Abstract
Poor inhibitory control is a known risk factor for substance use disorders, making it a priority to identify the determinants of these deficits. The aim of the current study was to identify genetic associations with inhibitory control using the stop signal task in a large sample (n=934) of healthy young adults of European ancestry. We genotyped the subjects genome-wide and then used a hierarchical approach in which we tested: 1) 7 a priori single nucleotide polymorphisms (SNPs) previously associated with stop signal task performance; 2) ~9,000 SNPs designated as high-value addiction (HVA) markers by the SmokeScreen© array; and 3) ~5M genotyped and imputed SNPs, followed by a gene-based association analysis utilizing the resultant p-values. A priori SNP analyses revealed nominally significant associations between response inhibition and one locus in HTR2A (rs6313; p = 0.04, dominance model, uncorrected) in the same direction as prior findings. A nominally significant association was also found in one locus in ANKK1 (rs1800497; p = 0.03, uncorrected), although this was in the opposite direction of previous reports. After accounting for multiple comparisons, the HVA, genome-wide, and gene-based analyses yielded no significant findings. This study implicates variation in serotonergic and dopaminergic genes, but also underscores the difficulty of detecting the influence of individual SNPs, even when biological information is used to prioritize testing. While such small effect sizes suggest limited utility of individual SNPs in predicting risk for addiction or other impulse control disorders, they may nonetheless shed light on complex biological processes underlying poor inhibitory control.
Keywords: inhibitory control, genetics, drug abuse, stop signal task, response inhibition
Introduction
Poor inhibitory control, defined broadly as an impaired ability to inhibit maladaptive or inappropriate behavior, is a well-established risk factor for substance abuse (de Wit, 2009; Perry & Carroll, 2008; Weafer, Mitchell, & de Wit, 2015). Longitudinal studies indicate that children who exhibit high levels of disinhibited behavior are more likely to use drugs as adolescents (Iacono, Carlson, Taylor, Elkins, & McGue, 1999; Sher, Bartholow, & Wood, 2000; Tarter, Kirisci, Reynolds, & Mezzich, 2004), and in laboratory animals, poor inhibitory control is a predictor of rapid acquisition, escalation and dysregulation of drug self-administration (Beckwith & Czachowski, 2016; Belin, Mar, Dalley, Robbins, & Everitt, 2008; Dalley et al., 2007; Diergaarde et al., 2008; Perry & Carroll, 2008). Associations between poor inhibitory control and risk for abuse have been observed across a number of drug classes, including alcohol, stimulants, and cannabis (Weafer et al., 2015; although perhaps less so for opiate drugs: Ahn & Vassileva, 2016; Badiani, Belin, Epstein, Calu, & Shaham, 2011). As such, there is a strong interest in determining the neurobiological factors, including genetic influences, underlying deficits in inhibition (Bevilacqua & Goldman, 2013; Iacono et al., 1999; Mackillop et al., 2016). Twin studies indicate that inhibitory control is heritable (31% – 50%) (Crosbie et al., 2013; Gagne & Saudino, 2010; Schachar, Forget-Dubois, Dionne, Boivin, & Robaey, 2011), yet the specific genetic influences on inhibitory control are not yet known. Identifying genes that contribute to poor inhibition might help to identify individuals at risk for developing drug and alcohol problems and to develop strategies to prevent, diagnose, and treat drug and alcohol abuse. Moreover, the genetic influences on inhibitory control should theoretically be less complex and more easily detectible than genetic influences on the more diffuse and heterogeneous construct of substance abuse, thus making inhibition a useful intermediary construct for understanding genetic influences on risk for substance abuse.
Several laboratory measures have been developed to assess inhibitory control in humans, including the stop signal task (Logan, Schachar, & Tannock, 1997) and the go/no-go task. These tasks require the quick execution of a motor response (i.e., finger press) following ‘go’ signals, and rapid inhibition of this response when a ‘stop’ or ‘no-go’ signal is presented. Poor inhibitory control is indicated by a greater number of inhibitory failures, or a longer amount of time needed to inhibit the response. For the current study, we focused on the stop signal task, because drug users exhibit poorer inhibitory control on this task compared to healthy controls (Crunelle, Veltman, van Emmerik-van Oortmerssen, Booij, & van den Brink, 2013; Fillmore & Rush, 2002; Joos et al., 2013; Lawrence, Luty, Bogdan, Sahakian, & Clark 2009; Moreno et al., 2012), and poorer inhibition on this task in childhood and adolescence predicts development of drug and alcohol problems later in life (Fernie et al., 2013; Nigg et al., 2006; Rubio et al., 2008).
Several candidate gene studies have reported associations between polymorphisms in specific genes and the stop signal task. These studies have identified associations in loci of monoaminergic genes, including noradrenergic: ADRA2B (Lei et al., 2012); dopaminergic: SLC6A3 (Cummins et al., 2012), ANKK1 (Rodriguez-Jimenez et al., 2006; White, Morris, Lawford, & Young, 2008), and DRD2 (Colzato, van den Wildenberg, & Hommel, 2013; Colzato, van den Wildenberg, van der Does, & Hommel, 2010); and serotonergic: HTR2A (Jakubczyk et al., 2012) and TPH2 (Stoltenberg et al., 2006) genes. However, these studies are limited by small sample sizes and heterogeneous populations varying in age, ethnicity, and substance use history. Additionally, it is now widely appreciated that candidate gene studies are prone to false positive errors due to their small sample sizes, failure to correct for multiple testing, and the potential for publication bias often noted in such studies (Duncan & Keller, 2011; Duncan, Pollastri, & Smoller, 2014; Hart, de Wit, & Palmer, 2013). As such, these initial findings require replication.
The aim of the current study was to examine genetic associations with inhibitory control using the stop signal task in a large sample of healthy young adults (N = 934). We aimed for a sample large enough to detect effects accounting for approximately 1.5% of the variance, as common variants rarely exceed 2% in effect size (Park et al., 2010; Visscher et al., 2017). We excluded individuals who were heavy substance users to avoid the potential confound of the deleterious effects of chronic drug use on frontal cognitive function, including inhibition (Goldstein & Volkow, 2011; Jentsch & Taylor, 1999). We employed a hierarchical analytic approach, in which groups of SNPs with higher priority were analyzed in three stratified subsets in order to maximize power to detect associations with response inhibition (Lin & Lee, 2012). First, we examined response inhibition in relation to a priori SNPs, providing a test of previously reported associations in the literature. Second, we tested the relationship between response inhibition and high-value addiction (HVA) markers prioritized in the SmokeScreen© array by BioRealm (Baurley, Edlund, Pardamean, Conti, & Bergen, 2016). The high-value SNP list is a compilation of loci from peer-reviewed publications and research consortia, such as the NeuroSNP project (Bergen et al., 2013; Saccone et al., 2007). This allowed for a formal assessment of loci that are biologically relevant to addiction that was intended to increase our power. Third, we conducted an exploratory genome-wide association study (GWAS) that included ~5M SNPs, and a gene-based association analysis that included 18,942 genes. Thus, the study provided a strong test of a priori relationships, a principled examination of high-value addiction markers, and an atheoretical genome-wide scan and gene-based analyses.
Material and Methods
Procedure
Participants were recruited at two sites (Athens, GA and Chicago, IL) through online and printed advertisements. Inclusion criteria were English fluency, age between 18 and 30 years, and self-reported Caucasian race and non-Hispanic ethnicity (to control for population stratification; Hutchison, Stallings, McGeary, & Bryan, 2004), and scores of 11 or below on the Alcohol Use Disorders Identification Test (AUDIT) or Drug Use Disorders Identification Test (DUDIT), to minimize confounding effects of substantial substance use. The AUDIT/DUDIT cut-off score of 11 was chosen to screen out individuals with heavy substance use or substance use disorders, while still allowing for low to moderate levels of substance use, given the high normative prevalence of substance use among young adults (Aertgeerts et al., 2000; SAMHSA, 2014). Participants were excluded if they reported any lifetime substance dependence, any current psychiatric prescriptions, or treatment in the last 12 months for depression, bipolar disorder, general anxiety, social anxiety, post-traumatic stress disorder, obsessive compulsive disorder, panic attacks/disorder, phobia, schizophrenia or related conditions, anorexia, bulimia, or binge eating. The study was approved by the Institutional Review Boards of the University of Chicago and the University of Georgia, and all participants provided informed consent.
Participants attended an experimental session during which they completed, in counterbalanced order, several self-report and behavioral measures, including the stop signal task (described below). Participants were instructed to abstain from alcohol and drugs other than their usual amounts of caffeine and nicotine for 24 hours before the visit, and breath and urine samples were obtained upon arrival to verify compliance. Participants were given two five-minute breaks during the 4-hour session. Participants provided a saliva sample in an Oragene DNA kit (DNA Genotek Inc., Kanata, ON, Canada) to acquire DNA samples. Most of the participants received compensation for their time ($10/hour) while a minority (n=281) were undergraduates who received research credit. Additionally, participants could receive additional compensation from one of the tasks. Full details of the phenotypic assessment are reported in MacKillop et al. (2016).
Measures
Demographics
Demographic characteristics were assessed including sex, age, race, household income, and education.
World Health Organization (WHO) Attention-Deficit/Hyperactivity Disorder Adult Self-Report Scale (ASRS)
The ASRS is an 18-item self-report screening scale for ADHD (Kessler et al., 2005). The analyses in this study examined the total ADHD score (comprised of all 18 items across inattention and hyperactivity-impulsivity domains).
Alcohol and drug abuse
As part of the exclusion criteria described above, problematic alcohol and drug use were measured using the Alcohol Use Disorders Identification Test (AUDIT; Saunders, Aasland, Babor, de la Fuente, & Grant, 1993) and Drug Use Disorders Identification Test (DUDIT; Berman, Bergman, Palmstierna, & Schlyter, 2005), respectively. These scales assess quantity, frequency, and consequences associated with drinking and drug use.
Inhibitory Control
Inhibitory control was assessed using a modified stop signal task (adapted from Logan et al., 1997), which measures the ability to inhibit a motor response. The task was administered on a desktop computer using E-prime software (E-prime 2.0.8.22, 2007). Participants were instructed to respond to ‘go’ signals (circles or squares) by pressing a key on the keyboard as quickly as possible, and to inhibit responses on 25% of trials when a ‘stop’ signal (auditory tone) was presented shortly after the ‘go’ signal. The task consisted of three blocks, each with 48 go trials and 16 stop trials. The stop signal delay was set at 250ms for the first stop trial of each block, and was adjusted downward by 50ms following each failed inhibition. Once a stop signal delay of 50ms was reached, the delay was held constant at 50ms until the end of the block. The primary outcome measure of response inhibition was the number of inhibitory failures on stop trials1.
Power Analysis
GWAPower (Feng, Wang, Chen, & Lan, 2011) was used to generate power estimates for 934 individuals. A Bonferroni rather than FDR approach was utilized in the power analyses because FDR requires the empirical p-values to generate the appropriate correction rate. For the 10 a priori loci, we were adequately powered (power > .8) to detect SNPs with effects accounting for ≥ 1.3% of variance. For the 8,762 HVA loci, we were adequately powered to detect SNPs with effects accounting for ≥ 2.8% of variance. The GWAS and gene-based analyses were exploratory and therefore no power analyses were conducted.
SNP Genotyping and Quality Control
Genotyping was performed using the Illumina PsychArray BeadChip platform, which includes ~600,000 SNPs; this platform has been optimized to capture the maximum amount of information about common variation. Quality control filtering was implemented in PLINK v1.9 (Chang et al., 2015). SNPs were filtered for call rates < 98%, Hardy-Weinberg Equilibrium (HWE) violations of p < 1 × 10−6, and MAF < 5%. After filtering we had 437,652 SNPs that were used for imputation. Imputation of missing genotypes and of new SNPs was performed with IMPUTE2 v.2.3.1 (Howie, Donnelly, & Marchini, 2009) using the 1000 Genomes Phase 3 b37 reference panel (1000 Genomes Project Consortium, 2015). Imputed SNPs were excluded for exhibiting an information score of < .3, MAF < 5%, HWE violations of p < 1 × 10−6, missingness > 5%, and multiallelic status. Imputed SNPs with confidence < .9 were set to missing for individuals. Four a priori loci were excluded for excessive missing values (> 5%). Finally, to ensure limited redundancy in the analysis of HVA SNPs via linkage disequilibrium and accurate implementation of false discovery rate (FDR) correction, the HVA loci were pruned using the plink command “–indep 50 5 5” (i.e., window size of 50 SNPs, shift the window at every 5 SNPs, and a variance inflation factor of 5, which implies R2 of .8). Following quality control, 10 a priori loci previously associated with stop signal task performance, 8,762 of the 20,652 HVA SNPs, and 4,873,750 genome-wide SNPs were available for analysis.
Participant Quality Control
1,000 participants had valid genotyping data (call rates ≥ 98%, inbreeding coefficient absolute value ≤ .02, concordant self-reported sex and X-chromosome determined sex) and satisfied the inclusion/exclusion criteria. To correct any self-reported race that was misclassified as Caucasian, principal components analysis (PCA; Price et al., 2006) was conducted. Two population outliers were identified and removed by visual inspection of the principal components plot (see Figure S2). 10 participants were excluded for missing data and 52 participants were excluded for invalid performance on the stop signal task, defined as < 5 correct ‘stop’ trials and < 116 correct ‘go’ trials. Finally, participants were assessed for cryptic relatedness (see equation 3 in Yang, Lee, Goddard, & Visscher, 2011) and two were removed for relatedness > .05, leaving a final sample of 934 European-ancestry participants.
Statistical Analysis
Stop signal task inhibitory failures were square root transformed to improve the distribution (before transformation: skewness = 1.04, kurtosis = 1.11; after transformation: skewness = .32; kurtosis = .19). Covariates for the study were ascertained via a univariate linear mixed model of inhibitory failures on the stop signal task with four candidate covariates: sex, age, income, and site (i.e., Athens or Chicago). Each covariate was tested as a fixed factor while the other three candidate covariates were entered into the model as covariates. Only variables which were significantly associated in the combined models were included as covariates in subsequent analyses. Additionally, ADHD total score was explored as a covariate separately in a bivariate correlation analysis with stop signal task performance. PLINK v1.9 software was utilized to conduct univariate linear mixed model associations between the loci from each strata (10 a priori loci, 8,762 HVA loci, and 4,873,750 genome-wide SNPs) and response inhibition, adjusting for the significant candidate covariates and the top three genetic principal components from the PCA. Of the 10 a priori loci, 3 offered largely redundant information (r2 > .99; all other loci exhibited r2 < .8) and were removed from the primary analyses (the locus with the least missing values was retained). All analyses employed an additive model; dominance models were also tested for the a priori loci because some previous studies have reported results based on dominance models. The Versatile Gene-based Association Study 2 (VEGAS2) software was utilized to conduct gene-based association tests utilizing the resultant p-values from the GWAS (Mishra & Macgregor, 2015). We utilized the top 10% SNP test for optimal sensitivity and specificity of true positives (Wojcik, Kao, & Duggal, 2015). Given the recent report of an erratum in the original VEGAS2 method for generating empirical p-values leading to increased type I error rate, we utilized the updated script (for details see Hecker, Prokopenko, Fier, & Lange, 2017). For the a priori loci, statistical significance was set at p < .05. For the prioritized HVA loci and the gene-based analyses, a Benjamini-Hochberg FDR correction was applied to the resultant p-values from the analyses (Benjamini & Hochberg, 1995). For atheoretical genome-wide tests, SNPs were examined at p < 5 × 10−8 (e.g., Pe’er et al., 2008).
Results
Sample Characteristics and Covariate Analyses
Participant characteristics and performance measures on the stop signal task are provided in Table 1, according to site (Chicago or Georgia). Mean number of inhibitory failures, go accuracy, and go reaction on the stop signal task were all correlated (rs > 0.23, ps < 0.001). In the univariate linear mixed model, age and site were significantly associated with inhibitory failures [F (4, 927) = 1.57, p = .03, partial ηp2 = .03; F (4, 927) = 6.00, p = .01, ηp2 = .01, respectively], and were thus included as covariates. Specifically, younger age and testing in Georgia were associated with more inhibitory errors. Income and sex were not significantly related to inhibitory failures [F (4, 927) = 1.13, p = .34, partial ηp2 = .005; F (4, 927) = .25, p = .62, ηp2 = .0003, respectively] and therefore were not covaried. Furthermore, ADHD total score was not associated with inhibitory failures (r = .040, p = .226), likely due to the small range of ADHD scores in this sample of healthy young adults, and therefore was not included as a covariate.
Table 1.
Participant characteristics (N = 934)
| Variable | Mean (SD), %, median | |
|---|---|---|
|
| ||
| University of Chicago (n=614) | University of Georgia (n=320) | |
| Age (years) | 22.87 (3.3) | 19.4 (1.8) |
| Sex | 61.6% Female | 63.6% Female |
| AUDIT | 4.93 (2.90) | 3.04 (3.24) |
| DUDIT | 1.74 (2.38) | 0.55 (1.35) |
| Years of education2 | 15.4 (2.12) | 12.97 (1.44) |
| SST inhibition errors3 | 3.61 (0.92) | 3.93 (0.94) |
| SST go accuracy (%) | 97.5 (2.5) | 97.1 (2.8) |
| SST go reaction time (ms) | 435.4 (61.2) | 430.6 (61.8) |
Note.
N = 932;
N = 933;
square root transformed
Primary Analyses
A priori loci (tier 1)
Of the seven loci tested for additive effects, one locus (rs1800497) was nominally significantly associated with response inhibition (p = .03, uncorrected), however the direction of effects was opposite of that observed in previous reports. Additionally, in the dominance tests, one locus (rs6313) was nominally significant (p = .04, uncorrected), and here the direction of the association was the same as previous reports using a dominance model. The associations are presented in Table 2 (the full 10 a priori loci are presented in Table S1).
Table 2.
Associations between a priori loci and stop signal task performance
| Minor | Add | Dom | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | Locus | Gene | Missing | Allele | MAF | B | p | B | p |
| 2 | rs2229169 | ADRA2B | 1 | T | .326 | .353 | .318 | .393 | .412 |
| 5 | rs37020 | SLC6A3 | 16 | C | .443 | .517 | .130 | .722 | .162 |
| 5 | rs460000 | SLC6A3 | 0 | T | .231 | .584 | .147 | .664 | .168 |
| 11 | rs1800497 | ANKK1 | 0 | A | .184 | −.934 | .031 | −.874 | .082 |
| 11 | rs6277 | DRD2 | 21 | G | .467 | −.279 | .410 | −.561 | .289 |
| 12 | rs1386483 | TPH2 | 0 | T | .374 | .439 | .203 | .506 | .296 |
| 13 | rs6313 | HTR2A | 0 | A | .423 | −.421 | .218 | −1.058 | .036 |
Note. Add = additive model, Dom = dominance model.
High-value addiction markers (tier 2)
Of the 8,762 HVA loci, none were significant using an FDR correction (p < 5.7*10−6). The strongest association was rs2288557 in the Methyltransferase Like 2B (METTL2B) gene on chromosome 7 (p = .00009; FDR q = 0.782). The next two strongest associations were rs9829009 (p = .0005; FDR q = 0.808) in the WD Repeat Domain 49 (WDR49) gene on chromosome 14 and rs4426337 (p = .0005; FDR q = 0.808) in an intergenic region on chromosome 11. The 50 most significant hits are included in Table S2.
Genome-wide association (tier 3)
The genome-wide scan did not yield any significant associations. The most significant association was rs879665 in the Maestro Heat Like Repeat Family Member 2A (MROH2A) gene on chromosome 2 (p = 4.52E-7). The next two strongest associations were rs11195620 and rs5787915 in intergenic regions on chromosome 10 (p = 1.78E-6, p = 1.89E-6, respectively). Figure 1 depicts the results of the GWAS using both Manhattan and Quantile-Quantile (Q-Q) plots. As can be seen in the Q-Q plots, the majority of markers fit null expectations and no markers show evidence for a true association. Furthermore, as can be seen in the Manhattan plot, rs879665 is a lone SNP, rather than the peak amongst many suggestive loci, indicating an increased likelihood of it being a false positive. The 50 most significant hits are included in Table S3.
Figure 1.
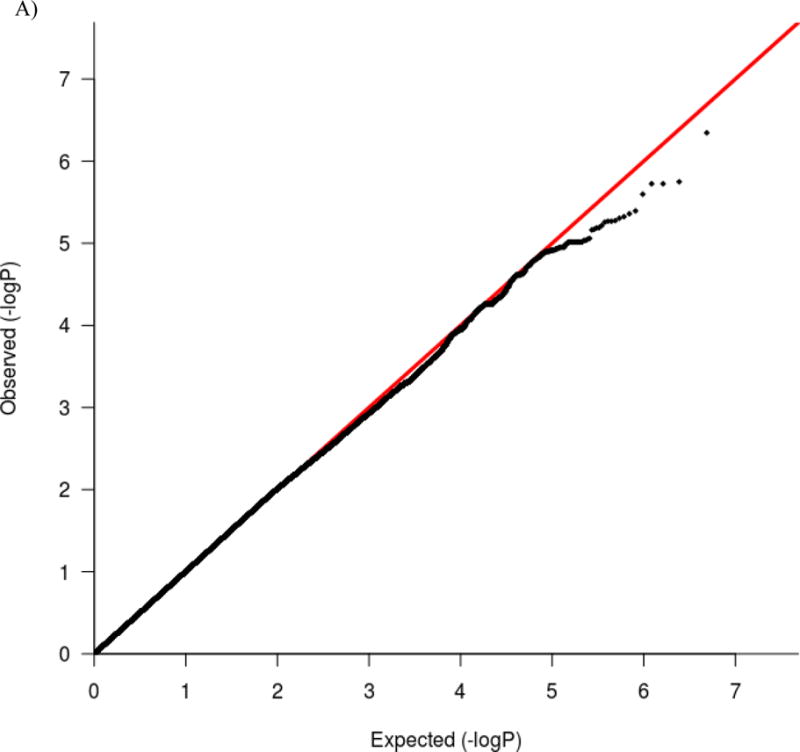
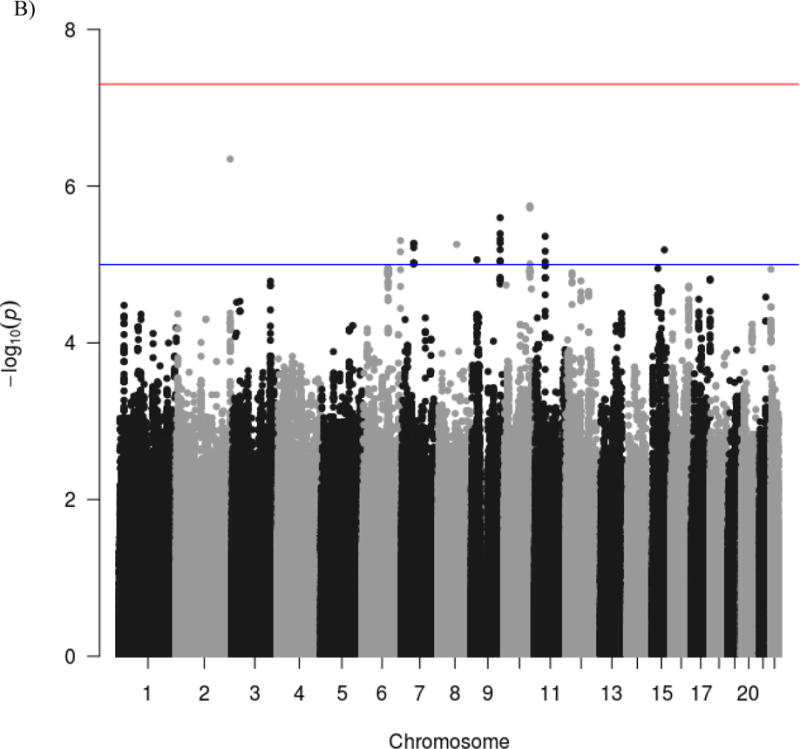
A) Q-Q plot and B) Manhattan plot of genome-wide associations for stop signal task performance. Significance values were –log10 transformed in order to display the smaller p-values as larger in the figures. The Manhattan plot displays level of significance for each SNP, organized by chromosomal position from chromosomes 1–22. The blue line indicates suggestive significance (1E-5). No SNPs achieved genome-wide significance (p < 5E-8), indicated by the red line. The Q-Q plot depicts the observed and expected p-values. The genomic control inflation factor was close to 1 (λ = 1.001), suggesting no systematic bias in the distribution of p-values.
Gene-based Analyses
None of the gene-based associations were significant after FDR correction (p < 2.64E-6). The strongest gene-based associations were solute carrier family 16 member 10 (SLC16A10; p = .00008), developmental pluripotency-associated protein 3 (DPPA3; p = .0004), and stimulated by retinoic acid 6 (STRA6; p = .0004). The top 10 genes are included in Table S4.
Discussion
This study examined genetic associations with response inhibition, assessed as the ability to inhibit a motor response using the stop signal task. We used a hierarchical approach in which 3 tiers of SNPs were analyzed: tier 1) a priori loci that have been previously associated with response inhibition on the stop signal task; tier 2) HVA markers; and tier 3) a genome-wide scan, followed by gene-based association analyses utilizing the resultant p-values from the GWAS. To our knowledge, this is the first study to attempt to identify genome-wide loci associated with a behavioral measure of response inhibition. Two a priori loci were nominally significantly associated with stop signal task performance: one in HTR2A and one in ANKK1. However, we discuss implications of these associations with caution given that: 1) these associations did not survive correction for multiple testing; 2) we did not statistically correct for additional phenotypes ascertained in the parent study (MacKillop et al., 2016) in the current analyses; and 3) the ANKK1 finding was in the opposite direction as previous findings. The HVA, genome-wide, and gene-based analyses yielded no statistically significant findings.
Our finding that response inhibition on the stop signal task was associated with rs6313 in HTR2A (serotonin 2A receptor gene) is in line with previous reports examining this SNP in relation to response inhibition. Specifically, Jakubczyk et al. (2012) found that the minor allele of rs6313 was associated with better response inhibition on the stop signal task in a sample of alcohol-dependent individuals. Similarly, Bjork et al. (2002) found that the minor allele was associated with better response inhibition on a continuous performance task in a sample of adults with histories of depression or substance abuse. We replicated those findings here, showing that in the same dominance model, the minor allele is also associated with better response inhibition in a large sample of healthy young adults. These findings are consistent with preclinical studies demonstrating that serotonin 5-HT2A receptors play an influential role in motor inhibition. For example, 5-HT2A agonists impair and antagonists enhance inhibition (Anastasio et al., 2011; Fletcher, Tampakeras, Sinyard, & Higgins, 2007; Koskinen, Haapalinna, & Sirvio, 2003), and greater density of cortical 5-HT2A receptors is associated with poorer response inhibition (Fink et al., 2015). Importantly, serotonin’s role in inhibition has been previously implicated in increased risk for drug abuse (Cunningham & Anastasio, 2014; Kirby, Zeeb, & Winstanley, 2011). In line with this, the minor allele of rs6313 in HTR2A (identified in this study as predictive of better response inhibition) was associated with less risk for relapse in alcohol dependent individuals (Jakubczyk et al., 2013), and a recent meta-analysis reported that the minor allele was protective across studies of opioid and alcohol abuse and dependence (Cao et al., 2014). Although necessarily speculative, taken together these findings suggest that the association between HTR2A and risk for drug abuse could be due in part to genetic influence on inhibitory control.
We also found an association between response inhibition on the stop signal task and rs1800497 in ANKK1 (ankyrin repeat and kinase domain containing 1 gene), which is proximal to the DRD2 (dopamine D2 receptor) gene and has been associated with differences in dopamine D2 receptors (Neville, Johnstone, & Walton, 2004). Two previous studies reported an association between the minor allele and poorer response inhibition on the stop signal task (Rodriguez-Jimenez et al., 2006; White, Morris, Lawford, & Young, 2008). By contrast, we found the opposite association, in that the minor allele was associated with better response inhibition, and as such this cannot be viewed as a replication of previous findings. Although it is not clear why the direction of effects differs between the current study and previous reports, it is important to note that both of the previous studies had small sample sizes (ns < 75). Moreover, the association between ANKK1 and response inhibition observed here is consistent with a body of literature linking both ANKK1 and D2 receptor signaling to inhibitory control and, more broadly, to drug and alcohol addiction. There is a well-established link between D2 receptor signaling and response inhibition on the stop signal task in both animals and humans (Eagle et al., 2011; Ghahremani et al., 2012; Nandam et al., 2013; Robertson et al., 2015). Additionally, as with serotonin, the link between D2 receptors and inhibitory control is thought to play a role in risk for drug abuse (Jentsch et al., 2014; London, 2016). As ANKK1 has long been associated with risk for drug abuse (for reviews, see Ma, Yuan, Jiang, Cui, & Li, 2015; Wang, Simen, Arias, Lu, & Zhang, 2013), this association could also be mediated in part by genetic influence on inhibitory control.
The current study failed to replicate associations with any of the 5 additional SNPs (tier 1) that have been associated with response inhibition on the stop signal task in previous studies, further suggesting that candidate gene finding should be interpreted with caution. Differences in sample populations could explain some of the discrepancies in findings. Specifically, we tested only healthy young adults of European ancestry with no history of drug or alcohol dependence, whereas previous samples have been comprised of participants with a wide range of ages, ethnicities, and substance use. It is possible that our exclusion of heavy substance users limited variability in inhibitory control within the sample, thus reducing our power to replicate previously reported associations. It is also possible that the age range of participants (18–30) limited our power to detect associations, given the on-going brain maturation during this period in prefrontal regions implicated in inhibitory control (Silveri, 2012). However, as mentioned in the introduction, such failure to replicate previous findings could also be due to the potential for false positives in under-powered candidate gene studies (Duncan & Keller, 2011; Duncan et al., 2014; Hart et al., 2013). Our results might suggest that some of the previously reported associations between specific loci and stop signal task performance may be specific to certain samples (e.g., among individuals with substance use disorders) or may have indeed been false positives, although of course neither possibility is definitive by any means.
It is possible that our use of number of inhibitory errors on the modified stop signal task (as opposed to the use of SSRT on the traditional task) as the measure of inhibitory control limited our ability to replicate previous findings. However, we believe this is unlikely given that the task modifications were subtle, and performance on the modified task correlated strongly with performance on the traditional task. One additional limitation of the accuracy measure used in the present study is that it does not rule out the possible influence of nonspecific performance impairment. Indeed, in the present study the accuracy in inhibiting responses was correlated with both Go reaction time and Go accuracy. It will be important in future studies to include measures that exclude this possibility.
Despite our careful assessment of inhibition within a comparatively large sample (n=934) of healthy, non-substance abusing young adults of European ancestry, we did not detect any significant HVA or genome-wide loci after implementing a false discovery rate correction. Although clinical and psychological phenotypes are likely to be highly polygenic, with numerous loci exerting very small effects (Robinson, Wray, & Visscher, 2014), we originally expected that our use of response inhibition would be a genetically simpler construct that might be amenable to genome wide significant hits within a relatively smaller sample. By contrast, our findings suggest that this is not the case, and that large samples will be needed for well-characterized behavioral tasks as well. This is consistent with the literature advising collection of much larger samples, perhaps through consortia or other collaborative endeavors, for sufficient power to observe very small magnitude effects at genome-wide significance (Agrawal, Edenberg, & Gelernter, 2016).
In sum, this study further advances the understanding of genetic variation and behavioral inhibition. Specifically, findings replicated previous associations between variation in the HTR2A and ANKK1 genes and inhibitory control as measured by a stop signal task. The study also highlights the potential for false positives in candidate gene studies, as we failed to replicate 5 of 7 previously reported associations between specific loci and response inhibition on the stop signal task, despite our larger sample size, which should have afforded us greater power compared to the original candidate gene studies. Finally, our findings underscore the difficulty in detecting the influence of individual SNPs from large panels of tests, even when those tests are biologically informed. Future studies may benefit from combining datasets across research sites to increase sample size, as well as from utilizing alternate approaches to measure genetic influence.
Supplementary Material
Public Significance Statement.
These findings provide important information regarding genetic influences on inhibitory control. Additionally, this study highlights the need for very large sample sizes in genetic studies, perhaps through combining datasets across research sites.
Acknowledgments
Disclosures
This research was partially supported by National Institute on Drug Abuse grant R01 DA032015 (HdW), National Institute on Alcohol Abuse and Alcoholism grant K01 AA024519 (JW), and the Peter Boris Chair in Addictions Research (JM). The funding sources had no other role other than financial support.
We would like to thank Dr. James Baurley for providing a list of Smokescreen HVAs.
Footnotes
Prior Dissemination: None of the findings appearing in this manuscript have been presented elsewhere, including conferences, meetings, or websites.
We used a variant of the stop signal task that did not provide the traditional measure of stop signal reaction time (SSRT; Logan et al., 1997), but instead provided a measure of accuracy in the ability to inhibit a response. Our task utilized an adaptive stop signal delay model, but because it only adjusted downward, it did not provide the stop signal delay time at which participants could inhibit at a rate of 50% (necessary to calculate the stop signal reaction time). To confirm that the measure of inhibitory control used here is comparable to SSRT obtained from the traditional task, we tested a new sample of healthy young adults (n=20) who completed both tasks to compare the outcome measures. As expected, inhibitory errors on the stop signal task used in the current study were significantly correlated with SSRT on the traditional stop signal task (r = 0.56; p = 0.010; see Figure S1 for a scatterplot of this correlation).
All authors significantly contributed to the manuscript and have read and approved the final manuscript.
All authors report no conflicts of interest.
References
- 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aertgeerts B, Buntinx F, Bande-Knops J, Vandermeulen C, Roelants M, Ansoms S, Fevery J. The value of CAGE, CUGE, and AUDIT in screening for alcohol abuse and dependence among college freshmen. Alcoholism: Clinical and Experimental Research. 2000;24:53–57. [PubMed] [Google Scholar]
- Agrawal A, Edenberg HJ, Gelernter J. Meta-analyses of genome-wide association data hold new promise for addiction genetics. Journal of Studies on Alcohol and Drugs. 2016;77:676–680. doi: 10.15288/jsad.2016.77.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn WY, Vassileva J. Machine-learning identifies substance-specific behavioral markers for opiate and stimulant dependence. Drug and Alcohol Dependence. 2016;161:247–257. doi: 10.1016/j.drugalcdep.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasio NC, Stoffel EC, Fox RG, Bubar MJ, Rice KC, Moeller FG, Cunningham KA. Serotonin (5-hydroxytryptamine) 5-HT2A receptor: association with inherent and cocaine-evoked behavioral disinhibition in rats. Behavioral Pharmacology. 2011;22:248–261. doi: 10.1097/FBP.0b013e328345f90d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani A, Belin D, Epstein D, Calu D, Shaham Y. Opiate versus psychostimulant addiction: the differences do matter. Nature Reviews Neuroscience. 2011;12:685–700. doi: 10.1038/nrn3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurley JW, Edlund CK, Pardamean CI, Conti DV, Bergen AW. Smokescreen: a targeted genotyping array for addiction research. BMC Genomics. 2016;17:145. doi: 10.1186/s12864-016-2495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith SW, Czachowski CL. Alcohol-preferring P rats exhibit elevated motor impulsivity concomitant with operant responding and self-administration of alcohol. Alcoholism: Clinical and Experimental Research. 2016;40:1100–1110. doi: 10.1111/acer.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin D, Mar AC, Dalley JW, Robbins TW, Everitt BJ. High impulsivity predicts the switch to compulsive cocaine-taking. Science. 2008;320:1352–1355. doi: 10.1126/science.1158136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B Methodology. 1995;57:289–300. [Google Scholar]
- Bergen AW, Javitz HS, Krasnow R, Nishita D, Michel M, Conti DV, Liu JH, Lee W, Edlund CK, Hall S, Kwok PY, Benowitz NL, Baker TB, Tyndale RF, Lerman C, Swan GE. Nicotinic acetylcholine receptor variation and response to smoking cessation therapies. Pharmacogenetics and Genomics. 2013;23:94–103. doi: 10.1097/FPC.0b013e32835cdabd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AH, Bergman H, Palmstierna T, Schlyter F. Evaluation of the Drug Use Disorders Identification Test (DUDIT) in criminal justice and detoxification settings and in a Swedish population sample. European Addiction Research. 2005;11:22–31. doi: 10.1159/000081413. [DOI] [PubMed] [Google Scholar]
- Bevilacqua L, Goldman D. Genetics of impulsive behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368 doi: 10.1098/rstb.2012.0380. 20120380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork JM, Moeller FG, Dougherty DM, Swann AC, Machado MA, Hanis CL. Serotonin 2a receptor T102C polymorphism and impaired impulse control. American Journal of Medical Genetics. 2002;114:336–339. doi: 10.1002/ajmg.10206. [DOI] [PubMed] [Google Scholar]
- Cao J, Liu XT, Han SZ, Zhang CK, Liu ZZ, Li DW. Association of the HTR2A gene with alcohol and heroin abuse. Human Genetics. 2014;133:357–365. doi: 10.1007/s00439-013-1388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colzato LS, van den Wildenberg WPM, Hommel B. The genetic impact (C957T-DRD2) on inhibitory control is magnified by aging. Neuropsychologia. 2013;51:1377–1381. doi: 10.1016/j.neuropsychologia.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Colzato LS, van den Wildenberg WPM, van der Does AJW, Hommel B. Genetic markers of striatal dopamine predict individual differences in dysfunctional, but not functional impulsivity. Neuroscience. 2010;170:782–788. doi: 10.1016/j.neuroscience.2010.07.050. [DOI] [PubMed] [Google Scholar]
- Crosbie J, Arnold P, Paterson A, Swanson J, Dupuis A, Li X, Shan J, Goodale T, Tam C, Strug LJ, Schachar RJ. Response inhibition and ADHD traits: correlates and heritability in a community sample. Journal of Abnormal Child Psychology. 2013;41:497–507. doi: 10.1007/s10802-012-9693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunelle CL, Veltman DJ, van Emmerik-van Oortmerssen K, Booij J, van den Brink W. Impulsivity in adult ADHD patients with and without cocaine dependence. Drug and Alcohol Dependence. 2013;129:18–24. doi: 10.1016/j.drugalcdep.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Cummins TDR, Hawi Z, Hocking J, Strudwick M, Hester R, Garavan H, Wagner J, Chambers CD, Bellgrove MA. Dopamine transporter genotype predicts behavioural and neural measures of response inhibition. Molecular Psychiatry. 2012;17:1086–1092. doi: 10.1038/mp.2011.104. [DOI] [PubMed] [Google Scholar]
- Cunningham KA, Anastasio NC. Serotonin at the nexus of impulsivity and cue reactivity in cocaine addiction. Neuropharmacology. 2014;76:460–478. doi: 10.1016/j.neuropharm.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalley JW, Fryer TD, Brichard L, Robinson ES, Theobald DE, Laane K, Pena Y, Murphy ER, Shah Y, Probst K, Abakumova I, Aigbirhio FI, Richards HK, Hong Y, Baron JC, Everitt BJ, Robbins TW. Nucleus accumbens D2/3 receptors predict trait impulsivity and cocaine reinforcement. Science. 2007;315:1267–1270. doi: 10.1126/science.1137073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit H. Impulsivity as a determinant and consequence of drug use: a review of underlying processes. Addiction Biology. 2009;14:22–31. doi: 10.1111/j.1369-1600.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diergaarde L, Pattij T, Poortvliet I, Hogenboom F, de Vries W, Schoffelmeer AN, De Vries TJ. Impulsive choice and impulsive action predict vulnerability to distinct stages of nicotine seeking in rats. Biological Psychiatry. 2008;63:301–308. doi: 10.1016/j.biopsych.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. American Journal of Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Pollastri AR, Smoller JW. Mind the gap: why many geneticists and psychological scientists have discrepant views about gene-environment interaction (GXE) research. American Psychologist. 2014;69:249–268. doi: 10.1037/a0036320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle DM, Wong JCK, Allan ME, Mar AC, Theobald DE, Robbins TW. Contrasting roles for dopamine D1 and D2 receptor subtypes in the dorsomedial striatum but not the nucleus accumbens core during behavioral inhibition in the stop-signal task in rats. Journal of Neuroscience. 2011;31:7349–7356. doi: 10.1523/JNEUROSCI.6182-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Wang SC, Chen CC, Lan L. GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genetics. 2011;12:12. doi: 10.1186/1471-2156-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie G, Peeters M, Gullo MJ, Christiansen P, Cole JC, Sumnall H, Field M. Multiple behavioural impulsivity tasks predict prospective alcohol involvement in adolescents. Addiction. 2013;108:1916–1923. doi: 10.1111/add.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore MT, Rush CR. Impaired inhibitory control of behavior in chronic cocaine users. Drug and Alcohol Dependence. 2002;66:265–273. doi: 10.1016/s0376-8716(01)00206-x. [DOI] [PubMed] [Google Scholar]
- Fink LHL, Anastasio NC, Fox RG, Rice KC, Moeller FG, Cunningham KA. Individual differences in impulsive action reflect variation in the cortical serotonin 5-HT2A receptor system. Neuropsychopharmacology. 2015;40:1957–1968. doi: 10.1038/npp.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher PJ, Tampakeras M, Sinyard J, Higgins GA. Opposing effects of 5-HT2A and 5-HT2C receptor antagonists in the rat and mouse on premature responding in the five-choice serial reaction time test. Psychopharmacology. 2007;195:223–234. doi: 10.1007/s00213-007-0891-z. [DOI] [PubMed] [Google Scholar]
- Gagne JR, Saudino KJ. Wait for it! A twin study of inhibitory control in early childhood. Behavioral Genetics. 2010;40:327–337. doi: 10.1007/s10519-009-9316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahremani DG, Lee B, Robertson CL, Tabibnia G, Morgan AT, De Shetler N, Brown AK, Monterosso JR, Aron AR, Mandelkern MA, Poldrack RA, London ED. Striatal dopamine D(2)/D(3) receptors mediate response inhibition and related activity in frontostriatal neural circuitry in humans. Journal of Neuroscience. 2012;32:7316–7324. doi: 10.1523/JNEUROSCI.4284-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RZ, Volkow ND. Dysfunction of the prefrontal cortex in addiction: neuroimaging findings and clinical implications. Nature Reviews Neuroscience. 2011;12:652–669. doi: 10.1038/nrn3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, de Wit H, Palmer AA. Candidate gene studies of a promising intermediate phenotype: failure to replicate. Neuropsychopharmacology. 2013;38:802–816. doi: 10.1038/npp.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker J, Prokopenko D, Fier H, Lange C. Reporting correct p-values in VEGAS analyses. bioRxiv. 2017 doi: 10.1017/thg.2017.16. 101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison KE, Stallings M, McGeary J, Bryan A. Population stratification in the candidate gene study: Fatal threat or red herring? Psycholical Bulletin. 2004;130:66–79. doi: 10.1037/0033-2909.130.1.66. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M. Behavioral disinhibition and the development of substance-use disorders: findings from the Minnesota Twin Family Study. Development and Psychopathology. 1999;11:869–900. doi: 10.1017/s0954579499002369. [DOI] [PubMed] [Google Scholar]
- Jakubczyk A, Klimkiewicz A, Kopera M, Krasowska A, Wrzosek M, Matsumoto H, Burmeister M, Brower KJ, Wojnar M. The CC genotype in the T102C HTR2A polymorphism predicts relapse in individuals after alcohol treatment. Journal of Psychiatry Research. 2013;47:527–533. doi: 10.1016/j.jpsychires.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczyk A, Wrzosek M, Lukaszkiewicz J, Sadowska-Mazuryk J, Matsumoto H, Sliwerska E, Glass J, Burmeister M, Brower KJ, Wojnar M. The CC genotype in HTR2A T102C polymorphism is associated with behavioral impulsivity in alcohol-dependent patients. Journal of Psychiatry Research. 2012;46:44–49. doi: 10.1016/j.jpsychires.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch JD, Ashenhurst JR, Cervantes MC, Groman SM, James AS, Pennington ZT. Dissecting impulsivity and its relationships to drug addictions. Annals of the New York Academy of Sciences. 2014;1327:1–26. doi: 10.1111/nyas.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch JD, Taylor JR. Impulsivity resulting from frontostriatal dysfunction in drug abuse: implications for the control of behavior by reward-related stimuli. Psychopharmacology. 1999;146:373–390. doi: 10.1007/pl00005483. [DOI] [PubMed] [Google Scholar]
- Joos L, Schmaal L, Goudriaan AE, Fransen E, Van den Brink W, Sabbe BG, Dom G. Age of onset and neuropsychological functioning in alcohol dependent inpatients. Alcoholism: Clinical and Experimental Research. 2013;37:407–416. doi: 10.1111/j.1530-0277.2012.01949.x. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Adler L, Ames M, Demler O, Faraone S, Hiripi E, Howes MJ, Jin R, Secnik K, Spencer T, Ustun TB, Walters EE. The World Health Organization Adult ADHD Self-Report Scale (ASRS): a short screening scale for use in the general population. Psychological Medicine. 2005;35:245–256. doi: 10.1017/s0033291704002892. [DOI] [PubMed] [Google Scholar]
- Kirby LG, Zeeb FD, Winstanley CA. Contributions of serotonin in addiction vulnerability. Neuropharmacology. 2011;61:421–432. doi: 10.1016/j.neuropharm.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen T, Haapalinna A, Sirvio J. Alpha-adrenoceptor-mediated modulation of 5-HT2 receptor agonist induced impulsive responding in a 5-choice serial reaction time task. Pharmacology and Toxicology. 2003;92:214–225. doi: 10.1034/j.1600-0773.2003.920504.x. [DOI] [PubMed] [Google Scholar]
- Lawrence AJ, Luty J, Bogdan NA, Sahakian BJ, Clark L. Impulsivity and response inhibition in alcohol dependence and problem gambling. Psychopharmacology. 2009;207:163–172. doi: 10.1007/s00213-009-1645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei XM, Chen CS, He QH, Moyzis R, Xue G, Chen CH, Cao ZY, Li J, Li H, Zhu B, Zhang MX, Li J, Dong Q. Haplotype polymorphism in the alpha-2B-adrenergic receptor gene influences response inhibition in a large chinese sample. Neuropsychopharmacology. 2012;37:1115–1121. doi: 10.1038/npp.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lee WC. Improving power of genome-wide association studies with weighted false discovery rate control and prioritized subset analysis. PLoS One. 2012;7:e33716. doi: 10.1371/journal.pone.0033716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan GD, Schachar RJ, Tannock R. Impulsivity and inhibitory control. Psychological Science. 1997;8:60–64. [Google Scholar]
- London ED. Impulsivity, stimulant abuse, and dopamine receptor signaling. Advances in Pharmacology. 2016;76:67–84. doi: 10.1016/bs.apha.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Ma YL, Yuan WJ, Jiang XZ, Cui WY, Li MD. Updated findings of the association and functional studies of DRD2/ANKK1 variants with addictions. Molecular Neurobiology. 2015;51:281–299. doi: 10.1007/s12035-014-8826-2. [DOI] [PubMed] [Google Scholar]
- MacKillop J, Weafer J, Gray JC, Oshri A, Palmer A, de Wit H. The latent structure of impulsivity: impulsive choice, impulsive action, and impulsive personality traits. Psychopharmacology. 2016;233:3361–3370. doi: 10.1007/s00213-016-4372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Research Human Genetics. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- Moreno M, Estevez AF, Zaldivar F, Montes JM, Gutierrez-Ferre VE, Esteban L, Sanchez-Santed F, Flores P. Impulsivity differences in recreational cannabis users and binge drinkers in a university population. Drug and Alcohol Dependence. 2012;124:355–362. doi: 10.1016/j.drugalcdep.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Nandam LS, Hester R, Wagner J, Dean AJ, Messer C, Honeysett A, Nathan PJ, Bellgrove MA. Dopamine D(2) receptor modulation of human response inhibition and error awareness. Journal of Cognitive Neuroscience. 2013;25:649–656. doi: 10.1162/jocn_a_00327. [DOI] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Human Mutation. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Nigg JT, Wong MM, Martel MM, Jester JM, Puttler LI, Glass JM, Adams KM, Fitzgerald HE, Zucker RA. Poor response inhibition as a predictor of problem drinking and illicit drug use in adolescents at risk for alcoholism and other substance use disorders. Journal of the American Academy of Child and Adolescent Psychiatry. 2006;45:468–475. doi: 10.1097/01.chi.0000199028.76452.a9. [DOI] [PubMed] [Google Scholar]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nature Genetics. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic Epidemiology. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Perry JL, Carroll ME. The role of impulsive behavior in drug abuse. Psychopharmacology. 2008;200:1–26. doi: 10.1007/s00213-008-1173-0. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Robertson CL, Ishibashi K, Mandelkern MA, Brown AK, Ghahremani DG, Sabb F, Bilder R, Cannon T, Borg J, London ED. Striatal D1- and D2-type dopamine receptors are linked to motor response inhibition in human subjects. Journal of Neuroscience. 2015;35:5990–5997. doi: 10.1523/JNEUROSCI.4850-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MR, Wray NR, Visscher PM. Explaining additional genetic variation in complex traits. Trends in Genetics. 2014;30:124–132. doi: 10.1016/j.tig.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Jimenez R, Avila C, Ponce G, Ibanez MI, Rubio G, Jimenez-Arriero MA, Ampuero I, Ramos JA, Hoenicka J, Palomo T. The TaqIA polymorphism linked to the DRD2 gene is related to lower attention and less inhibitory control in alcoholic patients. European Psychiatry. 2006;21:66–69. doi: 10.1016/j.eurpsy.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Rubio G, Jimenez M, Rodriguez-Jimenez R, Martinez I, Avila C, Ferre F, Jimenez-Arriero MA, Ponce G, Palomo T. The role of behavioral impulsivity in the development of alcohol dependence: a 4-year follow-up study. Alcoholism: Clinical and Experimental Research. 2008;32:1681–1687. doi: 10.1111/j.1530-0277.2008.00746.x. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human Molecular Genetics. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption–II. Addiction. 1993;88:791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Schachar RJ, Forget-Dubois N, Dionne G, Boivin M, Robaey P. Heritability of response inhibition in children. Journal of the International Neuropsychological Society. 2011;17:238–247. doi: 10.1017/S1355617710001463. [DOI] [PubMed] [Google Scholar]
- Sher KJ, Bartholow BD, Wood MD. Personality and substance use disorders: a prospective study. Journal of Consulting and Clinical Psychology. 2000;68:818–829. [PubMed] [Google Scholar]
- Silveri MM. Adolescent brain development and underage drinking in the United States: identifying risks of alcohol use in college populations. Harvard Review of Psychiatry. 2012;20:189–200. doi: 10.3109/10673229.2012.714642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration. Results from the 2013 National Survey on Drug Use and Health: summary of national findings. Rockville, MD: 2014. [PubMed] [Google Scholar]
- Stoltenberg SF, Glass JM, Chermack ST, Flynn HA, Li S, Weston ME, Burmeister M. Possible association between response inhibition and a variant in the brain-expressed tryptophan hydroxylase-2 gene. Psychiatric Genetics. 2006;16:35–38. doi: 10.1097/01.ypg.0000176528.30362.34. [DOI] [PubMed] [Google Scholar]
- Tarter RE, Kirisci L, Reynolds M, Mezzich A. Neurobehavior disinhibition in childhood predicts suicide potential and substance use disorder by young adulthood. Drug and Alcohol Dependence. 2004;76(Suppl):S45–52. doi: 10.1016/j.drugalcdep.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 years of GWAS discovery: biology, function, and translation. The American Journal of Human Genetics. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Simen A, Arias A, Lu QW, Zhang HP. A large-scale meta-analysis of the association between the ANKK1/DRD2 Taq1A polymorphism and alcohol dependence. Human Genetics. 2013;132:347–358. doi: 10.1007/s00439-012-1251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weafer J, Mitchell SH, de Wit H. Recent translational findings on impulsivity in relation to drug abuse. Current Addiction Reports. 2015;1:289–300. doi: 10.1007/s40429-014-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Morris CP, Lawford BR, Young RM. Behavioral phenotypes of impulsivity related to the ANKK1 gene are independent of an acute stressor. Behavioral and Brain Functions. 2008;4:54. doi: 10.1186/1744-9081-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik GL, Kao WHL, Duggal P. Relative performance of gene- and pathway-level methods as secondary analyses for genome-wide association studies. BMC Genetics. 2015;16:34. doi: 10.1186/s12863-015-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


