Abstract
Codon usage biases are found in all eukaryotic and prokaryotic genomes and have been proposed to regulate different aspects of translation process. Codon optimality has been shown to regulate translation elongation speed in fungal systems, but its effect on translation elongation speed in animal systems is not clear. In this study, we used a Drosophila cell-free translation system to directly compare the velocity of mRNA translation elongation. Our results demonstrate that optimal synonymous codons speed up translation elongation while non-optimal codons slow down translation. In addition, codon usage regulates ribosome movement and stalling on mRNA during translation. Finally, we show that codon usage affects protein structure and function in vitro and in Drosophila cells. Together, these results suggest that the effect of codon usage on translation elongation speed is a conserved mechanism from fungi to animals that can affect protein folding in eukaryotic organisms.
INTRODUCTION
Due to the redundancy of triplet genetic codons, most amino acids can be encoded by two to six synonymous codons. Synonymous codons are not used with equal frequencies, a phenomenon called codon usage bias that is observed in all organisms (1–4). Selection for efficient and accurate translation was proposed to be a major cause of codon usage bias (5–8). On the other hand, studies using protein expression in Escherichia coli suggested that translation rate and synonymous codon usage can affect protein folding and functions (9–15). Strongly supporting this hypothesis, we previously demonstrated that the codon usage of the Neurospora circadian clock gene, frequency (frq), is critical for the structure and function of FREQUENCY (FRQ) protein in vivo (8). More recently, we showed that the codon usage plays a similar role for the Drosophila period gene in vivo (16). Also importantly, these in vivo studies showed that codon usage influences protein structures in a location-specific manner. Consistent with this conclusion, bioinformatic analyses have revealed correlations between codon usage and protein structural motifs in different organisms (17–21). Together, these studies led to the hypothesis that there is a codon usage ‘code’ within genetic codons that regulates translation elongation speed to permit optimal co-translational protein folding.
Most of these proposed roles of codon usage are based on its effect on translation elongation speed. However, earlier studies addressing this issue based on protein overexpression in E. coli and indirect measurement of ribosome movement led to conflicting conclusions (22–25). Moreover, ribosome profiling, a powerful method that uses deep sequencing of the ribosome-protected fragments (RPF), initially found no correlations between codon usage and levels of RPF in different organisms (25–29). Ribosome profiling results are now known to be influenced by experimental conditions, sequencing depth, cloning/sequencing biases and the bioinformatic methods used (30–34). Furthermore, ribosome profiling relies on precise enzymatic 5′ end cleavage of the RPF to allow accurate A site determination, which are often difficult due to digestion biases and different experimental conditions (35). These results suggest that although ribosome profiling has codon-level resolution, it might not have codon-level sensitivity to detect the effect of codon usage on translation elongation speed.
It should be noted that although the implicated role of codon usage in regulating protein expression levels led to the hypothesis that codon usage impacts protein expression levels by affecting translation efficiency (36–39), a role for codon usage in translation elongation speed does not necessarily affect translation efficiency. In fact, recent studies suggest that translation efficiency is mainly determined by the efficiency of translation initiation, a process that is mostly affected by RNA structure but not codon usage near the start codon (40–42). In the current study, we only focus on the role of codon usage in regulating translation elongation speed.
To determine the effect of codon usage on translation elongation speed, we previously used Neurospora and yeast in vitro translation systems to directly monitor the speed of protein translation elongation (43). Our results demonstrate that codon usage plays an important role in regulating translation elongation speed on mRNA in vitro and in vivo in these fungal systems. However, the effect of codon usage on translation elongation rate in animal systems is still not known. Although single synonymous SNPs in human genes have been shown to associate with altered protein conformation and function (44,45), there is no clear evidence showing that these SNPs regulate translation elongation. In addition, although codon manipulation has also been shown to affect KRas expression and oncogenesis in mice, the role of codon usage in regulating gene expression in mammalian cells can also be explained by difference in GC contents in genes (46–49). More recently, we demonstrated that the codon usage of Drosophila Period gene is important for protein structure and function in vivo (16). These results raise the possibility that codon usage is a conserved mechanism from fungi to animals that influences translation elongation speed.
Similar to mammals and Neurospora, the Drosophila melanogaster genome has a strong codon bias for G/C at the wobble positions (5,8,50–52). Changing codon usage of Drosophila genes is known to alter protein expression levels (16,53) but the mechanism of such an effect is not clear. In addition, codon optimality was also suggested to play a potential role in splicing regulation in Drosophila (54). Previous ribosome profiling using Drosophila cells could not allow robust A site assignment due to enzyme cleavage biases (35). In this study, we used Drosophila in vitro translation system to determine the effect of codon usage on translation elongation. Our results demonstrate that codon usage affects translation elongation speed and local ribosome movement on mRNA. In addition, we show that codon usage influences protein structure and activity in vitro and in Drosophila cells. Together, our results suggest that the effect of codon usage on translation elongation is a conserved mechanism regulating protein structure and function from fungi to animals.
MATERIALS AND METHODS
Cell culture and preparation of S2 cell-free translation extract
The preparation of translation extract was modified from a protocol previously described (55). Drosophila Schneider 2(S2) cells (kindly provided by Dr Jin Jiang) were cultured in Schneider's Drosophila Medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Sigma) and 1% penicillin-streptomycin (10 000 units penicillin and 10 mg streptomycin/ml, Sigma) at 27°C. After reaching confluence, S2 were harvested by centrifugation at 1000 × g for 4 min and washed by 1 × phosphate bufferedsaline for three times. Cell pellets were resuspended in 2× volumes of hypotonic buffer (10 mM HEPES-KOH pH 7.6, 10 mM potassium acetate, 0.5 mM magnesium acetate, 5 mM Dithiothreitol) and incubated on ice for 40 min to 1 h. Cells were then homogenized in a Dounce homogenizer by 20–30 strokes on ice and the final concentration of potassium acetate was adjusted to 50 mM. The cell extract was centrifuged at 16 000 × g for 10 min at 4°C. The supernatant was aliquoted, snap frozen in liquid nitrogen and stored at −80°C before use.
Plasmid construction
OPT and dOPT luciferase constructs were designed based on the Drosophila codon usage table (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=7227). Except for the first 10 codons, all other codons are either the most preferred or the least preferred codons in the OPT or dOPT construct, respectively. Gene synthesis was carried out by Genescript and the resulting genes were cloned into pJI 204 vector containing a T7 promoter and 30-nt poly-A sequence (56). The pJI 204 vector was modified to include a NheI site following the poly-A sequence to enable plasmid linearization for in vitro transcription and a 5 c-Myc tag at the N terminus to allow detection of nascent peptides. N-OPT, M-OPT and C-OPT constructs were created by replacing the N-terminal part (codon 11–223, count from the first ATG of luciferase gene), middle part (codon 224–423) and C-terminal part (codon 424–550) of dOPT sequence with the corresponding optimized sequence, respectively. Constructs for S2 cell expression were created by cloning different versions of luciferase sequences into pUAST vector using NotI/XbaI sites. The pUAST vector and an ubiquitin-Gal4 construct for co-transfection were kindly provided by Dr Jin Jiang. The sequence information of different luciferase genes used in this study are provided in the supplemental information.
In vitro transcription assay
To obtain mRNA for in vitro translation, the plasmids were linearized by NheI and then transcribed using HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB). 3′-0-Me-m7G (5′)ppp (5′)G anti-reverse cap structure analog (NEB) was added following the manufacturer's instruction. The mRNA products were subsequently purified by LiCl precipitation, quantified by Nanodrop (Thermo Scientific), aliquoted and stored at −80°C before use. Denaturing agarose gel electrophoresis was used to determine the quality of the mRNA products.
Cell-free translation assay
To monitor the kinetics of luciferase activity in real time, a reaction mixture (20 mM HEPES-KOH pH 7.6, 0.5 mM spermidine, 8 mM creatine phosphate, 0.2 mM GTP, 1 mM ATP, 20 μM complete amino acids [Promega], 100 mM potassium acetate, 1 mM magnesium acetate, 0.13U/μl creatine phosphate kinase, 50 μM luciferin, 0.2U/μl SUPERase•In RNase Inhibitor [Invitrogen], 3 μl total volume per reaction) was prepared and aliquoted into 96-well plate. A total of 1 μl of the mRNA template (∼60–180 ng) and 8 μl cell-free translation extract were successively added to the reaction mixture. Afterward, the luminescence signal of the reaction was recorded every 30 s using FLUOstar Optima (BMG) at 26°C. The concentrations of potassium acetate and magnesium acetate in the reaction mixture were optimized for different batches of cell-free translation extracts.
To examine the protein products of the cell-free translation assays, the reactions were incubated in a 26°C water bath and stopped at the indicated time points by adding sodium dodecyl sulphate (SDS) sample buffer, followed immediately by heating at 90°C. The samples were subsequently denatured and analyzed by western blot.
For the harringtonine chase experiment, 20 ng/ml harringtonine was added to in vitro translation assay after 6 min of reaction to inhibit translation initiation. Afterward, the reaction mixture was withdrawn at the indicated time points and subjected to western blot analysis.
Isolation of ribosome-associate nascent chains
The method was modified from previously described (57). In vitro translation reaction was terminated at 15 min by adding cycloheximide to a final concentration of 0.5 mg/ml. Translation products are carefully layered on top of sucrose cushion (0.5M sucrose, 25 mM HEPES, pH 7.5, 80 mM KOAc, 1 mM Mg(OAc)2) and then centrifuged in a TL 100.3 rotor (Beckman Coulter) at 100 000 rpm for 5 min at 4°C. The ribosomal pellets are washed once with 25 mM HEPES (pH 7.5), 80 mM KOAc and 1 mM Mg(OAc)2 and then resuspended in 1× SDS sample buffer for western blot analysis. The supernatant is concentrated and mixed with SDS sample buffer to the same volume as pellet samples for western blot analysis.
S2 cell transfection
S2 cell transfection was carried out following the standard calcium phosphate transfection protocol in six-well plates. A total of 1.5 μg luciferase construct and 1.5 μg ubiquitin-Gal4 construct were co-transfected for each transfection. Cells were harvested 48–60 h afterward and lysed in 1× Passive Lysis Buffer (Promega) according to the manufacturer's instruction. Cell lysate was obtained by centrifugation at top speed for 1 min at 4°C and stored at −80°C.
To determine Luc mRNA stability, 10 μg/ml actinomycin D was added to stop transcription and cell samples were collected at indicated time points for RNA purification. Purified RNA samples were examined by northern blot analysis to determine Luc mRNA levels.
Limited trypsin digestion assay
For trypsin digestion of in vitro translated protein products, 0.5 mg/ml cycloheximide was added to terminate reaction after 20 min of in vitro translation. A total of 5 μg/ml trypsin was then added to the reaction and samples were withdrawn at indicated time points for western blot analysis. For trypsin digestion of S2 cell extract, total cell extract was used. Total protein concentrations of each sample was adjusted to 1 μg/μl. The trypsin digestion was carried out at 26°C by adding 300 ng/ml trypsin. Digestion products were withdrawn from the reaction at indicated time points and the denatured protein samples were analyzed by western blot.
RESULTS
Optimal codons speed up translation elongation while non-optimal codons slow it down in Drosophila cell-free system
To determine the role of codon usage on translation elongation speed, we established a cell-free translation system using Drosophila S2 cells to compare translation speeds of firefly luciferase (Luc) mRNAs with different codon preferences. All Luc mRNAs have identical 5′ and 3′ untranslated regions (UTR) and the same poly-A length. The first ten codons of the Luc open reading frame were also identical in these mRNAs to ensure the same translation initiation context. The use of the cell-free system allows the separation of transcription and translation processes so that translation can be synchronized. Because luciferase is known to be rapidly folded (within a few seconds) after translation (58), the time when luminescence signal is first detected after start of translation (time of first appearance (TFA)) should reflect the speed of translation process. Because different Luc mRNAs have the same translation initiation context, the difference in TFA values should mainly reflect the difference in translation elongation speed of different Luc mRNAs.
We first generated two Luc mRNAs by in vitro transcription: OPT (the entire Luc ORF codon optimized according to the Drosophila codon usage table) and dOPT (the least preferred codon is used for every amino acid). Afterward, they were separately translated in the S2 translation extracts supplemented with luciferin and luciferase activity was monitored in real-time. As shown in Figure 1A and B, the TFA of the OPT mRNA was ∼4 min earlier than that of the dOPT mRNA. To confirm this result, we performed Western blot analysis using a luciferase antibody to detect the production of full-length luciferase protein at different time points (Figure 1C and Supplementary Figure S1). As expected, the appearance of full-length luciferase for the OPT mRNA was also about 4 min earlier than that of the dOPT mRNA. It should be noted that the cell-free translation extract contained endogenous mRNA from S2 cells and could compete with luciferase mRNA for translation. By determining the total level of mRNA, we estimated that the luciferase mRNA added in the reaction is about 1–2% of total mRNA.
Figure 1.
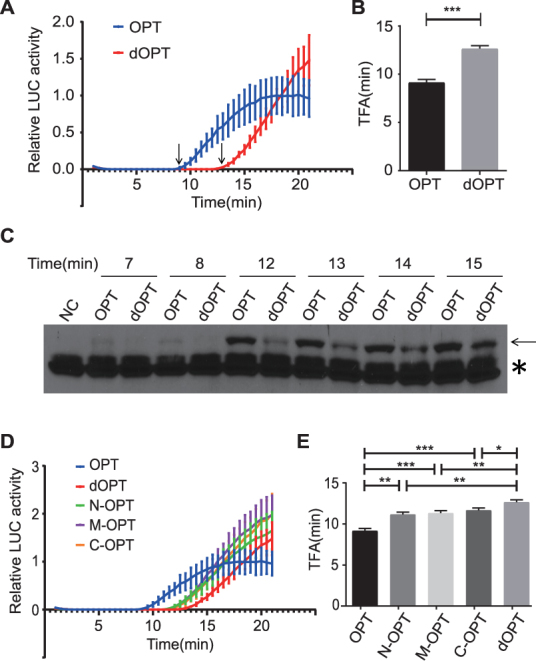
Codon usage affects translation elongation rate in S2 cell-free extract. (A) Real-time measurement of firefly luciferase activity using S2 cell-free translation system and the indicated Luc mRNA templates. TFA values are indicated by arrows. Luciferase signals were normalized to the signal level of the OPT mRNA at 21 min. Error bars represent standard deviations of three replicates. (B) Quantification of the TFA values of OPT and dOPT Luc mRNAs. ***P < 0.001. (C) Western blot analysis using luciferase antibody showing the production of the full length luciferase (indicated by the arrow) in the in vitro translation assays. The asterisk indicates the non-specific bands recognized by the luciferase antibody. NC: Negative Control. (D) Real-time measurement of firefly luciferase activity using the S2 cell-free translation system and the indicated Luc mRNA templates. mRNA concentrations are adjusted to produce similar activity levels. Error bars represent standard deviations of three replicates. (E) Quantification of the TFA values of the indicated Luc mRNAs. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether the effect observed is due to accumulative effect of codon usage along the ORF and not due to change of a specific mRNA structure, we generated three additional Luc mRNAs based on the dOPT mRNA. In the N-OPT, M-OPT and C-OPT mRNAs, the N-terminal region (codons 11–223), middle region (codons 224–423) and C-terminal region (codons 424–550) of the luciferase ORF was replaced by preferred codons, respectively. If the effect we observed above is due to change of mRNA structure at a specific location, one of these Luc mRNAs should have the same TFA as the OPT mRNA and the other two Luc mRNAs should be similar to the dOPT mRNA. Instead, we found that the TFA values of the N-OPT, M-OPT and C-OPT mRNAs were all significantly higher than that of the OPT mRNA but lower than that of the dOPT mRNA (Figure 1D and E). These results suggest that codon usage regulates the speed of translation elongation. In addition, the effect of codon usage on elongation rate is due to cumulative effects of codon usage in the coding sequence rather than changes in specific RNA structure.
Codon usage influences local ribosome stalling on mRNA
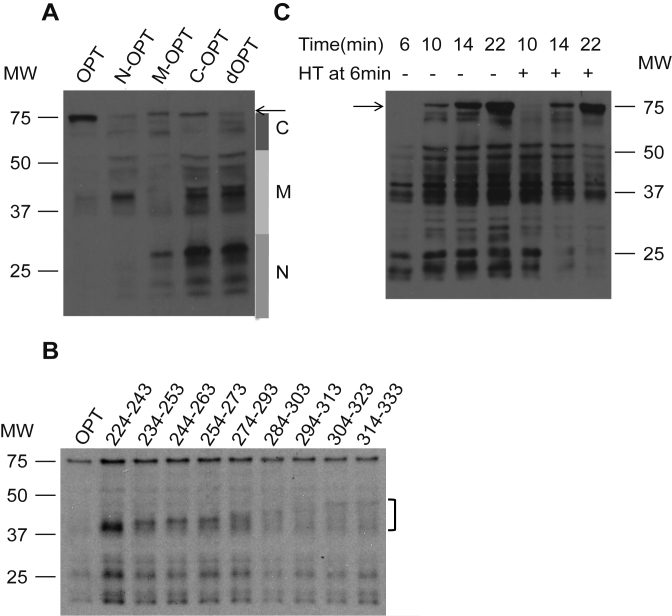
To further determine the effect of codon usage on ribosome movement on mRNA, we generated Luc mRNAs in which the luciferase protein is tagged by five consecutive c-Myc tags at the N-terminus so that nascent luciferase peptides can be detected by Western blot analysis using a c-Myc monoclonal antibody. After 15 min of translation in the S2 system, the production of luciferase protein from different mRNAs was examined by western blot analysis. Translation of the OPT and dOPT mRNAs resulted in dramatically different profiles of nascent luciferase peptides (Figure 2A). For the dOPT mRNA, most of the peptides seen are intermediate LUC species with a low level of the full-length protein. In contrast, translation of OPT mRNA resulted in mostly the full-length protein and much less intermediate LUC species. Remarkably, when N-OPT, M-OPT and C-OPT mRNAs, which each has a different region (codon 11–223, 224–423 or 424–550) of Luc codon-optimized in the dOPT background, are individually translated, we observed specific disappearance or reduction of the protein intermediate species within the expected molecular weight ranges. Furthermore, when the codons between 224–333 in the OPT luciferase gene were replaced by the least preferred codons in a series of 20 codon-windows, luciferase intermediates corresponding to the predicted molecular sizes appeared in the in vitro translated luciferase products (Figure 2B). Together, these results further demonstrate the role of codon usage in regulating elongation speed and show that codon usage regulates local ribosome movement on mRNA. Preferred codons speed up ribosome movement on mRNA and non-optimal codons cause ribosome stalling on mRNA.
Figure 2.
Non-optimal codon usage results in local ribosome stalling on mRNA. (A) Western blot analysis of denatured protein samples using c-Myc antibody showing the production of full-length and nascent luciferase peptides from the S2 cell-free extract translated Luc mRNAs. A five c-Myc tag was added to the N-terminal end of the Luc ORF for all constructs so that all nascent peptides could be detected by c-Myc antibody. The bars on the right represent approximate location of the N-terminal, middle part and C-terminal regions of LUC that were codon optimized, respectively. The arrow indicates the full-length LUC protein. The loadings for different samples were adjusted to produce similar full-length luciferase protein levels. (B) Western blot analysis showing the accumulation of nascent luciferase peptides at the locations of codon de-optimization of the Myc-OPT mRNA. The numbers on the top indicate the 20-amino-acid windows in which luciferase codons were de-optimized. (C) Western blot analysis showing the profiles of luciferase nascent peptides at the indicated time points with/without harringtonine treatment. For samples treated by harringtonine, the drug was added after six min of in vitro translation reaction.
To determine the fate of stalled ribosomes on mRNA, we added harringtonine, a translation initiation inhibitor, 6 min after the start of translation of the OPT mRNA and the translation of luciferase was monitored at different time points by western blot analysis. At 6 min of translation, only luciferase intermediates were observed (Figure 2C). After the addition of harringtonine, the luciferase intermediates gradually disappeared or reduced and the full-length luciferase appeared. In addition, by isolation of ribosome-associate nascent chains, we found that in contrast to the full-length protein, these low molecular weight luciferase species were mostly associated with the ribosomes (Supplementary Figure S2). Furthermore, these species are not degradation products of the full-length protein since the in vitro translated luciferase protein species were very stable (Supplementary Figure S3). Together, these results indicate that the low molecular weight luciferase species are nascent peptides associated with paused ribosomes that can continue to translate after temporary pausing on mRNA.
Codon usage affects protein structure in vitro and in S2 cells
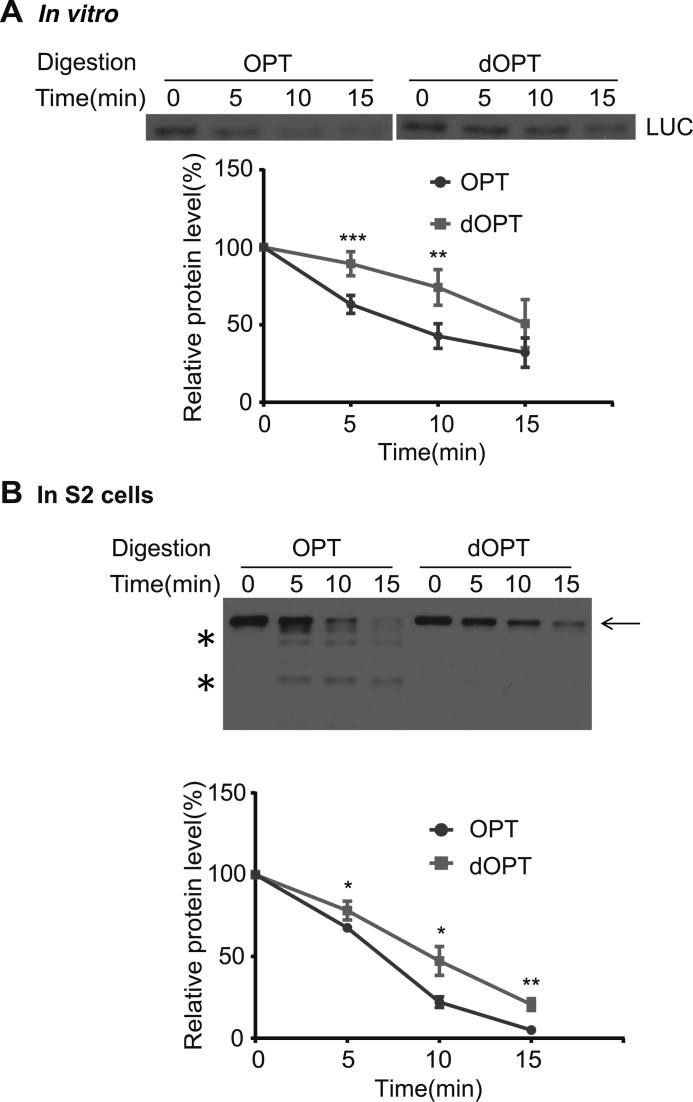
Since protein folds co-translationally, we predicted that the effect of codon usage on translation elongation speed should affect the time available for co-translational folding process, thus affecting protein structure. To test this hypothesis, we used limited trypsin digestion assay to probe the structural difference of the full-length luciferase proteins produced from OPT and dOPT Luc mRNAs by in vitro translation. As shown in Figure 3A, the OPT luciferase protein was found to be significantly more sensitive to trypsin digestion than the dOPT protein. To further confirm this conclusion in vivo, OPT and dOPT luciferase expressing constructs were individually transfected into S2 cells and we examined the trypsin sensitivity of the expressed luciferase in the S2 total cell extract. As expected, the S2 cell-expressed full-length OPT luciferase was found to be more sensitive to trypsin digestion than the dOPT protein (Figure 3B). It is worth noting that no luciferase intermediates could be observed for the S2 expressed luciferase, likely due to their rapid degradation in cells. Importantly, the trypsin digestion of the full-length OPT luciferase resulted in luciferase fragments that could not be detected for the digestion of dOPT protein (Figure 3B). Together, these in vitro and in vivo results indicate that codon usage affects protein structure.
Figure 3.
Codon usage affects trypsin sensitivity of luciferase protein produced in vitro and in S2 cells. (A) Limited trypsin digestion assay for the full-length OPT and dOPT luciferase proteins translated by the S2 cell-free system for 20 min. A representative western blot was shown in the upper panel. The lower panel shows the densitometric analysis results of LUC levels from independent experiments. **P < 0.01, ***P <0.001. n = 5 for OPT and n = 4 for dOPT mRNAs. Area under curve (AUCs) for OPT and dOPT samples are 860.1 ± 42 and 1195 ± 62.64, respectively. (B) Limited trypsin digestion assay for luciferase protein produced in S2 cells after transfecting the OPT or dOPT expressing construct. A representative western blot was shown in the upper panel. The lower panel shows the densitometric analysis results of LUC levels from three independent experiments. *P < 0.05, **P < 0.01. AUCs for OPT and dOPT samples are 710.7 ± 13.73 and 927.8 ± 38.47, respectively.
Codon usage affects protein activity and mRNA stability in cells
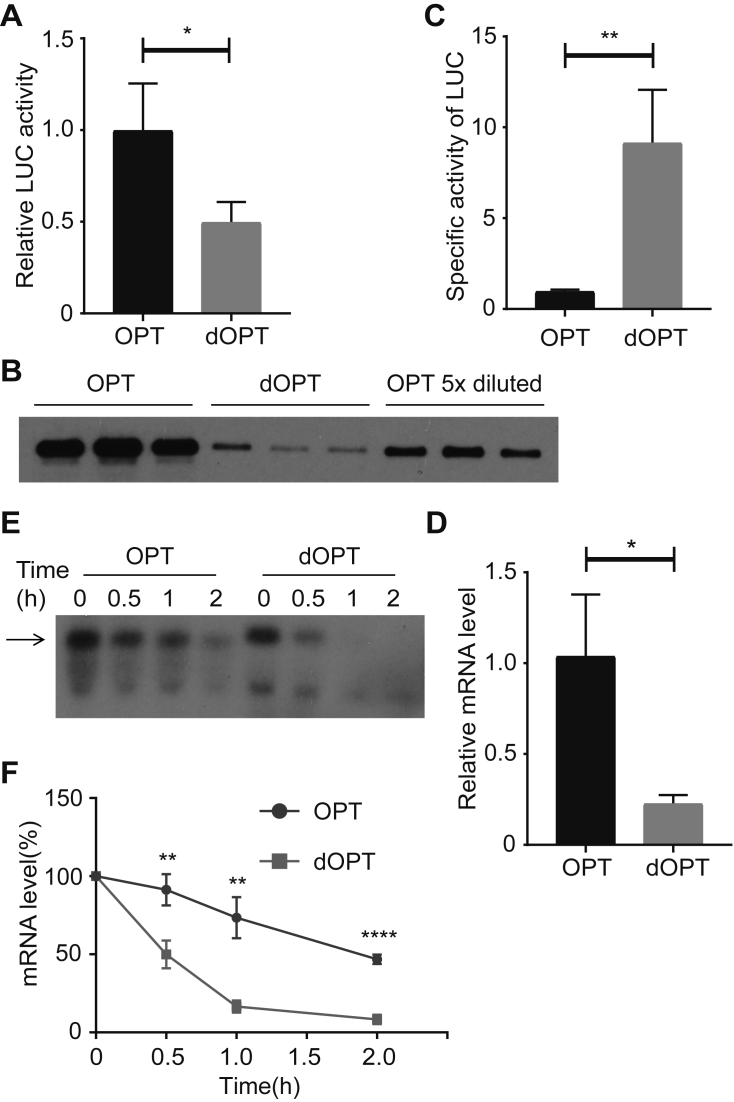
The altered luciferase structure shown above suggests that codon usage may affect luciferase activity in cells. To confirm this, we transfected OPT and dOPT Luc constructs into S2 cells and measured luciferase activity and luciferase protein levels. As shown in Figure 4A and B, although the luciferase activity from cells expressing dOPT Luc was about 50% of the OPT Luc, the luciferase protein level from the OPT Luc was more than 20-fold higher than that of dOPT Luc. Thus, the specific activity of dOPT luciferase is about 10-fold of that of OPT luciferase (Figure 4C). Thus, codon usage of luciferase mRNA greatly affects luciferase activity in Drosophila cells.
Figure 4.
Codon usage affects luciferase protein activity and mRNA stability in S2 cells. (A) Relative luciferase activity of S2 cells transfected with OPT and dOPT constructs. Same amount of protein extracts was used for each sample. Data were obtained from three independent experiments. (B) Western blot analysis showing the levels of the full-length luciferase in the OPT or dOPT transfected S2 cells from three replicates. (C) Specific activity of OPT and dOPT luciferase proteins calculated by dividing luciferase activity level by corresponding protein level. **P < 0.01. The signals from the OPT 5×diluted samples in (B) were used to determine the levels of the OPT LUC protein. (D) qRT-PCR analysis showing the relative levels of Luc mRNA from the OPT or dOPT transfected S2 cells. Error bars represent standard deviations from three independent replicates. *P < 0.05. (E) Representative northern blot analysis result showing the level of Luc mRNAs at the indicated time points after the addition of actinomycin D. The arrow indicates the full-length luciferase mRNA. (F) Quantification of three independent results of the experiment in (E) was shown. **P < 0.01. ****P < 0.0001. AUCs for OPT and dOPT samples are 149.1 ± 8.306 and 66.5 ± 4.128, respectively.
In addition to the difference in protein levels, the level of OPT Luc mRNA was much higher than that of the dOPT mRNA (Figure 4D), which largely explains the low level of luciferase protein in cells expressing dOPT construct. To determine whether the difference in mRNA levels is due to altered mRNA stability, we measured mRNA degradation rate after the addition of actinomycin D, a transcription inhibitor. As expected, the OPT Luc mRNA was significantly more stable than the dOPT mRNA after adding actinomycin D (Figure 4E and F). Therefore, similar to the reports in other organisms (59–63), codon usage also affects mRNA stability.
DISCUSSION
Although codon usage has been previously shown to regulate co-translational protein folding in fungi by affecting translation elongation speed, its effect in animal systems is not known. By using a cell-free Drosophila S2 translation system, we demonstrated here that codon usage plays an important role in regulating the speed of translation elongation: optimal codons enhance the rate of elongation while non-optimal codons slows it down (Figure 1). In addition, as shown by the accumulation of nascent intermediate peptides during translation (Figure 2), we showed that ribosome stalling occurs at the sites of non-optimal codons, indicating that codon usage regulates local ribosome movement speed on mRNA. Furthermore, we showed in vitro and in S2 cells that different codon usage biases led to structural differences of proteins with the same amino acid sequence, resulting in proteins with different functional activity (Figures 3–4). Together with our previous study showing the importance of codon usage for the function of a circadian clock protein in Drosophila (16), these results demonstrate that the role of codon usage in regulating protein structure and function by affecting translation elongation speed is conserved in fungi and Drosophila.
Bioinformatic analyses have uncovered correlations between codon usage and protein structural motifs in different fungal organisms (17–21). In addition, we recently showed that non-optimal codons are preferentially used in intrinsically disordered regions, and optimal codons are more frequently used in structured domains in yeast, Neurospora, Caenorhabditis elegans and Drosophila systems (20). Importantly, the functional importance of such correlations in vivo was confirmed by structure-based codon manipulation in the Neurospora and Drosophila circadian clock genes (16,20). Together with our current study, these studies suggest the existence of a codon usage code within genetic codons that generates rhythms of translation elongation rate optimal for protein structure and function in eukaryotic organisms.
Recently, codon optimality has been shown to affect mRNA stability due to its role in affecting translation elongation in budding yeast (59,63). A similar role for codon usage was also proposed in zebrafish and Xenopus systems (61,62). Consistent with these studies, we showed that codon usage manipulation of the luciferase gene altered Luc mRNA stability in Drosophila cells (Figure 4E and F). Therefore, codon usage may also affect mRNA stability due to its role in regulating translation elongation rate in animals.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Jin Jiang, Shuang Li, Youxing Jiang and Rolf A. Brekken and members of our laboratory for technical assistance and discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R35 GM118118]; Cancer Prevention and Research Institute of Texas [RP160268]; Welch Foundation [I-1560 to Y.L.]. Funding for open access charge: National Institutes of Health [R35 GM118118].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985; 2:13–34. [DOI] [PubMed] [Google Scholar]
- 2. Sharp P.M., Tuohy T.M., Mosurski K.R.. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986; 14:5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Comeron J.M. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004; 167:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plotkin J.B., Kudla G.. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011; 12:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994; 136:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drummond D.A., Wilke C.O.. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008; 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu Y., Ma P., Shah P., Rokas A., Liu Y., Johnson C.H.. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature. 2013; 495:116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J.M., Sachs M.S., Liu Y.. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013; 495:111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Komar A.A., Lesnik T., Reiss C.. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999; 462:387–391. [DOI] [PubMed] [Google Scholar]
- 10. Spencer P.S., Siller E., Anderson J.F., Barral J.M.. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012; 422:328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang G., Hubalewska M., Ignatova Z.. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009; 16:274–280. [DOI] [PubMed] [Google Scholar]
- 12. Siller E., DeZwaan D.C., Anderson J.F., Freeman B.C., Barral J.M.. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol. 2010; 396:1310–1318. [DOI] [PubMed] [Google Scholar]
- 13. Sander I.M., Chaney J.L., Clark P.L.. Expanding Anfinsen's principle: contributions of synonymous codon selection to rational protein design. J. Am. Chem. Soc. 2014; 136:858–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buhr F., Jha S., Thommen M., Mittelstaet J., Kutz F., Schwalbe H., Rodnina M.V., Komar A.A.. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell. 2016; 61:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Purvis I.J., Bettany A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J.. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol. 1987; 193:413–417. [DOI] [PubMed] [Google Scholar]
- 16. Fu J., Murphy K.A., Zhou M., Li Y.H., Lam V.H., Tabuloc C.A., Chiu J.C., Liu Y.. Codon usage affects the structure and function of the Drosophila circadian clock protein PERIOD. Genes Dev. 2016; 30:1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pechmann S., Chartron J.W., Frydman J.. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014; 21:1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pechmann S., Frydman J.. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013; 20:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou T., Weems M., Wilke C.O.. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 2009; 26:1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou M., Wang T., Fu J., Xiao G., Liu Y.. Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol. Microbiol. 2015; 97:974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warnecke T., Hurst L.D.. GroEL dependency affects codon usage–support for a critical role of misfolding in gene evolution. Mol. Syst. Biol. 2010; 6:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sorensen M.A., Kurland C.G., Pedersen S.. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989; 207:365–377. [DOI] [PubMed] [Google Scholar]
- 23. Bonekamp F., Dalboge H., Christensen T., Jensen K.F.. Translation rates of individual codons are not correlated with tRNA abundances or with frequencies of utilization in Escherichia coli. J. Bacteriol. 1989; 171:5812–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chevance F.F., Le Guyon S., Hughes K.T.. The effects of codon context on in vivo translation speed. PLoS Genet. 2014; 10:e1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charneski C.A., Hurst L.D.. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013; 11:e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qian W., Yang J.R., Pearson N.M., Maclean C., Zhang J.. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012; 8:e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Artieri C.G., Fraser H.B.. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014; 24:2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lareau L.F., Hite D.H., Hogan G.J., Brown P.O.. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014; 3:e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gardin J., Yeasmin R., Yurovsky A., Cai Y., Skiena S., Futcher B.. Measurement of average decoding rates of the 61 sense codons in vivo. Elife. 2014; 3:e03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakahigashi K., Takai Y., Shiwa Y., Wada M., Honma M., Yoshikawa H., Tomita M., Kanai A., Mori H.. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics. 2014; 15:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weinberg D.E., Shah P., Eichhorn S.W., Hussmann J.A., Plotkin J.B., Bartel D.P.. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016; 14:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hershberg R., Petrov D.A.. Selection on codon bias. Annu. Rev. Genet. 2008; 42:287–299. [DOI] [PubMed] [Google Scholar]
- 37. Quax T.E., Claassens N.J., Soll D., van der Oost J.. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015; 59:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duret L., Mouchiroud D.. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hiraoka Y., Kawamata K., Haraguchi T., Chikashige Y.. Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009; 14:499–509. [DOI] [PubMed] [Google Scholar]
- 40. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pop C., Rouskin S., Ingolia N.T., Han L., Phizicky E.M., Weissman J.S., Koller D.. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014; 10:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y.. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010; 141:344–354. [DOI] [PubMed] [Google Scholar]
- 43. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M.. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007; 315:525–528. [DOI] [PubMed] [Google Scholar]
- 45. Liu M.L., Zang T., Zhang C.L.. Direct lineage reprogramming reveals disease-specific phenotypes of motor neurons from human ALS patients. Cell Rep. 2016; 14:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lampson B.L., Pershing N.L., Prinz J.A., Lacsina J.R., Marzluff W.F., Nicchitta C.V., MacAlpine D.M., Counter C.M.. Rare codons regulate KRas oncogenesis. Curr. Biol. 2013; 23:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pershing N.L., Lampson B.L., Belsky J.A., Kaltenbrun E., MacAlpine D.M., Counter C.M.. Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Invest. 2015; 125:222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rudolph K.L., Schmitt B.M., Villar D., White R.J., Marioni J.C., Kutter C., Odom D.T.. Codon-driven translational efficiency is stable across diverse mammalian cell states. PLoS Genet. 2016; 12:e1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newman Z.R., Young J.M., Ingolia N.T., Barton G.M.. Differences in codon bias and GC content contribute to the balanced expression of TLR7 and TLR9. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1362–E1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanaya S., Yamada Y., Kinouchi M., Kudo Y., Ikemura T.. Codon usage and tRNA genes in eukaryotes: Correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001; 53:290–298. [DOI] [PubMed] [Google Scholar]
- 51. Hambuch T.M., Parsch J.. Patterns of synonymous codon usage in Drosophila melanogaster genes with sex-biased expression. Genetics. 2005; 170:1691–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heger A., Ponting C.P.. Variable strength of translational selection among 12 Drosophila species. Genetics. 2007; 177:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carlini D.B., Stephan W.. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics. 2003; 163:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Warnecke T., Hurst L.D.. Evidence for a trade-off between translational efficiency and splicing regulation in determining synonymous codon usage in Drosophila melanogaster. Mol. Biol. Evol. 2007; 24:2755–2762. [DOI] [PubMed] [Google Scholar]
- 55. Brasey A., Lopez-Lastra M., Ohlmann T., Beerens N., Berkhout B., Darlix J.L., Sonenberg N.. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003; 77:3939–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wei J., Zhang Y., Ivanov I.P., Sachs M.S.. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J. Biol. Chem. 2013; 288:9549–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCallum C.D., Do H., Johnson A.E., Frydman J.. The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. J. Cell Biol. 2000; 149:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kolb V.A., Makeyev E.V., Spirin A.S.. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994; 13:3631–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R. et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boel G., Letso R., Neely H., Price W.N., Wong K.H., Su M., Luff J.D., Valecha M., Everett J.K., Acton T.B. et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016; 529:358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mishima Y., Tomari Y.. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell. 2016; 61:874–885. [DOI] [PubMed] [Google Scholar]
- 62. Bazzini A.A., Del Viso F., Moreno-Mateos M.A., Johnstone T.G., Vejnar C.E., Qin Y., Yao J., Khokha M.K., Giraldez A.J.. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 2016; 35:2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Radhakrishnan A., Chen Y.H., Martin S., Alhusaini N., Green R., Coller J.. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016; 167:122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.