore, it is possible that the patterns detected in the synthetic polyploids may be effects remaining from the application of anti-mitotic substances. To test this, Husband et al. (2016) included in their comparison the offspring of diploid plants that were treated by colchicine but that did not become polyploid. They demonstrated that these plants do not differ from natural diploids and concluded that the colchicine does not affect the second generation. As this is the only indication that an effect was absent, further testing of this assumption is needed. In addition, the plants that did not become polyploid may not experience the same colchicine effect as those that became polyploid, so the absence of an effect in those plants may not signify that these effects do not exist. A potential approach to explore this issue may be to compare the offspring of synthetic polyploids with the offspring of synthetic polyploids that have reverted to the diploid state. To date, the author is not aware of any study that has attempted to do this.
Previous studies comparing natural diploids and polyploids with synthetic polyploids usually used plants from only a single population or did not distinguish between them (Husband et al., 2008, 2016; Maherali et al., 2009; Griffin et al., 2012). In an exceptional study, Oswald and Nuismer (2011) compared diploids and synthetic polyploids from two different populations and demonstrated that the differences were partly population specific. Their study, however, did not include any natural polyploids. Differences between synthetic polyploids of a different origin can be expected, as studies comparing natural diploids and polyploids demonstrate that the differences may be largely population specific (Eliášová and Münzbergová, 2014). These differences may be driven by the different habitat conditions of the origin (Černá and Münzbergová, 2013) or by different phylogenetic origins of the polyploids (e.g. Segraves and Thompson, 1999; Meimberg et al., 2009). To support this, Matsushita et al. (2012), Cohen et al. (2013) and Dar et al. (2013) demonstrated that different synthetic polyploids may indeed follow different evolutionary trajectories. Therefore, the effects of polyploidization should be studied in multiple populations to assess the generality of the observed patterns.
Polyploid seeds are often larger than diploid seeds (Maceira et al., 1993; Bretagnolle et al., 1995; Hoya et al., 2007; Cohen et al., 2013). As this is probably a direct consequence of large cell size in polyploids (e.g. Van Dijk and Van Delden, 1990; Otto, 2007), it can be expected that synthetic polyploids will also produce larger seeds (Hosseini et al., 2013). Seed size has direct consequences for plant performance (e.g. Vera, 1997; Benard and Toft, 2007, 2008; Münzbergová and Plačková, 2010; Skogen et al., 2010), so initial seed size should be included in models used to test between cytotype differences, but this has rarely been done (Eliášová and Münzbergová, 2014, 2017).
In a previous study on Vicia cracca, it was demonstrated that natural tetraploids differ significantly from diploids in a wide range of plant performance measures (Eliášová and Münzbergová, 2014, 2017). The tetraploids did not originate recently in situ. Instead, the two cytotypes met in a secondary contact zone (Eliášová et al., 2014), so is not clear whether the observed differences are due to polyploidization or to subsequent independent evolution of the two cytotypes. In a previous study, the methodology of the development of synthetic polyploids was optimized by using colchicine (Pavlíková et al., 2017), and a large number of synthetic polyploid seeds originating from plants from four mixed ploidy populations were produced.
In this current study, answers were sought to the following questions. (1) What are the differences in seed size, performance and flowering phenology between natural and synthetic tetraploids and natural diploids? (2) Do the differences vary between source populations? (3) Can the differences between diploids and tetraploids be explained by initial variations in seed size? As a high frequency of diploid seeds was detected in the synthetic tetraploid mother plants, it was also possible to test the effect of colchicine application in the second generation without the effects of polyploidization; thus, a further question was asked: (4) what are the effects of colchicine application on mother plants in regard to offspring performance?
MATERIALS AND METHODS
Study species
Vicia cracca, Fabaceae, was used as the model taxon. It is a self-compatible herb with a mixed mating system but with prevailing outcrossing and reduced fitness after selfing (Eliášová et al., 2014). Occurrence of apomixes is unlikely (Asker and Jerling, 1992). The species is autopolyploid (Eliášová et al., 2014) with diploid (2n = 2x = 14) and tetraploid (2n = 4x = 28) cytotypes forming a contact zone with mixed-ploidy populations in Central Europe (Trávníček et al. 2010). In a previous study, Eliášová and Münzbergová (2014, 2017) found that the performance of young and adult plants between the cytotypes was greatly different, indicating that polyploidization theoretically plays an important role in the performance of this species. On average, a single diploid individual produces 200–300 seeds, and a single tetraploid individual produces 150–450 seeds, depending on the watering regime (Eliášová and Münzbergová, 2017). The seed material used for this study was collected from four different mixed-ploidy populations occurring at the contact zone between the diploids and tetraploids along the border between the Czech Republic and Slovakia. These populations correspond to populations 6, 7, 19 and 20 in Eliášová et al. (2014). A previous study on the genetic diversity of these populations did not suggest any clear genetic differences between these populations (Eliášová et al., 2014).
Creation of synthetic polyploids
The procedure to develop synthetic polyploids in V. cracca using colchicine was optimized in a previous study (Pavlíková et al., 2017). To do this, seeds from diploid mothers of four mixed-ploidy populations that occurred at the contact zone between the two cytotypes in the Czech Republic were used. Plants that survived the polyploidization and became polyploid (confirmed by flow cytometry) were considered C1 synthetic polyploids. The plants were individually planted to pots (10 × 10 × 10 cm, 1:1 mixture of sand and soil) and kept in the experimental garden of the Institute of Botany, Czech Academy of Sciences in Průhonice. At the stage of flowering buds, the ploidy level of each flowering branch of the plant was individually measured using flow cytometry (using one leaf from the upper part of the flowering branch). Only the plants that were confirmed to be tetraploid were selected. In the case of a diploid flowering branch on a tetraploid plant, the diploid branch was removed at the base to ensure that all the flowering branches on the plant were tetraploid. Identifying the branch as chimeric may not be possible using a single leaf per branch (see the Discussion).
The tetraploid plants were then moved to an isolated location in the experimental garden to ensure that no V. cracca were flowering within a radius of at least 300 m; plants originating from different populations were maintained at least 50 m apart. This distance was assumed to be sufficient for preventing pollen flow between populations, as most pollen in Fabaceae can be dispersed up to 10 m away (Matter et al., 2013). In the same way, natural diploid and natural tetraploid plants were cultivated in the same experimental garden. The plants were left to cross-fertilize naturally over the course of flowering (approx. 12 weeks). Hand pollination of the plants was not conducted due to the complicated structure of the zygomorphic flowers of the species that contributes to a very low success of hand pollination. During the whole season, the ploidy levels of new flowering branches appearing on the C1 synthetic polyploid plants were assessed, and all diploid branches were removed. All ripe seeds, which represented C2 synthetic polyploids and C2 generations of the natural diploids and polyploids, were collected from each mother plant at the end of the field season. For the subsequent experiment, seeds from 3–5 mother plants per population for synthetic polyploids, 8–10 mother plants per population for natural diploids and 7–9 mother plants per population for natural tetraploids were available.
Growth experiment
To set up the growth experiment, 50 C2 seeds originating from natural diploids and natural polyploids and 100 C2 seeds from synthetic polyploids from each of the four original populations were used. All the seeds were individually weighed and then scarified. As the germination rate was of interest here and in order to reduce the possibility of different scarification intensity, the whole seed coat was removed from the seeds, resulting in an equal starting point for all seeds. Each seed was individually planted in one cell of a multipot (cells 2 × 2 × 5 cm, 1:1 mixture of sand and soil) at the end of March 2015. The multipots were placed in a growth chamber and regularly watered. The regime of the growth chamber was 12 h of light and 12 h of dark at temperatures of 20 and 10 °C, respectively. The multipots were inspected every other day for germination, and the germination date was recorded for each seed. A seed was considered germinated if the above-ground part of the plant was easily detectable at 1 cm in height. To describe plant growth, plant height was measured at the age of 1 and 2 weeks (plants had little leaf growth at this stage).
After a month, the plants were individually transplanted into pots (10 × 10 × 10 cm, 1:1 mixture of sand and soil) and cultivated in an unheated greenhouse. The total number of leaves and number of branches in plants were counted at 4 and 8 weeks old. At this stage, the plants were branching, so plant height was not a good measure of plant size. Ploidy level for all plants was assessed at the age of 8 weeks using flow cytometry.
At the age of 8 weeks, the plants were individually transplanted to large circular pots (19 cm in diameter, 19 cm deep, 1:1 mixture of sand and soil) and placed in the experimental garden. The plants were allowed to grow in the garden without interference for the whole growing season of 2015. When the plants began to flower, flowering was recorded on a weekly basis by counting the number of flowering inflorescences and it was visually estimated whether the flowers were one-third, two-thirds or fully bloomed. These data were used to calculate the start, end, duration and mean flowering for each plant. All the pods with ripe seeds were continuously collected, and the number of developed seeds in these pods were counted. Pod and seed production were used as the only two size measures of the plants after the age of 8 weeks because >80 % of plants had flowered by this time and information on reproductive output is a more appropriate measure of plant fitness than plant biomass. Due to a high correlation among the different plant characteristics, only the following variables on the mature plants were used in the subsequent tests: flowering (yes/no), fruiting of flowering plants (yes/no), proportion of developed seeds, number of seeds per pod, number of pods per plant, mean flowering time and flowering duration.
To estimate the width and length of the stomata, one composite leaf from each plant was selected, and the first and third leaflet were then collected from the composite leaf. The leaves were placed into Petri dishes filled with a mixture of 96 % ethanol and acetic acid in a 3:1 ratio. After 3 h, the leaves were moved to a solution of lactoglycerol (lactic acid, glycerol and water in a 1:1:1 ratio) for 2 h. The resulting samples were placed on a filter paper and then stored in an Eppendorf tube in lactoglycerol until they were measured for length and width of stomata. An Olympus BX60 microscope with a magnification of × 200 was used to observe the leaves. For each leaflet, the length of ten stomata in the centre of the abaxial side of the leaf was measured using QuickPHOTO Micro 3.0, so 20 stomata were measured per plant. For data analysis, the mean length and width of stomata were calculated for each plant. As the two size measures were largely correlated (R2 = 0·75), only data for the length of stomata are presented in the subsequent text. At the time of flowering, the specific leaf area (SLA) of one fully developed composite leaf was estimated for each plant. Size of the stomata and SLA were estimated using seven plants per population and plant type.
Ploidy level estimations using flow cytometry
For the flow cytometric analyses, the protocol described in Castro et al. (2012) was used. Pisum sativum ‘Ctirad’ was used as the internal standard for all measurements.
Data analysis
After having detected abundant diploid offspring on the synthetic tetraploid mothers (further referred to as synthetic diploids; see the Results), all subsequent analyses focused on distinguishing plants by their ploidy level (diploid and tetraploid), origin (natural and synthetic, i.e. offspring of the synthetic tetraploid mothers), locality and their interactions. All analyses only addressed germinated seeds, i.e. seeds with available ploidy level information.
First, the effect of ploidy level (diploid and tetraploid), plant origin (natural and synthetic), locality and their interactions on seed weight were tested using analysis of variance (ANOVA). Then, the effect of ploidy level, origin, locality, their interactions and seed weight on plant size were tested using ANOVA. Specifically, data were available on plant height at the age of 1 and 2 weeks and number of leaves and branches at the age of 4 and 8 weeks. The results of plant height at 1 and 2 weeks were similar, so only data at the age of 2 weeks are shown. Similarly, the number of leaves at 4 and 8 weeks also provided very similar results, so only data measured at 8 weeks are shown. The data on number of branches are not presented due to low variation in the data.
In addition, the effect of ploidy level, plant origin, locality and their interactions on flowering [yes/no, generalized linear model (GLM) with a binomial distribution], fruiting of flowering plants (yes/no, GLM with a binomial distribution), proportion of developed seeds (number of developed and undeveloped seeds linked using the cbind function, GLM with a binomial distribution), number of seeds per pod (GLM with a Poisson distribution), number of pods per plant (GLM with a Poisson distribution), mean flowering time (ANOVA) and flowering duration (ANOVA) were tested. Seed size was not used in these tests, as the plants were large and thus expected to be independent of seed size. This was also confirmed by results from preliminary analyses on the effect of seed size on these variables. Finally, the effect of ploidy level, plant origin, locality and their interactions on stomata length and SLA were tested with ANOVA. In each case, the model providing the best fit to the tested data was used. All statistical analyses were conducted using R software (R Development Core Team, 2011).
To compare all data across the ploidy levels, plant origins and localities, a redundancy analysis (RDA) using Canoco 5 was performed (ter Braak and Šmilauer, 1998). RDA is a direct gradient analysis that tests for joint structure by using multivariate data (Lepš and Šmilauer, 2003). This analysis was used to test if the overall phenotype differences are associated with ploidy (2× vs. 4×), mode of origin (natural vs. synthetic) and locality. All plant performance measurements represented dependent variables, and the dependent variables were standardized before analysis. Only individuals that had flowered and whose data had been collected on their SLA and size of stomata entered this analysis. These data were selected because missing values cannot be handled easily in an RDA. In total, 108 individuals were included in this analysis.
RESULTS
Ploidy levels and seed weight
Flow cytometric analysis of the seedlings germinated from seeds produced by the synthetic tetraploid mothers indicated that the seedlings may be not only tetraploid but also diploid, triploid or aneuploid (Table 1). The relative genome size was 0·716 ± 0·011 (mean ± s.e.) for diploids and 1·408 ± 0·023 for tetraploids, with the values being the same for the natural and synthetic plants. For triploids, the values of relative genome size were 1·069 ± 0·019. The aneuploids were plants that did not fit any of these types. The highest proportion of tetraploids was found in population 3, and the lowest was found in population 4 (Table 1). There were at least seven synthetic tetraploid individuals of the C2 generation available for the experiments in each population. In addition, there were at least nine diploid individuals originating from synthetic tetraploid mothers from each population (Table 1; further referred to as synthetic diploids). The triploid and aneuploid plants, which were very rare (Table 1), were discarded from further analyses, and the diploid and tetraploid offspring of the synthetic tetraploids were further distinguished.
Table 1.
Number of germinated seeds (out of 100) and ploidy level of the resulting seedlings originating from seeds produced by the synthetic tetraploid C1 mother plants from the four studied populations
| Population | Germinated | 2x | 3x | 4x | Aneuploid |
|---|---|---|---|---|---|
| 1 | 51 | 0·61 | 0 | 0·25 | 0·14 |
| 2 | 48 | 0·65 | 0 | 0·29 | 0·06 |
| 3 | 65 | 0·14 | 0 | 0·83 | 0·03 |
| 4 | 44 | 0·75 | 0·02 | 0·16 | 0·07 |
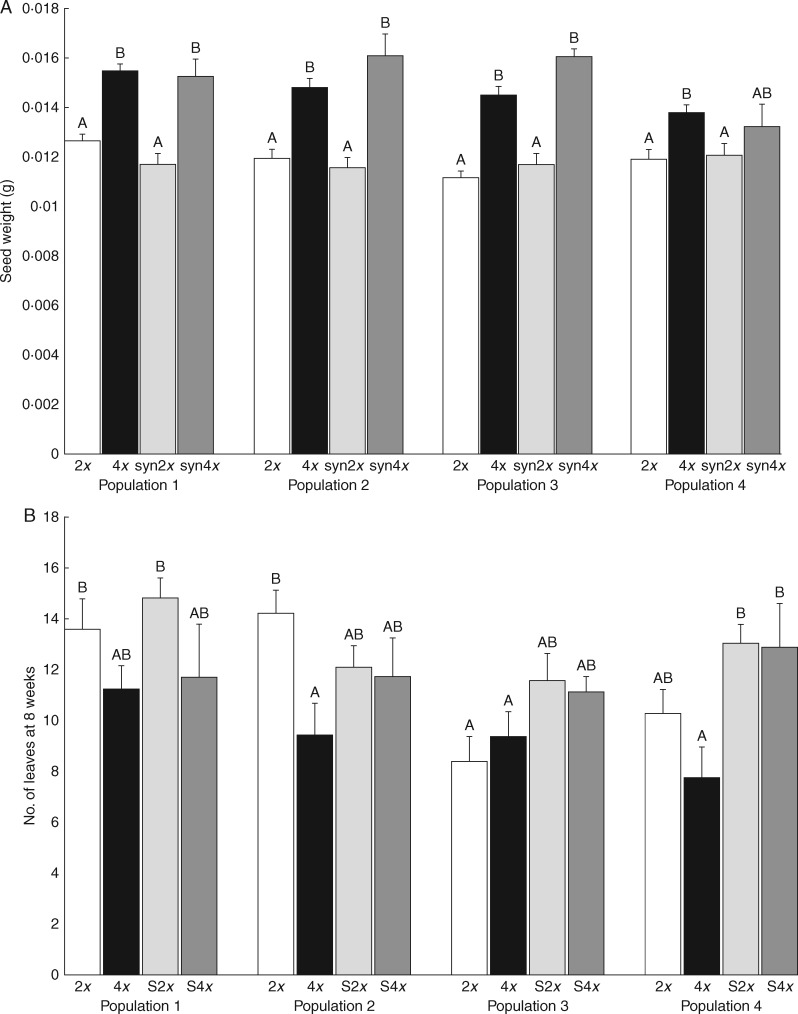
Seed weight of the germinated plants was significantly affected by locality, plant origin (natural vs. synthetic) and ploidy level (diploid vs. tetraploid). There was also a significant interaction between locality and plant origin, and locality and ploidy level (Table 2). Specifically, seeds of diploids were significantly lighter than seeds of tetraploids, and synthetic plants had heavier seeds than natural plants. The effect of ploidy level, however, was much stronger than the effect of plant origin (Fig. 1A).
Table 2.
The effect of locality, plant origin (natural, synthetic) and ploidy level on seed weight, plant height at the age of 2 weeks, the number of leaves at the age of 8 weeks, length of stomata and specific leaf area (SLA)
| Seed weight |
Height 2 weeks |
Leaves 8 weeks |
Length of stomata |
SLA |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| d.f. | F | P | F | P | F | P | F | P | F | P | |
| Seed weight | 1 | NT | NT | 27·10 | <0·001 | 2·72 | 0·100 | NT | NT | NT | NT |
| Locality | 3 | 8·78 | <0·001 | 0·73 | 0·536 | 7·98 | <0·001 | 2·50 | 0·064 | 0·51 | 0·679 |
| Plant origin | 1 | 7·16 | 0·001 | 29·43 | <0·001 | 6·47 | 0·002 | 0·88 | 0·352 | 11·19 | 0·001 |
| Ploidy level | 1 | 187·22 | <0·001 | 2·16 | 0·142 | 20·18 | <0·001 | 172·95 | <0·001 | 2·91 | 0·091 |
| Locality × origin | 3 | 7·73 | <0·001 | 3·66 | 0·002 | 1·22 | 0·295 | 3·95 | 0·011 | 1·65 | 0·184 |
| Locality × ploidy | 3 | 2·88 | 0·035 | 1·64 | 0·179 | 1·49 | 0·216 | 1·57 | 0·201 | 1·44 | 0·236 |
| Origin × ploidy | 1 | 2·05 | 0·152 | 12·64 | <0·001 | 0·50 | 0·478 | 0·00 | 0·968 | 0·47 | 0·493 |
| Locality × origin × ploidy | 3 | 0·76 | 0·515 | 4·71 | 0·003 | 1·39 | 0·244 | 0·17 | 0·914 | 0·64 | 0·591 |
| Residual d.f. | 804 | 357 | 365 | 94 | 93 | ||||||
Seed weight was used as a covariate for plant size.
NT, not tested.
Significant values (P ≤ 0·05) are in bold.
Fig. 1.
The effect of ploidy level (2x, 4x), plant origin (natural, synthetic) and population on (A) seed weight, (B) number of leaves at the age of 8 weeks, (C) number of pods per plant and (D) mean flowering time. S2x and S4x are diploid and tetraploid offspring of synthetic tetraploid mothers, respectively. Only germinated seeds are included in (A) because their ploidy level could be estimated. Columns sharing the same letter are not significantly different (P > 0·05). The graphs show means ± s.e.
Early plant performance
Plant height at the age of 2 weeks was significantly affected by seed weight, plant origin and the interaction of plant origin and locality. In addition, there was a significant interaction between ploidy level and plant origin, and a triple interaction between ploidy level, plant origin and locality (Table 2; Supplementary Data Table S1). Overall, the synthetic tetraploids were the tallest, followed by synthetic diploids, natural diploids and natural tetraploids. The ranking, however, partly varied between populations (Table S1).
The number of leaves at 8 weeks was independent of seed size (Table 2) but was significantly affected by locality, plant origin and ploidy level. No interactions were significant (Table 2). Synthetic diploids had the highest number of leaves, followed by natural diploids, synthetic tetraploids and natural tetraploids (Fig. 1B).
Flowering and plant fitness
Significant interactions between population, ploidy and origin make it difficult to describe general patterns for all variables except flowering duration (Table 1; Table S1). Still, several patterns can be seen from the figures, although their interpretation should account for the interactions. Diploids flowered more often, earlier and longer, and were more likely to produce pods when flowering. They also produced more pods in total and more developed seeds per pod than tetraploids. In contrast, tetraploids had a higher proportion of developed seeds than diploids (Table 3; Table S1). Synthetic plants produced pods more often when they flowered, produced more pods and seeds per pod and had a higher proportion of developed seeds (Fig. 1C). Synthetic and natural plants did not differ in flowering time and duration (Table 3; Fig. 1D).
Table 3.
The effect of locality, plant origin (natural, synthetic) and ploidy level on plant flowering (yes/no), fruiting of flowering plants (yes/no), proportion of developed seeds, number of seeds per pod, number of pods per plant, mean flowering time and flowering duration
| Flowering |
Fruiting when flowering |
Proportion of developed seeds |
Seeds per pod |
Pods |
Mean flowering time |
Flowering duration |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| d.f. | Deviance | P | Deviance | P | Deviance | P | Deviance | P | Deviance | P | F | P | F | P | |
| Locality | 3 | 20·76 | <0·001 | 14·72 | 0·002 | 401·1 | <0·001 | 5938 | <0·001 | 1872 | <0·001 | 779·70 | 0·010 | 6758 | <0·001 |
| Plant origin | 1 | 3·17 | 0·075 | 14·88 | <0·001 | 903·0 | <0·001 | 4008 | <0·001 | 2505 | <0·001 | 1·90 | 0·866 | 867 | 0·087 |
| Ploidy level | 1 | 22·39 | <0·001 | 19·85 | <0·001 | 55·2 | <0·001 | 3935 | <0·001 | 2915 | <0·001 | 723·90 | 0·001 | 1705 | 0·017 |
| Locality × origin | 3 | 8·67 | 0·034 | 6·80 | 0·078 | 120·9 | <0·001 | 900 | <0·001 | 232 | <0·001 | 1125·70 | 0·001 | 166 | 0·904 |
| Locality × ploidy | 3 | 1·17 | 0·761 | 7·88 | 0·048 | 32·0 | <0·001 | 73 | <0·001 | 42 | <0·001 | 591·00 | 0·035 | 820 | 0·427 |
| Origin × ploidy | 1 | 0·00 | 0·947 | 2·59 | 0·107 | 35·8 | <0·001 | 216 | <0·001 | 348 | <0·001 | 0·20 | 0·957 | 117 | 0·528 |
| Locality × origin × ploidy | 3 | 9·00 | 0·029 | 1·76 | 0·625 | 27·5 | <0·001 | 234 | <0·001 | 48 | <0·001 | 1177·60 | <0·001 | 1192 | 0·257 |
| Residual d.f. | 350 | 297 | 246 | 297 | 297 | 297 | 297 | ||||||||
Significant values (P ≤ 0·05) are in bold.
For many dependent variables, ploidy level interacted with plant origin. This was the case for number of pods, developed seeds per pod and proportion of developed seeds (Table 3; Table S1). For all variables, the difference between natural and synthetic plants was higher in diploids than in tetraploids (Fig. 1D).
Locality also interacted with ploidy level. It was significant for pod production in flowering plants, number of pods, proportion of developed seeds, seeds per pod and mean flowering time (Table 3). The effect of ploidy level on all the variables had the same direction for all localities, but localities differed in regards to the intensity of the differences between the cytotypes (Fig. 1C, D;Table S1).
Locality also interacted with plant origin. It was significant for number of pods, proportion of developed seeds, seeds per pod and mean flowering time (Table 3). The effect of origin on all variables, however, had the same direction for all localities, but localities differed in the intensity of the difference between the cytotypes (Fig. 1C). The only exception was for mean flowering time, which was higher in synthetic plants than in natural plants in some localities and vice versa for other localities (Fig. 1D). For several variables, there was also a triple interaction between ploidy level, plant origin and locality. This was the case for flowering, number of pods, proportion of developed seeds, seeds per pod and mean flowering time (Table 3; Table S1).
Size of stomata and specific leaf area
As expected, the length of stomata was significantly affected by ploidy level, with larger stomata in tetraploids, and was independent of locality and plant origin (Fig. 2, Table 2). Unexpectedly, there was also a significant interaction between locality and plant origin. This interaction was caused by the synthetic plants having shorter stomata than the natural plants in two populations, while the other two populations showed an opposite pattern. However, the difference between synthetic and natural plants was not significant for any of the populations. Specific leaf area was independent of ploidy level. The only significant effect was that of plant origin, with synthetic plants having a larger SLA than natural plants (Table 2, Tables S1).
Fig. 2.
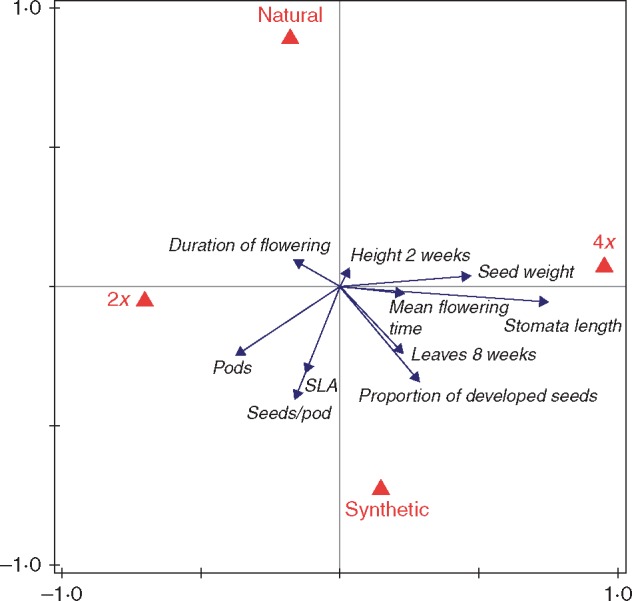
The results of the redundancy analysis of the effect of ploidy level (2x, 4x) and plant origin (natural, synthetic) on plant performance. The variables flowering (yes/no) and fruiting when flowering (yes/no) are not shown due to their low variation in the analysed sub-set of the data (see the Materials and Methods for details). The first (horizontal) axis explains 10·4 % and the second (vertical) axis explains 4·4 % of the total variation in the data set. The effect of plant origin and ploidy level is significant (Table 4).
Summary of plant performance
The redundancy analysis, which combined data for all the performance measures, demonstrated that ploidy level was the factor that explained most of the variation in the system (Table 4). Tetraploids were primarily characterized by longer stomata and heavier seeds (Fig. 2). In contrast, diploids were characterized mainly by higher number of seeds per pod and higher number of pods (Fig. 2). Plant performance was also affected by plant origin (Table 4), with the offspring of synthetic tetraploid mothers having more seeds per pod, a higher proportion of developed seeds and a higher SLA (Fig. 2). In contrast, natural plants had lower values for most of the performance measures. Locality alone did not have a significant effect on plant performance (Fig. 2; Table 4). However, locality significantly interacted with ploidy level and plant origin. Ploidy level interacted significantly with plant origin. The triple interaction between ploidy level, plant origin and locality was not significant (Table 4).
Table 4.
The results of the redundancy analysis of the effects of locality, plant origin (natural, synthetic) and ploidy level on all the plant traits studied
| Locality | Plant origin | Ploidy level | Locality × origin | Locality × ploidy | Origin × ploidy | Locality × origin × ploidy | |
|---|---|---|---|---|---|---|---|
| Prop. var. | 0·036 | 0·044 | 0·097 | 0·051 | 0·040 | 0·039 | 0·032 |
| P-value | 0·112 | 0·002 | 0·002 | 0·01 | 0·048 | 0·002 | 0·094 |
Significant values (P ≤ 0·05) are in bold.
Prop. var. indicates the proportion of total variance explained.
DISCUSSION
The synthetic tetraploids used as mother plants for this experiment produced a high proportion of diploid seeds, and this proportion was largely population specific. In addition, triploid and aneuploid individuals were found among the offspring of these plants. Husband et al. (2008) also detected the production of diploid and triploid offspring by their synthetic polyploids. However, they only excluded these from the experiments and did not report the frequencies. Other studies (e.g. Oswald and Nuismer, 2011) also screened their offspring for ploidy levels but did not report the results.
The occurrence of diploid seeds on the tetraploid C1 plants may be due to the flowering branches being detected as tetraploid (using a single leaf) on the C1 plants and used to produce the seeds, although they could be chimeric instead. In this study the seeds were collected separately from each maternal plant but no distinction was made between flowering branches and pods. Therefore, it is unknown if the diploid seeds were produced on a specific part of the plants (possibly from diploid tissue within an otherwise tetraploid branch, supporting the occurrence of chimeric tissue) or scattered throughout the whole individual. If they appeared within otherwise tetraploid pods or the diploid pods were intermixed among the tetraploid pods, this could indicate origin via haploid parthenogenesis (i.e. development of the diploid product of meiosis into seed without fertilization; Asker and Jerling, 1992). Alternatively, occurrence of different cytotypes could be explained by frequent chromosome pairing failures in the synthetic polyploids (Hassan and Jones, 1994; Srivastava and Srivastava, 2002).
Occurrence of haploid parthenogenesis was shown in a range of different taxa (e.g. Skalinska, 1971; Krahulcová and Krahulec, 2000; Krahulcová et al., 2004; Froelicher et al., 2007). Haploid parthenogenesis was suggested to be induced in sexual taxa by temperature shocks, particular kinds of pollination and also colchicine application (Yudin, 1970; Asker, 1980; Singsit and Hanneman, 1991; Asker and Jerling, 1992).
The effects of chromosome pairing failure may lead to multivalent formation during meiosis, supporting the production of haploid or aneuploid gametes. Multivalent formation is expected in polyploid taxa (Stebbins, 1940), but a range of polyploids developed different mechanisms to avoid this (e.g. Sanchez-Moran et al., 2001). The frequency of multivalent formation is much higher in synthetic than in natural tetraploids (e.g. McCollum, 1958; Ellis, 2013; Wu et al., 2014), although multivalents are also more frequent in odd polyploids (e.g. triploids; Loidl, 1995; Ellis, 2013). Even in the case of frequent multivalent formation, the multivalents may be resolved into bivalent associations at metaphase I in some taxa (Jones et al., 1996). Finally, the occurrence of aneuploid, and possibly triploid and diploid, offspring from the C1 synthetic polyploid mothers could be a result of chromosome elimination prior to meiosis or during mitosis (e.g. Bennett et al., 1976; Laurie and Bennett, 1989). Further studies on the meiotic behaviour of our model are necessary to understand the possibilities regarding the mode of diploid seed formation.
Seed weight (e.g. Maceira et al., 1993; Bretagnolle et al., 1995; Hoya et al., 2007; Cohen et al., 2013; Hosseini et al., 2013; Eliášová and Münzbergová, 2014) and size of stomata (e.g. Warner and Edwards, 1993; Bretagnolle and Lumaret, 1995; Maherali et al., 2009; Balao et al., 2011; Pavlíková et al., 2017) followed the general expectation, as they were larger in tetraploids than in diploids. The patterns held for both natural and synthetic plants. This is in line with the expectation that the sizes of seed and stomata are a function of cell size, which is larger in polyploids (Van Dijk and Van Delden, 1990; Comai, 2005; Otto, 2007). The significant effect of plant origin (natural vs. synthetic) on seed size also suggests that colchicine application has some, although weak, effect on seed size. In line with Eliášová and Münzbergová (2014), who worked on the same model species, the differences in seed size according to the cytotypes and population origin were partly population specific. This supports the need to consider plant material from multiple populations to obtain a general idea of the effects of polyploidization.
Growth of the plants in the first 2 weeks, but not later, was strongly affected by seed size. This is because seeds of V. cracca are relatively large and fully support plant growth over the first weeks before the plant develops leaves and a proper root system, and starts to acquire its own resources. A previous study (Eliášová and Münzbergová, 2014) looking at the growth of natural diploids and tetraploids in V. cracca indicated that seed weight has a significant effect on seedling size over the entire first growing season in tetraploids but not in diploids. Inclusion of seed weight in the model did not affect the significance of ploidy level for either the 2-week-old plants or the 8-week-old plants. This suggests that in spite of large differences in seed weight between cytotypes, seed weight differences are not the only factor affecting differential performance of the cytotype (Eliášová and Münzbergová, 2014). Therefore, other mechanisms, e.g. related to differences in cell size and thus growth rate of the tissue, should be considered to explain the cytotype differences (Comai, 2005).
Synthetic plants were often larger than the natural plants, produced more seeds per pod, produced more pods, had a larger proportion of developed seeds and had a larger SLA. However, there were exceptions to this pattern as plant origin often interacted with ploidy level and population. Overall, this still indicates that the offspring of synthetic polyploids gain some advantage from the application of colchicine in the maternal generation that is independent of their ploidy level and unrelated to seed size. This suggests that the application of anti-mitotic substances to the plants has many potential direct consequences for plant phenotype for both the maternal (C1; e.g. Francis et al., 1990; Hassan and Wazuddin, 2000; Jaskani et al., 2005; Contreras et al., 2007; Cohen et al., 2013) and the offspring (C2) generation. A possible explanation of the colchicine effects on the plants is linked to strong direct effects of colchicine on the production of various plant hormones and thus on cell functioning (e.g. Wang et al., 2001; Dudits et al., 2016), leading to strong differences in plant physiology and morphology (e.g. Francis et al., 1990; Hassan and Wazuddin, 2000, Jaskani et al., 2005; Cohen et al., 2013).
Other studies using synthetic polyploids only compare synthetic polyploids with natural diploids and polyploids. These studies often show that the synthetic polyploids, but not the natural polyploids, differ from the diploids (e.g. Maherali et al., 2009; Oswald and Nuismer, 2011; Pavlíková et al., 2017). The common interpretation of this result is that polyploidization directly affects the trait, and subsequent evolution brings the trait to its original value. The present study contradicts this interpretation by demonstrating that these effects may only be a carry-over of the substance used to create the synthetic polyploids in the second generation. Future studies exploring the consequences of polyploidization should attempt to use both the second- and third-generation polyploids to ensure that they do not merely study these adverse effects.
The strong positive effect of colchicine application on plant performance could also be due to the colchicine application exerting strong selection pressure on the plants. A previous study to optimize the protocol of creating the synthetic polyploids (Pavlíková et al., 2017) showed up to 98 % mortality of the plants after colchicine application. Thus, it is possible that the process of creating synthetic polyploids selects the fittest genotypes. In such a case, increased performance of the offspring of the synthetic polyploid mother is due to this selection. In contrast to any direct effect of colchicine, such an effect will persist over many generations, and using the third or any subsequent generation will not solve the issue. This may further reduce the usefulness of employing synthetic polyploids in ecological studies, so it must be considered when interpreting the results. However, data to test this are not available at present. Future studies may attempt to test this by recording information on seed size and/or seedling size for the plants due to undergo the colchicine treatments in order to determine if only the largest survive. This would confirm the hypothesis under the assumption that seed or seedling size is a good proxy of plant vigour in the later stages of its development.
In addition, it is possible that colchicine application affects the epigenome of the plants. Such an effect has already been shown in several previous studies (e.g. Hegarty et al., 2013; Zhang et al., 2014; Gao et al., 2016). As these changes are likely to persist over multiple generations, this may represent another mechanism that leads to strong differences between the offspring of synthetic and natural mothers, further complicating efforts to assess the consequences of polyploidization.
Interestingly, plant origin (natural vs. synthetic) also interacted with locality for plant size at 2 weeks, the measures of seed production and for size of stomata, indicating that the effects of colchicine application are partly population specific. This suggests that the colchicine effects in the C2 generation are context dependent and are probably due to varying genetic composition of the different populations. These results are similar to results of previous studies suggesting that different lineages of synthetic polyploids follow different trajectories (Matsushita et al., 2012; Dar et al., 2013). These differences were caused by differential gains and losses of chromosomes across the different generations (Matsushita et al., 2012) or changes in DNA methylation profiles (Dar et al., 2013).
Independent of origin, ploidy level had a significant effect on plant size in 8-week-old plants and all measures of flowering and seed production. In all cases, the values of the performance measures were higher in diploids than in tetraploids. This contrasts with a range of previous studies suggesting that polyploids are larger than diploids (e.g. Lumaret, 1988; Husband and Schemske, 2000; Ramsey and Schemske, 2002; Münzbergová, 2006; Mandáková and Münzbergová, 2008). In line with some other studies (e.g. Comai, 2005; Riddle et al., 2006, Münzbergová, 2007; Riddle and Birchler, 2008; Cohen et al., 2013), these results suggest that larger size in polyploids is far from general.
A possible explanation for the smaller size in polyploids could be the slower growth of polyploids than diploids (e.g. Otto and Whitton, 2000; Oswald and Nuismer, 2011), due to difficulties in the cell cycle and slower cell division (Comai, 2005). Although the study lasted for a full year and was longer than most similar studies, there is still the possibility that the polyploids will not be larger and have higher fitness in the subsequent years of life of this long-lived perennial.
Conclusions
The comparison of seed weight and size of stomata between natural and synthetic diploids and polyploids suggests that these variables are strongly affected by polyploidization and change very little during subsequent evolution. This is in line with a common expectation of studies that have compared diploids and polyploids. Most other performance measures were higher for diploids than for tetraploids, but were also higher for the offspring of synthetic tetraploids than for the offspring of natural plants. This suggests that plant performance is affected by colchicine application, even in the second generation. Therefore, the adverse effects of colchicine and possibly of other substances used to create synthetic polyploids in the second generation of plants must be considered in studies using synthetic polyploids. It is also possible that the strong differences between the natural and synthetic plants are caused by the fact that the colchicine treatment represents a strong selection pressure that only the fittest plants survive. The inclusion of diploid offspring of synthetic tetraploid mothers may be a useful way to detect which trait differences are due to polyploidization and which are primarily due to colchicine application, as was done in the present study. The high variation in responses between the populations suggests that future studies should also work with multiple populations to account for the possible variation between populations in the results.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of Table S1: mean and s.e. for the different traits studied.
Supplementary Material
ACKNOWLEDGEMENTS
I would like to thank to I. Chmelařová, I. Jarošincová and Z. Pavlíková for their help with the creation of the synthetic polyploids and maintaining and recording data for the experiment, L. Cosentino for measuring the stomatal size, K. Skácelová for measuring specific leaf area, and the POPEKOL discussion group and two anonymous reviewers for useful comments on previous versions of the manuscript. The study was supported by GAČR P505-13-32048S. Institutional research projects RVO 67985939, and a project of MŠMT also partly supported the work.
LITERATURE CITED
- Amiri S, Kazemitabaar S, Ranjbar K, Azadbakht M.. 2010. The effect of trifluralin and colchicine treatments on morphological characteristics of jimsonweed. Trakia Journal of Sciences 8: 47–61. [Google Scholar]
- Asker S. 1980. Gametophytic apomixis – elements and genetic regulation. Hereditas 93: 277–293. [Google Scholar]
- Asker S, Jerling L.. 1992. Apomixis in plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Balao F, Herrera J, Talavera S.. 2011. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach. New Phytologist, 192: 256–265. [DOI] [PubMed] [Google Scholar]
- Benard RB, Toft CA.. 2007. Effect of seed size on seedling performance in a long-lived desert perennial shrub (Ericameria nauseosa: Asteraceae). International Journal of Plant Sciences 168: 1027–1033. [Google Scholar]
- Benard RB, Toft CA.. 2008. Fine-scale spatial heterogeneity and seed size determine early seedling survival in a desert perennial shrub (Ericameria nauseosa: Asteraceae). Plant Ecology 194: 195–205. [Google Scholar]
- Bennett M, Finch R, Barclay I.. 1976. The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 54: 175–200. [Google Scholar]
- ter Braak C, Šmilauer P.. 1998. Canoco reference manual and users guide to Canoco forWindows: Software for canonical community ordination (version 4). Ithaca: Microcomputer Power. [Google Scholar]
- Bretagnolle F, Lumaret R.. 1995. Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica – occurence, morphological and genetic characteristics of first polyploids. Euphytica 84: 197–207. [Google Scholar]
- Bretagnolle F, Thompson JD, Lumaret R.. 1995. The influence of seed size variation on seed germination and seedling vigor in diploid and tetraploid Dactylis glomerata L. Annals of Botany 76: 607–615. [Google Scholar]
- Castro S, Loureiro J, Prochazka T, Münzbergová Z.. 2012. Cytotype distribution at a diploid–hexaploid contact zone in Aster amellus (Asteraceae). Annals of Botany 110: 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černá L, Münzbergová Z.. 2013. Comparative population dynamics of two closely related species differing in ploidy level. PLoS One 8: e75563. doi: 10.1371/journal.pone.0075563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H, Fait A, Tel-Zur N.. 2013. Morphological, cytological and metabolic consequences of autopolyploidization in Hylocereus (Cactaceae) species. BMC Plant Biology 13: 173. doi:10.1186/1471-2229-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Contreras RN, Ranney TG, Tallury SP.. 2007. Reproductive behavior of diploid and allotetraploid Rhododendron L. ‘fragrant affinity’. Hortscience 42: 31–34. [Google Scholar]
- Dar TH, Raina SN, Goel S.. 2013. Molecular analysis of genomic changes in synthetic autotetraploid Phlox drummondii Hook. Biological Journal of the Linnean Society 110: 591–605. [Google Scholar]
- Dudits D, Torok K, Cseri A, et al. 2016. Response of organ structure and physiology to autotetraploidization in early development of energy willow Salix viminalis. Plant Physiology 170: 1504–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliášová A, Münzbergová Z.. 2014. Higher seed size and germination rate may favour autotetraploids of Vicia cracca L. (Fabaceae). Biological Journal of the Linnean Society 113: 57–73. [Google Scholar]
- Eliášová A, Münzbergová Z.. 2017. Factors influencing distribution and local coexistence of diploids and tetraploids of Vicia cracca: inferences from a common garden experiment. Journal of Plant Research 130: 677–687. [DOI] [PubMed] [Google Scholar]
- Eliášová A, Trávníček P, Mandák B, Münzbergová Z.. 2014. Autotetraploids of Vicia cracca show a higher allelic richness in natural populations and a higher seed set after artificial selfing than diploids. Annals of Botany 113: 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MM. 2013. Evidence for transient dynamics in plant populations based on long-term demographic data. Journal of Ecology 101: 734–742. [Google Scholar]
- Francis A, Jones RN, Parker JS, Posselt UK.. 1990. Colchicine-induced heritable variation in cell-size and chloroplast numbers in leaf mesophyll-cells of diploid ryegrass (Lolium perenne L). Euphytica 49: 49–55. [Google Scholar]
- Froelicher Y, Bassene JB, Jedidi-Neji E, et al. 2007. Induced parthenogenesis in mandarin for haploid production: induction procedures and genetic analysis of plantlets. Plant Cell Reports 26: 937–944. [DOI] [PubMed] [Google Scholar]
- Gao R, Wang HB, Dong B, et al. 2016. Morphological, genome and gene expression changes in newly induced autopolyploid Chrysanthemum lavandulifolium (Fisch ex Trautv.) Makino. International Journal of Molecular Sciences 17: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin AR, Vuong TD, Vaillancourt RE, et al. 2012. The breeding systems of diploid and neoautotetraploid clones of Acacia mangium Willd. in a synthetic sympatric population in Vietnam. Sexual Plant Reproduction 25: 257–265. [DOI] [PubMed] [Google Scholar]
- Hassan L, Jones RN.. 1994. Long-range effects of colchicine sensitivity on meiosis in Lolium multiflorum L (italian ryegrass). Heredity 73: 65–71. [Google Scholar]
- Hassan L, Wazuddin M.. 2000. Colchicine-induced variation of cell size and chloroplast number in leaf mesophyll of rice. Plant Breeding 119: 531–533. [Google Scholar]
- Hegarty M, Coate J, Sherman-Broyles S, Abbott R, Hiscock S, Doyle J.. 2013. Lessons from natural and artificial polyploids in higher plants. Cytogenetic and Genome Research 140: 204–225. [DOI] [PubMed] [Google Scholar]
- Hosseini H, Chehrazi M, Sorestani M, Ahmadi D.. 2013. Polyploidy and comparison of diploid and autotetraploid seedling of Madagascar periwinkle (Catharanthus roseus cv. alba). International Research Journal of Applied and Basic Sciences 4: 402–406. [Google Scholar]
- Hoya A, Shibaike H, Morita T, Ito M.. 2007. Germination characteristics of native Japanese dandelion autopolyploids and their putative diploid parent species. Journal of Plant Research 120: 139–147. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW.. 2000. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. Journal of Ecology 88: 689–701. [Google Scholar]
- Husband BC, Ozimec B, Martin SL, Pollock L.. 2008. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. International Journal of Plant Sciences 169: 195–206. [Google Scholar]
- Husband BC, Baldwin SJ, Sabara HA.. 2016. Direct vs. indirect effects of whole-genome duplication on prezygotic isolation in Chamerion angustifolium: implications for rapid speciation. American Journal of Botany 103: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Jaskani MJ, Kwon SW, Kim DH.. 2005. Comparative study on vegetative, reproductive and qualitative traits of seven diploid and tetraploid watermelon lines. Euphytica 145: 259–268. [Google Scholar]
- Jones GH, Khazanehdari KA, FordLloyd BV.. 1996. Meiosis in the leek (Allium porrum L) revisited. 2. Metaphase I observations. Heredity 76: 186–191. [Google Scholar]
- Krahulcová A, Krahulec F.. 2000. Offspring diversity in Hieracium subgen. Pilosella (Asteraceae): new cytotypes from hybridization experiments and from open pollination. Fragmenta Floristica et Geobotanica 45: 239–255. [Google Scholar]
- Krahulcová A, Papoušková S, Krahulec F.. 2004. Reproduction mode in the allopolyploid facultatively apomictic hawkweed Hieracium rubrum (Asteraceae, subgen. Pilosella ). Hereditas 141: 19–30. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD.. 1989. The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32: 953–961. [Google Scholar]
- Lepš J, Šmilauer P.. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge: Cambridge University Press. [Google Scholar]
- Levin D. 1983. Polyploidy and novelty in flowering plants. American Naturalist 122: 1–25. [Google Scholar]
- Levin D. 2002. The role of chromosomal change in plant evolution. Oxford: Oxford University Press. [Google Scholar]
- Lignowski EM, Scott EG.. 1972. Effect of trifluralin on mitosis. Weed Science 20: 267–270. [Google Scholar]
- Loidl J. 1995. Meiotic chromosome-pairing in triploid and tetraploid Saccharomyces cerevisiae. Genetics 139: 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumaret R. 1988. Adaptive strategies and ploidy levels. Acta Oecologica-Oecologia Plantarum 9: 83–93. [Google Scholar]
- Maceira NO, Jacquard P, Lumaret R.. 1993. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytologist 124: 321–328. [DOI] [PubMed] [Google Scholar]
- Madon M, Clyde MM, Hasim H, Yusuf M, Mat H, Saratha S.. 2005. Polyploidy induction of oil palm through colchicine and oryzalin treatments. Journal of Oil Palm Research 17: 110–123. [Google Scholar]
- Maherali H, Walden AE, Husband BC.. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Münzbergová Z.. 2008. Morphometric and genetic differentiation of diploid and hexaploid populations of Aster amellus agg. in a contact zone. Plant Systematics and Evolution 274: 155–170. [Google Scholar]
- Matsushita SC, Tyagi AP, Thornton GM, Pires JC, Madlung A.. 2012. Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics 191: 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter P, Kettle CJ, Ghazoul J, Hahn T, Pluess AR.. 2013. Evaluating contemporary pollen dispersal in two common grassland species Ranunculus bulbosus L. (Ranunculaceae) and Trifolium montanum L. (Fabaceae) using an experimental approach. Plant Biology 15: 583–592. [DOI] [PubMed] [Google Scholar]
- McCollum GD. 1958. Comparative studies of chromosome pairing in natural and induced tetraploid Dactylis. Chromosoma 9: 571–605. [DOI] [PubMed] [Google Scholar]
- Meimberg H, Rice KJ, Milan NF, Njoku CC, McKay JK.. 2009. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). American Journal of Botany 96: 1262–1273. [DOI] [PubMed] [Google Scholar]
- Münzbergová Z. 2007. No effect of ploidy level in plant response to competition in a common garden experiment. Biological Journal of the Linnean Society 92: 211–219. [Google Scholar]
- Münzbergová Z. 2006. Ploidy level interacts with population size and habitat conditions to determine the degree of herbivory damage in plant populations. Oikos 115: 443–452. [Google Scholar]
- Münzbergová Z, Plačková I.. 2010. Seed mass and population characteristics interact to determine performance of Scorzonera hispanica under common garden conditions. Flora 205: 552–559. [Google Scholar]
- Oswald BP, Nuismer SL.. 2011. Neopolyploidy and diversification in Heuchera grossulariifolia. Evolution 65: 1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. 2007. The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J.. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Pavlíková Z, Paštová L, Münzbergová Z.. 2017. Synthetic polyploids in Vicia cracca: methodology, effects on plant performance and aneuploidy. Plant Systematics and Evolution. doi:10.1007/s00606-017-1414-y. [Google Scholar]
- Ramsey J, Ramsey TS.. 2014. Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society B: Biological Sciences 369: pii: 20130352. doi:10.1098/rstb.2013.0352.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW.. 2002. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics 33: 589–639. [Google Scholar]
- R Development Core Team. 2011. R: a language and environment for statistical computing .Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Riddle NC, Birchler JA.. 2008. Comparative analysis of inbred and hybrid maize at the diploid and tetraploid levels. Theoretical and Applied Genetics 116: 563–576. [DOI] [PubMed] [Google Scholar]
- Riddle NC, Kato A, Birchler JA.. 2006. Genetic variation for the response to ploidy change in Zea mays L. Theoretical and Applied Genetics 114: 101–111. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Benavente E, Orellana J.. 2001. Analysis of karyotypic stability of homoeologous-pairing (ph) mutants in allopolyploid wheats. Chromosoma 110: 371–377. [DOI] [PubMed] [Google Scholar]
- Segraves KA, Thompson JN.. 1999. Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution 53: 1114–1127. [DOI] [PubMed] [Google Scholar]
- Semeniuk P, Arisumi T.. 1968. Colchicine-induced tetraploid and cytochimeral roses. Botanical Gazette, 129: 190–193. [Google Scholar]
- Singsit C, Hanneman RE.. 1991. Haploid induction in Mexican polyploid species and colchicine-doubled derivatives. American Potato Journal 68: 551–556. [Google Scholar]
- Skalinska M. 1971. Experimental and embryological studies in Hieracium aurantiacum L. Acta Biologica Cracoviensia, Series Botanyo 14: 139–152 [Google Scholar]
- Skogen KA, Senack L, Holsinger KE.. 2010. Dormancy, small seed size and low germination rates contribute to low recruitment in Desmodium cuspidatum (Fabaceae). Journal of the Torrey Botanical Society 137: 355–365. [Google Scholar]
- Srivastava R, Srivastava GK.. 2002. Autopolyploids of Helianthus annuus L. var. Morden. Cytologia 67: 213–220. [Google Scholar]
- Stebbins G. 1940. The significance of polyploidy in plant evolution. American Naturalist 74: 54–66. [Google Scholar]
- Tate JA, Symonds VV, Doust AN, et al. 2009. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 years after Ownbey’s discovery. American Journal of Botany 96: 979–988. [DOI] [PubMed] [Google Scholar]
- Trávníček P, Eliášova A, Suda J.. 2010. The distribution of cytotypes of Vicia cracca in Central Europe: the changes that have occurred over the last four decades. Preslia 82: 149–163. [Google Scholar]
- Van Dijk P, Van Delden W.. 1990. Evidence for autotetraploidy in Plantago media and comparisons between natural and artificial cytotypes concerning cell-size and fertility. Heredity 65: 349–357. [Google Scholar]
- Vera ML. 1997. Effects of altitude and seed size on germination and seedling survival of heathland plants in North Spain. Plant Ecology 133: 101–106. [Google Scholar]
- Wang ZN, You RL, Chen-Zhu XZ.. 2001. Effects of colchicine on the accumulation of vacuolar H+-pyrophosphatase and H+-ATPase in germinating Acacia mangium seeds and the recovery effects by sucrose, indole butyric acid and 6-benzyladenine. Plant Growth Regulation 34: 293–303. [Google Scholar]
- Warner DA, Edwards GE.. 1993. Effects of polyploidy on photosynthesis. Photosynthesis Research 35: 135–147. [DOI] [PubMed] [Google Scholar]
- Wu JH, Datson PM, Manako KI, Murray BG.. 2014. Meiotic chromosome pairing behaviour of natural tetraploids and induced autotetraploids of Actinidia chinensis. Theoretical and Applied Genetics 127: 549–557. [DOI] [PubMed] [Google Scholar]
- Yudin BF. 1970. Capacity for parthenogenesis and effectiveness of selection on the basis of this character in diploid and autotetraploid maize. Genetika 6: 13–22. [Google Scholar]
- Zhang XS, Deng MJ, Fan GQ.. 2014. Differential transcriptome analysis between Paulownia fortunei and its synthesized autopolyploid. International Journal of Molecular Sciences 15: 5079–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.