Abstract
Mutations in the spliceosomal RNA binding protein RBM10 cause TARP syndrome and are frequently observed in lung adenocarcinoma (LUAD). We have previously shown that RBM10 enhances exon skipping of its target genes, including its paralog RBM5. Here, we report that RBM10 negatively regulates its own mRNA and protein expression and that of RBM5 by promoting alternative splicing-coupled nonsense-mediated mRNA decay (AS-NMD). Through computational analysis and experimental validation, we identified RBM10-promoted skipping of exon 6 or 12 in RBM10 and exon 6 or 16 in RBM5 as the underlying AS-NMD events. Importantly, we showed that LUAD-associated mutations affecting splice sites of RBM10 exons 6 or 12 abolished exon inclusion and correlated with reduced expression of RBM10 RNA. Together, our investigations have revealed novel molecular mechanisms underlying RBM10 autoregulation and cross-regulation of RBM5, thereby providing insights concerning the functions of RBM10 under various physiological and pathological conditions. Our combined computational and experimental approach should be useful for elucidating the role of AS-NMD in auto- and cross-regulation by other splicing regulators.
INTRODUCTION
Alternative splicing (AS) is a key molecular mechanism underlying transcriptomic and proteomic diversity in eukaryotic cells (1–3). AS plays critical roles in both normal and disease contexts (4–7). In addition to generating protein isoforms with distinct functions (8) and functionally important non-coding RNAs (9,10), AS also produces mRNA variants that are substrates for nonsense-mediated mRNA decay (NMD) (11). AS is largely regulated by interactions between cis-regulatory elements in pre-mRNA and trans-acting splicing regulators, which are primarily RNA binding proteins (RBPs) (12–14). Although the molecular mechanisms of many splicing regulators have been explored, it is not fully understood how their own expression is controlled to produce specific RNA splicing patterns under different physiological conditions (15).
AS events that generate premature termination codons (PTCs) are often coupled with NMD, which is important not only for eliminating aberrant mRNA transcripts containing PTCs, but also for post-transcriptional tuning of gene expression (11,16). mRNA transcripts containing a PTC located more than 50–55 nt upstream from the last exon-exon junction or a long 3’-untranslated region (3’UTR) are often substrates for NMD (16–20). mRNA degradation by this pathway depends on translation and subsequent recruitment of essential NMD factors, including the central factor, up frameshift 1 (UPF1) (16–18). Intriguingly, it has been observed that some splicing regulators maintain homeostasis via negative autoregulation mediated by AS-NMD (15). Splicing regulators that utilize this mechanism include SR and hnRNP family members (21,22), TDP43 (23), MBNL (24,25) and core spliceosomal proteins (26,27). Moreover, a number of splicing regulators have been found to regulate cellular concentrations of their family members and/or other splicing modulators (27–29). Examples of cross-regulating splicing factors include hnRNPL/hnRNPLL (30), PTBP1/PTBP2 (31) and Rbfox2/Ptbp2 (27). Notably, AS-NMD for these splicing regulators is often associated with regulatory regions containing highly evolutionarily conserved, or even ultra-conserved elements, as reported for SR, hnRNP (32–34) and MBNL family proteins (24). Disruption of auto- or cross-regulation of splicing regulators has been linked to various diseases, including neurodegenerative diseases and cancer (25,35–37). Understanding auto- and cross-regulatory mechanisms of splicing regulators may yield new clues to their functions under normal and disease conditions. The detection of AS-NMD, however, is often challenging due to the instability of PTC-containing transcripts.
RBM10 is an RNA binding protein and a component of the spliceosome, where it has been shown to interact with multiple spliceosomal proteins (38). Truncation mutations of RBM10 have been reported to cause the X chromosome-linked recessive genetic disorder, TARP syndrome (Talipes equinovarus, atrial septal defect, Robin sequence and persistent left superior vena cava, MIM #311900) (39–42). In addition, RBM10 mutations are frequently observed in lung adenocarcinomas (LUAD) (43,44) and have also been detected in colorectal carcinomas (45), intraductal papillary mucinous neoplasms (IPMN) (46) and pancreatic ductal adenocarcinomas (PDA) (47).
Our previous integrative analysis of transcriptome-wide RBM10-RNA interactions and RBM10-mediated splicing revealed that RBM10 suppresses exon inclusion by binding to flanking intronic regions adjacent to splice sites, including sites in its own pre-mRNA, and regulates AS of several RBPs, including its closest paralog RBM5 (42). Other studies have revealed RBM10-RNA interaction profiles (48,49) similar to those described in our previous study (42). We also identified an in-frame deletion in RBM10 that contributes to TARP syndrome, presumably via loss of its splicing function (42). Our recent functional analysis suggests that LUAD-associated RBM10 mutations contribute to LUAD pathogenesis by deregulating splicing (50). It has also been shown that RBM10 can suppress lung adenocarcinoma cell proliferation, at least partially by inhibiting inclusion of exon 9 in the NUMB long isoform, thereby negatively regulating NOTCH signaling activity (49,51). Collectively, those studies suggest that RBM10 functions as a crucial node in splicing regulatory networks that play important roles in normal development and disease. Whether RBM10 regulates its own expression and cross-regulates that of RBM5, and the molecular mechanisms underlying this regulation, however, remain unexplored.
In this study, we show that RBM10 negatively regulates its own expression and that of RBM5 via AS-NMD. By analyzing RBM10 binding sites using photoactivatable ribonucleotide-enhanced crosslinking and immunoprecipitation (PAR-CLIP) and measuring splicing changes following perturbations of RBM10, we identified and experimentally validated previously unknown RBM10-controlled AS-NMD events in RBM10 and RBM5. We also found that several LUAD-associated RBM10 splice site mutations disrupt splicing and thus significantly decreased RBM10 expression. Overall, our results reveal new molecular mechanisms underlying RBM10 autoregulation and cross-regulation of RBM5. The methods established here can readily be extended to investigate potential roles of AS-NMD in regulating other splicing factors.
MATERIALS AND METHODS
Constructs
An expression vector encoding RBM10-EGFP was constructed by subcloning the RBM10 coding sequence (NM_005676.4) from the plasmid pFRT-TO-RBM10 (42) into pEGFP-N3 (Clontech). A tet-on lentiviral plasmid encoding RBM10-EGFP (Tet-RBM10-EGFP) was constructed by subcloning RBM10-EGFP into pLVX-Tight-Puro (Clontech). Minigene splicing reporters for test exons (RBM10-E6M/E12M/E14M/E21M for RBM10 exon 6, 12, 14 or 21, pZW2C-E6M/E12M for RBM10 exon 6 or 12, and RBM5-E6M/E8M/E16M for RBM5 exon 6, 8 or 16, respectively) were constructed. For each exon, a chromosomal segment extending from the adjacent upstream exon to the adjacent downstream exon was amplified from HEK293 genomic DNA and inserted into restriction enzyme-linearized pcDNA3.1 vector (Invitrogen). For RBM10 exons 6 and 12, a DNA segment including the exon and its flanking introns was PCR-amplified from RBM10-E6M/E12M and inserted into the GFP coding segment of PCR-linearized pZW2C (52) respectively. In both cases, cloning was carried out using Clone Express II reagent (Vazyme) following the manufacturer's protocol. Mutations were introduced into the RBM10-E6M and RBM10-E12M minigene using Mut Express II Fast Mutagenesis Kit V2 (Vazyme) following the manufacturer's instructions. Primers used are listed in Supplementary Table S2.
Cell culture
HEK293 cells were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum (FBS, Invitrogen). 293FT (Thermo Fisher) cells were cultured in DMEM supplemented with 10% FBS, 6 mM L-glutamine, 1 mM sodium pyruvate (Sigma-Aldrich) and 0.1 mM non-essential amino acids (NEAA, Invitrogen). Mouse neuroblastoma N2a cells were cultured in MEM (Invitrogen) supplemented with 10% FBS. All cultures were maintained under standard culture conditions (37 °C, 5% CO2).
Generation of tet-on RBM10-EGFP HEK293 cells
Inducible overexpression of RBM10-EGFP in HEK293 cells (tet-RBM10-EGFP) was achieved using the Lenti-X Tet-On Advanced Inducible Expression System (Clontech) according to the manufacturer's manual. Briefly, the lentivirus constructs pLVX-Tet-On or pLVX-tight-RBM10-EGFP were transfected into 293FT cells together with three packaging plasmids (pLP1, pLP2, and pLP/VSVG, Invitrogen). Medium containing the lentiviruses was collected 48 and 72 h after transfection, filtered and used to infect cells in the presence of 10 μg/ml polybrene (Sigma-Aldrich). HEK293 cells were first infected with Tet-On lentivirus and selected by culturing infected cells in the presence of 500 μg/ml G418. The G418 selected cells were subsequently infected with pLVX-tight-RBM10-EGFP lentiviruses and selected by culturing the infected cells in the presence of 2 μg/ml puromycin. After selection, stably transfected cells were maintained in normal culture medium without antibiotics. RBM10-EGFP expression was induced by exposing the cells to 10, 100 or 1000 ng/ml doxycycline (Dox, Sigma-Aldrich).
Minigene splicing reporter assay
To study target exon splicing following RBM10 overexpression (OE) or knockdown (KD), HEK293 cells were co-transfected with minigene splicing reporters and the RBM10-EGFP expression plasmid or siRNA oligos targeting the RBM10 coding sequence (siRBM10), respectively. For RBM10 OE, HEK293 cells were seeded in six-well plates (4 × 105 cells per well), cultured for 24 h and transfected with 500 ng minigene reporter together with 500 ng RBM10-EGFP expression or EGFP-N3 control plasmids using Lipofectamine 3000 (Thermo Scientific). For RBM10 KD, siRNAs against RBM10 (siRBM10) or negative control oligos (siNC) were transfected into HEK293 cells at a final concentration of 20 nM with Lipofectamine RNAiMAX (Thermo Scientific), using the reverse transfection method described in the manufacturer's manual. The medium was replaced with complete growth medium 8 h after the transfection. Minigene plasmids were transfected into the cells 24 h after siRNA transfection using Lipofectamine 3000 (Thermo Scientific). Cells were harvested 48 h after siRNA transfection and used for isolation of total RNA, cDNA synthesis and PCR analysis. Sequences of siRNAs used in this study are listed in Supplementary Table S2.
Nonsense-mediated mRNA decay (NMD) inhibition and RBM10 overexpression
We carried out UPF1 depletion to directly block NMD or cycloheximide (CHX) treatment to indirectly block NMD under the control or RBM10 overexpression (OE) conditions in HEK293 cells inducibly expressing RBM10-EGFP. For UPF1 depletion, cells were transfected with siRNA oligos targeting UPF1 (siUPF1) or negative control oligos (siNC) at final concentration of 20 nM using Lipofectamine RNAiMAX (Thermo Scientific). Forty eight hours after the first transfection, the cells were again transfected with siRNAs at the same concentrations. RBM10 OE was induced with 1000 ng/ml doxycycline (Dox, Sigma-Aldrich), which was added to the culture media with first set of siRNAs. Cells were harvested 96 hrs after the first transfection. For CHX treatment, HEK293 cells were treated for 12 h with 10 μg/ml cycloheximide (Sigma-Aldrich) or the vehicle dimethylsulphoxide (DMSO) as a control. RBM10 overexpression was induced by addition of 1000 ng/ml doxycycline (Dox, Sigma-Aldrich) to the culture medium 48 h prior to addition of CHX or DMSO.
Total RNA preparation, reverse transcription followed by PCR amplification (RT-PCR) and quantitative real-time PCR (qPCR)
Total RNA was extracted from cell lysates in TRIzol (Invitrogen) according to the manufacturer's instructions. Genomic DNA was removed using TURBO DNA-free kits (Ambion). Purified total RNA (1 μg for 20 μl reaction) was reverse transcribed using HiScript II Reverse Transcriptase (Vazyme) and oligo dT20 primers. Splicing outcomes were assessed by PCR using specific primers binding to the flanking-exons, which detect both exon-included and skipped transcripts. PCR products were mixed with loading buffer containing GelRed fluorescent DNA stain (GENEray), resolved in 1.5 or 2% agarose gels and visualized using a UV image system. Signal intensity of bands was quantified using ImageJ software (http://rsb.info.nih.gov/ij/). We defined percent-spliced-in (PSI) as: intensity of the exon-included band/ (intensity of the exon-included band + intensity of the exon-excluded band) × 100%. When one product was amplified, relative expression levels of mRNA transcripts were measured by quantitative real-time PCR (qPCR) using AceQ qPCR SYBR Green Master Mix (Vazyme) and the ABI Step One Plus system (Applied Biosystems). Three technical replicates were included for each reaction condition. Quantification of qPCR results was performed using the ΔΔCt method. Sequences of PCR primers are listed in Supplementary Table S2.
Western blotting
Western blot analysis was performed as previously described (42). Briefly, cells were lysed in Laemmli lysis buffer (63 mM Tris–HCl, 10% glycerol, 2 % SDS, pH 6.8), followed by sonication, centrifugation and heat denaturation. Cell lysates were resolved by SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were blocked with 5% milk in TBST and sequentially probed with the primary antibodies and secondary antibodies coupled to horseradish peroxidase (HRP). Specific protein bands were detected using an enhanced chemiluminescence (ECL) system (Millipore) and visualized by ChemiDoc™ XRS+ image system (BioRad). The following antibodies were used: RBM10 (cat. No. HPA034972, 1:10 000, Sigma), RBM5 (cat. no. HPA017335, 1: 500, Sigma), α-tubulin (cat. no.11224–1-AP, 1:5000, Proteintech), Goat anti-rabbit IgG-HRP (cat. no. 458, 1:5000, MBL).
Fluorescence microscopy
Tet-RBM10-EGFP HEK293 cells were grown on poly-d-lysine coated coverslips and treated with 1 μg/ml doxycycline for 24 h. Cells were fixed with 4% paraformaldehyde, stained with 1 μg/ml DAPI and mounted on the glass slides. Fluorescent images were acquired using a Leica DM IL LED fluorescent microscope with a 40× objective and DFC450 C camera (Leica). Brightness and contrast were adjusted within the linear range using ImageJ software.
UV crosslinking (CL), immunoprecipitation (IP) and RT-PCR (CLIP-PCR)
HEK293 cells (2 × 107 cells per IP) were irradiated with 254 nm UV light (300 mJ/cm2) in a UV cross-linker (HL-2000 HybriLinker, UVP) and immediately lysed with CLIP buffer (50 mM Tris–HCl pH 7.4, 100 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate and Protease Inhibitor Cocktail III) on ice for 15 min. Lysates were sonicated (5 min, 30 s on/30 s off), treated with Turbo DNase I (Ambion) and cleared by centrifugation. The supernatants were incubated with anti-RBM10 antibody or IgG-bound beads (10 μg antibodies conjugated to 50 μl beads; anti-RBM10, cat. no. HPA034972, Sigma Aldrich; rabbit IgG, cat. no. I5006, Sigma Aldrich; Dynabeads Protein G, Invitrogen) and rotated at 4°C for 2 h. Beads were collected and sequentially washed six times with high salt wash buffer (50 mM Tris–HCl pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS and 0.5% sodium deoxycholate) and wash buffer (20 mM Tris–HCl pH 7.4, 10 mM MgCl2 and 0.2% Tween-20). Precipitated RNA fragments were released from beads by Protein K (NEB) digestion and extracted by acid phenol/chloroform/isoamyl alcohol. IP efficiency and specificity were confirmed by Western blot analysis. RBM10 bound RNA fragments were and reverse-transcribed by random primer and detected by PCR using primers listed in Supplementary Table S2. The following negative controls were used: IgG for RBM10 IP, no reverse transcriptase (RT-) for DNA contamination and non-template control (NTC) for PCR artifacts.
RBM10 mutations and RNA expression in the TCGA database
RBM10 mutation and expression data in lung adenocarcinoma patient samples were downloaded from The Cancer Genome Atlas (TCGA) website (http://cancergenome.nih.gov/). RNA expression levels in TCGA were estimated from RNA-Seq data using the RSEM algorithm (53) and presented as ‘normalized counts’ following upper quartile normalization (54).
PAR-CLIP data analysis
PAR-CLIP DNA sequencing reads were downloaded from the NCBI GEO database (GSE44976). After trimming-off adapter sequences, the sequencing reads were aligned to the human genome and transcriptome (GRCh37, Ensemblv71) allowing at most two mismatches and/or indels using Tophat2 (55). Potential binding clusters were defined by the overlapping reads as previously described (42). For each cluster, the crosslinking position was defined as the position with the highest number of T to C conversions.
RNA-Seq data analysis
RNA-Seq reads were downloaded from the NCBI GEO database (GSE44976) and aligned to the human genome and transcriptome (GRCh37, Ensemblv71), allowing at most 6 mismatches and/or indels using Tophat2 (55). We searched for all possible single and multiple-exon skipping events in RBM10 and RBM5. For each internal exon E, we counted the number of reads for inclusion events (reads mapped to E or exon junctions containing E with at least 6 nt overlap) and for exclusion events (reads mapped to exon junctions skipping E and both exons with at least 6 nt overlap).
We calculated the ratio between the average read coverage for two contiguous fragments in the last exon of RBM10: the last 100 nt of the RBM10 coding sequence and the first 100 nt of its 3’UTR. The former fragment derives from both exogenous and endogenous RBM10, while the latter derives from only endogenous RBM10. Under conditions of RBM10 overexpression, the number of sequencing reads supporting exon inclusion derived from endogenous RBM10 was estimated as total number of reads supporting exon inclusion divided by the calculated ratio.
Statistical analysis
For comparison of RBM10 RNA levels in LUAD samples, Welch's t-test was performed using R (https://www.r-project.org/). For all other experiments, statistical tests were carried out using GraphPad Prism 6 (GraphPad software). For comparisons between sample pairs, two-tailed Student's t-tests were used. For experiments with more than two conditions, one-way ANOVA followed by Dunnett's tests were used.
RESULTS
RBM10 negatively regulates its own expression
To investigate whether RBM10 regulates its own RNA expression, we quantified levels of 5’-untranslated region (5’UTR) and 3’UTR derived from endogenous RBM10 following overexpression (OE) of its coding sequence, using our previously acquired RNA sequencing (RNA-Seq) data in HEK293 cells (42). This analysis revealed that RBM10 OE dramatically and reproducibly reduced levels of both 5’UTR and 3’UTR sequences (Figure 1A), indicating that RBM10 negatively regulates its own RNA expression.
Figure 1.
RBM10 overexpression (OE) downregulates endogenous RBM10 RNA and protein expression. (A) Relative levels of 5’-untranslated region (5’UTR) and 3’UTR derived from endogenous RBM10 mRNA in the absence (Ctrl) and presence of overexpressed RBM10 (RBM10 OE) in HEK293 cells. Expression levels were estimated using RNA-Seq data and normalized with respect to GAPDH expression (n = 2; error bar: range). (B) RT-PCR analysis of total and endogenous RBM10 mRNA expression in tet-on HEK293 cells inducibly expressing RBM10-EGFP in response to doxycycline (Dox), using primers designed to amplify RBM10 coding region (CDS), 5’UTR and 3’UTR sequences, respectively. Left panel: representative agarose gel images of RT-PCR products. Right panel: qPCR results. Data are presented as mean ± SEM for n = 4 biological replicates. **P < 0.01, ***P < 0.001 (one-way ANOVA followed by Dunnett's tests). (C) Western blot analysis of RBM10 protein expression under conditions in (B), using an antibody that recognizes both endogenous and overexpressed RBM10-EGFP. α-Tubulin served as a loading control. A representative result from three independent experiments is shown. The faint band above the endogenous RBM10 band in RBM10-EGFP overexpressed samples, indicated by the open arrowhead, is likely an artifact resulting from RBM10-EGFP overexpression (see also Supplementary Figure S1D).
To confirm this observation and determine whether RBM10 OE also reduces protein levels of endogenous RBM10, we generated a tet-on HEK293 cell line that inducibly expresses an RBM10 fusion protein containing a C-terminal enhanced green fluorescent protein (EGFP) reporter. Expression of RBM10-EGFP mRNA and protein following induction with two different concentrations of doxycycline (Dox) was confirmed by reverse transcription followed by quantitative real-time PCR (RT-qPCR) and Western blot analysis, respectively (Figure 1B and C and Supplementary Figure S1A). To assess the functionality of the RBM10-EGFP fusion protein, we examined its subcellular localization and effects on RNA splicing for several RBM10 target genes. As expected, these measurements showed that RBM10-EGFP predominantly localized to the nucleus (Supplementary Figure S1B) and enhanced exon skipping for 6 RBM10 target genes (Supplementary Figure S1C), indicating that fusing EGFP to the C-terminus of RBM10 does not interfere with its function.
Using PCR primers designed to amplify RBM10 5’UTR and 3’UTR sequences, we observed that OE of RBM10-EGFP decreased levels of endogenous RBM10 RNA by approximately 50% (Figure 1B, P < 0.001), a result consistent with the trend observed in our RNA-Seq data (Figure 1A). Using an antibody that recognizes both endogenous RBM10 and RBM10-EGFP, we showed that endogenous RBM10 protein levels were also considerably reduced following overexpression of RBM10-EGFP (Figure 1C and Supplementary Figure S1D). Together, these results demonstrate that RBM10 autoregulates its own RNA and protein levels via negative feedback.
Computational analysis identifies candidate AS-NMD events underlying RBM10 autoregulation
Based on our previous findings on RBM10-mediated splicing inhibition (42), we hypothesized that RBM10 exerts negative autoregulation by promoting exon skipping by binding to splice site-adjacent regions in flanking introns within its own pre-mRNA, thereby producing mRNA isoforms that are degraded by NMD.
Previous studies have shown that RNA-Seq reads of sufficient depth can be used to characterize unstable splice variants, including putative substrates for NMD resulting from exon skipping/inclusion (56). To quantify exon skipping events promoted by RBM10, we estimated levels of individual RBM10 splice variants lacking one or multiple-coding exons and their proportions within endogenous RBM10, using splice junction reads in RNA-Seq data obtained from HEK293 cells overexpressing RBM10 (42). Under conditions of RBM10 overexpression (OE), levels of endogenous RBM10 were estimated from the ratio of the average read coverage for the last 100 nt of the coding sequence (present in both endogenous and exogenous mRNAs) versus the first 100 nt of the 3’UTR (present in endogenous mRNA only) (see Materials and Methods). This analysis showed that RBM10 OE increased levels of the exon 6-skipped splice variant about 2.5-fold (Figure 2A and Supplementary Figure S2A and S2B) and considerably increased its estimated proportion within endogenous RBM10 (Supplementary Figure S3). All other single exon skipping events occurred at very low levels, except for exon 4, an in-frame alternative exon, which showed reduced levels of exon skipping that largely reflect decreases in total RBM10 mRNA (Supplementary Figure S2A and Figure 1A). Examination of open reading frames revealed that skipping each of the coding exons 3, 5, 6, 8, 10–16 and 22 produces a stop codon >50 nt upstream from the last exon-exon junction and/or creates a long 3’UTR, which would be expected to trigger mRNA degradation via NMD. Through our RNA-Seq data analysis, we also detected multiple-exon skipping spanning exons 8–12, corresponding to a 585 nt deletion in the RBM10 coding sequence, and showed that its expression level increased under RBM10 OE (Supplementary Figure S2C). Because this multiple-exon skipping is in-frame and occurs at low levels (Supplementary Figure S2C), it is unlikely to significantly contribute to RBM10 autoregulation via NMD.
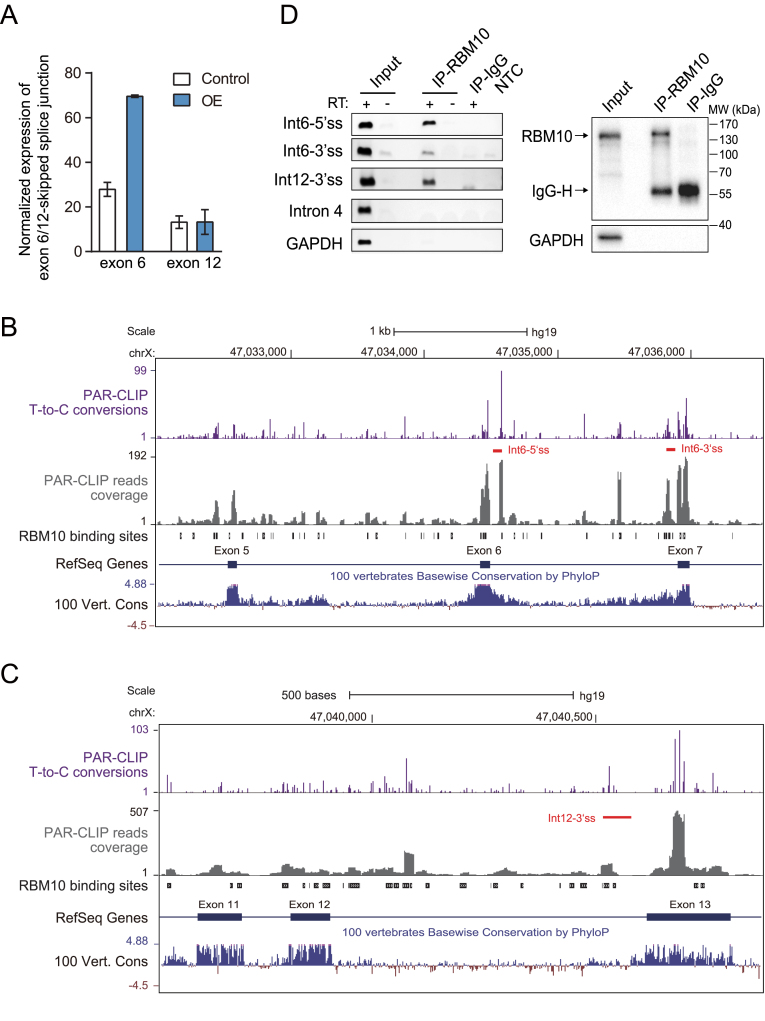
Figure 2.
Identification of candidate AS-NMD events underlying RBM10 autoregulation via computational analysis and confirmation of RBM10 PAR-CLIP binding via CLIP-PCR. (A) Expression levels of RBM10 exon 6- or 12-skipped splice junctions in HEK293 cells under control (Ctrl) and RBM10 overexpression (OE) conditions, respectively. Expression levels of the splice junction were estimated from numbers of aligned RNA-Seq reads after normalization to GAPDH expression. Error bar: range, n = 2 biological replicates. (B and C) Genome browser views of RBM10 PAR-CLIP binding sites within genomic regions containing upstream and downstream exons flanking exon 6 (A) or exon 12 (B). Shown are intensities of T-to-C conversions at reproducible crosslinking sites (purple), aligned PAR-CLIP reads (gray), and RBM10 binding sites (black boxes), as well as sequence conservation across vertebrates. Red lines below T-to-C conversion peaks mark the region amplified by CLIP-PCR in (D). (D) CLIP-PCR validation of RBM10 PAR-CLIP binding in RBM10 pre-mRNA. Left panel: gel image of CLIP-PCR products using three oligonucleotide primers targeting RBM10 binding regions within introns 6 and 12. Primers targeting sequences of RBM10 intron 4 and GAPDH, which lack of RBM10 binding sites, were included as negative controls. RT + and – indicate with and without addition of reverse transcriptase, respectively (IP: immunoprecipitation; Int: intron; NTC: non-template control). Right panel: Western blot assessment of the IP specificity. GAPDH was included as a negative control.
To obtain additional evidence for RBM10-enhanced exon skipping events within its own pre-mRNA, we analyzed RBM10 binding sites in its own pre-mRNA using our previous PAR-CLIP data obtained in HEK293 cells inducibly over-expressing RBM10 (42). Because we previously found that RBM10 binding to flanking introns, but not binding to exons, generally correlates with exon skipping (42), we focused on intronic regions within 300 nt of splice sites, where RBM10 binding sites are enriched and most known splicing regulatory elements are located (57). After filtering out background crosslinking positions (58,59), we defined reproducible RBM10 binding sites as those with crosslinking positions detected in both biological replicates (42). We also examined T-to-C conversion profiles of the reproducible RBM10 binding sites located in those intronic regions, which are an indication of genuine PAR-CLIP binding (60). Because RBM10 likely interacts with multiple spliceosomal RNA binding proteins (38) and such interactions may create many transient or weak RBM10 binding sites, we focused on strong RNA binding sites in our analyses. We found that introns 6 and 12 exhibited strong T-to-C conversion with intensities above the upper quartile of all observed intronic binding regions (Supplementary Figure S4 and Figure 2B and C), and thus are likely to contain true RBM10 binding sites (60).
We further verified this binding by RNA crosslinking immunoprecipitation followed by RT-PCR (CLIP-PCR) in HEK293 cells using an anti-RBM10 antibody. This analysis showed that RBM10 interacted specifically with identified RBM10 binding sites in introns 6 and 12 of its own pre-mRNA, but not with sequences in RBM10 intron 4 or GAPDH, which lack RBM10 PAR-CLIP binding signals (Figure 2D). Based on our previous finding that RBM10 promotes exon skipping by binding to flanking introns at regions near splice sites (42), the observed binding patterns in intron 6 and 12 (Figure 2B and C) suggest that RBM10 may cause skipping of their adjacent exons. For the exons flanking intron 6 and 12, our RNA-Seq analysis showed that RBM10 OE dramatically increased the level of exon 6 skipping (Figure 2A). In addition, exon 12 skipped splice variant were represented by more than 10 sequencing reads in all the samples (Figure 2A and Supplementary Figure S2A). By contrast, splice variants with skipped exon 7 or 13 were rarely detected (Supplementary Figure S2A), suggesting that they are constitutive exons. Taken together, the results from our computational analysis and CLIP-PCR validation demonstrate that RBM10 likely enhances skipping of exons 6 and 12 by binding to splice site-adjacent regions in their flanking introns and thereby promotes NMD of the corresponding mRNA transcripts.
Confirmation that RBM10 enhances skipping of its own exons 6 and 12
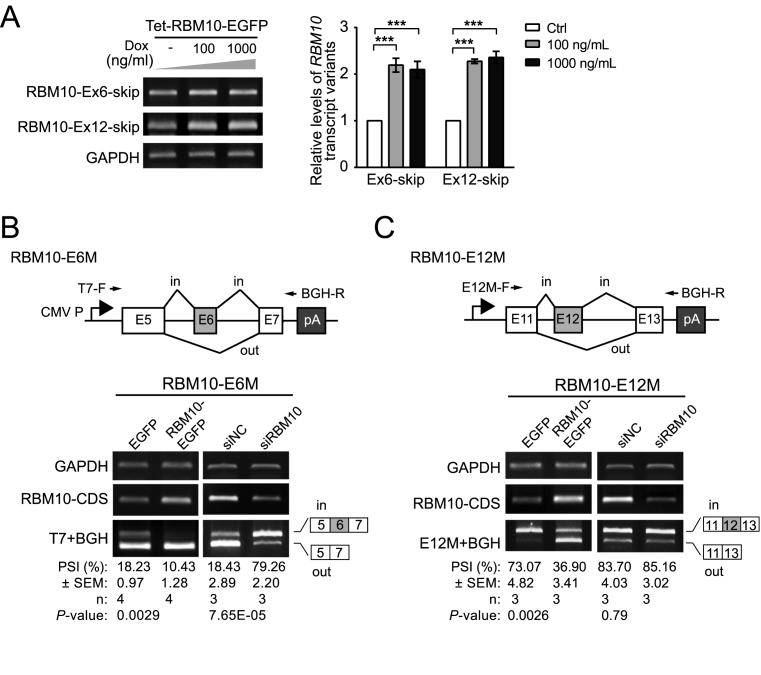
To confirm that RBM10 enhances skipping of its own exons 6 and 12, we performed RT-PCR using splice junction-specific primers to examine levels of transcript variants lacking exon 6 or 12 (RBM10-Ex6/12-skip) in HEK293 cells overexpressing RBM10-EGFP (primer sequences are listed in Supplementary Table S2). This analysis showed that levels of both exon 6-skipped and exon 12-skipped splice variants increased following RBM10-EGFP OE (Figure 3A). By contrast, levels of splice variant lacking exon 4 (RBM10-Ex4-skip) decreased to an extent similar to the observed decrease in total RBM10 mRNA (Supplementary Figure S5 and Figure 1B), consistent with results obtained from RNA-Seq data (Supplementary Figure S2A and Figure 1A). To further verify RBM10-mediated enhancement of skipping of its own exons 6 and 12, we generated minigene splicing reporters for each of the two exons (RBM10-E6M and RBM10-E12M, respectively) containing the complete genomic sequence from the adjacent upstream exon to the adjacent downstream exon (Figure 3B and C). Each of the two minigenes was then co-transfected with RBM10-EGFP or EGFP (control) expression plasmids into HEK293 cells. We found that RBM10-EGFP OE promoted the skipping of both exon 6 and exon 12 (Figure 3B and C, P = 0.0029 and 0.0026 for RBM10-E6M and -E12M, respectively). By contrast, RBM10 depletion using siRNAs significantly increased the inclusion of exon 6 (Figure 3B, P = 7.65E–05 and Supplementary Figure S6), but not of exon 12 (Figure 3C). The lack of a significant increase in exon 12 inclusion following RBM10 knockdown is likely due to its high basal level of inclusion (PSI: ∼79%) and/or possible antagonizing effects of other splicing factors regulated by RBM10. Taken together, these results confirm that RBM10 promotes skipping of its own exons 6 and 12, producing transcript variants that are predicted to be substrates for NMD.
Figure 3.
RBM10 enhances skipping of its own exons 6 and 12. (A) Changes in expression of RBM10 transcript variants lacking exons 6 or 12 (RBM10-Ex6/12-skip) following induction of RBM10-EGFP expression with 100 or 1000 ng/ml doxycycline (Dox) in HEK293 cells assessed by RT-PCR. Representative agarose gel images of RT-PCR products are shown in the left panel. qPCR results are shown in right panel. Error bars represent ±SEM from four biological replicates. ***P < 0.001 (one-way ANOVA followed by Dunnett's tests). (B and C) Changes in splicing of minigene reporters for exons 6 and 12 (RBM10-E6M and -E12M) in HEK293 cells following RBM10-EGFP overexpression (OE) or RBM10 knockdown (KD) assessed by RT-PCR. Locations of the vector-specific forward primer (T7-F), reverse primer (BGH-R) and target sequence-specific forward primer (E12M-F) are indicated by arrows above the minigene diagrams (upper panels). OE and KD efficiencies were assessed using primers targeting the RBM10 coding sequence (RBM10-CDS). GAPDH served as a loading control. PSI: percent-spliced-in. Error bars represent ±SEM from three biological replicates (Student's t-test).
RBM10-mediated skipping of its exons 6 and 12 was further confirmed using heterologous minigene reporters, in which exons 6 or 12 and their flanking introns were inserted within the GFP coding sequence to exclude possible effects of RBM10 binding to neighboring exons (Supplementary Figure S7). Deletion of the two strongest RBM10 binding sites within intron 12 in minigene reporter RBM10-E12M attenuated RBM10-meditated exon 12 skipping (Supplementary Figure S8). In addition, minigene reporters containing RBM10 exons that have internal RBM10 binding sites, but lack strong binding sites within flanking introns, did not show splicing changes following RBM10 OE (Supplementary Figure S9). Together, these observations support the importance of RBM10 binding sites within flanking introns for exon skipping and the usefulness of information concerning the RBM10 intronic binding for predicting exon splicing outcomes.
RBM10 transcripts lacking exon 6 or 12 are substrates for NMD
Skipping of RBM10 exon 6 or 12 is predicted to introduce premature stop codons and induce NMD (16,20). To confirm the predicted effects and the contribution of these exon skipping events to RBM10 autoregulation, we inhibited NMD alone and in combination with RBM10 overexpression (OE). We first directly inhibited NMD by depleting the NMD pathway regulator UPF1 using a previously verified siRNA (Supplementary Figure S10) (34). As predicted, expression levels of RBM10 transcript variants lacking exon 6 or exon 12 significantly increased following NMD inhibition alone and synergistically increased following NMD inhibition together with RBM10 OE (Figure 4A). To rule out possible artifacts of UPF1 depletion unrelated to NMD (16), we also indirectly inhibited NMD by blocking translation with cycloheximide (CHX) and obtained similar results (Figure 4B). Consistent with the observed accumulation of the NMD-susceptible transcripts, expression levels of total endogenous RBM10 mRNA significantly increased following NMD inhibition and decreases in mRNA levels observed under RBM10 OE were reversed by NMD inhibition (Supplementary Figure S10). Together, these results demonstrate that RBM10-promoted skipping of its exon 6 or 12 produces RNA transcripts that are substrates for NMD, and thereby contributes to autoregulation.
Figure 4.
RBM10 splice variants lacking exon 6 or exon 12 are substrates for nonsense-mediated decay. (A and B) RT-PCR analysis of the expression of RBM10 transcript variants lacking exon 6 or exon 12 (RBM10-Ex6/12-skip) following NMD pathway inhibition via siRNA-mediated depletion of the regulator UPF1 (A) or cycloheximide (CHX)-mediated inhibition of protein synthesis (B) with or without RBM10 overexpression (OE) in HEK293 cells using splice variant-specific primers. Dox: doxycycline; NC: negative control. Left panel: representative agarose gel images of RT-PCR products. Right panel: qPCR results. Error bars: ±SEM, n = 5 biological replicates in (A) and n = 4 biological replicates in (B). *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant, compared to control sample (siNC/Dox- in (A) and DMSO/Dox- in (B), respectively) and between sample pairs indicated by lines (Student's t-test).
RBM10 negatively regulates its paralog RBM5 via AS-NMD
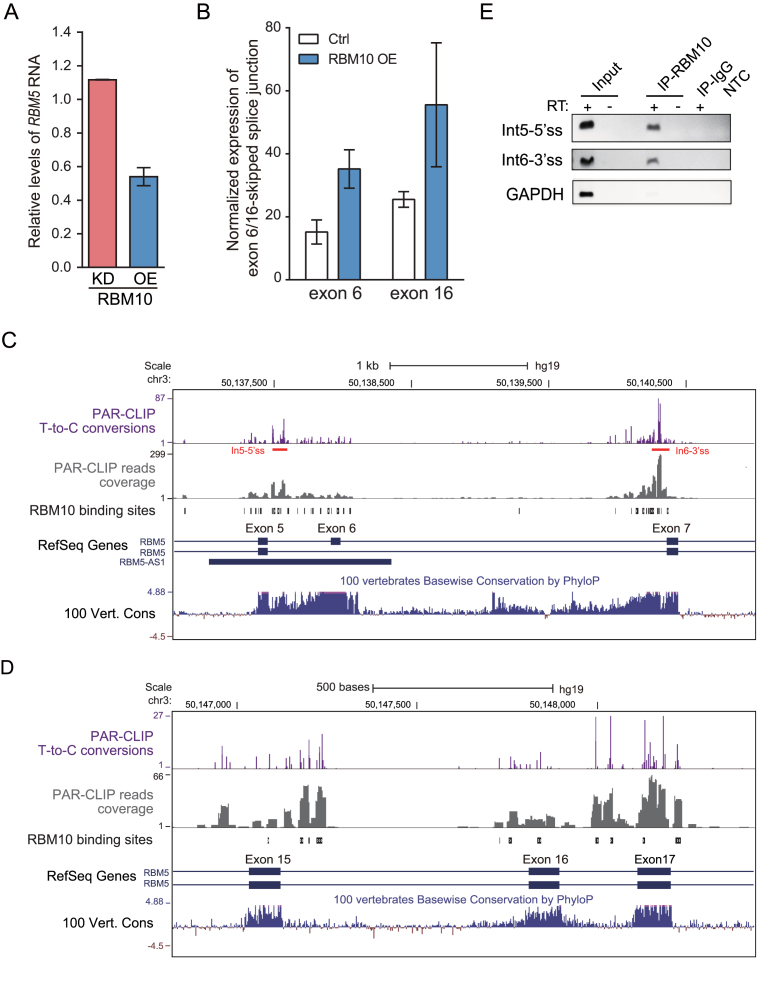
RBM5 is the closest paralog of RBM10 with 46% amino acid identity. Our previous results showed that RBM10 OE downregulated the expression of RBM5 mRNA, while RBM10 knockdown (KD) upregulated its expression (Figure 5A) (42). Given the observed cross-regulation between other paralogous splicing factors (15), we postulated that RBM10 negatively modulates RBM5 expression through AS-NMD. Using the computational method described above, we found that: (i) RBM10 promoted skipping of RBM5 exons 6 and 16, and exon 8 to a lesser extent (Figure 5B and Supplementary Figure S11A); (ii) RBM5 transcripts lacking one of the coding exons 3, 5, 6, 8, 10–12, 14–17 or 23 are predicted to trigger NMD; and (iii) RBM10 binds to RBM5 introns 4, 5, 6, 16, 17 and 20 at regions near the splice sites (Figure 5C and D and Supplementary Figure S12). The observed RBM10 PAR-CLIP binding sites in RBM5 intron 5 and 6 were confirmed by CLIP-PCR (Figure 5E). Together, these data suggest that RBM10 promotes skipping of RBM5 exon 6 and 16 by binding to flanking intronic regions and likely promotes skipping of RBM5 exon 8 via indirect effects, resulting in transcript variants that are subject to NMD. Notably, splice site adjacent regions in flanking introns of exon 6 and 16 that contain RBM10 binding sites are evolutionarily conserved across species (Figure 5C and D).
Figure 5.
Computational analysis identifies RBM10 binding sites and regulated AS-NMD events within RBM5. (A) Changes in RBM5 mRNA expression in HEK293 cells, estimated by RNA-Seq data following RBM10 overexpression (OE) or knockdown (KD), respectively. Ctrl: Control. (B) Changes in levels of splice junctions specific for RBM5 transcript variants lacking exon 6 or 16, respectively, following RBM10 OE, as described in the legend of Figure 2A. Error bar: range, n = 2 biological replicates. (C and D) Genome browser view of RBM10 PAR-CLIP binding sites and sequence conservation across vertebrates in genomic regions spanning the proximal upstream and downstream exons of RBM5 exon 6 and 16, respectively, with panels as described in the legend of Figure 2B and C. (E) Confirmation of the RBM10 binding sites in RBM5 intron 5 and 6 using CLIP-PCR, under conditions described in Figure 2D. GAPDH served as a negative control.
Several lines of experimental evidence support our computational predictions. First, both mRNA and protein levels of RBM5 were reduced following RBM10-EGFP OE (Figure 6A and B). Second, expression levels of RBM5 splice variants lacking exon 6 or 16, from both endogenous pre-mRNA and the corresponding minigene splicing reporters, increased following RBM10-EGFP OE (Figure 6A and C, respectively). By contrast, skipping of exon 8, for which there is only very low intensity of RBM10 binding within flanking introns (Supplementary Figure S14A), did not significantly change under RBM10-EGFP OE (S14B–D, compare DMSO/Dox– with DMSO/Dox+ levels in S14C and S14D), suggesting that skipping of this exon is not regulated by the binding of RBM10. Third, expression levels of RBM5 splice variants lacking exon 6, 8 or 16 significantly increased following NMD inhibition via UPF1 depletion or CHX treatment (Figure 6D and E and Supplementary Figure S13 and S14C). Skipping of exon 6 or 16, but not exon 8, further increased following NMD inhibition under RBM10 OE (Figure 6D and E and Supplementary Figure S13, S14C and S14D). Consistently, expression levels of total RBM5 mRNA increased following NMD inhibition, and their decreases under RBM10 OE were reversed by NMD inhibition (Supplementary Figure S10). Collectively, these observations strongly support our findings that RBM10 negatively regulates mRNA and protein expression of its closest paralog RBM5 via AS-NMD.
Figure 6.
RBM10 negatively regulates the expression of its paralog RBM5 via AS-NMD. (A) Changes in levels of RBM5 total mRNA and splice variants lacking exon 6 or 16 (RBM5-Ex6/16-skip) following RBM10-EGFP overexpression (OE) assessed using RT-PCR as described in the legend of Figure 3A. n = 3 biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant (one-way ANOVA followed by Dunnett's tests). (B) Western blot analysis of RBM5 protein expression levels under conditions in (A). (C) RT-PCR analysis of the expression levels of RBM5 splice variants lacking exon 6 or 16 from minigene splicing reporters (RBM5-E6M, -E16M) co-transfected with EGFP control and RBM10-EGFP, respectively, as in Figure 3B. n = 4 biological replicates for RBM5-E6M and n = 3 for RBM5-E16M (Student's t-test). (D and E) RT-PCR analysis of expression changes of RBM5 splice variants lacking exon 6 or 16 following NMD inhibition via UPF1 depletion (D) or CHX treatment (E) with or without RBM10 OE, respectively, as described in the legend of Figure 4. Upper panels: representative gel images; lower panels: qPCR results. n = 5 biological replicates in (D) and n = 4 biological replicates in (E). *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant, compared to control sample (siNC/Dox– in (A) and DMSO/Dox- in (B), respectively) and between sample pairs indicated by lines (Student's t-test).
Lung adenocarcinoma (LUAD)-associated mutations affecting RBM10 exons 6 and 12 splice sites disrupt splicing and correlate with reduced RBM10 RNA expression
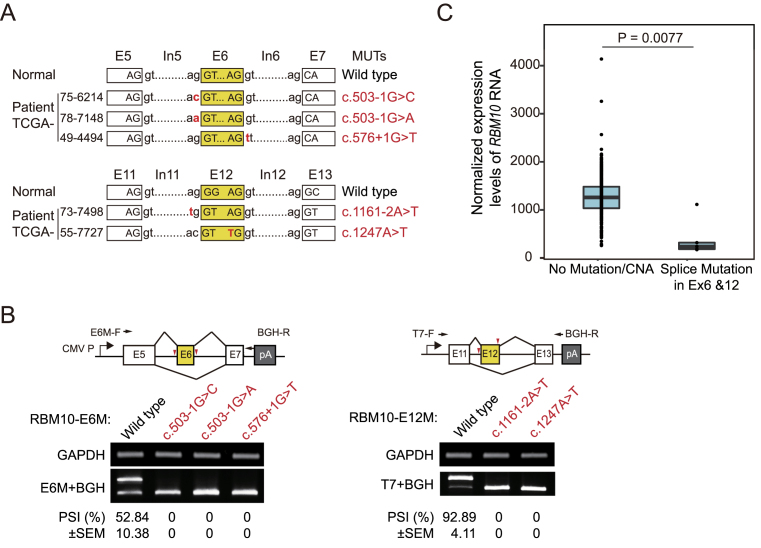
RBM10 mutations have been frequently observed in LUAD patients (43,44). To examine whether these cancer-associated mutations occur within splice sites of RBM10 and subsequently affect splicing of its pre-RNA, we analyzed RBM10 mutations and RNA expression data for LUAD patients catalogued in The Cancer Genome Atlas (TCGA) database. In total, 9 of 43 RBM10 mutations found in 546 TCGA LUAD samples are located within splicing sites. Of particular interest, 5 of these 9 mutations are located within splice sites for exons 6 or 12 (Figure 7A and Supplementary Table S1). To determine the effects of these mutations on exon inclusion, we introduced them into the RBM10-E6M/E12M minigene reporters and transfected the wild type and mutant RBM10 minigene reporters into two cell lines with different genetic backgrounds: N2a, a mouse neuroblastoma cell line, and HEK293. Analysis of minigene splicing in these cells revealed that each of the five mutations completely disrupted the inclusion of their affected exons in both cell lines (Figure 7B and Supplementary Figure S15). Interestingly, exon 6 of RBM10-E6M reporter has higher inclusion levels in N2a cells compared to HEK293 cells, suggesting cell type specific regulation of RBM10 via AS-NMD (compare Figure 7B and Supplementary Figure S15).
Figure 7.
LUAD-associated splice site mutations in RBM10 exons 6 and 12 disrupt splicing and correlate with significantly reduced RBM10 RNA expression. (A) Locations of splice site mutations for RBM10 exon 6 and 12 identified in TCGA LUAD patients. Ex: exon; In: intron. (B) RT-PCR analysis of exon 6 and 12 inclusion levels derived from wild type and splice site mutants of minigene reporters RBM10-E6M and -E12M transfected in N2a cells. PCR primers are indicated by black arrows and positions of mutations are marked by red arrowheads in minigene diagrams (upper panel). PSI: percent-spliced-in. n = 2 biological replicates. (C) RBM10 RNA expression levels in four LUAD patients harboring splice site mutations for RBM10 exons 6 or 12 compared to individuals lacking RBM10 mutations or copy number alternation (CNA). Data from TCGA, see Materials and Methods for details. P = 0.0077 (Welch's t-test).
Since RBM10 transcript variants lacking exon 6 or 12 were predicted to be substrates for NMD, we expected that splice site mutations for these exons would greatly reduce RNA expression levels of RBM10. To rule out the potential influence of copy number alteration (CNA) on gene expression, we excluded LUAD samples with RBM10 CNA from our analysis, including one case carrying an RBM10 mutation in an exon 6 splice site (Supplementary Table S1). As expected, we observed that RNA expression levels of RBM10 in the remaining four patients carrying splice site mutations for exon 6 or 12 as the only RBM10 mutations were significantly lower compared with LUAD patients without RBM10 mutations or CNA (Figure 7C, P = 0.0077). Of note, one case among the four patient samples showed higher RBM10 RNA expression than the others. After careful inspection, we found that the frequency of the mutant allele in the tumor tissue excised from this individual is much lower than the others (Supplementary Table S1), indicating that wild type RBM10 constitutes the majority of this sample (44,61,62). The difference in RBM10 RNA expression levels is even more significant after removing this case (P = 7.41e–07), strongly supporting our finding that skipping of RBM10 exon 6 or 12 activates NMD and consequently decreases levels of RBM10 RNA.
DISCUSSION
In this study, we uncovered a novel molecular mechanism underlying RBM10 negative autoregulation and cross-regulation of its paralog RBM5. Taking advantage of our RNA sequencing data following RBM10 perturbations and published RBM10 PAR-CLIP results (42), we identified RBM10-enhanced exon skipping events that trigger NMD for four exons and RBM10 binding sites in RBM10 and RBM5. Notably, alternative splicing (AS) of these exons has not previously been linked to NMD, thus highlighting the advantage of our methods for characterizing AS-NMD-mediated auto- and cross-regulation of splicing regulators. Together, our findings provide new insights regarding the functions of RBM10 in normal and disease contexts. The approach established here should be useful for elucidating the mechanisms of auto- or cross-regulation by other splicing factors.
Our study began with an integrative analysis of RNA-seq and CLIP data to systematically search for candidate AS-NMD events in RBM10 and RBM5, which were then thoroughly validated by experiments. RNA sequencing is a powerful and extensively used method for the systematic identification of new splicing events, including those couple to NMD (56). NMD transcripts are often difficult to detect due to rapid degradation. However, we successfully identified changes in three of four exon skipping events that trigger NMD in RBM10 and RBM5 using RNA-Seq data under control and RBM10 OE conditions (Supplementary Figure S2A, S3 and S11). RNA-Seq data is also useful for detecting highly unstable mRNA transcripts, for example RBM10 transcripts lacking exon 12, although additional experiments such as those carried out in our study may be required for confirmation. Notably, the fact that splice site-adjacent intronic regions flanking RBM10 exon 12 lack high sequence conservation implies that, although often correlated with splicing regulatory functions (15), sequence conservation is not necessarily required for exon skipping/inclusion-coupled NMD events that underlie auto- or cross- regulation of splicing regulators.
In cases where splicing regulator-RNA interaction data are available, binding sites in target RNAs can be precisely identified, thereby providing evidence for regulation. Caution should to be taken, however, concerning identification of the biologically meaningful binding sites using CLIP, because CLIP is a very sensitive method that can capture both stable and transient binding. In addition, UV crosslinking, immunoprecipitation and deep sequencing procedures may produce artifacts, even when carried out in a stringently controlled manner (59). Control CLIP experiments characterizing background binding have been shown to be helpful for excluding experimental artifacts, but have not been routinely included in most published CLIP datasets (59,63).
Because our previous study found that RBM10 binding to splice site-adjacent regions in flanking introns, but not binding to exons, globally correlates with exon skipping (42), we analyzed RBM10 intronic binding to its own pre-mRNA and that of RBM5. Results from our study support a role for RBM10 binding to flanking introns near splice sites in exon skipping and the usefulness of RBM10 intronic binding information for predicting exon splicing outcomes (Supplementary Figure S7-S9 and S14). Because several independent CLIP studies identified RBM10 binding to exons (42,48,49) and a very recent study showed that RBM10 promotes skipping of TNRC6A exon7 partly by binding to this exon (64), we do not preclude possible functions of exonic binding sites in individual cases. It should be noted that the observed RBM10 binding to its own exons in our study may be overrepresented because the PAR-CLIP data was obtained in HEK293 cells inducibly overexpressing FLAG-tagged RBM10, where its coding sequence were overexpressed.
After filtering out common PAR-CLIP background binding signals (59), we prioritized intronic RBM10 PAR-CLIP binding regions within RBM10 and RBM5 based on T-to-C conversions intensities, an indication of genuine PAR-CLIP binding (60), to identify candidate binding sites. Because potential interactions of RBM10 with other spliceosomal RNA binding proteins (RBPs) (38) may give rise to many transient or indirect weak RBM10 binding sites, we focused on strong binding sites that are more likely to be functional. In this way, we discovered that RBM10 binding sites are enriched in introns flanking RBM10 exons 6 and 12 and introns flanking RBM5 exon 6 and 16 (Figures 2B, C, 5C and D and Supplementary Figure S4 and S12), suggesting that RBM10 promotes skipping of the these exons. Notably, combined with the analysis of CLIP data, we found that skipping of RBM10 exon 12 is promoted by RBM10, but the magnitude of change is too low to be precisely quantified by RNA-Seq alone (Figure 2A and Supplementary Figure S2A). We also found that skipping of RBM5 exon 8, which lacks RBM10 binding in adjacent introns, is likely not significantly promoted by RBM10 (Supplementary Figure S14), even though RNA-Seq data suggests that skipping of this exon is weakly promoted by RBM10 (Supplementary Figure S11A).
We also used our computational method to analyze the splicing regulators PTBP1 and PTBP2, a well-characterized example of autoregulation and cross-regulation mediated by AS-NMD (31,65). Skipping of PTBP1 exon 11 and PTBP2 exon 10 was previously identified as the underlying AS-NMD events by analyzing expressed sequence tags (ESTs) and/or sequence conservation (31,65). To determine if our method confirms these results, we analyzed publically available RNA-seq data following PTBP1 depletion and PTBP1 CLIP data in Hela cells (66,67). Using this RNA-Seq data, we found that expression levels of PTBP2 exon 10 skipping and the proportion of the resulting splice variant dramatically decreased (Supplementary Figure S16A and S16B), and PTBP2 expression dramatically increased (Supplementary Figure S16E), following PTBP1 depletion, consistent with previous findings (31). By contrast, skipping of other PTBP2 exons showed no change or very small decreases (Supplementary Figure S16A and S16B). It is difficult to estimate changes of specific PTBP1 exons under conditions of PTBP1 depletion, because the siRNA-mediated depletion affects levels of all PTBP1 transcripts (Supplementary Figure S16C and S16D). Instead, we observed that the proportion of exon 11 skipping is very low under control conditions (Supplementary Figure S16C and S16D). As described for RBM10 in this study, RNA-Seq data obtained under conditions of PTBP1 overexpression should be helpful for identifying candidate AS-NMD events underlying negative autoregulation. Using PTBP1 PAR-CLIP data with high sequencing depth (67), we detected PTBP1 binding to flanking intronic regions close to splice sites for both PTBP1 exon 11 and PTBP2 exon 10 (Supplementary Figure S17). Based on a proposed model for PTBP1-mediated splicing regulation (66), the observed binding patterns suggest that PTBP1 suppresses inclusion of these exons.
These results demonstrate the usefulness of our method for identifying candidate AS-NMD events underlying auto- and cross-regulation of splicing regulators. It should be noted, however, that we observed differences in PTBP1 binding between the two CLIP datasets (Supplementary Figure S17). This may be due to large differences in sequencing depth and distinct methods used in corresponding CLIP experiments (66,67). Again, as discussed above, highly reproducible and well-controlled CLIP datasets are essential for identifying true binding sites and interpreting their functions.
The RBM10 transcript lacking exon 6 is predicted to encode a truncated protein in the UCSC database (uc004dhe.2, GRCh37) and RBM10 exon 12 is not annotated as an alternative exon in the Refseq and UCSC databases. Similar to RBM10, the RBM5 transcript variant lacking exon 6 is listed in the UCSC database (uc011bdk.2, GRCh37) and predicted to encode an alternative protein using a different downstream start codon. By contrast, this transcript is annotated as a non-coding transcript in Refseq (NR_036627, GRCh37). Based on similar RBM10 binding patterns and high conservation in flanking intronic sequences for RBM5 exon 6 and RBM10 exon 6, we speculate that skipping of the two exons induces NMD in a similar manner. Although exogenous overexpression of the predicted coding sequence of RBM5 transcript variant lacking exon 6 was shown to accelerate cell cycle and suppress apoptosis (68), the full-length and functions of this transcript have not been characterized in vivo. RBM5 exon 8 and 16 are not annotated as alternative exons in the Refseq and UCSC databases. Although RBM5 exon 8 is likely not regulated via direct binding of RBM10, our results identify it as a new alternative exon for which skipping results in NMD (Supplementary Figure S14).
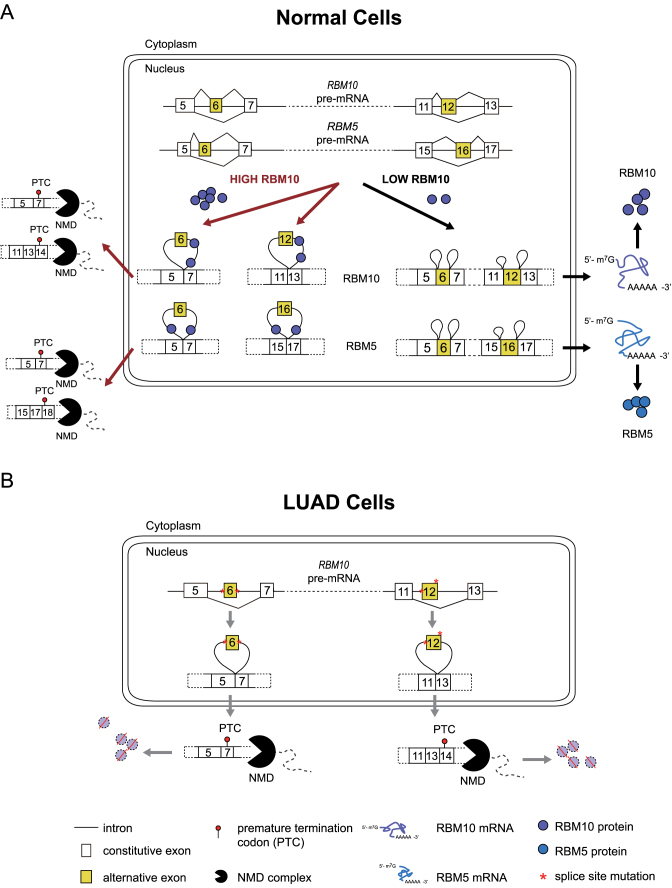
Theoretically, multiple AS-NMD events can be used by splicing regulators to increase the efficiency of auto- and cross-regulation. As illustrated in Figure 8A, AS-NMD events in RBM10 or RBM5 can increase the magnitude of auto- and cross-regulation in an additive manner. Specifically, when cellular concentrations of RBM10 increase, skipping of exon 6 also increases, thus activating NMD and decreasing levels of RBM10 mRNA and protein. RNA transcripts that contain exon 6, however, may undergo exon 12 skipping, thereby further reducing mRNA and protein levels and increasing the effectiveness of negative autoregulation. A similar mechanism holds true for negative cross-regulation of RBM5, where RBM10 enhances skipping of RBM5 exons 6 and 16. Conversely, when cellular concentrations of RBM10 decrease, inclusion levels of RBM10 exons 6/12 and RBM5 exons 6/16 increase, thereby increasing mRNA and protein levels of both splicing regulators (Figure 8A).
Figure 8.
(A, B) Proposed model for RBM10 autoregulation and cross-regulation of RBM5 in normal cells (A) and possible mechanisms for disruption of RBM10 expression due to specific splice site mutations in lung adenocarcinoma (LUAD) cells (B).
Additionally, given that different cofactors likely contribute to the regulation of splicing of the four identified NMD exons in RBM10 and RBM5 (12), the AS-NMD events individually and in combination could provide RBM10 with multiple ways to differentially modulate its own expression and that of RBM5 under different normal and pathological contexts. We hypothesize that negative autoregulation of RBM10 can be used by the cells to maintain homeostasis. Meanwhile, cross-regulation of RBM5 can give rise to varying combinations of RBM10 and RBM5 concentrations, thereby potentially expanding functional diversity.
Importantly, the circuit for RBM10 autoregulation and cross-regulation of RBM5 established in this study (Supplementary Figure S18) has also been observed in other splicing regulators (15), and thus may represent a common mechanism for controlling splicing regulatory networks. The functional significance of such regulatory circuits is clearly demonstrated by PTBP1 and PTBP2. PTBP1 is expressed in most types of cells, while PTBP2 is primarily expressed in neurons (69). PTBP1 can negatively autoregulate its own expression and cross-regulate that of PTBP2 by triggering AS-NMD (31,65). During neuronal differentiation, expression of PTBP1 is repressed, alleviating the negative cross-regulation of PTBP2 and leading to a switch to PTBP2 expression (69,70). In addition, depletion of PTBP1 in fibroblasts has been shown to be sufficient to induce PTBP2 expression and neuronal trans-differentiation (71), strongly supporting the functional importance of these splicing regulators in determining neuronal cell states.
Since RBM10 regulates alternative splicing of hundreds of target genes, including its paralog RBM5 and several other RBPs (42), it is crucial for cells to tightly regulate its expression. In this regard, it is not surprising that RBM10 mutations cause multifaceted developmental abnormalities in patients with TARP syndrome (39–42) and contribute to molecular changes in LUAD (43,44) and other types of cancer (45–47).
LUAD-associated splice site mutations of RBM10 exon 6 or 12 correlated with significantly reduced RNA expression of RBM10 (Figures 7 and 8B), strongly supporting our finding that skipping of one of these exons triggers NMD (Figure 8B). We hypothesize that the occurrence of mutations that block inclusion of these exons is not a coincidence, but instead the consequence of positive selection of tumor clonal evolution process. This hypothesis is consistent with reported suppressive role of RBM10 in LUAD (49,51). More broadly, splice site mutations that promote transcript degradation via NMD may be one of the strategies used by tumor cells to block the expression of tumor suppressive genes and consequently promote tumor development and maintenance.
Because RBM5 shares 46% identify in amino acid sequence and high similarity in conserved protein domains with RBM10, it is conceivable that there are functional redundancies between them. Indeed, compensation at the protein level between RBM10 and RBM5 has been reported in mouse brain and neuroblastoma-derived SH-SY5Y cells, suggesting possible functional compensation (72). Notably, emerging evidence suggests that RBM10 and RBM5 target different sets of RNAs and exhibit distinct regulatory and biological functions, at least with respect to regulating splicing and apoptosis (49,72). For these reasons, possible confounding effects resulting from cross-regulation of RBM5 by RBM10 should be taken into consideration when investigating the functions of RBM10 under various physiological conditions. It should also be pointed out that RBM10-mediated cross-regulation may play an important role in the regulation of RBM5 levels and functions in different cell types, such as shaping the high levels of RBM5 expression in brain and testis cells (72,73). Elucidating the various possible roles of auto- and cross-regulation by RBM10 remains an important goal for further investigations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Zhen Shao at Key Lab of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Chinese Academy of Science, Shanghai, China, for discussions regarding TCGA data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31371299 to Y.W.]; Fudan University [EZF101528, EZF101600/03 and JIF101011 to Y.W.]. Funding for open access charge: National Natural Science Foundation of China [31371299 to Y.W.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Roy B., Haupt L.M., Griffiths L.R.. Review: alternative splicing (AS) of genes as an approach for generating protein complexity. Curr. Genomics. 2013; 14:182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Merkin J., Russell C., Chen P., Burge C.B.. Evolutionary dynamics of gene and isoform regulation in mammalian Ttissues. Science. 2012; 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Çolak R. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012; 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 4. Cooper T.A., Wan L., Dreyfuss G.. RNA and disease. Cell. 2009; 136:777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalsotra A., Cooper T.A.. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011; 12:715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scotti M.M., Swanson M.S.. RNA mis-splicing in disease. Nat. Rev. Genet. 2016; 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J., Weiss W.A.. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015; 34:1–14. [DOI] [PubMed] [Google Scholar]
- 8. Yang X., Coulombe-Huntington J., Kang S., Sheynkman Gloria M., Hao T., Richardson A., Sun S., Yang F., Shen Yun A., Murray Ryan R. et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016; 164:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 10. Sibley C.R., Blazquez L., Ule J.. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016; 17:407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGlincy N.J., Smith C.W.J.. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense. Trends Biochem. Sci. 2008; 33:385–393. [DOI] [PubMed] [Google Scholar]
- 12. Fu X.-D., Ares M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M., Manley J.L.. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009; 10:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee Y., Rio D.C.. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015; 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jangi M., Sharp P.A.. Building robust transcriptomes with master splicing factors. Cell. 2014; 159:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lykke-Andersen S., Jensen T.H.. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015; 16:665–677. [DOI] [PubMed] [Google Scholar]
- 17. Brogna S., Wen J.. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009; 16:107–113. [DOI] [PubMed] [Google Scholar]
- 18. Chang Y.-F., Imam J.S., Wilkinson M.F.. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007; 76:51–74. [DOI] [PubMed] [Google Scholar]
- 19. Peccarelli M., Kebaara B.W.. Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryotic Cell. 2014; 13:1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popp M.W., Maquat L.E.. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016; 165:1319–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long J.C., Caceres J.F.. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009; 417:15–27. [DOI] [PubMed] [Google Scholar]
- 22. Han S.P., Tang Y.H., Smith R.. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010; 430:379–392. [DOI] [PubMed] [Google Scholar]
- 23. Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.-C., Sun E., Wancewicz E., Mazur C. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011; 14:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gates D.P., Coonrod L.A., Berglund J.A.. Autoregulated splicing of muscleblind-like 1 (MBNL1) pre-mRNA. J. Biol. Chem. 2011; 286:34224–34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kino Y., Washizu C., Kurosawa M., Oma Y., Hattori N., Ishiura S., Nukina N.. Nuclear localization of MBNL1: splicing-mediated autoregulation and repression of repeat-derived aberrant proteins. Hum. Mole. Genet. 2015; 24:740–756. [DOI] [PubMed] [Google Scholar]
- 26. Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J.. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008; 28:4320–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jangi M., Boutz P.L., Paul P., Sharp P.A.. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014; 28:637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huelga Stephanie C., Vu Anthony Q., Arnold Justin D., Liang Tiffany Y., Liu Patrick P., Yan Bernice Y., Donohue John P., Shiue L., Hoon S., Brenner S. et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012; 1:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calarco J.A., Superina S., O’Hanlon D., Gabut M., Raj B., Pan Q., Skalska U., Clarke L., Gelinas D., van der Kooy D. et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009; 138:898–910. [DOI] [PubMed] [Google Scholar]
- 30. Rossbach O., Hung L.-H., Schreiner S., Grishina I., Heiner M., Hui J., Bindereif A.. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell. Biol. 2009; 29:1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spellman R., Llorian M., Smith C.W.J.. Crossregulation and functional redundancy between the splicing regulator PTB and Its paralogs nPTB and ROD1. Mol. Cell. 2007; 27:420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E.. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007; 446:926–929. [DOI] [PubMed] [Google Scholar]
- 33. Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O’Brien G., Shiue L., Clark T.A., Blume J.E., Ares M. Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007; 21:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Valacca C., Bonomi S., Buratti E., Pedrotti S., Baralle F.E., Sette C., Ghigna C., Biamonti G.. Sam68 regulates EMT through alternative splicing–activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 2010; 191:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Y., Liu S., Liu G., Ozturk A., Hicks G.G.. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013; 9:e1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Budini M., Buratti E.. TDP-43 autoregulation: implications for disease. J. Mol. Neurosci. 2011; 45:473–479. [DOI] [PubMed] [Google Scholar]
- 37. Guo J., Jia J., Jia R.. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Scientific Rep. 2015; 5:14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hegele A., Kamburov A., Grossmann A., Sourlis C., Wowro S., Weimann M., Will C.L., Pena V., Lührmann R., Stelzl U.. Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell. 2012; 45:567–580. [DOI] [PubMed] [Google Scholar]
- 39. Johnston J.J., Sapp J.C., Curry C., Horton M., Leon E., Cusmano-Ozog K., Dobyns W.B., Hudgins L., Zackai E., Biesecker L.G.. Expansion of the TARP syndrome phenotype associated with de novo mutations and mosaicism. Am. J. Med. Genet. A. 2014; 0:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnston J.J., Teer J.K., Cherukuri P.F., Hansen N.F., Loftus S.K., Chong K., Mullikin J.C., Biesecker L.G.. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 2010; 86:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gripp K.W., Hopkins E., Johnston J.J., Krause C., Dobyns W.B., Biesecker L.G.. Long term survival in TARP syndrome and confirmation of RBM10 as the disease causing gene. Am. J. Med. Genet. A. 2011; 155:2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y., Gogol-Döring A., Hu H., Fröhler S., Ma Y., Jens M., Maaskola J., Murakawa Y., Quedenau C., Landthaler M. et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol. Med. 2013; 5:1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012; 150:1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giannakis M., Mu X.J., Shukla S.A., Qian Z.R., Cohen O., Nishihara R., Bahl S., Cao Y., Amin-Mansour A., Yamauchi M. et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016; 15:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Furukawa T., Kuboki Y., Tanji E., Yoshida S., Hatori T., Yamamoto M., Shibata N., Shimizu K., Kamatani N., Shiratori K.. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Scientific Rep. 2011; 1:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Witkiewicz A.K., McMillan E.A., Balaji U., Baek G., Lin W.-C., Mansour J., Mollaee M., Wagner K.-U., Koduru P., Yopp A. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015; 6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rodor J., FitzPatrick D.R., Eyras E., Cáceres J.F.. The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 2017; 14:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bechara E.G., Sebestyén E., Bernardis I., Eyras E., Valcárcel J.. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell. 2013; 52:720–733. [DOI] [PubMed] [Google Scholar]
- 50. Zhao J., Sun Y., Huang Y., Song F., Huang Z., Bao Y., Zuo J., Saffen D., Shao Z., Liu W. et al. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Scientific Rep. 2017; doi:10.1038/srep40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hernández J., Bechara E., Schlesinger D., Delgado J., Serrano L., Valcárcel J.. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016; 13:466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y., Ma M., Xiao X., Wang Z.. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 2012; 19:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bullard J.H., Purdom E., Hansen K.D., Dudoit S.. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010; 11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36–R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rivas M.A., Pirinen M., Conrad D.F., Lek M., Tsang E.K., Karczewski K.J., Maller J.B., Kukurba K.R., DeLuca D.S., Fromer M. et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015; 348:666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J.. Deciphering the splicing code. Nature. 2010; 465:53–59. [DOI] [PubMed] [Google Scholar]
- 58. Reyes-Herrera P.H., Speck-Hernandez C.A., Sierra C.A., Herrera S.. BackCLIP: a tool to identify common background presence in PAR-CLIP datasets. Bioinformatics. 2015; doi:10.1093/bioinformatics/btv442. [DOI] [PubMed] [Google Scholar]
- 59. Friedersdorf M.B., Keene J.D.. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014; 15:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Collins K.M., Kainov Y.A., Christodolou E., Ray D., Morris Q., Hughes T., Taylor I.A., Makeyev E.V., Ramos A.. An RRM-ZnF RNA recognition module targets RBM10 to exonic sequences to promote exon exclusion. Nucleic Acids Res. 2017; doi:10.1093/nar/gkx225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W.J.. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004; 13:91–100. [DOI] [PubMed] [Google Scholar]
- 66. Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y.-S., Zhang C., Yeo G., Black D.L., Sun H. et al. Genome-wide analysis of PTB-RNA Iinteractions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009; 36:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ling J.P., Chhabra R., Merran J.D., Schaughency P.M., Wheelan S.J., Corden J.L., Wong P.C.. PTBP1 and PTBP2 repress nonconserved cryptic exons. Cell Rep. 2016; 17:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mourtada-Maarabouni M., Sutherland L.C., Meredith J.M., Williams G.T.. Simultaneous acceleration of the cell cycle and suppression of apoptosis by splice variant delta-6 of the candidate tumour suppressor LUCA-15/RBM5. Genes Cells. 2003; 8:109–119. [DOI] [PubMed] [Google Scholar]
- 69. Boutz P.L., Stoilov P., Li Q., Lin C.-H., Chawla G., Ostrow K., Shiue L., Ares M., Black D.L.. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007; 21:1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T.. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007; 27:435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y. et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2012; 152:82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jackson T.C., Du L., Janesko-Feldman K., Vagni V.A., Dezfulian C., Poloyac S.M., Jackson E.K., Clark R.S., Kochanek P.M.. The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J. Cerebral Blood Flow Metab. 2015; 35:655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O’Bryan M.K., Clark B.J., McLaughlin E.A., D'Sylva R.J., O’Donnell L., Wilce J.A., Sutherland J., O’Connor A.E., Whittle B., Goodnow C.C. et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 2013; 9:e1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.