Abstract
Many animals with genetic sex determination harbor heteromorphic sex chromosomes, where the heterogametic sex has half the gene dose of the homogametic sex. This imbalance, if reflected in the abundance of transcripts or proteins, has the potential to deleteriously disrupt interactions between X-linked and autosomal loci in the heterogametic sex. Classical theory predicts that molecular mechanisms will evolve to provide dosage compensation that recovers expression levels comparable to ancestral expression prior to sex chromosome divergence. Such dosage compensating mechanisms may also, secondarily, result in balanced sex-linked gene expression between males and females. However, numerous recent studies addressing sex chromosome dosage compensation (SCDC) in a diversity of animals have yielded a surprising array of patterns concerning dosage compensation in the heterogametic sex, as well as dosage balance between sexes. These results substantially contradict longstanding theory, catalyzing both novel perspectives and new approaches in dosage compensation research. In this review, we summarize the theory, analytical approaches, and recent results concerning evolutionary patterns of SCDC in animals. We also discuss methodological challenges and discrepancies encountered in this research, which often underlie conflicting results. Finally, we discuss what outstanding questions and opportunities exist for future research on SCDC.
Keywords: sex chromosomes, dosage compensation, heterogamety
Introduction
From the book Sex Chromosomes and Sex-Linked Genes (Ohno 1967).
“During the course of evolution, an ancestor to the placental mammals must have escaped a peril resulting from the hemizygous existence of all the X-linked genes in the male by doubling the rate of product output of each X-linked gene. Once this step was accomplished, the female no longer needed two X’s in her somatic cells. Hence, the dosage compensation mechanisms by random inactivation of one or the other X evolved.
In the case of Drosophila, on the other hand, it appears that a needed increase of the rate of product output by the individual X-linked genes did not take place in their evolutional past. Thus, two alleles at each X-linked gene locus are still needed by the female. The presence of modifier genes is required primarily to raise the efficiency of individual X-linked genes in the hemizygous state as a means of minimizing a peril encountered by the male.” (p. 99)
“Although the two are rather similar in absolute size, the avian Z-chromosome makes up nearly 10% of the genome, while the original X of placental mammals comprises only 5%. It would appear that birds have an even greater need than mammals for developing an effective dosage compensation mechanism for the Z-linked genes. It is a great surprise to find that avian species apparently failed in developing an effective means for achieving the dosage compensation.” (p. 144)
Most discussions of sex chromosome dosage compensation (SCDC), including this one, start with Ohno’s “peril” of hemizygosity. Susumu Ohno was the first to concisely articulate the notion that dosage compensating mechanisms might be expected to evolve concomitantly with well-differentiated sex chromosomes (also called allosomes) (for a historical overview of the discovery and early research of SCDC, see Gartler 2014). From an historical perspective, this peril of hemizygosity was a formative concept that strongly shaped research and theory concerning SCDC for several decades (Charlesworth 1978, 1996; Vicoso and Bachtrog 2009; Mank etal. 2011). Yet even at the time, Ohno’s (1967) arguments did not easily or coherently explain known patterns of SCDC, as the above excerpts demonstrate. In recent years, genomic analyses have revealed a striking degree of variation in SCDC across taxa. These results have largely overturned Ohno’s once predominant hypothesis while also raising numerous further questions about the role of dosage compensation in sex chromosome evolution.
In a narrow sense, “Ohno’s hypothesis” is associated specifically with his assessment of SCDC in mammals: that degradation of the Y catalyzed up-regulation of the X, causing over-expression in females, which subsequently was mitigated through X-chromosome inactivation (XCI) (Nguyen and Disteche 2006; Xiong etal. 2010; Pessia etal. 2014; Graves 2016). While this proposed chain of evolutionary events form an intuitively appealing hypothesis that long appeared correct, recent genomic analyses have caused it to unravel (Julien etal. 2012; Lin etal. 2012; Chen and Zhang 2015). The broader hypothesis, that sex chromosome differentiation presents a “peril” to be mitigated via dosage compensation, has also long served as an important conceptual framework in sex chromosome research (Charlesworth 1978, 1996; Bachtrog 2006; Vicoso and Bachtrog 2009; Mank 2013). Yet this broader hypothesis is also eroding as genomic investigations of SCDC expand into increasingly diverse taxa. It now appears that evolving SCDC via chromosome-wide regulation is the exception, not the norm (Disteche 2016; Chandler 2017).
As the classical Ohnian paradigm crumbles, research in SCDC evolution faces major challenges in integrating a diverse and growing patchwork of observations into a coherent theoretical framework. This research ultimately aims to understand clearly why SCDC evolves or is absent, both for particular genes as well as for whole chromosomes. Recent empirical results and novel modeling efforts offer some clues; for instance it may depend on how dosage “sensitive” a gene is, as well as whether the degenerate sex chromosome resides in males or females (Pessia etal. 2012; Mullon etal. 2015). Nonetheless, a comprehensive understanding of SCDC evolution remains an elusive but alluring goal, with much work still required to provide broader taxonomic sampling, detailed dissection of mechanism, and creative application of theory.
Here, were present on overview of contemporary research on SCDC evolution. We begin by summarizing the relevant theory and connecting it to the criteria, methodologies, and specific vocabulary used in the context of studying SCDC. Next, we address technical issues and complications that are frequently encountered in genomic analysis of SCDC. We then provide a summary of empirical results currently available concerning SCDC across animals. Finally, we address several outstanding questions and opportunities that exist for future studies of dosage compensation. We focus our discussion on animals, as SCDC in plants is thoroughly addressed in the recent review from Muyle etal. (2017)
The Evolution of Sex Chromosomes and the Peril of Hemizygosity
The prevailing theory of sex chromosome evolution is that allosomes arise from a pair of homologous and recombining autosomes, in which one homolog acquires a sex-determining locus (Charlesworth 1991; Charlesworth etal. 2005). Subsequently, selection will favor tight linkage between the sex determining locus and sexually antagonistic alleles benefiting the heterogametic sex. This results in a loss of recombination between the proto-allosomes, allowing for processes like Muller’s ratchet or adaptive hitch-hiking to erode the gene content of one allosome, yielding an intact X chromosome and a degenerate Y chromosome in male-heterogametic taxa (or a Z and degraded W chromosomes in female-heterogametic species like birds and butterflies) (Charlesworth 1978; Rice 1996; Bachtrog 2013).
One important consequence of this process is that the X chromosome becomes increasingly monoallelic in the males, causing a difference in gene dose between sexes: females retain two copies of X-linked genes while males are left with only a single copy. (For the sake of simplicity we will assume male-heterogamety in our discussion, unless otherwise specified, though all arguments apply equally to female-heterogamety and ZW chromosomes.) This imbalance, if reflected in the abundance of transcripts or proteins, has the potential to deleteriously disrupt interactions between X-linked and autosomal loci in males (Birchler etal. 2001, 2005; Mank 2009). Proper function for much of development, cellular physiology, and metabolism requires tightly controlled stoichiometric ratios of interacting proteins (Birchler etal. 2005; Zhang and Oliver 2007). Grossly altering these ratios is expected to substantially reduce fitness. During allosome divergence, females presumably retain the ancestral gene dosage (diploid X chromosome) and experience no disruptions to protein–protein interactions between X and autosomes. In contrast, large monoallelic regions on the X in males will likely cause detrimental gene dosage effects. For this reason, it is expected that dosage compensating mechanisms should evolve that recapitulate the ancestral expression levels for the monosomic male X (i.e., comparable to XX females) (Ohno 1967; Rice 1987; Charlesworth 1996; Mank etal. 2011). In other words, dosage compensation mechanisms arise due to stabilizing selection to maintain ancestral expression levels on the X chromosome. Primarily this selection acts on the heterogametic sex, though potentially also on the homogametic sex as a secondary, sexually antagonistic step in the process of SCDC evolution (Ohno 1967; Rice 1987; Mank etal. 2011). Yet because most genes have comparable expression in both sexes, the mechanisms that evolve to achieve SCDC should also produce balanced expression on the X chromosome between males and females.
We can thus specifically define SCDC as the maintenance of ancestral expression levels of sex-linked genes relative to autosomal expression in the heterogametic sex (Ohno 1967; Mank etal. 2011). SCDC is often further qualified as “complete” versus “incomplete” or “partial.” SCDC is “complete” when sex-linked expression in the heterogametic sex is indistinguishable from the ancestral state. SCDC that is “partial” or “incomplete” refers to sex-linked expression in the heterogametic sex that is reduced relative to ancestral expression. Importantly, this specific definition of complete SCDC can exclude some cases where sex-linked expression is well-balanced between males and females. For example, this appears to be the case in placental mammals and in Lepidoptera (moths and butterflies), where the average X or Z expression is reduced relative to ancestral states in both sexes (Walters and Hardcastle 2011; Julien etal. 2012; Lin etal. 2012; Kiuchi etal. 2014; Chen and Zhang 2015; Gu etal. 2017 ). These patterns are notably inconsistent with theoretical expectations and are not easily explained at present, other than to suggest SCDC may not be universally required for allosome divergence as has long been believed. Nonetheless, gene-by-gene compensation of highly dosage sensitive genes seems to occur in several species (Julien etal. 2012; Lin etal. 2012; Pessia etal. 2012; Disteche 2016; Zimmer etal. 2016).
This definition of SCDC also includes cases where the X chromosome is over-expressed in the homogametic sex such that sexes are not balanced, even though the heterogametic sex is dosage compensated (Prince etal. 2010; Mank etal. 2011; Allen etal. 2013). This scenario can result from the spread of X-linked alleles that benefit males by up-regulating the X chromosome to achieve dosage compensation. If these alleles act similarly in females, they will cause over-expression of X-linked loci relative to the ancestral state, potentially reducing female fitness (Ohno 1967; Charlesworth 1978, 1996; Rice 1984, 1987; ). This sexual antagonism surrounding optimal X-linked expression levels should ultimately cause a secondary mechanism to evolve that down-regulates X-linked expression specifically in females, much as Ohno proposed as the reason for XCI in placental mammals (Ohno 1967; Engelstädter and Haig 2008; Mank etal. 2011). However, if overexpression in the homogametic sex is not deleterious, or if the process has not yet equilibrated, a pattern of female over-expression will result on the X (Mullon etal. 2015).
We emphasize that “SCDC” is a term that has long been loosely and variably applied in the context of gene dose and expression differences between sexes (Mank 2011; Disteche 2016). However, in light of evolutionary theory, we argue that SCDC should be limited specifically to mean the pattern and/or process in which heterogametic sex-linked expression recapitulates ancestral levels prior to allosome divergence. In adopting this definition of SCDC, additional terminology is needed to clarify the discourse surrounding this topic. In particular, one scenario that has been increasingly observed is equal sex-linked expression between sexes but without complete SCDC. We henceforth use dosage balance to refer to equal average expression between sexes on the sex chromosome regardless of ancestral expression levels. In other words, if the X-linked genes (or the whole chromosome on average) show no bias in male: female expression ratios, but expression has diverged substantially from ancestral levels of the proto-X, then there is dosage balance without dosage compensation.
Approaches, Challenges, and Caveats in SCDC Analysis
Methods and Criteria for Assessing SCDC
Ultimately, the goal of SCDC analysis is to determine whether sex chromosome gene expression has changed relative to the ancestral state before allosome divergence. The hypothetically ideal experiment would be to compare the focal species with differentiated allosomes to a progenitor with undifferentiated proto-sex chromosomes. Complete SCDC would be inferred if no difference were found between the ancestral diploid expression and contemporary monoallelic expression. However, since directly measuring expression from ancestral proto-sex chromosomes is generally not possible, the typical approach to assessing SCDC compares expression of current sex-linked genes to a reference gene set which serves as a proxy for ancestral expression levels of these same genes. There are three common approaches for this: (1) comparative analysis contrasts X-linked genes with autosomal orthologs in an outgroup species, (2) the X-to-autosome approach compares X-linked genes to autosomal loci in one species, and (3) male:female ratios contrasts X-linked expression between the heterogametic versus homogametic sex in one species.
Comparative Analysis
Perhaps the most convincing and informative approach to assessing SCDC is comparative analysis, in which orthologous loci are compared between species with distinct and independently evolved sex chromosomes, such that the loci are sex-linked in a focal species, but autosomal in a reference species. The diploid autosomal expression in the reference species serves as a proxy for the ancestral diploid expression levels in the focal species, against which the magnitude of dosage compensation in the heterogametic sex may be assessed. Complete compensation is inferred if expression of monoallelic X-linked loci in the heterogametic sex is comparable to expression levels of autosomal diploid orthologs in the reference species. Comparison of orthologs that remained autosomal in both species can serve as a control for expression divergence due to drift or selection (Julien etal. 2012; Lin etal. 2012; Nozawa etal. 2014; Vicoso and Bachtrog 2015; White etal. 2015; Gu etal. 2017). This comparative approach has the advantage of comparing dosage effects on the same genes in the same sex, so it is a relatively controlled comparison that is arguably the best way to assess dosage effects relative to ancestral expression. However, this approach may be limited when only a relatively small number of 1-to-1 orthologs can be identified, such that generalizing to the entire sex chromosome may not be robust. Thus, it is preferable to use closely-related species where possible in order to maximize available orthologs and minimize the effects of divergence in this analysis.
X-to-Autosome Differences
When a comparative analysis is impossible or impractical, the next best approach will often be to test for differences in average expression of X-linked versus autosomal loci. Complete SCDC is inferred when the heterogametic sex exhibits no difference in average expression between the X and autosomes (Mank 2009, 2013). Incomplete SCDC would cause X < Autosomes for the heterogametic sex. Testing for SCDC using X-to-autosome differences rests on the assumption that the average autosomal expression is a reasonable proxy for average expression of the ancestral diploid proto-X; in other words, it assumes that average chromosomal expression is equal in the absence of dosage effects. However, this is often not the case and average autosomal expression can vary considerably (Julien etal. 2012; Wheeler etal. 2016). Accordingly, it may be that the average X chromosome expression differs from the overall autosomal average, but is not unusual compared with other individual chromosomes.
Examining X-to-autosome differences in the homogametic sex is a necessary companion to analysis in the heterogametic sex. If no differences are detected, this supports the assumptions that average expression is equal across chromosomes and that autosomal expression is a reasonable proxy for the proto-X. Alternatively, X > Autosomes potentially indicates unresolved sexual antagonism in the evolution of SCDC (Engelstädter and Haig 2008; Prince etal. 2010; Mank etal. 2011; Allen etal. 2013). X < Autosomes in the homogametic sex has, to date, never been observed without a (nearly) comparable reduction the heterogametic sex. A comparable reduction in X expression relative to autosomes in both sexes may reflect two scenarios. In one scenario, it may be that SCDC is (nearly) complete, but the ancestral expression of the diploid proto-X was markedly lower than autosomes before allosome divergence (Vicoso etal. 2013). Alternatively, it may be that the homogametic sex has evolved a reduction in gene expression relative to the ancestral state, so as to achieve balanced expression with the heterogametic sex that exhibits incomplete SCDC. These two scenarios can be distinguished through comparative analyses, as described above (Julien etal. 2012; Lin etal. 2012; Gu etal. 2017), and also by experimental manipulation of dosage compensation mechanisms to expose the “unmodified” gene dosage (Kiuchi etal. 2014).
Male:Female Ratios
The distribution of male:female expression ratios on the sex chromosome is best regarded as an assessment of gene dosage effects (i.e., whether a species is dosage balanced). Although male:female ratios may be informative concerning SCDC in some cases, they are potentially problematic as such. In principle, if sex-linked expression has not changed in the homogametic sex in the course of allosome divergence, then diploid X expression in contemporary females is an accurate representation of ancestral expression levels. In this case, equal expression between sexes on the X is a good indicator of SCDC. However, if sex-linked expression has changed significantly in the homogametic sex, observing complete gene balance (i.e., equal average male: female expression ratios) does not convey information about SCDC. Rather, both the male and female sex-linked expression have equally diverged from the ancestral state, producing a scenario of dosage balance without compensation. Comparing male:female ratios on the X relative to autosomes is an important internal control in such analyses, where overall artifacts in the data can lead to spurious results if only the sex chromosome is assayed (Zha etal. 2009; Walters and Hardcastle 2011).
Analytical Challenges and Artifacts
There are numerous statistical and technical issues that must be navigated in pursuing an assessment of SCDC. If genes are not already mapped to chromosomes in the studied organism, then the effort will begin with the non-trivial task of identifying sex-linked loci. The traditional solution to this involves genetic crosses and linkage mapping, which is increasingly a component of genome sequencing projects (Werren etal. 2010; Heliconius Genome Consortium 2012; Ahola etal. 2014). Linkage mapping has the advantage of assigning genome scaffolds and genes to specific chromosomes, including the X or Z, but it is laborious and time-consuming. Fundamentally, simply partitioning genes as autosomal or sex-linked is sufficient for SCDC analysis. One recent approach to this uses RNA-seq to track SNPs in parent-offspring groups; this is particularly useful when allosomes are recently diverged and retain substantial homology (Muyle etal. 2016). For species with well-differentiated sex chromosomes, this can be readily accomplished by examining differences in DNA abundance between males and females, either via Illumina sequencing or microarrays (Baker and Wilkinson 2010; Martin etal. 2013; Mahajan and Bachtrog 2015; Vicoso and Bachtrog 2015). Autosomes will have equal amounts of DNA between sexes, while X- or Z-linked sequences will yield a 2-fold difference. Alternatively, when synteny and sex-chromosome identity is well-conserved between taxa, it is possible to use orthology to predict sex-linkage (Harrison etal. 2012; Gu etal. 2017).
After sex-linkage is established and measures of gene expression are obtained, typically as RNA-seq data normalized to reads per kilobase per million mapped reads (RPKM) (Oshlack etal. 2010; Conesa etal. 2016), the next step is to perform appropriate statistical tests for expression differences between chromosomes or sexes. There is a strong precedent in the literature for using a Mann–Whitney U (MWU) test for such inferences, either contrasting expression on the X versus autosomes within sex (requiring two tests) or contrasting expression ratios (male:female, or modern:ancestral in a comparative analysis) between X and autosomes (Vicoso and Bachtrog 2011; Walters and Hardcastle 2011; Harrison etal. 2012; Lin etal. 2012; Vicoso etal. 2013; Smith etal. 2014). Another similar approach is to employ bootstrap analysis to gauge statistical differences in X: autosome or male: female expression ratios (Uebbing etal. 2013; Gu etal. 2017).
While the use of MWU or bootstrap tests is intuitive and sufficient in most cases, these approaches also have limitations. First, when biological replicates are available, they are typically averaged together within sex for a single RPKM value per locus, which discards information on expression variance between replicates. Second, dosage and balance are not examined simultaneously in a unified statistical framework, since the former is in units of RPKM while the latter reflects ratios. This makes it difficult to meaningfully compare the magnitude of chromosome-specific effects on expression with the magnitude of any dosage effect between sexes. An alternative approach that circumvents these issues is to employ linear modeling of expression levels (Walters etal. 2015). Using linear modeling, reduced X expression can be captured as a primary effect of chromosome, while X-specific dosage effects that unbalance expression between sexes are modeled as an interaction between sex- and chromosome-specific effects. This provides an appealingly unified and flexible framework for SCDC analysis, which also happens to accommodate biological replicates without prior averaging. However, implementing this linear mixed-modeling framework is not as straightforward as the non-parametric approaches provided by MWU and bootstrapping.
Another noteworthy technical issue in SCDC analysis is whether and how best to exclude unexpressed genes, which are decisions that substantially influence results (Xiong etal. 2010; Castagné etal. 2011; Deng etal. 2011; He etal. 2011; Kharchenko etal. 2011; Jue etal. 2013). Including unexpressed genes when they are not evenly distributed across chromosomes can bias average expression levels between chromosomes. Furthermore, applying too stringent a threshold when discerning “active” transcription may disproportionately truncate the distribution of expression levels in one portion of the data, artificially compressing the averages of X and autosomal expression to mask true differences. Currently there is very little consensus on the appropriate method or criteria for discerning “unexpressed” loci. Some studies employ a uniform cutoff, for instance excluding genes with <1 RPKM. However, a uniform RPKM threshold is problematically arbitrary because different types of between-sample normalizations will produce dramatically different library-size scaling factors (e.g., TMM versus 75% quantile versus total reads [Bullard etal. 2010; Robinson and Oshlack 2010]). These scaling factors are in the denominator when calculating RPKM, thus RPKM values have little real biological relevance and applying the same threshold (e.g., <1 RPKM) may produce wildly different filtering stringencies across normalization methods. We suggest that a probabilistic approach to filtering is more sensible. This may simply reflect removing outlier loci with expression levels at the extremes of the observed distribution (Jue etal. 2013). Alternatively, employing likelihood or other inference-based approaches to discern loci that are providing signal, rather than noise, may also be a good option (Walters and Hardcastle 2011; Hart etal. 2013; George and Chang 2014; Hardcastle 2016). In any case, it is certainly a good practice to confirm that results do not qualitatively change under different filtering criteria.
Despite these numerous statistical challenges commonly encountered by researchers working on SCDC, currently there is no established bioinformatic software package available for performing SCDC analysis. Having a standard software tool for implementing a variety of relevant statistical tests and filtering would be a welcome advance toward improving consistency and reproducibility in analyses of dosage compensation.
Gonads Are Different, and Problematic
There is substantial evidence from diverse taxa that gonads show extreme and idiosyncratic patterns of gene expression relative to somatic tissues. For this reason, it is highly desirable to separate gonads from soma in studies of SCDC, since failing to do so may produce results that are misleading or difficult to interpret. There are at least three major factors underlying distinctive patterns of expression in gonads. The first is that mechanisms mediating dosage compensation or balance may often not operate in the gonads. This is apparently the case in Drosophila and Lepidoptera (Meiklejohn etal. 2011; Walters and Hardcastle 2011; Vicoso and Bachtrog 2015; Gu etal. 2017). Other examples include the reactivation of the silenced X chromosome in germ cells of female mammals, and the silencing of the X chromosome in germ cells of both sexes of nematodes (Kelly etal. 2002; Sugimoto and Abe 2007). The second factor is the often unequal complement of sex-biased genes on the sex chromosomes relative to autosomes, reflecting the fact that sex chromosomes are a “hotspot” for sexually antagonistic evolution (Rice 1984; Connallon and Clark 2010; Meisel etal. 2012; Parsch and Ellegren 2013). In particular, the X/Z chromosome is typically enriched for genes with expression biased toward the homogametic sex. As gonads normally have very extreme patterns of sex-biased expression relative to soma, this potentially interacts with the unequal distribution of sex-biased genes to produce a pattern that mimics a dosage effect. Another idiosyncrasy of germline tissues is meiotic sex chromosome inactivation, which silences the single male X expression in mammals (Turner 2007), Drosophila (Vibranovski 2014), and possibly chicken (Schoenmakers etal. 2009, but also see Guioli etal. 2012). Each of these factors may substantially skew sex-linked expression patterns in the same—or opposite—direction as a dosage effect, potentially obscuring or confounding accurate inferences about SCDC. For this reason, dosage compensation studies that include gonadal tissues should seek to isolate and analyze them separately from somatic tissues, which arguably provide a more accurate assessment of SCDC.
Separation of soma and gonad is of particular concern for invertebrates, where whole-body analysis is tractable and common, but where reproductive tissues constitute a substantial fraction of the adult body. Clear examples of this issue are known from flies and nematodes. In a survey of multiple species across dipteran lineages, Vicoso and Bachtrog (2015) showed that X-linked loci were hypertranscribed in the ovaries and hypotranscribed in the testis relative to ancestral expression levels, while the X expression in somatic tissues were both compensated and balanced. Yet whole-body samples yielded an intermediate pattern, with biases in the same direction as gonads, but reduced in magnitude, presumably mitigated by mixing with dosage compensated somatic tissues. The nematode Caenorhabditis elegans has an increasing fraction of germline cells relative to somatic cells as it develops. Since the X chromosome is inactivated in germ line cells, the stage-specific X:autosome ratio decreases over the course of development as the ratio of germline: somatic cells increases (Deng etal. 2011). These two examples highlight the potential pitfalls of analyzing a mixture of soma and gonadal tissues, as has unfortunately been done in a few other studies reporting unusual patterns of dosage compensation (see Type IV pattern, below) (Prince etal. 2010; Allen etal. 2013).
Observed Patterns of Sex Chromosome Dosage Compensation in Animals
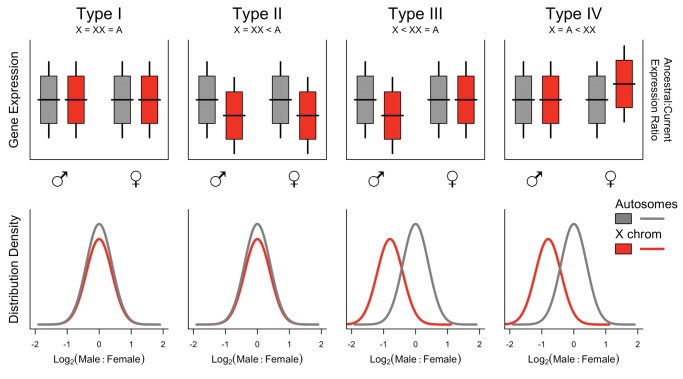
Having considered the theoretical and practical framework for addressing SCDC, what do we actually observe? Dozens of studies have examined SCDC across a wide range of animal taxa and have revealed a striking array of patterns, many of which do not fit well with theoretical predictions. Nonetheless, these observations can be readily grouped into four distinct types, based on a combination of “dosage compensation” and “dosage balance.” For the sake of facilitating discussion we arbitrarily label these patterns as Type I–IV. Hypothetical patterns of gene expression obtained from these four types of SCDC are presented in figure 1. An overview of taxonomic occurrences is given in table 1, which summarizes a detailed listing with references provided in supplementary table S1, Supplementary Material online.
Fig. 1.
—Hypothetical examples of gene expression levels/ratios encountered in Types I-IV of SCDC. The top row represents an assessment of complete dosage compensation, which emphasizes whether X-linked expression levels have changed relative to the ancestral state. In the subtitles, “X” and “XX” refer, respectively, to heterogametic and homogametic expression levels of sex-linked loci, and “A” may reflect diploid autosomal or ancestral expression levels, depending on the empirical approach used (e.g., comparative method or X-to-autosome). The bottom row represents an assessment of dosage balance, which emphasizes the presence of a dosage effect on gene expression resulting from heterogametic sex chromosomes.
Table 1.
A Summary of SCDC Patterns Observed in Animals
| Sex Determination | SCDC Pattern | Taxon (number of species surveyed) |
|---|---|---|
| Male heterogamety (XX/XY) | Type I (X = XX = Ancestral) | True bugs (Hemiptera) (4) |
| Strepsipteran (1) | ||
| Beetle (Coleoptera) (1) | ||
| Flies and mosquitoes (Diptera) (7) | ||
| Type II (X = XX < Ancestral) | Nematodes (2) | |
| Therian mammals (9) | ||
| Type III (X < XX = Ancestral) | Three-spined stickleback (1) | |
| Platypus (1) | ||
| Female heterogamety (WZ/ZZ) | Type II (Z = ZZ < Ancestral | Moths and butterflies (Lepidoptera) (5) |
| Type III (Z < ZZ = Ancestral) | Blood-fluke (Schistosoma) (1) | |
| Tonguefish (1) | ||
| Snakes (2) | ||
| Birds (5) |
Note.—A complete listing of individual species, with references, is provided in supplementary table S1, Supplementary Material online.
Type I: Complete Compensation with Balance (X = XX = Ancestral)
In this case, SCDC is complete in the heterogametic sex and X-linked expression is balanced with the homogametic sex. This pattern was long considered to be widespread because initial assays of SCDC in major model systems (i.e., flies, nematodes, humans) yielded results consistent with this pattern (Meller 2000; Gupta etal. 2006; Nguyen and Disteche 2006; Straub and Becker 2007; Zhang and Oliver 2007). However, SCDC analyses in a broadening spectrum of species have revealed that this pattern is far from common (Mank 2013). In fact, so far it is primarily observed among insects.
The most prominent and well-studied example is Drosophila melanogaster. D. melanogaster up-regulates the single X chromosome in males by roughly two folds, resulting in both “balance” and “compensation” without changes in female X expression (recently reviewed by Lucchesi and Kuroda 2015; Kuroda etal. 2016). Vicoso and Bachtrog (2015) surveyed a diversity of dipteran species and showed that this mode of dosage compensation is broadly conserved across flies, despite substantial turnover of sex chromosomes between lineages (including one conversion to ZW female-heterogamety). Separate studies on the stalk-eyed fly Teleopsis dalmanni (Wilkinson etal. 2013), Australian sheep blowfly Lucilia cuprina (Linger etal. 2015) and two mosquito species, Anopheles stephensi (Jiang etal. 2015), and Anopheles gambiae (Rose etal. 2016), all support the consistency of this pattern.
In D. melanogaster dosage compensation is mediated by Male-Specific Lethal (MSL) complex, which binds to the X chromosome in the male and is not assembled in females (Lucchesi and Kuroda 2015). The same mechanism is co-opted by the recently evolved neo-X chromosome in D. pseudoobscura and D. miranda (Marín etal. 1996; Steinemann and Steinemann 1998; Alekseyenko etal. 2013; Zhou etal. 2013) but does not operate on the retrogenes on the X chromosome in the Australian sheep blowfly L. cuprina (Linger etal. 2015). The MSL-dependent dosage compensation mechanism seems to be unique to the Drosophila genus, and even within Drosophila, alternative mechanisms of SCDC may exist (Park and Kuroda 2001). Despite multiple transitions in sex chromosome identity across Diptera the patterns of SCDC remain quite consistent, so it will be interesting to learn the extent to which various lineages employ molecular mechanisms similar to, or even homologous to, that found in Drosophila (Baker and Wilkinson 2010; Toups and Hahn 2010; Pease and Hahn 2012; Vicoso and Bachtrog 2015).
Outside Diptera, this pattern is also reported in two other insect orders: Hemiptera and Coleoptera. Four hemipteran species have been examined and all show patterns consistent with male-specific up-regulation of the X chromosome (Pal and Vicoso 2015). Three of them are completely compensated, or nearly so, but the fourth exhibits a notable dosage effect. While these results possibly reflect true variation among species, this study sampled whole adult bodies so results may be biased by inclusion of the gonads. Potential complications from including gonads may also plague an early study of SCDC using whole bodies of the beetle Tribolium castaneum (see section on Type IV) (Prince etal. 2010). Yet subsequent analyses of isolated somatic tissues indicated that expression of genes from the X and the autosomes is similar between males and females (Mahajan and Bachtrog 2015). Thus, T. castaneum most likely follows the Type I pattern of SCDC.
It is striking that, at least where described, the mechanisms for achieving a Type I pattern of complete dosage compensation with balance appear to operate by male-specific up-regulation of the haploid X chromosome. As for eutherian mammals and caenorhabditid nematodes that were long considered to have a Type I pattern of SCDC evolving via the Ohnian model (Marín etal. 2000; Nguyen and Disteche 2006; Straub and Becker 2007; Mank 2009), recent analyses indicate this is not the case, as we address next under Type II patterns.
Type II: Incomplete Compensation with Balance (X = XX < Ancestral)
The use of comparative genomics to infer ancestral expression levels for dosage compensation analysis has catalyzed substantial rethinking of long-standing assumptions concerning SCDC in several lineages, most prominently for eutherian mammals (Julien etal. 2012; Lin etal. 2012; Mank 2013; Pessia etal. 2014). This novel approach to analyzing dosage compensation has also been applied to nematodes (Albritton etal. 2014), with similar results: X chromosome expression is effectively balanced between sexes, but apparently without dosage compensation sufficient to recapitulate ancestral diploid expression levels. This is also a pattern found repeatedly among Lepidoptera (Walters and Hardcastle 2011; Kiuchi etal. 2014; Smith etal. 2014; Walters etal. 2015; Gu etal. 2017).
In the case of eutherian mammals, until quite recently the perspective on SCDC was consistent with “Ohno’s hypothesis”: XCI was a dosage compensating mechanism arising in response to the initial up-regulation of X-linked expression required in males to maintain ancestral expression levels as the Y degraded (Marín etal. 2000; Nguyen and Disteche 2006; Straub and Becker 2007; Mank 2009). In the last 5 years, this Ohnian paradigm has become surprisingly contentious. This body of work has been reviewed repeatedly and extensively in recent years (c.f. Birchler 2012; Disteche 2012, 2016; Mank 2013; Pessia etal. 2014; Veitia etal. 2015; Graves 2016), so here we provide only a brief overview. While various X-to-autosome comparisons provided more or less support for Ohno’s elusive “X-upregulation” (Xiong etal. 2010; Deng etal. 2011; He etal. 2011; Kharchenko etal. 2011), subsequent comparative analyses contrasting mammalian X-linked loci with autosomal loci in chickens yielded compelling evidence for the general absence of SCDC in eutherian mammals (Julien etal. 2012; Lin etal. 2012; Ka etal. 2016). This finding was further supported by quantitative proteomic analyses as well as expression in haploid versus diploid human parthenogenetic stem cells (Lin etal. 2012; Chen and Zhang 2015, 2016). Yet notable complexities persist in this issue. For example, the subset of X-linked genes that encode members of large protein complexes, thus likely to be highly dose-sensitive, do show effectively complete dosage compensation, indicating gene-by-gene up-regulation has occurred for ∼5% of X-linked loci (Lin etal. 2012; Pessia etal. 2012). There is also some evidence for chromosome-wide mechanisms that specifically enhance X transcription and translation, consistent with the notion of male-driven up-regulation. Examinations of RNA half-life, RNA polymerase occupancy, activating histone marks, and ribosome density point to a range of distinct regulatory patterns on the placental X chromosome that should increase transcription and translation (Yildirim etal. 2011; Deng etal. 2013; Faucillion and Larsson 2015). So it seems that complete chromosome-wide X dosage compensation is not the case for eutherian mammals, but at least some dosage-sensitive genes are fully compensated, and there are hints that some global mechanisms for up-regulation have evolved.
The status of SCDC in marsupials is ambiguous at the moment. The two primary studies addressing this topic in therian mammals both used the same data sets, and reported largely consistent results across taxa, except for opossum (Monodelphis domestica), the only marsupial represented in the studies (Julien etal. 2012; Lin etal. 2012). Both studies indicate equal average X-linked expression between sexes, suggesting opossum is dosage balanced. However, in assessing dosage compensation by comparison to autosomal chicken orthologs, Julien etal. (2012) reported substantial or complete dosage compensation while Lin etal. (2012) showed very little X compensation relative to ancestral expression. It is worth noting that the numbers of X-linked genes used differ considerably between the two studies. Julien etal. (2012) restricted their analysis to only 91 conserved 1:1 orthologs across 9 mammal species and chicken. Meanwhile, Lin etal. (2012) considered 197 genes on the opossum X with chicken autosomal orthologs. While this larger number of genes may well reflect a more reliable assessment of X chromosome dynamics, additional data and analyses are clearly needed in marsupials. Also of note, the differences between the gene sets and the observed degree of compensation could indicate that widely conserved genes also may be prone to better dosage compensation.
One corollary of recognizing that most X-linked genes in therian mammals are likely not completely dosage compensated is the need to explain XCI as something other than a female “response” to male-driven dosage compensation. It is not obvious why halving X expression in females to match males would be initially advantageous if X up-regulation has not already globally occurred, though it does solve the problem of dosage for any genes arising on the X after allosome divergence (Lin etal. 2012). An alternative explanation for XCI is offered by theory invoking paternal imprinting of the X in the context of sexual antagonism over fetal growth rates and maternal resources (Haig 2006; Engelstädter & Haig 2008; Pessia etal. 2014). Thus, it may be that any balancing of X-linked expression in mammals due to XCI is an outcome, not a cause, of this phenomenon. Therian sex chromosome divergence and the origins of XCI occurred quite close in time, if not coincidentally, so teasing this apart may prove difficult (Graves 2016; Whitworth and Pask 2016).
Relative to mammals, there has been less contention concerning SCDC in nematodes, particularly for the C. elegans X chromosome, where molecular mechanisms of balance, but not compensation, have been carefully dissected (Pferdehirt and Meyer 2013; Ferrari etal. 2014; Ercan 2015). Nonetheless, longstanding assumptions are being overturned for this group, much like with mammals. Nematodes, or at least C. elegans, were long regarded as another classic example Ohno‘s 2-step model of SCDC (Straub and Becker 2007; Zhang and Oliver 2007; Mank 2009). There is a well-described epigenetic mechanism operating in the homogametic sex (hermaphrodites) that halves expression from each X chromosome. Like XCI in mammals, this so-called “dosage compensation complex” (DCC) was considered the homogametic response to male-driven up-regulation of the X chromosome. Yet, again like in mammals, the argument for this up-regulation is based on X:autosome ratios near 1, while the mechanisms of the purported X up-regulation remain largely unknown (Deng etal. 2011). However, the relevance of this X:autosome comparison was called into question by the recognition that average expression varies between autosomes as much as 2-fold (Wheeler etal. 2016). Also, comparative analysis between C. elegans and Pristionchis pacificus indicates SCDC is incomplete, based on ∼350 orthologs that are reciprocally X-linked in one species and autosomal in the other (Albritton etal. 2014). Further evidence for incomplete SCDC, as well as dosage balance, in C. elegans comes from analysis of single-copy transgenes. Transgenes inserted on the X had equal expression in males (XO) versus hermaphrodites (XX), but half the expression of autosomal copies (Wheeler etal. 2016). So it now appears that allosome divergence may have occurred in nematodes without much compensation, leaving open the question of what underlies the evolution of the DCC if it was not male-driven X up-regulation.
While the uncertainties in both worms and mammals revolve around dosage compensation without any question of dosage balance, the situation in Lepidoptera (moths and butterflies) is quite different. This group of insects was initially considered to have incomplete dosage compensation without balance (i.e., Type III, see below), much like other ZW species. This early perspective was shaped by two reports both showing strong Z-linked male bias, one in silkworm (Bombyx mori), and the other from Indian meal moth (Plodia interpunctella) (Zha etal. 2009; Harrison etal. 2012). However, a subsequent reanalysis of the silkworm data demonstrated substantial analytical artifacts in the initial study and, further, demonstrated dosage balance with reduced Z:autosome ratios (Walters and Hardcastle 2011). The meal moth study used whole-body adults, including gonads, which (as explained above) is problematic, especially because Lepidopteran gonads tend to show patterns of unbalanced dosage (Walters and Hardcastle 2011; Gu etal. 2017). A later study in meal moth analyzed gonads separately from soma, confirmed this distinction, and yielded patterns in somatic tissues similar to silkmoth (Huylmans etal. 2017). In addition, several studies analyzing somatic tissues in other lepidopteran species consistently showed that moths and butterflies actually have equally reduced Z transcriptional output in both sexes compared with autosomal expression (Smith etal. 2014; Fukui etal. 2015; Sugimoto etal. 2015; Walters etal. 2015; Gu etal. 2017). Two additional lines of evidence support this pattern and further suggest dosage balance reflects male-specific down-regulation of the Z chromosome. The first is comparative analyses involving the codling moth, Cydia pomonella, which is the most ancestrally diverging Lepidopteran species yet investigated for dosage compensation. Codling moth harbors a neo-Z chromosome, arising from a translocation event that fused an autosome to the ancestral Z chromosome (Nguyen etal. 2013). Genes on the neo-Z segment were expressed on average about 30% less than their autosomal orthologs in other lepidopteran species (Gu etal. 2017). Second, in silkworm, feminizing male embryos via RNAi depletion of the male-determining protein, Masc, causes substantial and wide-spread Z-specific up-regulation (Kiuchi etal. 2014). Thus, it appears that Lepidoptera employ a molecular mechanism specific to the homogametic sex that reduces expression and mitigates Z dosage effects between sexes. This pattern of dosage balance without complete compensation is a striking parallel with mammalian XCI as well as the nematode DCC and it will be important to understand better the molecular details of this phenomenon in Lepidoptera.
Type III: Incomplete Compensation Without Balance (X< XX = Ancestral)
When sex-linked expression is substantially reduced in the heterogametic sex relative to the homogametic sex, the pattern is often referred to as incomplete or partial SCDC. In such cases, a substantial gene dosage effect is inferred to occur for sex-linked gene expression (yielding a scenario with Z < ZZ = AA or X < XX = AA) (Vicoso and Bachtrog 2011; Wolf and Bryk 2011; Mank 2013). Strikingly, this pattern is almost universally observed among female-heterogametic taxa examined to date. This includes species with Z chromosomes that are very old (e.g., birds [Ellegren etal. 2007; Itoh etal. 2007, 2010; Wolf and Bryk 2011; Uebbing etal. 2013]) and young (e.g., sole flatfish [Chen etal. 2014]), as well as from snakes and a parasitic trematode (Vicoso and Bachtrog 2011; Vicoso etal. 2013). As mentioned above, the only exception to this trend among ZW taxa thus far surveyed is Lepidoptera (Walters and Hardcastle 2011; Smith etal. 2014; Gu etal. 2017). In contrast, this Type III pattern is rarely found in male heterogametic species and is so far only reported in platypus (a monotreme), which shares homologous sex chromosomes with birds, and in the three-spined stickleback fish (Julien etal. 2012; Lin etal. 2012; White etal. 2015).
Chicken is the best studied species exhibiting a Type III pattern. Global expression analyses based on both microarray and RNA-seq, as well as comparison to inferred ancestral expression (i.e., comparative analysis), showed that transcriptional output of the Z chromosome is ∼30% lower in females than in males and that male Z expression is comparable to autosomal expression (Ellegren 2007; Itoh etal. 2007; Julien etal. 2012). While most loci exhibit intermediate dosage effects, some are fully compensated. In particular, ohnologs appear better compensated than other genes on the Z, in line with the idea that genes preserved after whole genome duplication are particularly dosage sensitive (Zimmer etal. 2016). Further, a recent proteomic analysis revealed that many Z-linked genes exhibit additional dosage compensation during protein translation (Uebbing etal. 2015). Therefore, it appears that in chicken, and likely all birds, SCDC operates through localized gene-by-gene mechanisms involving a mix of the transcriptional and translational modulation.
Among mammals and other male-heterogametic taxa, the platypus is noteworthy for its peculiar complement of sex chromosomes and that it exhibits a Type III pattern of SCDC. Platypus (like echidnas, their sister monotremes) have a complement of five distinct X chromosomes, each of which has a degraded Y counterpart and is largely homologous to the avian Z chromosome (Veyrunes etal. 2008; Graves 2016; Whitworth and Pask 2016). Complications of genome assembly have thus far allowed dosage analysis of only X1 and X5, but there is a clear dosage effect (e.g., female-biased expression) among genes outside of pseudoautosomal regions (Julien etal. 2012; Lin etal. 2012). Notably, comparative analyses in both chicken and platypus did show some upregulation of monosomic sex-linked expression in the heterogametic sex relative to ancestral levels, though it is insufficient to completely recover ancestral levels (Julien etal. 2012). Yet the unchanged sex-linked expression in the homogametic sex suggests either up-regulation being limited to the heterogametic sex or a secondary evolution of down-regulation in the homogametic sex (Whitworth and Pask 2016). Epigenetic marks typically associated with chromosome-wide transcriptional inactivation mechanisms (e.g., histone modifications) have not been detected on the platypus X chromosomes and the bird Z chromosomes (Itoh etal. 2010; Rens etal. 2010). However, RNA–FISH analysis of a couple dozen loci in both chicken and platypus fibroblast cells did reveal that a substantial fraction of homogametic cells inactivate one gene copy (Deakin etal. 2008; Livernois etal. 2013). Furthermore, the fraction of cells with inactivated copies correlates positively with the magnitude of expression in the heterogametic sex. These observations suggest homogametic gene-by-gene down-regulation that provides some degree of “balance.” Contradicting this, however, is an analysis of allele-specific expression in chicken that suggests inactivation is not widespread among Z-linked loci (Zimmer etal. 2016). Further work on X/Z inactivation is clearly needed in these species. If it does occur, it may have evolved to avoid allosome overexpression in the homogametic sex, which is a hallmark of the final (Type IV) category of SCDC we address.
Type IV: Complete Compensation Without Balance (X = Ancestral < XX)
A key component of theory concerning SCDC evolution is that selection on (heterogametic) males for increased expression of sex-linked loci should cause a correlated increase in (homogametic) female sex-linked expression (Ohno 1967; Rice 1987; Engelstädter and Haig 2008; Mullon etal. 2015). Without a mechanism secondarily reducing female expression, a pattern of dosage compensation without balance would be expected, where females exhibit X hypertranscription relative to males and autosomes (Mank etal. 2011). Such a pattern has been reported in three different taxa so far, but in each case there is also contradictory evidence, so whether this Type IV pattern actually occurs remains uncertain.
The first empirical report of this pattern was from the flour beetle, T. castaneum. Initial results from microarray analysis of whole adult bodies strongly indicated female X hypertranscription (Prince etal. 2010). Later analyses limited to somatic tissues using RNA-Seq contradicted this, indicating X-linked expression is comparable to autosomes in both sexes (Mahajan and Bachtrog 2015). This discrepancy could arise from the technology used; the DNA–hybridization dynamics of microarrays may be less accurate than RNA-Seq in providing a measure of absolute expression differences between loci (Carey and Gentleman 2005; Marioni etal. 2008). More likely, it reflects the inclusion of gonads in the initial whole-body experiments, which may be obscuring or confounding patterns in the soma, as we noted earlier.
Another report of female X hypertranscription was in Drosophila serrata, again from microarray analysis of whole-body adults including gonads (Allen etal. 2013). Although this study collected data from separated soma and gonads, these were analyzed only for sex-biased gene expression but not SCDC. Notably, the somatic tissues showed minimal differences between sexes, which seemingly contradicts the observation of X-hypertranscription in females observed in whole-body analyses. Reproductive tissues yielded substantial sex-biased expression, with the X chromosome showing both an excess of female-biased genes and a dearth of male-biased genes, relative to autosomes. Thus the apparent observation of female X hypertranscription might primarily reflect idiosyncrasies of gonadal gene expression causing higher X than autosomal expression in the ovary, calling into question the generality of female X hypertranscription across tissues.
Finally, three-spined stickleback fish have recently evolved heterogametic XY chromosomes that have three distinct strata of differentiation. At one end of the X is a pseudo-autosomal region (PAR) that is adjacent to a second, medial pericentric stratum of intermediate X–Y divergence. The third, distal stratum exhibits substantial Y degeneration. Two studies have independently analyzed patterns of dosage compensation across these three stickleback X chromosome strata (Schultheiß etal. 2015; White etal. 2015). Both reported the absence of a gene dosage effect in the PAR and medial, younger stratum, but also the presence of a notable dosage effect in the distal, older stratum, which is therefore not dosage balanced. However, the reports differ in their assessment of dosage compensation for the distal section. Using autosomal expression levels as a reference, Schultheiß etal. (2015) argued that substantial up-regulation of male expression occurs and concomitantly causes female hypertranscription, consistent with a Type IV scenario. In contrast, White etal. (2015) analyzed expression levels in a comparative framework to infer ancestral expression levels of the X chromosome and concluded no dosage compensation or female hypertranscription is occurring in the distal stratum, leading to a Type III pattern.
Despite the current lack of any unambiguous evidence for this Type IV pattern, such pattern of heterogametic compensation accompanied by homogametic hypertranscription remains an intriguing theoretical possibility (Mullon etal. 2015). It will be interesting to see whether any solid evidence for this pattern will emerge from future research.
Dosage and Balance: Why or Why Not?
Recent advances in SCDC research present a very complex set of observations to interpret and reconcile with theory. Importantly, despite theoretical predictions and known costs of large dosage imbalances (Mank 2009; Veitia and Potier 2015), it increasingly appears that complete dosage compensation (regardless of balance) is not very common. Why might this be? One simple answer may be that complete compensation for all loci is unnecessary; for many genes, partial upregulation may be sufficient to mitigate negative fitness effects, even if expression still falls significantly short of ancestral expression levels. Achieving this “incomplete but sufficient” level of expression may result from pre-existing buffering mechanisms that generally mitigate the effects of aneuploidy (Stenberg and Larsson 2011; Malone etal. 2012; Mank 2013); additional selection for compensatory up-regulation may be unnecessary for such loci.
Furthermore, besides directly increasing expression of X-linked loci, several other processes may contribute to reducing the cost of dosage imbalances and reduced X-linked expression due to Y degeneration. Rice (1987) suggested that the evolution of autosomal tolerance of X-linked dosage effects may be an important outcome of Y degeneration. Indeed, the Type III pattern of SCDC (X < XX = AA) may reflect some combination of incomplete compensation and autosomal dosage tolerance. When X-linked expression is balanced between sexes, and fitness costs still exist due to incomplete compensation (as might occur for Type II; X = XX < AA), the evolution of reduced expression in autosomal loci interacting with X-linked loci may occur. For example, Julien etal. (2012) report that the evolution of reduced expression among many autosomal loci with interacting X-linked loci was coincident with sex chromosome differentiation in therian mammals. This same scenario could also prompt increased rates of gene duplication on the X chromosome as a means of increasing expression of X-linked transcripts relative to autosomal loci, a pattern recently noted in Humans (Hurst etal. 2015). There is also the possibility that increased rates of protein translation on the X helps to recapitulate ancestral levels of protein abundance, even when transcription is not completely compensated (Uebbing etal. 2015). Finally, X-to-autosome translocations would allow dosage sensitive genes to avoid negative fitness consequences of incomplete compensation (Vicoso and Charlesworth 2009b; Mikhaylova and Nurminsky 2011; Albritton etal. 2014; Gu etal. 2017).
Whatever the reason, in many cases the evidence suggests that complete dosage compensation may evolve only among small subsets of genes which are most likely to be sensitive to dosage effects, for instance among loci with many interacting autosomal partners or among sex-linked ohnologs (Lin etal. 2012; Pessia etal. 2012; Zimmer etal. 2016). Many other loci may be compensated to some extent, but whether this reflects generalized buffering of aneuploidies or locus-specific evolution of increased expression is difficult to discern. Nonetheless, the fact that only some genes are completely compensated stands in contrast to the chromosome-wide nature of “balance” mechanisms operating in the homogametic sex of various taxa, including therian mammals (XCI), Caenorhabditis (DCC), and Lepidoptera. Why should the gene-by-gene evolution of complete dosage compensation for a minority of X-linked loci spur the evolution of a global regulatory mechanism that halves homogametic expression of most or all X-linked loci? Given the substantial evidence for sex-specific control of gene expression levels at individual loci (Grath and Parsch 2016), invoking XCI or DCC as the resolution to sexual antagonism over optimal expression of a minority of X-linked loci seemingly invites comparisons to driving nails with a sledgehammer. Of course, partial compensation of other X-linked loci may also cause sexual antagonism that, when integrated across loci, might be sufficient to promote a global balance mechanism.
The evolutionary dynamics of this situation have not been thoroughly modeled and it remains substantially uncertain the extent to which moderate sexual antagonism around expression levels, when occurring simultaneously at many X-linked loci, could promote the spread of a global balance mechanism (Engelstädter and Haig 2008). Perhaps the equilibrium point for a global balance mechanism to emerge is when most loci are only partially compensated, causing a Type II pattern (X = XX < AA). Similarly, perhaps it is sufficient for strong sexual antagonism at just a few dosage sensitive loci to drive a global balance mechanism, so long as other affected loci are not dosage sensitive or are otherwise accommodated via mechanisms noted above. In other words, could it be that Ohno’s 2-step hypothesis is essentially correct, but with a relaxed requirement for complete compensation of most loci before XCI evolves (or comparable global balance mechanisms in other taxa)? The dynamics of this process may also depend substantially on how the Y chromosome degrades, for instance via Muller’s ratchet or selective sweeps, which may determine whether dosage compensation evolves gene-by-gene or in a more block-wise fashion (Charlesworth 1996; Vicoso and Charlesworth 2009a). Clearly there is much opportunity for further theoretical work in this area. However, we should not ignore the distinct possibility that these balance mechanisms initially evolved for reasons unrelated to SCDC, as has been suggested for mammalian XCI (Haig 2006; Engelstädter and Haig 2008; Pessia etal. 2014). Such alternative hypotheses have yet to be introduced for other taxa that show global balance mechanisms but generally incomplete dosage compensation.
Another important dynamic in understanding when dosage compensation or balance will evolve seems to be male- versus female-heterogamety. As noted above, with the exception of Lepidoptera, ZW taxa exhibit Type III patterns (Z < ZZ = AA) of SCDC, raising questions about why this pattern is so common among female-heterogametic species. One suggested reason is that male mutation bias (due to increased DNA replication in spermatogenesis versus oogenesis) causes the Y to degenerate faster than the W, since the Y is always found in males but the W never is (Vicoso and Bachtrog 2009; Wilson Sayres and Makova 2011; Mank 2013). This slower-W phenomenon would allow more time for dosage-sensitive genes on the Z to compensate individually (e.g., through cis-regulatory evolution), thus mitigating selection for chromosome-wide mechanisms to evolve (Naurin etal. 2012). Alternatively, a smaller-Z phenomenon may explain the dichotomy with XY systems, suggesting that a reduced effective population size (Ne) on the Z leaves it less able to adapt to the eroding W as compared with XY systems (Mank 2013). This smaller-Z effect rests on how sexual selection influences Ne of the Z and X chromosomes relative to autosomes. Sexual selection typically causes high variance in male-mating success such that females genetically contribute more than males to each generation. This lowers NeZ but raises NeX relative to NeA, reducing the adaptive potential of the Z relative to X (Vicoso and Charlesworth 2009a; Mank etal. 2010). A reduced ability to adapt to the eroding W may explain the lack of completely dosage compensated Z chromosomes. This explanation involving sexual selection and Ne was modeled formally and was also supported by empirical studies in chicken (Mullon etal. 2015).
Future Directions in Studying the Evolution of SCDC
Much has been learned about SCDC in the last few years, though in many ways it has been one step back for two steps forward. While expanding genomic and comparative analyses of SCDC have uncovered a surprising diversity of patterns, this work has also demonstrated the limitations of our current theory in explaining this diversity. Continuing to broaden the scope of taxa in which dosage compensation has been assayed is one clear path forward; this would provide a broader comparative context of SCDC patterns across animals (and plants). Another important path forward is through further integrating studies of molecular mechanisms with characterizations of pattern. Deeper proximal knowledge of the epigenetic and transcriptional machinery governing dosage compensation and balance will substantially advance our ultimate theoretical understanding of evolutionary process by highlighting to what extent different organisms share, or have converged on, specific mechanisms.
Studying young sex chromosomes in the early stages of degeneration is one obvious area for future focus that has already proved fruitful (Bachtrog 2013; Zhou etal. 2013; Nozawa etal. 2014; White etal. 2015; Gu etal. 2017). However, an important distinction should be made among such systems, which may be broadly split into situations concerning de novo versus a priori sex chromosome evolution. In many ways, the most informative sex chromosome systems to study would be those where sex chromosomes have recently evolved de novo, for instance in the transition from environmental to genetic sex determination (Bachtrog etal. 2014). Presumably, in such cases, any dosage compensating system would evolve independently “from scratch.” This should be contrasted with an a priori scenario, such as a neo-sex chromosome resulting from an autosomal fusion with an existing allosome. In the case of a neo-sex chromosome, it seems most likely that the evolution of SCDC would be strongly influenced by pre-existing dosage compensation mechanisms that could be readily co-opted from the ancestral sex chromosome to the recently acquired neo-sex fragment (Vicoso and Bachtrog 2009). Such neo-sex chromosomes are unquestionably informative concerning how sex chromosomes and dosage compensation evolve, particularly for a given lineage already possessing genetic sex determination, but they cannot necessarily be considered as independent data points in our understanding of how dosage compensation evolved in the first place (e.g., de novo). Nonetheless, such a priori scenarios are going to be relatively more common than de novo origins of sex chromosomes, so in terms of understanding common events in sex chromosome biology, they are important to investigate. Between these two extremes (de novo versus a priori) are scenarios such as found in Diptera, with many independent transitions of sex chromosomes, which may or may not involve allosome–autosome fusion, but which indicate substantial turnover in sex chromosome identity (Vicoso and Bachtrog 2015). Dosage compensation and balance remain conserved in flies, but it is largely uncertain to what extent the same mechanisms are retained across these multiple changes in sex chromosomes.
Another poorly addressed issue in SCDC evolution is the extent to which dosage compensation (as well as balance) evolves via localized gene-by-gene mechanisms or, in contrast, through a “global” mechanism involving multi-gene blocks or even entire chromosomes. Whether one or the other is expected initially may depend on the manner by which the Y chromosome degrades, for instance via selective sweeps or Muller’s ratchet. It may also be influenced by the effective population size of the organism in question. And of course, it may be possible for localized compensation to precede “global” regulation. An extensive discussion of these issues is provided by Charlesworth (1996) and Vicoso and Bachtrog (2009), so we will not reiterate it further here. Yet it does not seem we are much closer to answering these questions now than in decades past.
It is clear that we are currently in the midst of a dynamic and revolutionary moment for both research and theory concerning SCDC. While the emerging diversity of patterns and occasionally inconsistent results can seem bewildering or frustrating at times, opportunity abounds for future investigations, insights, and discoveries.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by NSF-DEB 1457758 (to J.R.W.) and start-up funds provided to J.R.W. by the University of Kansas. Three anonymous reviewers made helpful suggestions for improving the manuscript. We are grateful to B. Vicoso and colleagues for sharing their results concerning P. interpunctella.
Literature Cited
- Ahola V. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun. 5:4737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albritton SE, et al. 2014. Sex-biased gene expression and evolution of the x chromosome in nematodes. Genetics 197(3):865–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, et al. 2013. Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 27:853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SL, Bonduriansky R, Chenoweth SF.. 2013. The genomic distribution of sex-biased genes in Drosophila serrata: X chromosome demasculinization, feminization, and hyperexpression in both sexes. Genome Biol Evol. 5(10):1986–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2006. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 16(6):578–585. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processesof Y-chromosome degeneration. Nat Rev Genet. 14(2):113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12(7):e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RH, Wilkinson GS.. 2010. Comparative genomic hybridization (CGH) reveals a neo-X chromosome and biased gene movement in stalk-eyed flies (genus Teleopsis). PLoS Genet. 6(9):e1001121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. 2012. Claims and counterclaims of X-chromosome compensation. Nat Publish Group 19(1):3–5. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL.. 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 234(2):275–288. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA.. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21:219–226. [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S.. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey V, Gentleman R.. 2005. Analysis overview In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer Science & Business Media; p. 183–185. [Google Scholar]
- Castagné R, et al. 2011. The choice of the filtering method in microarrays affects the inference regarding dosage compensation of the active X-chromosome. PLoS ONE 6(9):e23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler CH. 2017. When and why does sex chromosome dosage compensation evolve? Ann N Y Acad Sci. 1389(1):37–51. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1978. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 75(11):5618–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1996. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 6(2):149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1991. The evolution of sex chromosomes. Science 251(4997):1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais GAB.. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 46(3):253–260. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J.. 2015. No X-chromosome dosage compensation in human proteomes. Mol Biol Evol. 32(6):1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang J.. 2016. The X to autosome expression ratio in haploid and diploid human embryonic stem cells. Mol Biol Evol. 33(12):3104–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG.. 2010. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64(12):3417–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JA.. 2008. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 4(7):e1000140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 43(12):1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, et al. 2013. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev Cell 25(1):55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. 2016. Dosage compensation of the sex chromosomes and autosomes. Semin Cell Dev Biol. 56:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. 2012. Dosage compensation of the sex chromosomes. Annu Rev Genet. 46:537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2007. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc R Soc B: Biol Sci. 274(1606):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. 2007. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biology 5:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstädter J, Haig D.. 2008. Sexual antagonism and the evolution of X chromosome inactivation. Evolution 62(8):2097–2104. [DOI] [PubMed] [Google Scholar]
- Ercan S. 2015. Mechanisms of X chromosome dosage compensation. J Genomics 3:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucillion ML, Larsson J.. 2015. Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome Biol Evol. 7(4):1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F, Alekseyenko AA, Park PJ, Kuroda MI.. 2014. Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat Publish Group 21(2):118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, et al. 2015. The endosymbiotic bacterium wolbachia selectively kills male hosts by targeting the masculinizing gene. PLoS Pathog. 11(7):e1005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM. 2014. A brief history of dosage compensation. J Genet. 93(2):591–595. [DOI] [PubMed] [Google Scholar]
- George NI, Chang C-W.. 2014. DAFS: a data-adaptive flag method for RNA-sequencing data to differentiate genes with low and high expression. BMC Bioinformatics 15:92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grath S, Parsch J.. 2016. Sex-biased gene expression. Annu Rev Genet. 50:29–44. [DOI] [PubMed] [Google Scholar]
- Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 17(1):33–46. [DOI] [PubMed] [Google Scholar]
- Gu L, Walters JR, Knipple DC.. 2017. Conserved patterns of sex chromosome dosage compensation in the Lepidoptera (WZ/ZZ): insights from a moth neo-Z chromosome. Genome Biol Evol. 9(3):802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guioli S, Lovell-Badge R, Turner JMA.. 2012. Error-prone ZW pairing and no evidence for meiotic sex chromosome inactivation in the chicken germ line. PLoS Genet. 8(3):e1002560.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, et al. 2006. Global analysis of X-chromosome dosage compensation. J Biol. 5(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. 2006. Self-imposed silence: parental antagonism and the evolution of X-chromosome inactivation. Evolution 60(3):440–447. [PubMed] [Google Scholar]
- Hardcastle TJ. 2016. Generalized empirical Bayesian methods for discovery of differential data in high-throughput biology. Bioinformatics 32(2):195–202. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Mank JE, Wedell N.. 2012. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 4(11):1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DR.. 2013. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14:778.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, et al. 2011. He etal. reply. Nat Genet. doi:10.1038/ng.1010. [Google Scholar]
- Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Ghanbarian AT, Forrest ARR, FANTOM consortium, Huminiecki L.2015. The constrained maximal expression level owing to haploidy shapes gene content on the mammalian X chromosome Barton, NH, editor. PLoS Biol. 13:e1002315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huylmans AK, Macon A, Vicoso B.. 2017. Global dosage compensation is ubiquitous in Lepidoptera, but counteracted by the masculization of the Z chromosome. Mol Biol Evol. 10.1093/molbev/msx190. [DOI] [PMC free article] [PubMed]
- Itoh Y, et al. 2007. Dosage compensation is less effective in birds than in mammals. J Biol. 6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, et al. 2010. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 20(4):512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Biedler JK, Qi Y, Hall AB, Tu Z.. 2015. Complete dosage compensation in Anopheles stephensi and the evolution of sex-biased genes in mosquitoes. Genome Biol Evol. 7(7):1914–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue NK, et al. 2013. Determination of dosage compensation of the mammalian X chromosome by RNA-seq is dependent on analytical approach. BMC Genomics 14:150.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, et al. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation Barton, NH, editor. PLoS Biol. 10(5):e1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka S, et al. 2016. Status of dosage compensation of X chromosome in bovine genome. Genetica 144(4):435–444. [DOI] [PubMed] [Google Scholar]
- Kelly WG, et al. 2002. X-chromosome silencing in the germline of C. elegans. Development 129(2):479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi R, Park PJ.. 2011. correspondence. Nat Genet. 43(12):1167–1169. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509:633–636. [DOI] [PubMed] [Google Scholar]
- Kuroda MI, Hilfiker A, Lucchesi JC.. 2016. Dosage compensation in Drosophila—a model for the coordinate regulation of transcription. Genetics 204(2):435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X.. 2012. Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc Natl Acad Sci. 109(29):11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger RJ, Belikoff EJ, Scott MJ, Marais GAB.. 2015. Dosage compensation of X-linked Muller element F genes but not X-linked transgenes in the Australian sheep blowfly. PLoS ONE 10(10):e0141544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livernois AM, Waters SA, Deakin JE, Marshall Graves JA, Waters PD.. 2013. Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals. PLoS Genet. 9(7):e1003635.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kuroda MI.. 2015. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 7(5):a019398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Bachtrog D.. 2015. Partial dosage compensation in strepsiptera, a sister group of beetles. Genome Biol Evol. 7(2):591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, et al. 2012. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 13(4):r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29(12):677–683. [DOI] [PubMed] [Google Scholar]
- Mank JE. 2012. Small but mighty: the evolutionary dynamics of W and Y sex chromosomes. Chrom Res. 20(1):21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2009. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet. 25(5):226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N.. 2011. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution 65(8):2133–2144. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B.. 2010. Effective population size and the Faster-X effect: empirical results and their interpretation. Evolution 64(3):663–674. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y.. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18(9):1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín I, Franke A, Bashaw GJ, Baker BS.. 1996. The dosage compensation system of Drosophila is co-opted by newly evolved X chromosomes. Nature 383(6596):160–163. [DOI] [PubMed] [Google Scholar]
- Marín I, Siegal ML, Baker BS.. 2000. The evolution of dosage-compensation mechanisms. BioEssays 22(12):1106–1114. [DOI] [PubMed] [Google Scholar]
- Martin SH, et al. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Landeen EL, Cook JM, Kingan SB, Presgraves DC.. 2011. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation eisen, MB, editor. PLoS Biol. 9(8):e1001126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Malone JH, Clark AG.. 2012. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 22:1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH. 2000. Dosage compensation: making 1X equal 2X. Trends Cell Biol. 10(2):54–59. [DOI] [PubMed] [Google Scholar]
- Mikhaylova LM, Nurminsky DI.. 2011. Lack of global meiotic sex chromosome inactivation, and paucity of tissue-specific gene expression on the Drosophila X chromosome. BMC Biology 9:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullon C, Wright AE, Mank JE, Reuter M, Pomiankowski A.. 2015. Evolution of dosage compensation under sexual selection differs between X and Z chromosomes. Nat Commun. 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, et al. 2016. SEX-DETector: a probabilistic approach to study sex chromosomes in non-model organisms. Genome Biol Evol. 8(8):2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Shearn R, Marais G.. 2017. The evolution of sex chromosomes and dosage compensation in plants. Genome Biol Evol. 9:627–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naurin S, Hasselquist D, Bensch S, Hansson B.. 2012. Sex-biased gene expression on the avian Z chromosome: highly expressed genes show higher male-biased expression. PLoS ONE 7(10):e46854.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM.. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet. 38(1):47–53. [DOI] [PubMed] [Google Scholar]
- Nguyen P, et al. 2013. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc Natl Acad Sci. 110(17):6931–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Fukuda N, Ikeo K, Gojobori T.. 2014. Tissue- and stage-dependent dosage compensation on the Neo-X chromosome in Drosophila pseudoobscura. Mol Biol Evol. 31(3):614–624. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex-linked genes. Berlin: Springer. [Google Scholar]
- Oshlack A, Robinson MD, Young MD.. 2010. From RNA-seq reads to differential expression results. Genome Biol. 11(12):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Vicoso B.. 2015. The X chromosome of hemipteran insects: conservation, dosage compensation and sex-biased expression. Genome Biol Evol. 7:3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Kuroda MI.. 2001. Epigenetic aspects of X-chromosome dosage compensation. Science 293(5532):1083–1085. [DOI] [PubMed] [Google Scholar]
- Parsch J, Ellegren H.. 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 14(2):83–87. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW.. 2012. Sex chromosomes evolved from independent ancestral linkage groups in winged insects. Mol Biol Evol. 29(6):1645–1653. [DOI] [PubMed] [Google Scholar]
- Pessia E, Engelstädter J, Marais GAB.. 2014. The evolution of X chromosome inactivation in mammals: the demise of Ohno's hypothesis? Cell Mol Life Sci. 71(8):1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB.. 2012. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci. 109(14):5346–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pferdehirt RR, Meyer BJ.. 2013. SUMOylation is essential for sex-specific assembly and function of the Caenorhabditis elegans dosage compensation complex on X chromosomes. Proc Natl Acad Sci. 110(40):E3810–E3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince EG, Kirkland D, Demuth JP.. 2010. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol Evol. 2:336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC.. 2010. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci. 107(41):17657–17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38(4):735–742. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1996. Evolution of the Y sex chromosome in animals. Bioscience 46(5):331–343. [Google Scholar]
- Rice WR. 1987. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116(1):161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, et al. 2016. Dosage compensation in the African malaria mosquito Anopheles gambiae. Genome Biol Evol. 8(2):411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmakers S, et al. 2009. Female meiotic sex chromosome inactivation in chicken. PLoS Genet 5: e1000466. doi: 10.1371/journal.pgen.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiß R, Viitaniemi HM, Leder EH.. 2015. Spatial dynamics of evolving dosage compensation in a young sex chromosome system. Genome Biol Evol. 7(2):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD.. 2014. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol Evol. 6(3):526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinemann M, Steinemann S.. 1998. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica 102–103:409–420. [PubMed] [Google Scholar]
- Stenberg P, Larsson J.. 2011. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120(3):213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Becker PB.. 2007. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 8(1):47–57. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Abe K.. 2007. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 3(7):e116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto TN, Kayukawa T, Shinoda T, Ishikawa Y, Tsuchida T.. 2015. Misdirection of dosage compensation underlies bidirectional sex-specific death in Wolbachia-infected Ostrinia scapulalis. Insect Biochem Mol Biol. 66:72–76. [DOI] [PubMed] [Google Scholar]
- Toups MA, Hahn MW.. 2010. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 186(2):763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA. 2007. Meiotic sex chromosome inactivation. Development 134(10):1823–1831. [DOI] [PubMed] [Google Scholar]
- Uebbing S, et al. 2015. Quantitative mass spectrometry reveals partial translational regulation for dosage compensation in chicken. Mol Biol Evol. 32(10):2716–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing S, Kunstner A, Makinen H, Ellegren H.. 2013. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 5(8):1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, Potier MC.. 2015. Gene dosage imbalances: action, reaction, and models. Trends Biochem Sci. 40(6):309–317. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Veyrunes F, Bottani S, Birchler JA.. 2015. X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. J Mol Cell Biol. 7(1):2–11. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, et al. 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18(6):965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD. 2014. Meiotic sex chromosome inactivation in Drosophila. J Genomics 2:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 3:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13(4):e1002078.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D.. 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chrom Res. 17(5):585–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B.. 2009a. Effective population size and the faster-X effect: an extended model. Evolution 63(9):2413–2426. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B.. 2009b. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 68(5):576–583. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D.. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11(8):e1001643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JR, Hardcastle TJ.. 2011. Getting a full dose? Reconsidering sex chromosome dosage compensation in the silkworm, Bombyx mori. Genome Biol Evol. 3:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JR, Hardcastle TJ, Jiggins CD.. 2015. Sex chromosome dosage compensation in heliconius butterflies: global yet still incomplete? Genome Biol Evol. 7(9):2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid nasonia species. Science 327(5963):343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler BS, Anderson E, Frøkjær-Jensen C, Bian Q.. 2016. Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. Elife doi:10.7554/eLife.17365.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Kitano J, Peichel CL.. 2015. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol Biol Evol. 32(8):1981–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth DJ, Pask AJ.. 2016. The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Semin Cell Dev Biol. 56:117–121. [DOI] [PubMed] [Google Scholar]
- Wilkinson GS, Johns PM, Metheny JD, Baker RH.. 2013. Sex-biased gene expression during head development in a sexually dimorphic stalk-eyed fly. PLoS ONE 8(3):e59826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres MA, Makova KD.. 2011. Genome analyses substantiate male mutation bias in many species. BioEssays 33(12):938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Bryk J.. 2011. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics 12:91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, et al. 2010. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 42(12):1043–1047. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Sadreyev RI, Pinter SF, Lee JT.. 2011. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Publish Group 19(1):56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha X, et al. 2009. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem Mol Biol. 39(5–6):315–321. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Oliver B.. 2007. Dosage compensation goes global. Curr Opin Genet Dev. 17(2):113–120. [DOI] [PubMed] [Google Scholar]
- Zhou Q, et al. 2013. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11(11):e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer F, Harrison PW, Dessimoz C, Mank JE.. 2016. Compensation of dosage-sensitive genes on the chicken Z chromosome. Genome Biol Evol. 8(4):1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.