Abstract
Deep-sequencing reveals extensive variation in the sequence of endogenously expressed microRNAs (termed ‘isomiRs’) in human cell lines and tissues, especially in relation to the 3′ end. From the immunoprecipitation of the microRNA-binding protein Argonaute and the sequencing of associated small RNAs, we observe extensive 3′-isomiR variation, including for miR-222 where the majority of endogenously expressed miR-222 is extended by 1–5 nt compared to the canonical sequence. We demonstrate this 3′ heterogeneity has dramatic implications for the phenotype of miR-222 transfected cells, with longer isoforms promoting apoptosis in a size (but not 3′ sequence)-dependent manner. The transfection of longer miR-222 isomiRs did not induce an interferon response, but did downregulate the expression of many components of the pro-survival PI3K-AKT pathway including PIK3R3, a regulatory subunit whose knockdown phenocopied the expression of longer 222 isoforms in terms of apoptosis and the inhibition of other PI3K-AKT genes. As this work demonstrates the capacity for 3′ isomiRs to mediate differential functions, we contend more attention needs to be given to 3′ variance given the prevalence of this class of isomiR.
INTRODUCTION
MicroRNAs post-transcriptionally regulate the expression of genes by acting as sequence-specific guides, bringing effector proteins of the RNA-induced silencing complex (RISC) in proximity to target mRNAs where they suppress gene expression through a combination of translational inhibition and RNA destabilization (1). Typically, miRNAs are annotated as a single defined sequence, though many recent RNA sequencing studies (2–20) demonstrate miRNAs are actually expressed as a range of naturally occurring variants, termed ‘isomiRs’ (21). In the conventional biogenesis pathway, miRNA 5′ and 3′ ends are defined by the activities of the RNAses Drosha and Dicer, which can cause heterogeneity in length if cleavage occurs at variable positions. Factors such as secondary structure of the precursor miRNA, sequence around cleavage sites and the influence of accessory proteins can all modulate cleavage location (22–25). Length heterogeneity can also arise from the ‘nibbling’ activities of exonucleases (26–29). Both of these mechanisms result in ‘templated isomiRs’ where the sequence of the mature miRNA matches the genomically encoded sequence. Non-templated isomiRs (where the mature sequence differs from the genome) can arise from the post-transcriptional addition of nucleotides (primarily A and U) to the 3′ termini or, less commonly, from the activities of RNA editing enzymes (21).
The majority of endogenous mature miRNA sequences are in fact isomiRs that differ from the canonical sequence annotated in the miRNA sequencing repository, miRbase (30). Interestingly, the relative abundance of different isomiRs can vary between tissue and developmental stages (20,31), race and gender (14,32), pathogen exposure (6,16) or diseases including pre-eclampsia (7), alzheimer's (33) and cancer (13,18,34). Heterogeneity at the 5′ end is expected to have functional consequences because it results in ‘seed-shifting’, whereby the target-specifying seed region (nt 2–8) is altered, thereby changing the expected pool of target genes. Although functional consequences of seed-shifting have been demonstrated (9,10,12,17,18,35–37), the occurrence of 5′ variation is relatively rare. Far more common is 3′ heterogeneity, which has been shown to be of functional consequence with non-templated 3′ adenylation and uridylation having been linked to miRNA stability (27,38–40) (so called ‘tailing and trimming’). However, despite its frequent occurrence, there are few reports in which the functional consequence of templated 3′ variation have been assessed.
In this study, we catalogued endogenous length variation across breast cancer tumour samples and cell lines, and observed the frequent occurrence of templated 3′ isomiRs. We selected the 3′ variants of miR-222 for further study and found that the longer miR-222 isomiRs elicit a progressively more-pronounced apoptotic phenotype that is not attributable to the induction of an interferon response. Instead, we found that members of the pro-survival PI3K/AKT signalling pathway are differentially regulated by different miR-222 isomiRs. We also found that longer endogenous miRNAs, including longer miR-222 isomiRs, are more nuclear enriched than shorter forms. Due to the abundance of multiple endogenous miRNAs of longer size (≥24 nt) (10), these observations are likely to have wider consequence beyond miR-222 and suggest consideration should be given to 3′ heterogeneity, even though it is removed from the target-specifying seed-site.
MATERIALS AND METHODS
Cell culture
MCF10A (breast epithelial), MDA-MB-468, MCF7, T47D (breast cancer) and KGN (granulosa) cells were purchased from the ATCC (American Type Culture Collection). All cell lines were subjected to regular mycoplasma monitoring every 1–2 months using a MycoAlert mycoplasma detection kit (Lonza). MCF10A cells were cultured in DMEM/F12 medium (Sigma-Aldrich) supplemented with 5% horse serum (Invitrogen, Grand Island, NY, USA), 20 ng/ml EGF, 0.5 μl/ml Hydrocortisone, 100 ng/ml Cholera toxin, 10 μg/ml Insulin (Sigma-Aldrich). MB231, MB468 and KGN cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich), whereas MCF7 and T47D were cultured in RPMI-1640 (Sigma-Aldrich), both supplemented with 10% (v/v) fetal calf serum (Invitrogen). All media were supplemented with 2 mmol/l L-Glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. All cell lines were cultured at 37°C in 5% CO2.
miRNA and siRNA overexpression and inhibition
For miRNA overexpression studies, cells were seeded at 2.5 × 105 cells/well in 6-well plates or 0.5 × 105 cells/well in 24-well plates. Reverse-transfections were performed according to manufacturer’s instructions using Lipofectamine RNAiMAX (Invitrogen) and miRVana (Ambion, Grand Island, NY, USA) or Genepharma (Genepharma, Shanghai, China) miRNA mimics (20 nM), paired with negative control RNAs from the same manufacturers. Cells were harvested 72 h post-transfection, unless otherwise specified. Single-stranded miRNA mimics were obtained from Genepharma and transfected at 200 nM following the same procedure.
2′-Oxymethelated siRNAs and paired negative controls were purchased from Genepharma, and transfected following the same procedure (50 nM).
siPIK3R3: CAACCUCGUUUCCUUACAAAU.
Luciferase assay
Target 3′UTRs were cloned downstream of Renilla luciferase in the psiCHECK2 dual-luciferase vector (Promega). The Firefly luciferase gene expressed from the same vector was used as an internal reference. Approximately 0.5 × 105 MCF10A cells were plated in each well of 24-well plates. After 24 h, psiCHECK2 vectors (nM) were co-transfected with 10 nM miRNA mimics (or negative control RNA), using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Cells were harvested 72 h post-transfection and assessed using a GloMax 20/20 Luminometer (Promega). A ratio of Renilla/Firefly luminescence intensity was used to calculate the relative luciferase expression activity.
Immunoblotting and antibodies
Cells were rinsed with phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mmol/l Tris–HCl, pH 7.5, 250 mmol/l NaCl, 1% Triton X-100 and 1× protease inhibitors—Roche) on ice for 30 min. Insoluble components of cell lysates were removed by centrifugation (12 000 g for 10 min at 4°C) and protein concentration measured using a bicinchoninic acid protein assay kit (Pierce). Proteins were resolved using Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8–15% polyacrylamide gels, depending on the molecular weights of the target proteins and transferred to a Hybond- C Extra nitrocellulose membrane (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The membrane was then blocked for 30 min in blocking buffer, made by dissolving 5% skim milk in TBST (50 mM Tris–HCl, 150 mM NaCl, 0.05% Tween 20, pH = 7.6).
For quantification, the following proteins were probed in blocking buffer at 1:1000 - p53 (Santa Cruz sc-126), p21waf1/cip1 (Cell Signalling Technology #2947), PI3K p55 (CST #11889), phospo-Akt (CST #4051), Akt (CST #9272), PARP (CST #9542) and Casp3 (CST #9662). Puma (ProSci #41232) and BCL-xL (CST #2762) were probed at 1:500 and 1:1000. Equal loading was confirmed by using β-actin (Sigma-Aldrich #AC-15) or β-tubulin (Abcam ab6046), both at 1:2000. Chemiluminescent detection of protein was conducted using appropriate secondary antibodies conjugated with horseradish peroxidise (GE Healthcare) and the enhanced chemiluminescence kit (GE Healthcare) according to the manufacturer’s instructions.
RNA extraction and real-time PCR
RNA (>250 nt) was extracted from cells using a GenElute™ mRNA Miniprep Kit (Sigma) with on-column RNase-free DNase digestion (Sigma). Total RNA, including small RNAs, was extracted using Trizol (Invitrogen) as per manufacturer instruction. RNA was quantitated using a ND-1000 NanoDrop spectrometer (Thermo Scientific). Thereafter, 1 μg of total RNA was reverse-transcribed into cDNA using Moloney Murine Leukaemia Virus Reverse Transcriptase (Promega) with random 6′mer primers (Promega). Real-time polymerase chain reaction (PCR) was performed on a CFX Connect Real-time PCR Detection System (Bio-Rad) using a QuantiTect SYBR Green PCR Kit (QIAGEN). RT-PCR program: 1 × 95°C, 3 min/40× (95°C, 10 s/57°C 60 s). For quantification of small RNAs, TaqMan gene expression assays (Applied Biosystems) were performed following manufacturer's instructions. The 18 s rRNA was used for normalization.
Incucyte scanning and cell proliferation assay
Cells were plated in 24 or 96 well trays, reverse-transfected then scanned over 72 h by Incucyte (Essen BioScience). For proliferation assays, cells were reverse-transfected then cell numbers measured over various time points using either a CyQUANT proliferation kit (Invitrogen) or CellTiter 96 Aqueous MTS proliferation assay kit (Promega) according to manufacturer’s instructions.
Cell cycle assay, viability assay and flow cytometry
Cell cycle assay was performed using PI (propidium iodide) staining and flow cytometry. Briefly, cells were harvested, fixed in ice-cold ethanol (70%), and incubated overnight at 4°C. Cells were then stained with PI solution (50 μg/ml; Sigma-Aldrich) and 100 μg/ml RNase A (Sigma-Aldrich) for 45 min at 37°C. DNA content was determined using a FACS Calibur flow cytometer with cell cycle profiles analysed by FLOWJO software.
Cell viability assay was performed using 7AAD (7-aminoactinomycin D) staining. Briefly, cells were harvested 72 h post-treatment, centrifuged at 1300 × g, washed in PBS and stained with 7AAD solution (2 μg/ml) (Invitrogen) for 10 min at room temperature. Viable cells were determined by flow cytometry as above.
Nuclear fractionation
Nuclear/cytoplasmic fractionation was performed using a PARIS kit (Ambion). To enable small RNA purification, after fractionation the lysates were mixed with 5× volume of Trizol (Invitrogen) and total RNA extracted following manufacturer instruction.
RNA deep sequencing and HITS-CLIP (high-throughput sequencing of crosslinked and immunoprecipitated RNA)
MDA-MB-231 cells (20 × 10 cm dishes) were suspended in ice-cold PBS and crosslinked with UV at 254 nM (Stratalinker). Cell pellets were lysed (0.1% SDS, 0.5% deoxycholate, 0.5% NP-40 with protease inhibitors, Roche) for 10 min on ice followed by RQ1 DNAse (Promega) at 37°C for 15 min with shaking. RNAse A/T1 (Ambion) was then added for a further 8 min, prior to the addition of ethylenediaminetetraacetic acid (EDTA) (30 mM). Pellets were then spun (30 000 rpm) and the lysate subjected to immunoprecipitation for 2 h with a pan-anti-Ago antibody (2A8) conjugated to protein-A dynabeads (Invitrogen) using bridging rabbit anti-mouse IgG (Jackson Immunolabs). Pellets were then successively washed (0.1% SDS, 0.5% deoxycholate, 0.5% NP40 in 1 × PBS; 0.1% SDS, 0.5% deoxycholate, 0.5% NP40 in 5xPBS; 50 mM Tris pH 7.5, 10 mM MgCl2, 0.5% NP40) and on-bead phosphatase treatment performed for 30 min with antarctic phosphatase (New England Biolabs) in the presence of superasin RNAse inhibitor (Ambion). The 3′ RNA linker (CAGACGACGAGCGGG) was labelled with P32 using T4-PNK (NEB) and ligated on-bead for 1 h at 16°C with T4 RNA ligase (Fermentas). Beads were then washed as previous and treated with T4 Polynucleotide kinase to ligate the 5′ RNA linker (AGGGAGGACGAUGCGGxxxG, with ‘X’ representing different nucleotides for barcoding). Beads were resuspended in 4× Lithium dodecyl sulfate Novex loading buffer with 4% B-mercaptoethanol, incubated at 70°C for 10 min and the supernatant loaded on Novex NuPAGE 4–12% Bis-Tris acrylamide gels (Biorad). After running, the Ago–RNA complexes were then transferred to nitrocellulose and exposed to film at −80°C for 3 days. Complexes running at ∼110 kDa (AGO + miRNA) and ∼130 kDa (AGO + mRNA fragments) were then excised with a scalpel and resuspended (100 mM Tris pH 7.5, 50 mM NaCl, 10 mM EDTA, 4 mg/ml proteinase K) for 20 min at 37°C. The sample was incubated for an additional 20 min in the presence of 3.5 M urea and RNA isolated by a phenol–chloroform extraction. Sample was then run on a 10% denaturing (1:19) polyacrylamide gel and exposed to film with an intensifying screen at −80°C for 5 days. Thin bands corresponding to the Ago-miRNA (∼110 kD) and AGO mRNA fragments (∼130 kDa) were excised, crushed and eluted at 37°C for 1 h (1M NaOAc, pH 5.2, 1 mM EDTA). RNA was then precipitated overnight with ethanol, centrifuged and dried. RNA was then resuspended in 8 μl H2O, primer added (TCCCGCTCGTCGTCTG) and reverse transcription performed using SuperScriptIII (Invitrogen). PCR was then performed with the above primer and an additional primer (ACGGAGGACGATGCGG) for 25 cycles. PCR product was then run on a 10% native (1:29) polyacrylamide gel, stained with Sybr Gold (Qiagen) and bands excised over a UV light box. The DNA was then precipitated using isopropanol and a final 10 cycle PCR performed with the following primers: AATGATACGGCGACCACCGACTATGGATACTTAGTCAGGGAGGACGATGCGG, CAAGCAGAAGACGGCATACGATCCCGCTCGTCGTCTG. Reactions were then run on 2% metaphor agarose/Tris-Borate-EDTA gels and bands (∼115 and ∼135 bp) excised corresponding to the linker sequence + RNA CLIP tag. Samples were finally purified using quick-spin columns (Qiagen) and subjected to Illumina sequencing. In house Perl scripts were then used to filter reads for quality, trimmed of linker sequence, separated by barcode and aligned to the human genome using bowtie.
Immunofluorescence
MCF10A cells adhered to Thermanox coverslips (Nalge Nunc) were fixed in 4% PFA (paraformaldehyde) for 15 min then blocked in 2% BSA (bovine serum albumin) for 30 min. Primary antibodies (β-actin and β-tubulin) were added to cells overnight at 4°C, 1:500 dilution. Secondary antibodies (goat anti-Mouse Alexa Fluor 488 IgG and goat anti-rabbit Alexa Fluor 594 IgG) (Invitrogen), also at 1:500 were incubated for 1 h. Coverslips were mounted in Vectasheld Antifade Mounting Medium with DAPI (4′,6-Diamidino-2-Phenylindole, Dihydrochloride) (Vector Laboratories) and visualized using laser confocal microscopy.
Statistical analysis
Results are given as mean of at least three independent experiments ± standard deviation. Student’s t-test was performed using replicate values to indicate significance. Values of P < 0.05 were considered statistically significant (as labelled with ** in figures), while values of P < 0.1 were indicated by *.
RESULTS
Endogenous expression of miR-222 isoforms
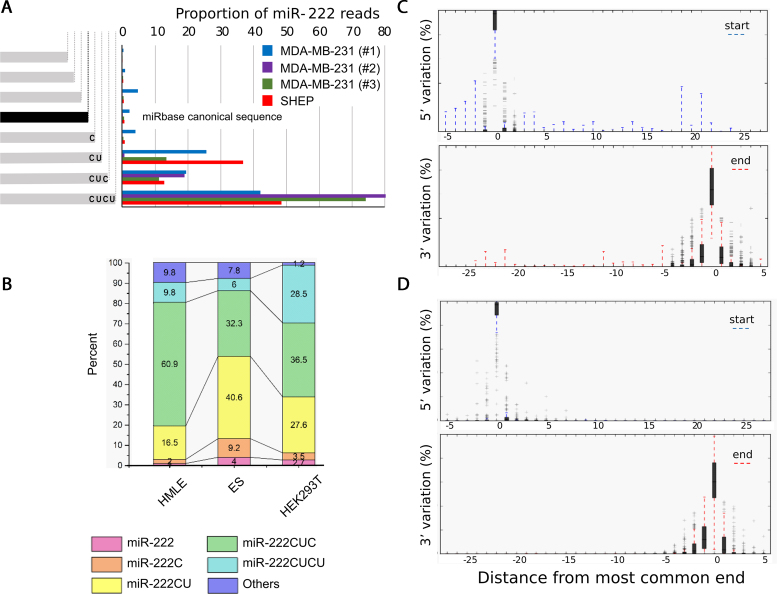
Prompted by reports that numerous miRNAs exist as length-differing isomiRs, we examined the degree of isomiR heterogeneity of Ago-bound miRNAs in MDA-MB-231 (and SHEP) cells and noticed in particular that miR-222, (which is abundant in these cells) had marked length heterogeneity at the 3′ end (Figure 1A). In fact most endogenous, AGO-bound miR-222 differed from the canonical miRBase definition with the majority of miR-222 isomiRs extended by between 2 and 4 nt at the 3; end. The 3′extension of CUCU corresponds to the natural sequence within the pri-miR, consistent with the variation in length being created by alternative sites of cleavage by Drosha.
Figure 1.
Endogenous miRNAs exhibit substantial 3′ variation. (A) Frequency of miR-222 mapping reads are shown from multiple independent AGO-CLIPs in MDA-MB-231 and SHEP cells. The sequence and abundance of canonical miR-222 from miRbase is shown. (B) Extensive miR-222 3′-heterogeneity is also seen in publicly available whole cell RNA sequencing from three cell lines. (C and D) The distribution of 5′ and 3′ variants of all miRNAs classified by distance from the most common end is shown by box plot. Shaded areas and whiskers indicate the 25–75th and 5–95th percentile ranges respectively. Data were obtained from (C) AGO-bound miRNAs from MDA-MB-231 cells and (D) the most abundant 227 miRNAs from the Cancer Genome Atlas (TCGA).
To assess whether this heterogeneity is a general feature of miR-222, and not a quirk of the MDA-MB-231 cell line, we examined the heterogeneity of miR-222 in other cell lines (Figure 1B) and again found marked heterogeneity in each dataset. To compare the length heterogeneity of miR-222 to that of miRNAs in general, we plotted the cumulative length distributions of miRNAs in MDA-MB-231 cells and TCGA data (Figure 1C and D) and observed that 3′ length heterogeneity, but not 5′ heterogeneity, is a common feature of miRNAs, as has been previously reported.
miR-222 isoforms significantly alter cell morphology
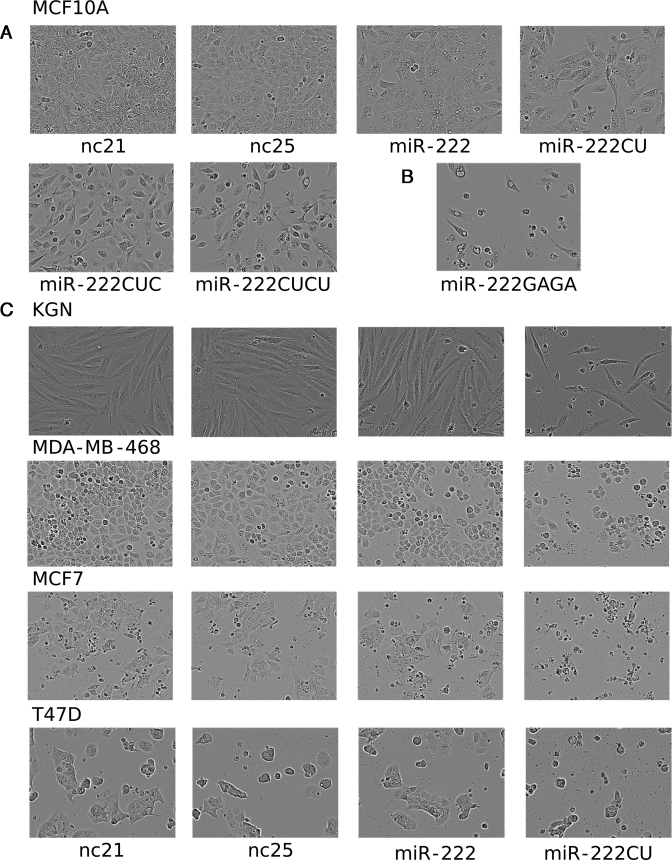
The fact that the most abundant forms of miR-222 are not the canonical form specified in miRbase raises the possibility that experiments performed in the past using miRNA mimics to examine the functions of miR-222 may have used an isomiR that is not the predominant form. Mindful of the fact that the literature contains conflicting conclusions regarding the functions of miR-222 (41–50), we decided to examine the effects of transfecting cells with each isoform. To investigate potential biological roles, we transfected miRNA mimics that match endogenous miR-222 isoforms from 21 to 25 nt in length, along with size-matched negative control RNAs (nc21 and nc25). Cells transfected with the larger isoforms displayed morphological alteration, adopting a rounded phenotype ∼48 h post-transfection, followed by blebbing and shrinkage at ∼72–96 h post-transfection (Figure 2). These effects were progressively more severe with the transfection of larger miR-222 isoforms. No morphological changes were seen with the transfection of either 21 or 25 nt scrambled controls (Figure 2A and Supplementary Figure S1). To ascertain if this was specific to the sequence of the 3′ extension, the 3′ end was mutated from CUCU to GAGA (miR-222GAGA), which caused similar morphological changes in MCF10A cells to the endogenous sequence (Figure 2B). Length-dependent, sequence-independent effects were further confirmed by the transfection of additional miR-222 mimics possessing additional, non-physiological 3′ extensions (Supplementary Figure S2). We therefore conclude that the changes in morphology are not due to the exact sequence of the 3′ extension, but occur in response to length. Further investigation revealed the effects of longer miR-222 isomiRs are not restricted to MCF10A cells, but are also present in multiple other cell lines including KGN, MDA-MB-468, MCF7 and T47D cells (Figure 2C).
Figure 2.
Longer miR-222 isomiRs alter cell morphology. (A and B) MCF10A cells are shown 72 h after the transfection of miR-222 isomiRs, the 21 or 25 nt controls (nc21 and nc25) or the miR-222GAGA mimic (in B). (C) Representative images of similarly transfected additional cell lines.
The miR-222 isomiRs all inhibit proliferation and canonical target gene expression
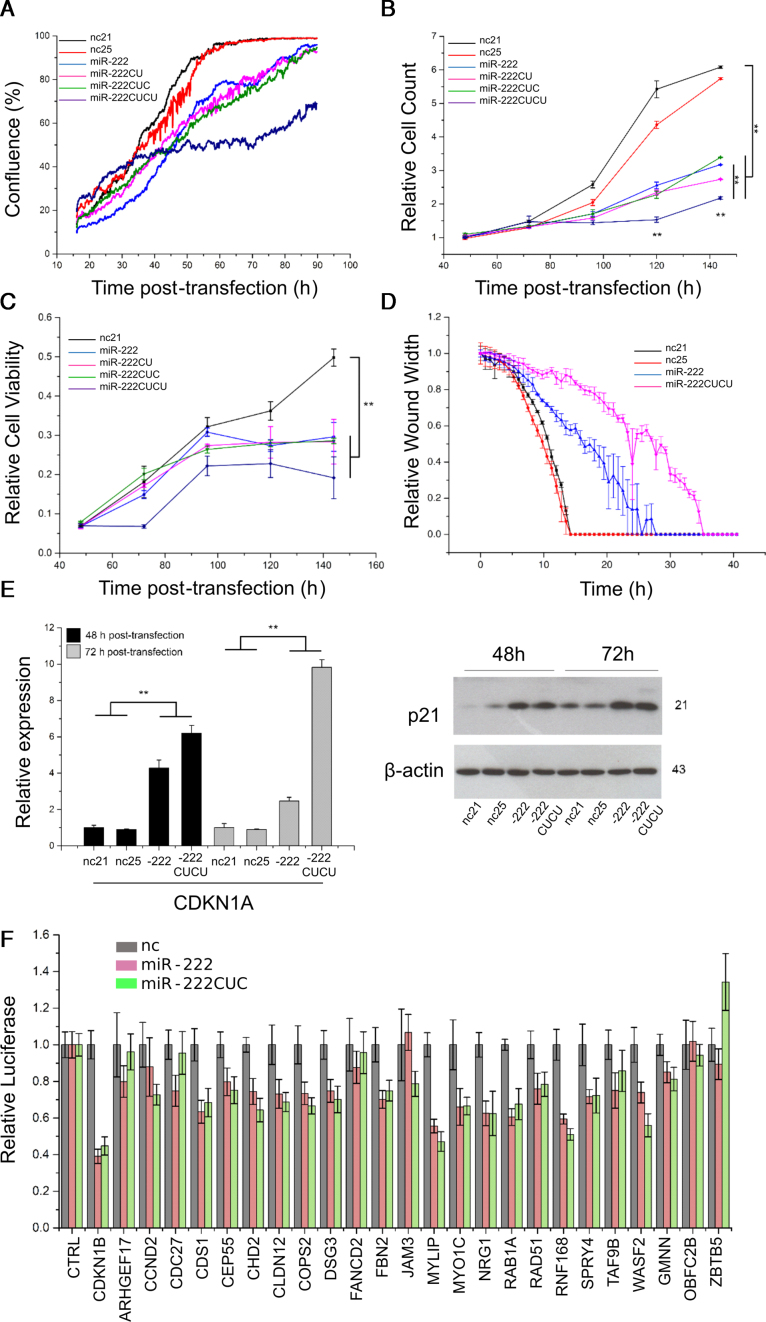
To further investigate the biological function of miR-222 isoforms, each of the four variants (21–25 nt in size) were transfected into MCF10A cells and proliferation (and/or migration) was assessed by confluence (Incucyte), cell count (Cyquant), viability (MTS assay) and scratch wound closure assay (Figure 3A–D). All miR-222 isomiRs effectively suppressed proliferation, with miR-222CUCU having the strongest effects (Figure 2A). Consistent with a generally anti-proliferative effect, both miR-222 and 222CUCU (but not nc21 and nc25), also induced the expression of CDKN1A (p21), a universal inhibitor of cyclin kinases and mediator of cell cycle delay (51), at both the mRNA and protein level (Figure 3E). Together, these data indicate all isomiRs of miR-222 play anti-proliferative roles in MCF10A cells.
Figure 3.
miR-222 and its isomiRs inhibit proliferation and share the same target recognition potential. (A–C) MCF10A cells were transfected as indicated and proliferation assessed by (A) confluence, (B) CyQuant and (C) MTS assay. (D) Scratch wound closure assay was also performed after transfection, though effects on proliferation versus migration are difficult to disentangle. (E) The expression of CDKN1A (p21) was determined at both the mRNA and protein level, 48 and 72 h post-transfection of miR-222, miR-222CUCU and the negative controls. (F) Relative luciferase activity was measured for 25 putative and established miR-222 target genes by cloning the respective 3′UTRs downstream of the reporter gene and co-transfecting with miR-222 isoforms or negative controls as indicated. The parental vector was used as a negative control.
MiRNAs function through the post-transcriptional suppression of target mRNAs, typically interacting with the 3′UTR of targets via their 5′ seed sequence, though internal and 3′ sequences may also confer target selection in some circumstances (52). To test whether 3′UTR extension alters targeting potential, 3′UTRs of 25 strongly predicted miR-222 targets were cloned downstream of luciferase reporters and the potential for differential targeting investigated with co-transfection of either miR-222, the 3′-CUC isomiR or the nc21 and nc25 negative controls. We saw no differential targeting capacity between miR-222 isoforms, indicating differential effects are not likely mediated through a difference in the general post-transcriptional targeting capacities of the isomiRs (Figure 3F). Consistent with the previous data (Figure 2B and Supplementary Figure S2), a range of miR-222 mimics possessing variable 3′ extensions all displayed similar repressive capacity on representative miR-222-responsive 3′UTR target gene reporters (Supplementary Figure S3). Combined with the fact that longer isomiRs are still capable of binding AGO (Figure 1A and C), these data suggest differential effects are unlikely to result from differences in the basic functional properties of miRNAs such as stability, AGO-loading or generalized target selection.
Longer miR-222 isomiRs induce apoptosis
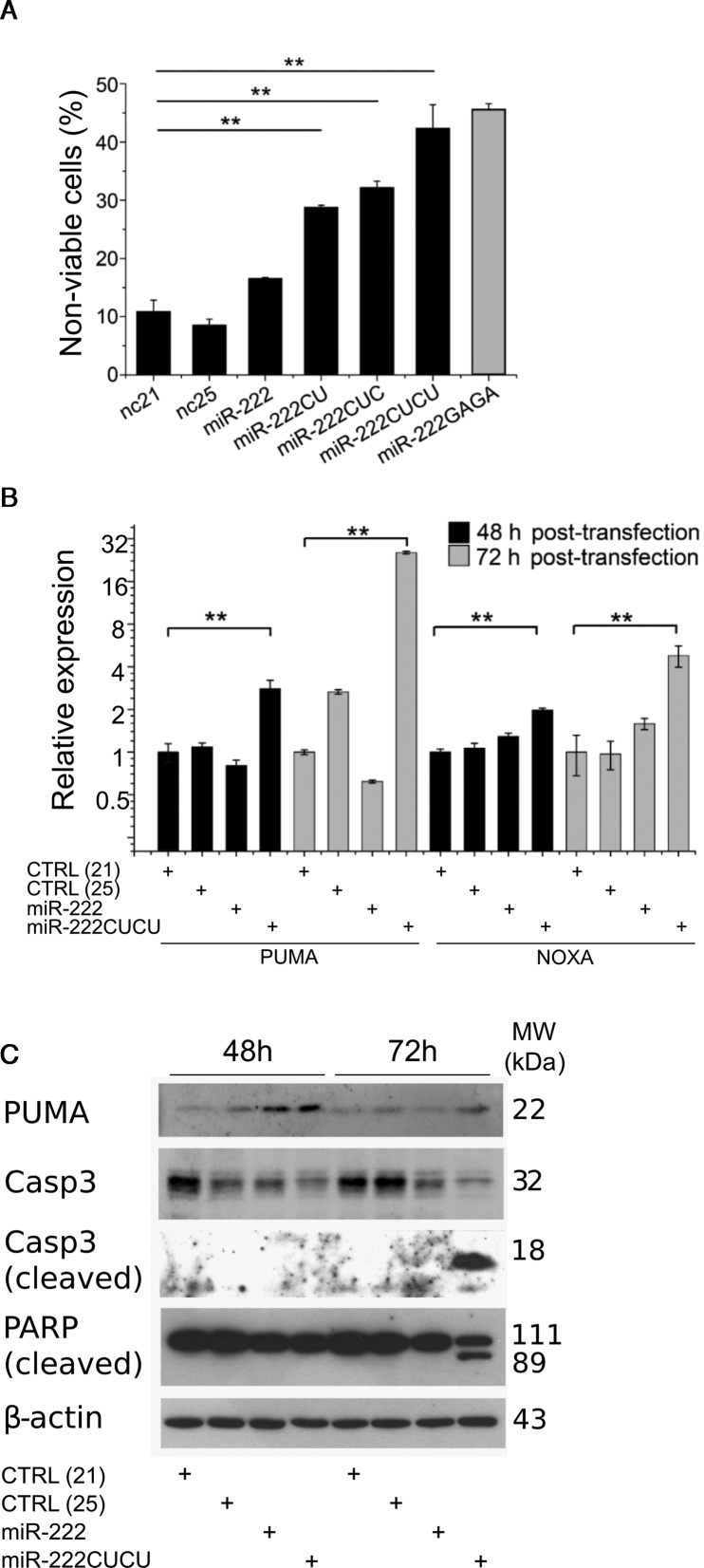
To further investigate the cell death caused by the expression of longer miR-222 isomiRs we performed flow cytometry to evaluate the proportion of non-viable cells after transfection. Consistent with morphological observations, progressively longer miR-222 isomiRs displayed a progressively stronger tendency to promote cell death, from 17% by miR-222, to 42% by miR-222CUCU (Figure 4A). Once again, miR-222GAGA (Figure 2B), exhibited an identical activity to that of miR-222CUCU, again suggesting apoptosis was a function of length and not sequence. In further confirmation of apoptotic cell death, miR-222CUCU-transfected cells displayed cleavage of both caspase 3 and PARP and prolonged activation of the apoptosis-promoter Puma (BBC3) 72 h post-transfection (Figure 4B). Further, both apoptosis promoters Puma and Noxa were specifically upregulated at the RNA level by miR-222CUCU (Figure 4C). Collectively, these observations indicate the triggering of the intrinsic apoptosis pathway upon expression of longer miR-222 isomiRs.
Figure 4.
Apoptosis is induced by longer miR-222 isomiRs. (A) The proportion of non-viable cells and the expression of apoptotic marker genes at the (B) RNA and (C) protein level were determined after transfection of the miR-222 isoforms or 21 and 25 nt controls (nc21 and nc25). MiR-222CUCU-mediated apoptosis is indicated by the induction of pro-apoptotic Puma and Noxa and the cleavage of Caspase 3 and PARP.
The differential apoptotic activity of longer 222 isomiRs is not attributable to interferon responses
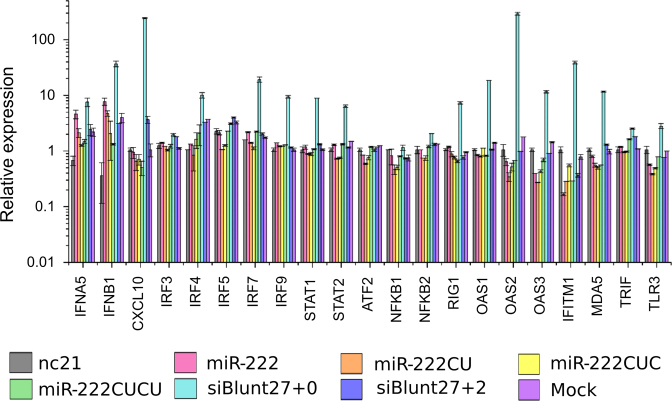
Longer double-stranded (ds-) RNAs are known to trigger an interferon immune response, evolved as a defensive mechanism against viral infection (53). Recently, longer double-stranded miRNA mimics were reported to inhibit cell proliferation and induce apoptosis independent of the seed sequence through the activation of an interferon response (54). Throughout our work, the inclusion of a size-matched 25 nt ds-negative control (nc25) failed to elicit anti-proliferative or pro-apoptotic responses which argues against a non-specific interferon response as a mechanism to explain the activities of miR-222CUCU. To further confirm this however, we performed RT-PCR of 21 key IFN-responsive genes and observed no activation after the transfection of any miR-222 isoform. A double-stranded positive control (siBlunt27) which promotes an interferon response through direct interaction with RIG-1 (retinoic acid-inducible gene 1) (55) successfully promoted the expression of the majority of these genes (Figure 5). A matched negative control (siBlunt27+2), known to not elicit an interferon response, accordingly failed to induce IFN-responsive gene expression (56).
Figure 5.
Interferon response genes are not induced by miR-222 transfection. MiR-222 isomiRs, control RNAs (nc21 and nc25), a known inducer of the interferon response (siBlunt27+0) or a non-inducing negative control (siBlunt27+2) were transfected into MCF10A cells and a series of IFN (interferon) pathway markers measured by RT-PCR.
Unfortunately, due to the high sequence similarity between miR-222 isomiRs, we are not able to inhibit them differentially, nor are we able to guide the cell to process certain miR-222 isomiRs over others by the construction and transfection of exogenous miRNA expression vectors. We did however recapitulate the effects of double-stranded miR-222 mimics from two separate sources (Ambion and GenePharma, data not shown). As interferon responses are triggered by the engagement of different cell surface receptors by RNAs of different size and structure, we sought to further test the separate functions of miR-222 isomiRs through the application of single-stranded (ss-) miRNA mimics, offering an alternate overexpression model and, presumably, the potential to engage different interferon pathways. These mimics re-capitulated previous observations, indicating reduced proliferation by all miR-222 isoforms and the specific promotion of apoptosis by miR-222CUCU but not canonical (21 nt) miR-222 or size-matched negative controls (Supplementary Figure S4).
The PI3K-AKT pathway is inhibited by miR-222CUCU
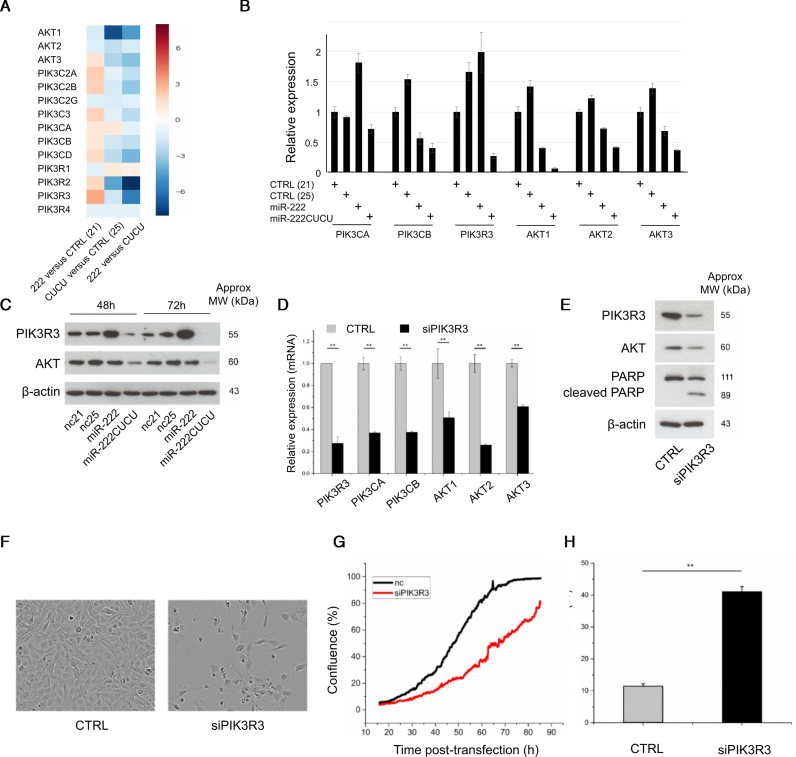
In order to identify genes and pathways through which miR-222 isomiRs might be exerting differential functions, mRNA-sequencing was performed on MCF10A cells transfected with control (nc21 and nc25), miR-222 or miR-222CUCU miRNA mimics. Among the genes upregulated by both miR-222 and miR-222CUCU, there was a clear and strong over-representation of gene ontology classes associated with the cell cycle and cell division (Supplementary Figure S5). There were also groups of genes differentially regulated between the miR-222 isomiRs, including cell signalling genes upregulated by the CUCU extended isomiR and small molecule metabolic processes downregulated by this same miRNA mimic. We utilized IMPaLA (Integrated Molecular Pathway Level Analysis) (57) to identify genetic networks that are consistently modulated by miR-222CUCU in comparison to miR-222 and found ECM-membrane and integrin associated pathways were enriched, consistent with the decreased adherence we observe in miR-222CUCU transfected cells prior to apoptosis. We also identified that components of a number of signalling pathways (namely the PI3K-Akt component of larger EGFR and TGF-B signalling networks) were enriched, which led us to investigate PI3K-Akt cell survival signalling in greater detail. We found that the expression of miR-222CUCU downregulated the expression of a number of pathway components, in sharp contrast to miR-222 which typically upregulated AKT-survival genes (Figure 6A). RT-PCR and western blotting confirmed many key components (including PIK3CA, PIK3CB, PIK3R3, AKT1, AKT2 and AKT3) were repressed upon miR-222CUCU transfection (Figure 6B and C).
Figure 6.
Longer miR-222 isomiRs suppress the PIK3/AKT pathway. (A–C) The expression of multiple PIK3/AKT pathway components in MCF10A cells in response to the transfection of miR-222 and miR-222CUCU are shown using (A) RNA sequencing, (B) qPCR or (C) western blotting. (D and E) PI3KR3 knockdown decreases PI3K/AKT pathway gene expression and promotes apoptosis as indicated by (E) PARP cleavage, (F) morphology, (G) cell confluence and (H) flow cytometry analysis for non-viable cells.
Among these genes, the AKT-regulator PIK3R3 (p55) was of particular interest as it is strongly differentially regulated by different miR-222 isomiRs and its knockdown phenocopied the effects of miR-222CUCU transfection in four aspects: (i) siPIK3R3 inhibited the expression of key PI3K-Akt genes, i.e. PIK3CA, PIK3CB, AKT1, 2 and 3 (Figure 6D and E); (ii) MCF10A cells displayed a blebbing morphology and PARP cleavage indicative of apoptosis (Figure 6E and F); (iii) cell confluency was significantly reduced after siPIK3R3 treatment (Figure 6F and G); and (iv) siPIK3R3 induced cell death to a similar extent as miR-222CUCU expression (Figure 6H).
The size of endogenous miRNAs correlates with their nuclear localization
To further explore miR-222 isomiR differences, we subjected our miR-222-affected gene expression profiles to EISA (Exon-Intron Split Analysis) (58) to differentiate genes regulated at the transcriptional and post-transcriptional levels. Of the AKT-pathway genes that are downregulated by miR-222CUCU, several (PIK3R3, PIK3C2A, PIK3C2B) were downregulated primarily at the transcriptional level (Supplementary Figure S6), suggesting the possibility of non-canonical transcriptional silencing (59,60).
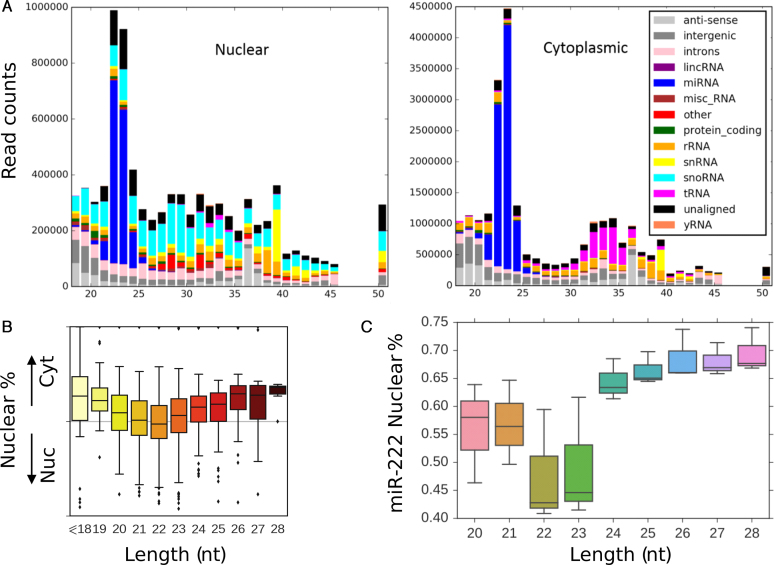
To investigate the possibility of direct transcriptional regulation, nuclear and cytoplasmic compartments from MCF10A cells were fractionated and the small RNA population was sequenced. We observed abundant mature miRNAs within nuclei (Figure 7A), with specific miRNAs preferentially enriched in one or the other cell compartment. Clean fractionation was demonstrated by the respective nuclear and cytoplasmic enrichment of snoRNAs and tRNAs respectively (Figure 7A) and through qPCR of known nuclear and cytoplasmic RNAs (Supplementary Figure S7). Although we did not observe any specific sequences correlating with sub-cellular localization (in keeping with some reports (61,62) but in disagreement with others (63)), we found a strong relationship between the size of endogenous miRNAs and their sub-cellular location. On a global scale, we observed a clear size-dependent trend for the nuclear enrichment of both shorter (<19 nt) and longer (>23 nt) miRNAs compared to miRNAs of typical 20–22 nt size (Figure 7B). With regard to endogenous miR-222, there was a dramatic nuclear enrichment of miR-222 isomiRs of >23 nt in size (Figure 7C), corresponding to the same isomiRs that promote apoptosis.
Figure 7.
Endogenous miRNAs are abundant within the nucleus and exhibit size-dependent nuclear enrichment. (A) MCF10A cells were separated into nuclear and cytoplasmic fractions and subjected to Illumina sequencing. Clean fractionation is indicated by the differential localization of nuclear snoRNAs and snRNAs and cytoplasmic tRNAs. Endogenous, mature miRNAs are both nuclear and cytoplasmic. The y-axis indicates total sequencing read counts and the x-axis, length of the associated small RNA. (B) Shorter (<20 nt) and longer (>23 nt) miRNAs are preferentially enriched within nuclei. (C) Proportion of the nuclear distribution of miR-222 isomiRs of varying size are indicated by box plot.
DISCUSSION
It is suggested that miR-222 is a valuable biomarker for cancer and cardiovascular disease (64,65) and its role as a regulator of such processes as proliferation, apoptosis, chemoresistance and epithelial-mesenchymal transition have been extensively examined (64–71). Despite this, the response of cells to miR-222 perturbation varies widely. Anti-apoptotic roles for miR-222 are often reported (20,72–86), with many of these studies also observing pro-proliferative (66,73,75,76,79–82,84,85,87–95) and pro-invasion/migration (73–76,80–82,85,87,89,90,94,96,97) effects. In direct contrast, pro-apoptotic effects (as we report here with the transfection of longer miR-222 isomiRs) are also observed (45,98–100). For reasons that remain to be defined, cell-specificity accounts for at least some of these differences, with varying effects on proliferation and chemosensitivity reported between different lung cancer cell lines in the same study (49). We speculate that differential functions of miR-222 isomiRs might also contribute to these apparently contradictory findings, as we demonstrate isomiR-specific effects on apoptosis and regulation of the AKT pathway within the one cell line. Very few studies specify the sequence of the transfected miRNA mimics, though it is noteworthy that of those which do, all utilize miR-222 mimics that correspond to the canonical 21 nt form, and all of these report anti-apoptotic effects (75,83,84,90)—in direct contrast to our findings using longer isomiRs.
MiRbase, a reference sequence repository for miRNAs, changed the canonical definition of human miR-222 in version 10 (August 2007) from the +CUC containing sequence to the shorter and less abundant 21 nt sequence still annotated today. Species-specific discrepancies also exist, with the shortened 21 nt version representing the canonical miR-222 sequence in most mammalian species (human, mouse, rat, cow, etc.), whilst in other species (spider monkey, zebrafish, chicken), it is the +CUC isomiR which is canonical. This has directly informed the production of Taqman (Applied Biosystems) qPCR probes, with human (shorter, RT002276) probes able to equally detect all major human miR-222 isomiRs (21 nt, +CU, +CUC, +CUCU), whilst probes designed against spider monkey miR-222 (+CUC, RT000525) preferentially detect the human +CUCU isomiR but completely fail to detect the shorter 21 nt canonical form (data not shown). Species-specific miR-222 annotation and the version of mirBase used for reference may therefore have a major effect on the isomiR of miR-222 that is both capable of being detected and that is chosen for use in expression studies. These differences in turn may have significant consequences for the investigation of miR-222 functional roles.
Although we have focused exclusively on the differential effects of miR-222 isomiRs, it is noteworthy that more than half of all endogenously expressed miRNAs vary in the sequence or position of their 3′ terminus. Our work suggests 3′ variance may have a wider effect on the diversification of miRNA function than has been previously thought, with functional consequences of 3′ variance having received little attention as it is removed from the 5′ ‘seed-site’ which is responsible for the majority of miRNA–target interaction.
It has been reported that the transfection of longer miRNA mimics (23 nt in size) decreases cell confluency and promotes apoptosis, in conjunction with the upregulation of interferon pathway genes and PARP cleavage (54). This occurred in response to both the length of the miRNA mimic, and the seed-sequence of the co-transfected passenger strand, and suggests that miRNA transfection can promote cell death non-specifically as a result of interferon pathway activation. We conclude that the phenomenon we report here is not a length-dependent interferon response for several reasons. All assays were accompanied by co-transfection with size-matched (21 and 25 nt) negative control miRNA mimics which behaved identically to each other with regard to cell morphology, confluency, survival, apoptosis and the expression of the apoptosis and AKT-associated proteins that we examined. Further, it was reported that the triggering of interferon-associated apoptosis was accompanied by the upregulation of key interferon pathway genes such as IFNB1, OAS1/OAS 2, CXCL10, IFITM1 and RIG1, but none of these genes were induced by any miR-222 isomiR or size-matched control sequence in our work, though they were strongly upregulated by the transfection of a positive control—a double stranded 27-nt sequence known to activate the interferon pathway. Further, the contribution of the passenger strand sequence is discounted by the capacity of single stranded longer miR-222 isomiRs to also trigger apoptosis.
Despite the markedly different outcome of miR-222 isomiR expression with regard to survival, most effects of miR-222 and miR-222CUCU are in fact common between isoforms. All isoforms inhibited cell proliferation and luciferase assays designed to examine the effect of miR-222 variation on canonical target genes, failed to demonstrate any functional differences between short and long isomiRs. This strongly argues against standard canonical targeting, or other generalized properties such as AGO loading or stability, as likely explanations for these differential effects. Mutation of the 3′-extended nucleotides also had no effect on the capacity of longer miR-222 isomiRs to promote apoptosis, indicating differential effects occur in a size and not-sequence dependent manner. The response of the pro-survival PI3K-AKT signalling pathway; however, was vastly different between isoforms, with miR-222 upregulating the expression of many components whilst miR-222CUCU downregulated expression, not only of PI3KR3 whose knockdown phenocopied miR-222CUCU, but also of many other signalling components as well.
We also find that longer miRNAs display an increased proclivity for nuclear localization. This is true for both endogenous miRNAs globally, and miR-222 specifically, where we observe a marked increase in the relative nuclear localization of endogenous miR-222 isomiRs >23 nt in length. Although AGO and RISC-associated proteins, along with an abundance of endogenous mature miRNAs, have been observed within the nuclei of various cells (61,62,101–105), the functional significance of these observations are often unclear. Nuclear roles for AGO, miRNAs and their isomiRs that are present within nuclei, include well established examples in both the positive and negative regulation of transcription and alternative splicing (reviewed in (59,60,106–108)). AGO, bound to small RNAs other than miRNAs, also has additional nuclear functions including DNA damage repair (small RNAs produced at sites of UV-induced DNA damage) (109,110), RNA processing (3′ tRNA trailer sequences) (111) and regulating nucleosome occupancy (small RNAs produced proximal to transcription start sites) (112,113).
Much as there remains uncertainty regarding the function of nuclear miRNAs, the mechanisms that govern nuclear import/retention are also unclear and there is little consensus between the identity of nuclear enriched miRNAs between studies. One early study reported a hexanucleotide motif at the 3′ terminus of miR-29b that governs its nuclear import (63), which was partially supported by the identification of a related 3′ A(C/G)U(C/G) motif that was over-represented within nuclear localized miRNAs in neural stem cells (102). Post-transcriptional 3′ guanylation was also reported to promote nuclear localization (61). Virtually all studies however fail to support the conclusions of others regarding factors that contribute to nuclear localization. Similarly, we find no sequence motifs that are enriched within nuclear localized miRNAs in MCF10A cells. Our observation of size-dependent nuclear enrichment however is supported by a separate study that also examines the sub-cellular distribution of endogenous miRNAs within breast cancer cells (MCF7, MCF10A, MDA-MB-231), that found a progressive nuclear enrichment of longer miRNAs (101).
Although 3′ variance has been associated with miRNA stability (reviewed in (21)) in both plants and animals, in each of these reports 3′ variance has involved 3′-tailing whereby nucleotides (mostly A and U) are added in a non-templated manner. This is the first report (to our knowledge) detailing functional consequences for differential processing of the miRNA hairpin. Given the extensive 3′ variability that exists with miRNAs leading to a variety of endogenous miRNA sizes, this work strongly suggests more careful consideration be given to the effects of 3′ variance and more careful reporting be detailed in future studies with regard to the sequence of the miRNA isoform being examined. It also highlights the potential importance of miRNA size in regulating nuclear uptake and/or retention, which may not only deplete the cytoplasmic pool of functional miRNAs, but which may actively take on additional functions.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [#GNT1069128]; NHMRC Fellowship [Goodall #GNT1026191]; Royal Adelaide Hospital Research Fund (Bracken, Florey Postdoctoral Fellowship); Australian Research Council (Gantier, Future Fellowship) [#140100594]. Funding for open access charge: Ongoing Grant Funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed F., Senthil-Kumar M., Lee S., Dai X., Mysore K.S., Zhao P.X.. Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol. 2014; 11:1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azuma-Mukai A., Oguri H., Mituyama T., Qian Z.R., Asai K., Siomi H., Siomi M.C.. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:7964–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baran-Gale J., Fannin E.E., Kurtz C.L., Sethupathy P.. Beta cell 5′-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One. 2013; 8:e73240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cloonan N., Wani S., Xu Q., Gu J., Lea K., Heater S., Barbacioru C., Steptoe A.L., Martin H.C., Nourbakhsh E. et al. . MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011; 12:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Etebari K., Osei-Amo S., Blomberg S.P., Asgari S.. Dengue virus infection alters post-transcriptional modification of microRNAs in the mosquito vector Aedes aegypti. Sci. Rep. 2015; 5:15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo L., Yang Q., Lu J., Li H., Ge Q., Gu W., Bai Y., Lu Z.. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PLoS One. 2011; 6:e21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo L., Zhao Y., Yang S., Zhang H., Chen F.. A genome-wide screen for non-template nucleotides and isomiR repertoires in miRNAs indicates dynamic and versatile microRNAome. Mol. Biol. Rep. 2014; 41:6649–6658. [DOI] [PubMed] [Google Scholar]
- 9. Hinton A., Hunter S.E., Afrikanova I., Jones G.A., Lopez A.D., Fogel G.B., Hayek A., King C.C.. sRNA-seq analysis of humanTh embryonic stem cells and definitive endoderm reveals differentially expressed microRNAs and novel IsomiRs with distinct targets. Stem Cells. 2014; 32:2360–2372. [DOI] [PubMed] [Google Scholar]
- 10. Humphreys D.T., Hynes C.J., Patel H.R., Wei G.H., Cannon L., Fatkin D., Suter C.M., Clancy J.L., Preiss T.. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012; 7:e30933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Humphreys D.T., Suter C.M.. miRspring: a compact standalone research tool for analyzing miRNA-seq data. Nucleic Acids Res. 2013; 41:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karali M., Persico M., Mutarelli M., Carissimo A., Pizzo M., Singh Marwah V., Ambrosio C., Pinelli M., Carrella D., Ferrari S. et al. . High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016; 44:1525–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S.C., Liao Y.L., Ho M.R., Tsai K.W., Lai C.H., Lin W.C.. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012; 13(Suppl. 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loher P., Londin E.R., Rigoutsos I.. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget. 2014; 5:8790–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGahon M.K., Yarham J.M., Daly A., Guduric-Fuchs J., Ferguson L.J., Simpson D.A., Collins A.. Distinctive profile of IsomiR expression and novel microRNAs in rat heart left ventricle. PLoS One. 2013; 8:e65809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siddle K.J., Tailleux L., Deschamps M., Loh Y.H., Deluen C., Gicquel B., Antoniewski C., Barreiro L.B., Farinelli L., Quintana-Murci L.. bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS Genet. 2015; 11:e1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan G.C., Chan E., Molnar A., Sarkar R., Alexieva D., Isa I.M., Robinson S., Zhang S., Ellis P., Langford C.F. et al. . 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014; 42:9424–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Telonis A.G., Loher P., Jing Y., Londin E., Rigoutsos I.. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 2015; 43:9158–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaz C., Ahmad H.M., Bharti R., Pandey P., Kumar L., Kulshreshtha R., Bhattacharya A.. Analysis of the microRNA transcriptome and expression of different isomiRs in human peripheral blood mononuclear cells. BMC Res. Notes. 2013; 6:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou H., Arcila M.L., Li Z., Lee E.J., Henzler C., Liu J., Rana T.M., Kosik K.S.. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012; 40:5864–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neilsen C.T., Goodall G.J., Bracken C.P.. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012; 28:544–549. [DOI] [PubMed] [Google Scholar]
- 22. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A.. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012; 151:900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma H., Wu Y., Choi J.G., Wu H.. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20687–20692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Starega-Roslan J., Witkos T.M., Galka-Marciniak P., Krzyzosiak W.J.. Sequence features of Drosha and Dicer cleavage sites affect the complexity of isomiRs. Int. J. Mol. Sci. 2015; 16:8110–8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han B.W., Hung J.H., Weng Z., Zamore P.D., Ameres S.L.. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr. Biol. 2011; 21:1878–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Katoh T., Hojo H., Suzuki T.. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015; 43:7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu N., Abe M., Sabin L.R., Hendriks G.J., Naqvi A.S., Yu Z., Cherry S., Bonini N.M.. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr. Biol. 2011; 21:1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandez-Valverde S.L., Taft R.J., Mattick J.S.. Dynamic isomiR regulation in Drosophila development. RNA. 2010; 16:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo L., Liang T., Yu J., Zou Q.. A comprehensive analysis of miRNA/isomiR expression with gender difference. PLoS One. 2016; 11:e0154955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang S., Xu Y., Li M., Tu J., Lu Z.. Dysregulation of miRNA isoform level at 5′ end in Alzheimer's disease. Gene. 2016; 584:167–172. [DOI] [PubMed] [Google Scholar]
- 34. Saito K., Inagaki K., Kamimoto T., Ito Y., Sugita T., Nakajo S., Hirasawa A., Iwamaru A., Ishikura T., Hanaoka H. et al. . MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One. 2013; 8:e71480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan Y.T., Lin Y.C., Lin R.J., Kuo H.H., Thang W.C., Chiu K.P., Yu A.L.. Concordant and discordant regulation of target genes by miR-31 and its isoforms. PLoS One. 2013; 8:e58169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Llorens F., Banez-Coronel M., Pantano L., del Rio J.A., Ferrer I., Estivill X., Marti E.. A highly expressed miR-101 isomiR is a functional silencing small RNA. BMC Genomics. 2013; 14:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manzano M., Forte E., Raja A.N., Schipma M.J., Gottwein E.. Divergent target recognition by coexpressed 5′-isomiRs of miR-142-3p and selective viral mimicry. RNA. 2015; 21:1606–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boele J., Persson H., Shin J.W., Ishizu Y., Newie I.S., Sokilde R., Hawkins S.M., Coarfa C., Ikeda K., Takayama K. et al. . PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11467–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katoh T., Sakaguchi Y., Miyauchi K., Suzuki T., Kashiwabara S., Baba T., Suzuki T.. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009; 23:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marcinowski L., Tanguy M., Krmpotic A., Radle B., Lisnic V.J., Tuddenham L., Chane-Woon-Ming B., Ruzsics Z., Erhard F., Benkartek C. et al. . Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012; 8:e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S. et al. . MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:18081–18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fuse M., Kojima S., Enokida H., Chiyomaru T., Yoshino H., Nohata N., Kinoshita T., Sakamoto S., Naya Y., Nakagawa M. et al. . Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J. Hum. Genet. 2012; 57:691–699. [DOI] [PubMed] [Google Scholar]
- 43. Gan R., Yang Y., Yang X., Zhao L., Lu J., Meng Q.H.. Downregulation of miR-221/222 enhances sensitivity of breast cancer cells to tamoxifen through upregulation of TIMP3. Cancer Gene Ther. 2014; 21:290–296. [DOI] [PubMed] [Google Scholar]
- 44. Goto Y., Kojima S., Nishikawa R., Kurozumi A., Kato M., Enokida H., Matsushita R., Yamazaki K., Ishida Y., Nakagawa M. et al. . MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer. 2015; 113:1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Medina R., Zaidi S.K., Liu C.G., Stein J.L., van Wijnen A.J., Croce C.M., Stein G.S.. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008; 68:2773–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Visone R., Russo L., Pallante P., De Martino I., Ferraro A., Leone V., Borbone E., Petrocca F., Alder H., Croce C.M. et al. . MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer. 2007; 14:791–798. [DOI] [PubMed] [Google Scholar]
- 47. Wang M., Ge X., Zheng J., Li D., Liu X., Wang L., Jiang C., Shi Z., Qin L., Liu J. et al. . Role and mechanism of miR-222 in arsenic-transformed cells for inducing tumor growth. Oncotarget. 2016; 7:17805–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wurz K., Garcia R.L., Goff B.A., Mitchell P.S., Lee J.H., Tewari M., Swisher E.M.. MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer. 2010; 49:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamashita R., Sato M., Kakumu T., Hase T., Yogo N., Maruyama E., Sekido Y., Kondo M., Hasegawa Y.. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015; 4:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao J.J., Lin J., Yang H., Kong W., He L., Ma X., Coppola D., Cheng J.Q.. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 2008; 283:31079–31086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51. Karimian A., Ahmadi Y., Yousefi B.. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst). 2016; 42:63–71. [DOI] [PubMed] [Google Scholar]
- 52. Seok H., Ham J., Jang E.S., Chi S.W.. MicroRNA target recognition: insights from transcriptome-wide non-canonical interactions. Mol. Cells. 2016; 39:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murira A., Lamarre A.. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front. Immunol. 2016; 7:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goldgraben M.A., Russell R., Rueda O.M., Caldas C., Git A.. Double-stranded microRNA mimics can induce length- and passenger strand-dependent effects in a cell type-specific manner. RNA. 2016; 22:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ishibashi O., Ali M.M., Luo S.S., Ohba T., Katabuchi H., Takeshita T., Takizawa T.. Short RNA duplexes elicit RIG-I-mediated apoptosis in a cell type- and length-dependent manner. Sci. Signal. 2011; 4:ra74. [DOI] [PubMed] [Google Scholar]
- 56. Gantier M.P., Williams B.R.. Monitoring innate immune recruitment by siRNAs in mammalian cells. Methods Mol. Biol. 2010; 623:21–33. [DOI] [PubMed] [Google Scholar]
- 57. Kamburov A., Cavill R., Ebbels T.M., Herwig R., Keun H.C.. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011; 27:2917–2918. [DOI] [PubMed] [Google Scholar]
- 58. Gaidatzis D., Burger L., Florescu M., Stadler M.B.. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2015; 33:722–729. [DOI] [PubMed] [Google Scholar]
- 59. Kalantari R., Chiang C.M., Corey D.R.. Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res. 2016; 44:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Salmanidis M., Pillman K., Goodall G., Bracken C.. Direct transcriptional regulation by nuclear microRNAs. Int. J. Biochem. Cell Biol. 2014; 54:304–311. [DOI] [PubMed] [Google Scholar]
- 61. Khudayberdiev S.A., Zampa F., Rajman M., Schratt G.. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013; 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H.. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010; 5:e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hwang H.W., Wentzel E.A., Mendell J.T.. A hexanucleotide element directs microRNA nuclear import. Science. 2007; 315:97–100. [DOI] [PubMed] [Google Scholar]
- 64. Ding S., Huang H., Xu Y., Zhu H., Zhong C.. MiR-222 in cardiovascular diseases: physiology and pathology. Biomed. Res. Int. 2017; 2017:4962426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wei T., Ye P., Peng X., Wu L.L., Yu G.Y.. Prognostic value of miR-222 in various cancers: a systematic review and meta-analysis. Clin. Lab. 2016; 62:1387–1395. [DOI] [PubMed] [Google Scholar]
- 66. Galardi S., Mercatelli N., Giorda E., Massalini S., Frajese G.V., Ciafre S.A., Farace M.G.. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007; 282:23716–23724. [DOI] [PubMed] [Google Scholar]
- 67. Hwang M.S., Yu N., Stinson S.Y., Yue P., Newman R.J., Allan B.B., Dornan D.. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013; 8:e66502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafre S.A. et al. . Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007; 26:3699–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stinson S., Lackner M.R., Adai A.T., Yu N., Kim H.J., O’Brien C., Spoerke J., Jhunjhunwala S., Boyd Z., Januario T. et al. . miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011; 4:pt5. [DOI] [PubMed] [Google Scholar]
- 70. Stinson S., Lackner M.R., Adai A.T., Yu N., Kim H.J., O’Brien C., Spoerke J., Jhunjhunwala S., Boyd Z., Januario T. et al. . TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011; 4:ra41. [DOI] [PubMed] [Google Scholar]
- 71. Wang D.D., Yang S.J., Chen X., Shen H.Y., Luo L.J., Zhang X.H., Zhong S.L., Zhao J.H., Tang J.H.. miR-222 induces Adriamycin resistance in breast cancer through PTEN/Akt/p27kip1 pathway. Tumour Biol. 2016; 37:15315–15324. [DOI] [PubMed] [Google Scholar]
- 72. Brognara E., Fabbri E., Montagner G., Gasparello J., Manicardi A., Corradini R., Bianchi N., Finotti A., Breveglieri G., Borgatti M. et al. . High levels of apoptosis are induced in human glioma cell lines by co-administration of peptide nucleic acids targeting miR-221 and miR-222. Int. J. Oncol. 2016; 48:1029–1038. [DOI] [PubMed] [Google Scholar]
- 73. Chun-Zhi Z., Lei H., An-Ling Z., Yan-Chao F., Xiao Y., Guang-Xiu W., Zhi-Fan J., Pei-Yu P., Qing-Yu Z., Chun-Sheng K.. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010; 10:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jiang F., Zhao W., Zhou L., Liu Z., Li W., Yu D.. MiR-222 targeted PUMA to improve sensitization of UM1 cells to cisplatin. Int. J. Mol. Sci. 2014; 15:22128–22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jiang F., Zhao W., Zhou L., Zhang L., Liu Z., Yu D.. miR-222 regulates the cell biological behavior of oral squamous cell carcinoma by targeting PUMA. Oncol. Rep. 2014; 31:1255–1262. [DOI] [PubMed] [Google Scholar]
- 76. Liu W., Song N., Yao H., Zhao L., Liu H., Li G.. miR-221 and miR-222 simultaneously target RECK and regulate growth and invasion of gastric cancer cells. Med. Sci. Monit. 2015; 21:2718–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu X., Yu J., Jiang L., Wang A., Shi F., Ye H., Zhou X.. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics. 2009; 6:131–139. [PMC free article] [PubMed] [Google Scholar]
- 78. Qin B., Cao Y., Yang H., Xiao B., Lu Z.. MicroRNA-221/222 regulate ox-LDL-induced endothelial apoptosis via Ets-1/p21 inhibition. Mol. Cell. Biochem. 2015; 405:115–124. [DOI] [PubMed] [Google Scholar]
- 79. Wang L., Liu C., Li C., Xue J., Zhao S., Zhan P., Lin Y., Zhang P., Jiang A., Chen W.. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene. 2015; 572:252–258. [DOI] [PubMed] [Google Scholar]
- 80. Yang F., Wang W., Zhou C., Xi W., Yuan L., Chen X., Li Y., Yang A., Zhang J., Wang T.. MiR-221/222 promote human glioma cell invasion and angiogenesis by targeting TIMP2. Tumour Biol. 2015; 36:3763–3773. [DOI] [PubMed] [Google Scholar]
- 81. Yang X., Yang Y., Gan R., Zhao L., Li W., Zhou H., Wang X., Lu J., Meng Q.H.. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One. 2014; 9:e98833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang Y., Zhao X., Li H.X.. MiR-221 and miR-222 simultaneously target ARID1A and enhance proliferation and invasion of cervical cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016; 20:1509–1515. [PubMed] [Google Scholar]
- 83. Zhang C.Z., Zhang J.X., Zhang A.L., Shi Z.D., Han L., Jia Z.F., Yang W.D., Wang G.X., Jiang T., You Y.P. et al. . MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer. 2010; 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang J., Han L., Ge Y., Zhou X., Zhang A., Zhang C., Zhong Y., You Y., Pu P., Kang C.. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int. J. Oncol. 2010; 36:913–920. [DOI] [PubMed] [Google Scholar]
- 85. Zhao X., Wang P., Liu J., Zheng J., Liu Y., Chen J., Xue Y.. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol. Ther. 2015; 23:1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhou Z., Zhou L., Jiang F., Zeng B., Wei C., Zhao W., Yu D.. Downregulation of miR-222 induces apoptosis and cellular migration in adenoid cystic carcinoma cells. Oncol. Res. 2017; 25:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li N., Yu N., Wang J., Xi H., Lu W., Xu H., Deng M., Zheng G., Liu H.. miR-222/VGLL4/YAP-TEAD1 regulatory loop promotes proliferation and invasion of gastric cancer cells. Am. J. Cancer Res. 2015; 5:1158–1168. [PMC free article] [PubMed] [Google Scholar]
- 88. Li Q., Shen K., Zhao Y., He X., Ma C., Wang L., Wang B., Liu J., Ma J.. MicroRNA-222 promotes tumorigenesis via targeting DKK2 and activating the Wnt/beta-catenin signaling pathway. FEBS Lett. 2013; 587:1742–1748. [DOI] [PubMed] [Google Scholar]
- 89. Li Y., Liang C., Ma H., Zhao Q., Lu Y., Xiang Z., Li L., Qin J., Chen Y., Cho W.C. et al. . miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014; 19:7122–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu B., Che Q., Qiu H., Bao W., Chen X., Lu W., Li B., Wan X.. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERalpha. PLoS One. 2014; 9:e87563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu X., Cheng Y., Zhang S., Lin Y., Yang J., Zhang C.. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009; 104:476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sun C., Li N., Zhou B., Yang Z., Ding D., Weng D., Meng L., Wang S., Zhou J., Ma D. et al. . miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27kip1. Oncol. Lett. 2013; 6:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang Y.F., Wang F., Xiao J.J., Song Y., Zhao Y.Y., Cao Y., Bei Y.H., Yang C.Q.. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int. J. Clin. Exp. Med. 2014; 7:893–902. [PMC free article] [PubMed] [Google Scholar]
- 94. Yu B., Zhou S., Wang Y., Qian T., Ding G., Ding F., Gu X.. miR-221 and miR-222 promote Schwann cell proliferation and migration by targeting LASS2 after sciatic nerve injury. J. Cell Sci. 2012; 125:2675–2683. [DOI] [PubMed] [Google Scholar]
- 95. Zhao Y., Wang Y., Yang Y., Liu J., Song Y., Cao Y., Chen X., Yang W., Wang F., Gao J. et al. . MicroRNA-222 controls human pancreatic cancer cell line capan-2 proliferation by P57 targeting. J. Cancer. 2015; 6:1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang C., Zhang J., Hao J., Shi Z., Wang Y., Han L., Yu S., You Y., Jiang T., Wang J. et al. . High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012; 10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Y., Yao J., Huan L., Lian J., Bao C., Li Y., Ge C., Li J., Yao M., Liang L. et al. . GNAI3 inhibits tumor cell migration and invasion and is post-transcriptionally regulated by miR-222 in hepatocellular carcinoma. Cancer Lett. 2015; 356:978–984. [DOI] [PubMed] [Google Scholar]
- 98. Chung H.K., Chen Y., Rao J.N., Liu L., Xiao L., Turner D.J., Yang P., Gorospe M., Wang J.Y.. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including frizzled-7. Mol. Med. 2015; doi:10.2119/molmed.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dai R., Li J., Liu Y., Yan D., Chen S., Duan C., Liu X., He T., Li H.. miR-221/222 suppression protects against endoplasmic reticulum stress-induced apoptosis via p27(Kip1)- and MEK/ERK-mediated cell cycle regulation. Biol. Chem. 2010; 391:791–801. [DOI] [PubMed] [Google Scholar]
- 100. Ihle M.A., Trautmann M., Kuenstlinger H., Huss S., Heydt C., Fassunke J., Wardelmann E., Bauer S., Schildhaus H.U., Buettner R. et al. . miRNA-221 and miRNA-222 induce apoptosis via the KIT/AKT signalling pathway in gastrointestinal stromal tumours. Mol. Oncol. 2015; 9:1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen B., Zhang B., Luo H., Yuan J., Skogerbo G., Chen R.. Distinct microRNA subcellular size and expression patterns in human cancer cells. Int. J. Cell Biol. 2012; 2012:672462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jeffries C.D., Fried H.M., Perkins D.O.. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011; 17:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Park C.W., Zeng Y., Zhang X., Subramanian S., Steer C.J.. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010; 7:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wong J.J., Ritchie W., Gao D., Lau K.A., Gonzalez M., Choudhary A., Taft R.J., Rasko J.E., Holst J.. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J. Hematol. Oncol. 2014; 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kalantari R., Hicks J.A., Li L., Gagnon K.T., Sridhara V., Lemoff A., Mirzaei H., Corey D.R.. Stable association of RNAi machinery is conserved between the cytoplasm and nucleus of human cells. RNA. 2016; 22:1085–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Huang V., Li L.C.. Demystifying the nuclear function of Argonaute proteins. RNA Biol. 2014; 11:18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ross J.P., Kassir Z.. The varied roles of nuclear argonaute-small RNA complexes and avenues for therapy. Mol. Ther. Nucleic Acids. 2014; 3:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schraivogel D., Meister G.. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014; 39:420–431. [DOI] [PubMed] [Google Scholar]
- 109. Schalk C., Cognat V., Graindorge S., Vincent T., Voinnet O., Molinier J.. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2965–E2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y.. A role for small RNAs in DNA double-strand break repair. Cell. 2012; 149:101–112. [DOI] [PubMed] [Google Scholar]
- 111. Couvillion M.T., Bounova G., Purdom E., Speed T.P., Collins K.. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell. 2012; 48:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Carissimi C., Laudadio I., Cipolletta E., Gioiosa S., Mihailovich M., Bonaldi T., Macino G., Fulci V.. ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human transcription start sites. Nucleic Acids Res. 2015; 43:1498–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zamudio J.R., Kelly T.J., Sharp P.A.. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014; 156:920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.