Abstract
Phosphorylated histone H2AX, termed ‘γH2AX’, mediates the chromatin response to DNA double strand breaks (DSBs) in mammalian cells. H2AX deficiency increases the numbers of unrepaired DSBs and translocations, which are partly associated with defects in non-homologous end joining (NHEJ) and contributing to genomic instability in cancer. However, the role of γH2AX in NHEJ of general DSBs has yet to be clearly defined. Here, we showed that despite little effect on overall NHEJ efficiency, H2AX deficiency causes a surprising bias towards accurate NHEJ and shorter deletions in NHEJ products. By analyzing CRISPR/Cas9-induced NHEJ and by using a new reporter for mutagenic NHEJ, we found that γH2AX, along with its interacting protein MDC1, is required for efficient classical NHEJ (C-NHEJ) but with short deletions and insertions. Epistasis analysis revealed that ataxia telangiectasia mutated (ATM) and the chromatin remodeling complex Tip60/TRRAP/P400 are essential for this H2AX function. Taken together, these data suggest that a subset of DSBs may require γH2AX-mediated short-range nucleosome repositioning around the breaks to facilitate C-NHEJ with loss of a few extra nucleotides at NHEJ junctions. This may prevent outcomes such as non-repair and translocations, which are generally more destabilizing to genomes than short deletions and insertions from local NHEJ.
INTRODUCTION
DNA double strand breaks (DSBs) are a form of highly mutagenic DNA lesions that can arise exogenously and endogenously, and pose a serious threat to the integrity of eukaryotic genomes. In order to prevent genome instability and cancer, DSBs have to be properly recognized and repaired (1). In mammalian cells, repair of DSBs is carried out primarily by either homologous recombination (HR) or non-homologous end joining (NHEJ). HR repairs DSBs generated in post-replicative cells where sister chromatids are available as a homologous template for repair (2). In contrast, NHEJ, including classical NHEJ (C-NHEJ) and alternative NHEJ (A-NHEJ), functions throughout the cell cycle and is the primary DSB repair pathway in G1-arrested somatic cells (3). C-NHEJ requires several core proteins including Ku70, Ku80, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), XRCC4 and DNA ligase 4, whereas A-NHEJ repairs DSBs without participation of either one of these core NHEJ factors and generates deletions frequently associated with short tracts of homology at repair junctions (4–6). In C-NHEJ, upon DNA breakage, the Ku70 and Ku80 heterodimer binds to ‘naked’ DNA ends of DSBs and helps recruit other core NHEJ factors to the same DNA ends. DNA-PKcs is activated to assist NHEJ upon recruitment to the Ku–DNA complex. XRCC4 and DNA ligase 4 form a complex in cells and function in the final steps of NHEJ to catalyze ligation of DNA ends.
DSBs in eukaryotes have to be detected and repaired within the context of chromatin (7), and in mammalian cells, a key response to DSBs in chromatin is rapid phosphorylation of H2AX, a histone H2A variant. This occurs on serine 139 (S139) of the H2AX carboxyl-terminal SQEY motif by ataxia telangiectasia mutated (ATM), DNA-PKcs and ataxia telangiectasia mutated- and Rad3-related (ATR) over large chromatin regions surrounding the DSBs (8–12). Phosphorylated H2AX, termed ‘γH2AX’, subsequently initiates a DNA damage signaling cascade on chromatin near the DSBs and establishes the ‘chromatin domain’ of the DNA damage response by providing a chromatin platform for protein interactions and modifications (13). It is expected that deletion of H2AX would disrupt this ‘chromatin domain’, induce aberrant DSB response and cause genomic instability. In fact, H2AX-deficient cells are sensitive to ionizing radiation (IR), have an elevated level of chromosomal and chromatid breaks as well as translocations, and exhibit genomic instability (14–18). When combined with p53 deficiency, H2AX-deficient mice are predisposed to cancer, in particular, T and B lineage lymphomas (15,16).
As DSB repair occurs in the context of chromatin, the γH2AX chromatin domain adds a layer of control to the regulation of DSB repair, and has been implicated in both HR and NHEJ repair pathways (13). Previous studies have shown that HR is impaired in H2AX (H2A in yeast)-deficient cells (14,19,20), but it remains unclear how H2AX facilitates HR, especially between sister chromatids. It has been proposed that the γH2AX chromatin domain may promote end resection for HR engagement or have a synapsis function in HR (21,22), but the roles of H2AX in NHEJ are more confusing. Studies thus far have mostly focused on V(D)J recombination and class switch recombination (CSR) in H2AX-deficient lymphocytes and H2AX-deficient mice (15–18,23–26), as both processes require programmed DSB induction by recombination activated gene (RAG) endonucleases and activation-induced cytidine deaminase (AID) respectively and joining of DSBs by NHEJ. A recent study has also indicated that end resection during NHEJ in G1 is distinct from that during HR (27); by extension, if this occurs, end resection promoted by the γH2AX chromatin domain in HR may not be applicable to NHEJ.
V(D)J recombination, the mechanism responsible for generating antigen receptor diversity in developing lymphocytes, is grossly normal in H2AX-deficient mice (15,16). Prior work has suggested that γH2AX is dispensable for end synapsis and end joining within the RAG–DSB synaptic complex in C-NHEJ-mediated V(D)J recombination (22,24). However, H2AX-deficient mice have an increased level of chromosomal breaks and translocations derived from RAG endonuclease-initiated DNA breaks (15,16,23–25,28). It is speculated that the γH2AX chromatin domain may help stabilize broken DNA strands, preventing DSBs from progressing into unrepaired chromosomal breaks and translocations (22,24). H2AX is also shown to suppress hairpin opening and subsequent end resection of RAG-initiated DSBs (23).
After V(D)J recombination, CSR further diversifies antigen receptors of antigen-stimulated B cells (29). In this NHEJ-mediated process, AID initiates DSBs on two switch (S) regions, which can be nearly 100kb apart from each other at IgH locus (15,26,29). Subsequent end joining of these DSBs can lead to either intra-S region deletion or productive CSR with intervening sequences deleted (15,26,29). In the absence of H2AX, CSR-associated NHEJ is deficient, resulting in modestly deficient CSR and increased level of chromosomal breaks and translocations (15–18,26,28). It is thought that the γH2AX chromatin domain promotes synapsis of two distantly located DSBs for efficient NHEJ during CSR (22,29,30).
Despite the understanding of the role of H2AX in specialized NHEJ during V(D)J recombination and CSR, the function of H2AX in NHEJ in general settings is poorly understood. In Saccharomyces cerevisiae, γH2A is required for efficient end joining in a plasmid NHEJ assay (31). In mammalian cells, the γH2AX chromatin domain is responsible for repairing ∼10–15% of DSBs induced by IR (32). H2AX deficiency causes genomic instability including an increased level of chromosomal breaks and translocations, indicating a defect in general NHEJ (15,17,18,26,29). Paradoxically, using a green fluorescent protein (GFP)-based NHEJ reporter assay, we found that H2AX loss has little effect on the efficiency of general NHEJ of two site-specific chromosomal DSBs tandemly induced by I-SceI meganuclease in mouse (ES) cells (33,34). This suggests that H2AX may not be needed for NHEJ of two adjacent, readily rejoined DNA ends. But it is also possible that H2AX deficiency may contribute to defects in a subtype of NHEJ, which are masked by largely unaltered overall NHEJ efficiency. In addition, DNA ends of many DSBs generated in chromatin are associated with nucleosomes that may hinder the binding of NHEJ proteins to DNA ends for NHEJ. Hence, the presence of γH2AX may help reposition nucleosomes around these DSBs, allowing efficient NHEJ. It has been shown that the γH2AX chromatin domain contains chromatin remodelers and has chromatin remodeling activities (21,31).
Given the development and potential applications of clustered regularly interspaced short palindromic repeats (CRISPR) technology and its unique requirement for DSB induction and repair (35–37), it is also of interest to determine whether H2AX plays a role in NHEJ of CRISPR/Cas9-induced DSBs, and by extension, in NHEJ-mediated CRISPR genome editing. Here, we report a role of H2AX in NHEJ that repairs a site-specific chromosomal DSB induced by I-SceI and by the CRISPR Cas9 nuclease, as well as epistasis analysis of this role.
MATERIALS AND METHODS
Plasmids
The sGEJ/vGEJ reporters were previously constructed (33). To generate the mNHEJ reporter BGN, the BSD gene was amplified by PCR (see Supplementary Table S7 for primer sequences) with a HindIII site in one primer and tandem I-SceI–EcoRI sites in the other primer. PCR product was digested with HindIII and EcoRI and inserted into the HindIII–EcoRI region between the PGK promoter and the EGFP cassette of pBigT previously constructed (19). pcDNA3β-Hyg-based expression vectors for human H2AX, human XRCC4, mouse MDC1 tandem BRCT domain (mBRCT) and its mutant (mBRCT-KM) were described previously (19,33,34,38). CRISPR/Cas9 plasmids px330 was originally obtained from Addgene (Cat #42230). The U6-sgRNA vector (pU6-gRNA) was derived from px330 by removing the CBh-hSpCas9 cassette. Individual sgRNAs cloning was performed as described previously (39). The sgRNA target sequences are listed in Supplementary Table S7. Plasmids newly constructed were confirmed by Sanger sequencing.
Antibodies and chemicals including small molecular inhibitors
Antibodies included anti-HA Tag (SC-805; 1:500) and XRCC4-C20 (SC-8285; 1:500) from Santa Cruz, anti-H2AX (07-627; 1:1000) from Millipore, anti-β-actin (A5441; 1:10 000) from Sigma, and peroxidase-conjugated goat anti-rabbit, donkey anti-goat and rabbit anti-mouse IgG (315-035-048; 1:10 000) from Jackson ImmunoResearch. Small molecule inhibitors included ATR inhibitor (VE821) from Axon MedChem, ATM inhibitor (KU60019), DNA-PKcs inhibitor (NU7441) from Tocris, and poly(ADP-ribose) polymerase 1 (PARP1) inhibitor (Olaparib) from Selleck. Olaparib was used at 2 μM final concentration and the others at 5 μM. Chloroquine (S4157) was purchased from Selleck and sodium butyrate (A510838) from Sangon Biotech. The hypotonic buffer contains phosphated buffered saline, 50 mM NaCl, 0.45% (w/v) glucose and 1% FBS.
Cell lines
The sGEJ/vGEJ reporter mouse ES cells were previously established and described (33). The BGN reporter cell lines were generated as previously described (19,33). Specifically, mouse H2AXflox/flox and XRCC4flox/flox ES cells were electroporated with linearized BGN reporter vector and seeded in 10 cm plates with neo+ feeders. Individual clones were selected in the presence of 400 μg/ml neomycin (Invitrogen). Clones were isolated and confirmed by Southern blot with GFP and ROSA26 probes as done previously (19), and only individual clones with single copy of intact reporter cassette were selected for this study. To generate mouse ES cell lines stably and ectopically expressing H2AX or XRCC4, 2 × 105 mouse ES cells were transfected with Lipofectamine 2000 (Invitrogen) in 24-well plate as previously described (19,33), and were seeded onto a 10 cm plate 3 days post-transfection. Stable clones were selected and pooled in the presence of 400 μg/ml hygromycin (Invitrogen). For Cas9-mediated gene knockout, 2 × 105 ES cells were transfected twice and then plated on MEF for single clones without antibiotics selection. Knockout clones were verified by PCR along with Sanger sequencing and/or Western blot. Primers are listed in Supplementary Table S7.
NHEJ reporter assays
Mouse ES cells harboring NHEJ reporter were transfected with pcDNA3β-I-SceI, the U6-sgRNA plasmids and pX330, and/or siRNA as previously described (33). SiRNAs were purchased from RiboBio Co with a ‘Scramble’ siRNA as control. SiRNA sequences are listed in Supplementary Table S7. For drug treatment, inhibitors were added at 6 h post-transfection, and replaced with fresh addition next day for a continued treatment for the rest of the experiment. Cells transfected and treated were analyzed for GFP+ frequencies using the BD LSRFortessa™ cell analyzer and the Beckman Coulter FC500 cell analyzer 3 days post-transfection. The NHEJ frequencies were calculated after being corrected with background readings and normalized with transfection efficiencies as described before (33,34). Statistical comparisons between two unpaired populations and between paired samples were analyzed by Anova and Student's two-tailed paired t test, respectively.
Western blotting
To analyze histones, cells were first lysed for 30 min using cytolysis buffer (10 mM HEPES, pH 7.9, 50 mM NaCl, 0.25 M sucrose, 0.1 mM EDTA, 0.5% Triton X-100). Histones were then acid-extracted with the buffer containing 0.25 M HCl and 5% glycerol from pellets of cell lysates as described previously (19), resolved by SDS-PAGE, and analyzed by Western blotting. To analyze non-histone proteins, cells were lysed in the NP buffer (150 mM NaCl, 20 mM Tris–HCl, pH 7.4, 5 mM EDTA and 0.5% Nonidet P-40). Cell extracts were resolved by SDS-PAGE, and analyzed by western blotting with corresponding antibodies.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNAs were extracted by TRIzol reagent (Invitrogen) and then reverse-transcribed to complementary DNA using the PrimeScript RT reagent kit (TaKaRa). qRT-PCR was performed using Power SYBR Green Master mix (Applied Biosystems) on an ABI Viia7 DX System (Applied Biosystems). Primers are listed in Supplementary Table S7.
gDNA extraction, PCR amplification and Illumina DNA sequencing
To analysis the deletion/insertion pattern in NHEJ products, I-SceI-induced and Cas9-induced GFP+ cells were sorted by flow cytometry (BD FACSAria II). For analysis of NHEJ or targeted genome editing at endogeneous loci, cells were directly collected after I-SceI-induced NHEJ or CRISPR/Cas9-induced NHEJ. gDNA was isolated from these cells using genomic DNA purification kit (Axygen). The genomic regions of 300–500 bp surrounding the I-SceI and CRISPR/Cas9 target sites were PCR amplified with respective primers (Supplementary Table S7). PCR products were purified using gel extraction kit (Axygen). As NHEJ in normal cells generates deletions mostly within ∼50 bp (27,33,40,41), PCR products of 300–500 bp are expected to cover most of NHEJ products. For next-generation sequencing at Annoroad Gene Technology (Beijing), PCR products of 4–20 different genomic target sites were mixed, end-repaired, adenylated at 3′-ends, ligated with adapters, purified, and amplified by the second round of PCR to incorporate the P7 and P5 Illumina adapters and a unique 8-mer barcode sequence according to the manufacturer's protocols. The final PCR products were cleaned, quantified, normalized into one library pool and sequenced on the Illumina Hiseq.
Quantification of NHEJ events in Illumina DNA sequencing data
After demultiplexing, forward and reverse paired-end reads were merged to generate a single consensus sequence per read pair using PEAR 0.9.8 (42). Merged sequences were analyzed using the ‘DeepSeq Pipeline’, a self-analysis platform, which implemented a pairwise local sequence alignment algorithm called DBS-Aligner (Double Breakpoints Sequence Aligner) to align two genomic sequences with one or two breakpoints. DBS-Aligner uses Smith–Waterman algorithm to compute the basic alignments and makes refinements on alignments in breakpoint regions (43,44). ‘Insertion’ sequences were identified by taking continuous letters in read sequence that paired up with gaps (dashes) in reference sequence, and ‘Deletion’ sequences determined by gaps (dashes) in read sequence. The length of ‘Deletion’ and ‘Insertion’ defined by the distance from both ends of a DSB were counted.
RESULTS
H2AX deficiency causes increased level of accurate NHEJ and a bias toward shorter deletions in NHEJ
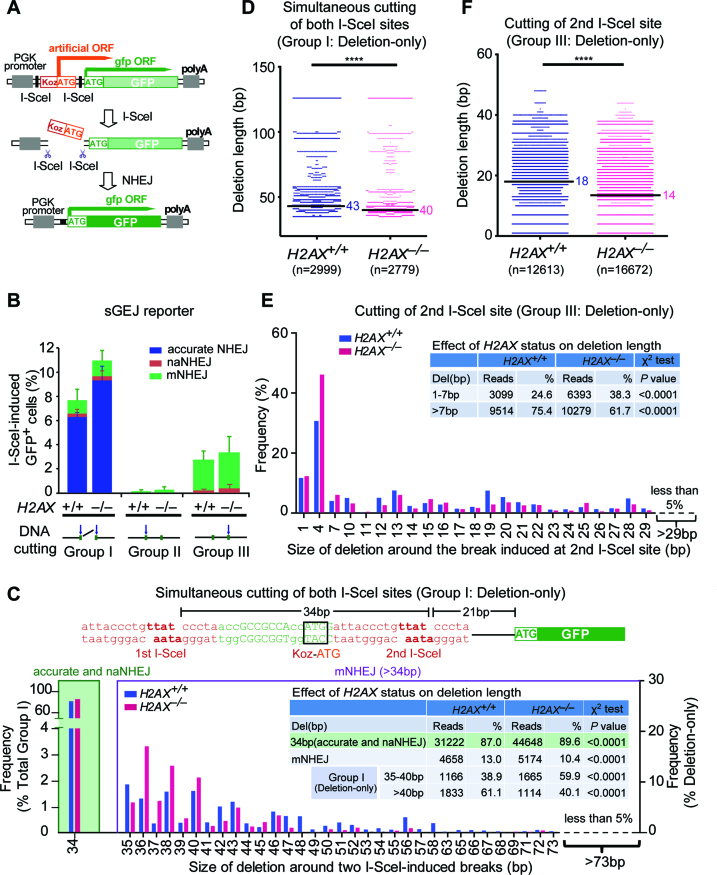
We previously developed a reporter (sGEJ or vGEJ) for quantitatively measuring I-SceI-induced NHEJ in mammalian cells (Figure 1A) (33). In this reporter, due to an upstream, out-of-frame translation start site (‘Koz-ATG’), no wild-type GFP is translated. When DSBs are induced at either or both of two closely positioned, tandem I-SceI sites by I-SceI meganuclease (45), repair by NHEJ corrects the reading frame of GFP, making cells positive for GFP expression (GFP+). The frequency of I-SceI-induced GFP+ cells reflects relative NHEJ efficiency. Using mouse ES cells harboring the NHEJ reporter sGEJ or vGEJ that we previously established (33), we re-analyzed the effect of H2AX deficiency on I-SceI-induced NHEJ. Consistent with previous findings (33,34), H2AX deletion causes little change in overall NHEJ efficiency (Figure 1B, Supplementary Figure S1A).
Figure 1.
H2AX deficiency causes a bias towards accurate NHEJ and shorter length of deletion. (A) Schematic of the NHEJ reporter sGEJ/vGEJ. Tandem I-SceI sites are either sequentially (sGEJ) or invertedly positioned (vGEJ). With no I-SceI-induced DSB, GFP is out-of-frame, resulting no GFP+ cells. Repair of I-SceI-induced DSB by NHEJ generates in-frame GFP, making cells GFP+. (B) Percentage of I-SceI-induced GFP+ cells with distributions of accurate NHEJ (blue), naNHEJ (red) and mNHEJ (orange) from H2AX+/+ and H2AX–/– sGEJ reporter cells transfected with the I-SceI expression plasmid. I-SceI-induced GFP+ cells were categorized into three groups according to I-SceI-induced break site detected by junction analysis. Group I, II and III respectively represent NHEJ of two simultaneous I-SceI cuts, NHEJ of the first I-SceI cut and NHEJ of the second I-SceI cut as indicated. Three independent experiments were performed for I-SceI-induced GFP+ cells. Bars represent the mean ± S.D. of three independent experiments. In each group between H2AX+/+ and H2AX–/– cells, accurate NHEJ, naNHEJ and mNHEJ characterized from deep sequencing are proportionated to total NHEJ reads and distributed accordingly in each bar as indicated in color. (C) Frequency of accurate NHEJ/naNHEJ in NHEJ of two simultaneous I-SceI cuts (Group I; left), and frequency of deletions with different deletion length in ‘Del’ events of Group I NHEJ (right) between H2AX+/+ and H2AX–/– sGEJ reporter cells. These NHEJ events were also grouped into 34 bp (accurate and naNHEJ), 35–40 bp and >40 bp, and the reads and frequencies were summarized in inset and compared between H2AX+/+ and H2AX–/– cells by a χ2 test with P values indicated. The sequence around two I-SceI sites is shown on the top with two I-SceI sites in red and the ‘Koz-ATG’ start codon highlighted with black box. (D) Deletion distributions of ‘Del’ events in NHEJ of two simultaneous I-SceI cuts (Group I) between H2AX+/+ and H2AX–/– sGEJ reporter cells. The median deletion length is indicated, and deletion distributions between H2AX+/+ and H2AX–/– cells here and below were compared by a two-tailed Mann–Whitney test, indicating a shift toward shorter deletions in H2AX–/– cells. ****P <0.0001. (E) Frequency of deletions with different deletion length in ‘Del’ events among NHEJ of the second I-SceI cut between H2AX+/+ and H2AX–/– sGEJ reporter cells. These NHEJ events grouped into 1–7 and >7 bp were summarized in inset according to their reads and frequencies. P values from a χ2 test are indicated. (F) Deletion distributions of ‘Del’ events in NHEJ of the second I-SceI cut (Group III) between H2AX+/+ and H2AX–/– sGEJ reporter ES cells demonstrate a shift towards shorter deletions in H2AX–/– cells. ****P < 0.0001.
However, it is unclear whether H2AX modulates the accuracy of NHEJ junctions, which could be revealed by deep sequencing. We thus sorted I-SceI-induced GFP+ cells from H2AX+/+ and H2AX–/– sGEJ reporter cells by fluorescence-activated cell sorting (FACS), isolated genomic DNA (gDNA), and amplified repair junctions by PCR for Illumina sequencing. As expected, sequence analysis reveals that I-SceI induces a DSB at either or both of tandem I-SceI sites in the reporter. Upon simultaneous induction of DSBs at two I-SceI sites, end joining through full pairing of complementary 4-protruding nucleotides (4Pnt) generated by I-SceI could delete the intervening sequence between two I-SceI-induced DSBs, but cause no additional junctional deletions, and is therefore defined as accurate NHEJ used hereafter. This type of accurate NHEJ is different from accurate NHEJ that restores the original sequence. We also define end joining through partial pairing of complementary 4Pnt generated by I-SceI as near accurate NHEJ (naNHEJ) because this end joining does not cause additional deletions at junctions either. However, naNHEJ may or may not cause the loss of the intervening sequence between two I-SceI sites. It is notable that, in all sorted GFP+ products, based on the combined reads from three independent experiments, the efficiency of accurate NHEJ and naNHEJ elevates from 61.1 ± 5.2% (31 918 of 52 231) in H2AX+/+ cells to 64.6 ± 7.5% (45 934 of 71 075) in H2AX–/– cells (χ2 test: P < 0.0001) (Figure 1B, Supplementary Table S1). Conversely, mutagenic NHEJ (mNHEJ) that generates deletions and insertions extended beyond 4Pnt decreases from 38.9 ± 3.2% (20 313 of 52 231) to 35.4 ± 7.5% (25 141 of 71 075) (χ2 test: P< 0.0001) by H2AX deletion (Figure 1B, Supplementary Table S1).
To better define the effect of H2AX on NHEJ, we must compare NHEJ of the same type of DNA breakage between H2AX+/+ and H2AX–/– cells. Based on the sites of DNA breakage, we divided I-SceI-induced DSBs into three groups: (i) simultaneous cutting of two I-SceI sites that pops out Koz-ATG (Group I); (ii) cutting of the first I-SceI site that destroys Koz-ATG (Group II); (iii) cutting of the second I-SceI site that either destroys Koz-ATG or introduces additional ‘3n+1’ frame-shift to correct the reading frame of GFP with Koz-ATG retained as the translation start site (Group III). About 70% of GFP+ cells arise from Group I and ∼25% from Group III in both H2AX+/+ cells and H2AX–/– cells whereas the contribution of Group II is minimal (5.3% and 4.2% respectively) (Figure 1B, Supplementary Table S1).
We then examined major types of NHEJ events for possible functions of H2AX. In Group I, compared with H2AX+/+ cells, in which 84.3 ± 13.3% (30 255 of 35 880) of GFP+ cells is derived from accurate NHEJ, H2AX deficiency slightly increases the frequency of this type of NHEJ (87.6 ± 9.9%, 43 640 of 49 822) (χ2 test: P< 0.0001; Supplementary Table S1). In addition, H2AX–/– cells have a lower frequency of mNHEJ than H2AX+/+ cells in Group I (10.4 ± 6.2% versus 13.0 ± 9.5%, χ2 test: P< 0.0001) and Group III (93.0 ± 15.5% versus 94.9 ± 4.0%, χ2 test: P< 0.0001) (Figure 1B, Supplementary Table S1). This paradoxically suggests that H2AX deficiency may tilt NHEJ towards accurate NHEJ and naNHEJ.
Of note, besides the whole Group II, NHEJ events associated with insertions in both Group I and Group III are rare (Supplementary Table S1); therefore, these types of NHEJ events are ignored in further analysis of H2AX’s role. Thus, we only analyzed the deletion length of deletion-only (‘Del’) NHEJ products in Group I and III in order to gain a better view on the role of H2AX in controlling the accuracy of NHEJ. Over 95% of ‘Del’ NHEJ events show deletion length <29 bp, in addition to the pop-out of 34 bp between two I-SceI sites if it occurs. In Group I’s ‘Del’ events, H2AX–/– cells have short-range deletions (1–6 bp + 34 bp pop-out) at the frequency of 59.9%, significantly higher than H2AX+/+ cells at 38.9% (Figure 1C and inset). In contrast, for deletions over 6 bp in addition to 34 bp pop-out, H2AX–/– cells have a lower frequency (40.1%) than H2AX+/+ cells (61.1%) (Figure 1C and inset). In fact, compared with H2AX+/+ cells, deletions in H2AX–/– cells are shifted toward shorter length with the median deletion length of 43 bp (i.e. 9 bp + 34 bp pop-out) in H2AX+/+ cells and 40 bp (i.e. 6 bp + 34 bp pop-out) in H2AX–/– cells (Mann–Whitney test: P< 0.0001; Figure 1D). In Group III’s ‘Del’ events, 38.3% of H2AX–/– cells have deletions of 1–7 bp but only 24.6% of H2AX+/+ cells do. Conversely, fewer H2AX–/– cells (61.7%) have deletions over 7 bp than H2AX+/+ cells (75.4%) (Figure 1E and inset). Compared to H2AX+/+ cells, deletions in H2AX–/– cells are biased towards shorter length with the median deletion length of 18 bp in H2AX+/+ cells and 14 bp in H2AX–/– cells (Mann–Whitney test: P< 0.0001; Figure 1F). Taken together, these data demonstrate that H2AX deficiency reduces the level of mNHEJ, shifts deletion of mNHEJ products to a shorter length by a few base pairs and appears to cause a bias towards accurate NHEJ.
Bias towards shorter deletions caused by H2AX deficiency is bi-directional
DNA end processing, which affects the composition of NHEJ junctions, occurs at I-SceI-induced DSBs bi-directionally before end ligation during NHEJ, generating deletion on both directions from breakage sites in NHEJ products. We therefore analyzed deletions in both directions to assess whether the effect of H2AX deficiency on deletion length is bi-directional. In Group I’s deletions, H2AX–/– cells have a higher frequency of short-range deletions (0–4 bp) at 48.6% to the upstream (versus 34.5% in H2AX+/+ cells) and 90.1% to the downstream (0–5 bp) (versus 81.4% in H2AX+/+ cells) (χ2 test: P< 0.0001; Supplementary Figure S1B and inset). In contrast, the frequency of deletions over 4 bp is lower in H2AX-deficient cells at 51.4% to the upstream (versus 65.5% in H2AX+/+ cells) and 9.9% (versus 18.6% in H2AX+/+ cells) to the downstream (over 5 bp) than in H2AX+/+ cells (χ2 test: P< 0.0001; Supplementary Figure S1B and inset). Similarly, in Group III’s deletions, the frequency of short-range deletions is elevated in H2AX–/– cells (<7 bp upstream from 28.0% to 41.4% and <4 bp downstream from 68.6% to 73.8%) and that of long-range deletions (over 6 bp upstream and over 3 bp downstream) suppressed as compared to H2AX+/+ cells (χ2 test: P< 0.0001; Supplementary Figure S1C and inset). This suggests that H2AX deficiency reduces the length of deletions during NHEJ in a bi-directional manner.
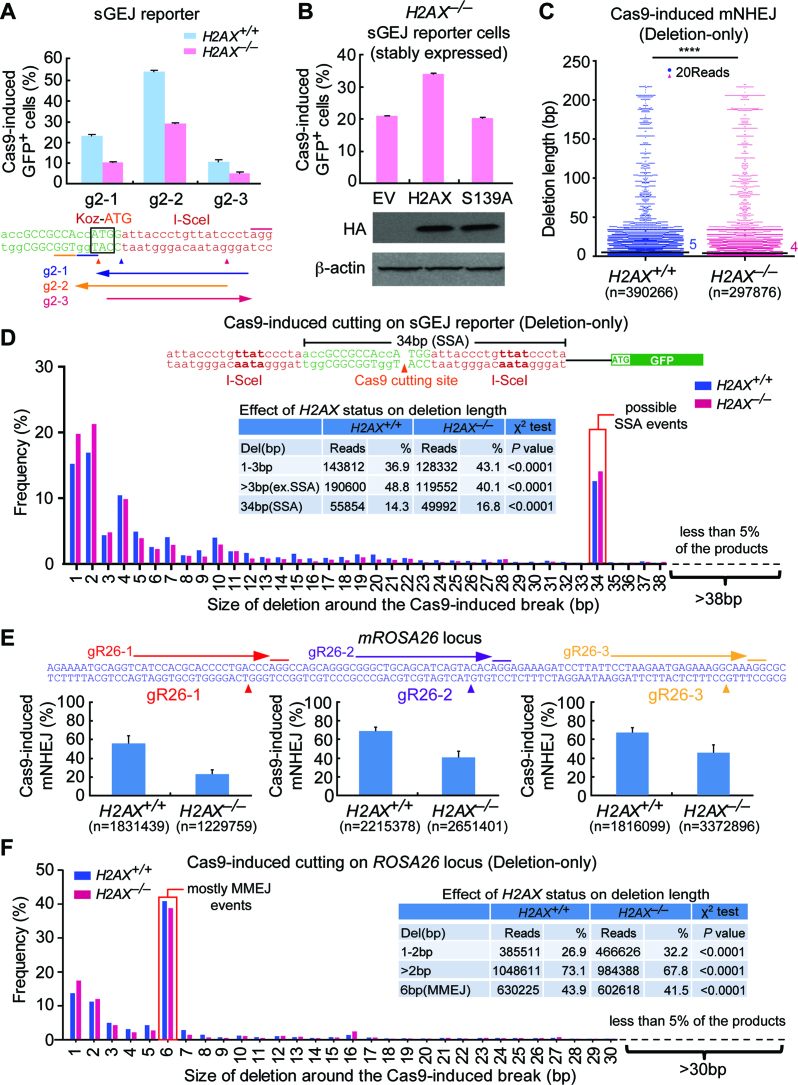
H2AX deficiency reduces the efficiency and deletion length of mNHEJ-mediated CRISPR/Cas9 genome editing
In the presence of fully complementary 4Pnt 3′-overhangs at two DNA ends of an I-SceI-induced chromosomal DSB, H2AX deficiency surprisingly causes a slight increase of accurate NHEJ and a reduction of mNHEJ along with a bias towards shorter deletions. Given that some of CRISPR/Cas9 genome editing is mediated by mNHEJ, we reasoned that H2AX would facilitate mNHEJ-mediated CRISPR/Cas9 genome editing. We thus used Streptococcus pyogenes CRISPR/Cas9, in place of I-SceI, to induce a blunt-end DSB around the second I-SceI site of the sGEJ reporter in mouse ES cells. We generated three gRNAs (g2–1, g2–2 and g2–3) targeting the vicinity of the second I-SceI site of the sGEJ reporter (Figure 2A). Since DSBs generated either lie within ATGG of ‘Koz-ATG’ or 15 bp downstream of ‘Koz-ATG’ in this sGEJ reporter, end processing during repair of these DSBs by mNHEJ can either destroy ATG or cause a ‘3n+1’ frame-shift in NHEJ products to generate GFP+ cells. We found that mNHEJ, induced by three Cas9/gRNA, is reduced by nearly a half in H2AX–/– cells as compared to H2AX+/+ cells (Figure 2A). The percentage of Cas9/g2–2-induced GFP+ cells is much higher than that of the other two gRNAs, likely due to the DNA breakage between A and TG of the ‘Koz-ATG’ induced by Cas9/g2–2; any modifications would abolish this ATG start codon and initiate in-frame translation of the GFP gene. The efficiency of mNHEJ induced by Cas9/g2–2 is increased in H2AX–/– cells by stable complementation of wide-type (wt) H2AX (Student's paired t-test: P = 0.0008), but not by the mutant S139A or empty vector control (EV) (Figure 2B), indicating S139 phosphorylation of H2AX is required for efficient CRISPR/Cas9 genome editing by mNHEJ.
Figure 2.
H2AX deficiency reduces the efficiency and deletion length in mNHEJ-mediated CRISPR/Cas9 genome editing. (A) Percentage of gRNA/Cas9-induced GFP+ cells from H2AX+/+ and H2AX–/– sGEJ reporter cells. Three gRNAs (g2–1, g2–2 and g2–3) targeting the regions surrounding the second I-SceI site are indicated. Values are the mean ± S.D. of three independent experiments, each in triplicates. Student paired t-test between ‘H2AX+/+’ and ‘H2AX–/–’: P = 0.03 for g2–1; P = 0.047 for g2–2; P = 0.007 for g2–3. (B) Percentage of g2–2/Cas9-induced GFP+ cells from H2AX–/– sGEJ reporter cells stably transfected with expression plasmids for HA-tagged mouse H2AX and its mutant S139A as well as empty vector control (EV). Values are the mean ± S.D. of three independent experiments, each in triplicates. Student paired t-test between ‘EV’ and ‘H2AX’: P = 0.0008; between ‘EV’ and ‘S139A’: not significant (NS). HA-tagged H2AX and S139A detected by anti-HA antibody are indicated under the bar chart with β-actin as the loading control. (C) Deletion distributions of ‘Del’ events in g2–2/Cas9-induced mNHEJ between H2AX+/+ and H2AX–/– sGEJ reporter ES cells demonstrate a shift toward shorter deletions in H2AX–/– cells. Each blue or red dot represents 20 reads. ****P < 0.0001 (two-tailed Mann–Whitney test). (D) Frequency of deletions with different deletion length in ‘Del’ events of g2–2/Cas9-induced mNHEJ between H2AX+/+ and H2AX–/– sGEJ reporter cells. The NHEJ events were grouped into 1–3 bp, >3 bp (excluding SSA or MMEJ) and 34 bp (SSA or MMEJ events) according to the deletion length. The combined reads and frequencies were summarized in inset with P values from a χ2 test indicated. (E) The relative efficiency of Cas9-induced mNHEJ at three different sites of endogenous ROSA26 locus in H2AX+/+ and H2AX–/– mouse ES cells. Three gRNAs (gR26–1, gR26–2 and gR26–3) targeting three different sites of endogenous ROSA26 locus are indicated. Cas9-induced mNHEJ was identified by Illumina sequencing, and the relative efficiency was calculated as ratios of mNHEJ reads to total reads and normalized by transfection efficiency. Student's paired t-test between ‘H2AX+/+’ and ‘H2AX–/–’: P = 0.004 for gR26–1; P = 0.003 for gR26–2; P= 0.019 for gR26–3. (F) Frequency of deletions with different deletion length in ‘Del’ events of gR26–3/Cas9-induced mNHEJ at the ROSA26 locus between H2AX+/+ and H2AX–/– mouse ES cells. The NHEJ events were grouped into deletions of 1–2 bp, deletions over 2 bp (>2 bp) and deletions of 6 bp (MMEJ). The combined reads and frequencies were summarized in inset with P values from a χ2 test indicated.
To determine the effect of H2AX on repair junctions of CRISPR/Cas9-induced mNHEJ, we next sorted Cas9/g2–2-induced GFP+ cells from H2AX+/+ and H2AX–/– sGEJ reporter cells by FACS, isolated gDNA, and analyzed repair junctions by deep sequencing. Among CRISPR/Cas9-induced mNHEJ in H2AX+/+ cells, 83.7 ± 4.7% is ‘Del’ events, 11.0 ± 3.7% is insertion-only (‘Ins’) events and 5.3 ± 1.0% carries both deletion and insertion (‘InDel’) (Supplementary Table S2). H2AX deficiency significantly decreases the percentage of ‘Del’ events (79.6 ± 5.8%) and InDel (5.2 ± 3.7%), but elevates ‘Ins’ events to 15.2 ± 2.2% (Supplementary Table S2). Consistent with I-SceI-induced NHEJ, further examination of ‘Del’ events reveals that, compared with H2AX+/+ cells, H2AX–/– cells are ‘biased’ toward shorter deletions in CRISPR/Cas9-induced mNHEJ. The median length of deletion is 5 bp in H2AX+/+ cells and 4 bp in H2AX–/– cells (Mann–Whitney test: P< 0.0001; Figure 2C). In addition, 1–3 bp deletion is observed in 36.9% of H2AX+/+ cells and 43.1% of H2AX–/– cells (χ2 test: P< 0.0001; Figure 2D and inset). However, over 48.8% of H2AX+/+ cells show deletion exceeding 3 bp, and only 40.1% of H2AX–/– cells have such length of deletion (χ2 test: P< 0.0001; Figure 2D and inset). Due to Cas9/g2–2-induced single strand annealing (SSA) or microhomology-mediated end joining (MMEJ) mediated by 18 bp homology of two tandem I-SceI sites, mNHEJ with 34 bp deletion is observed in both H2AX+/+ and H2AX–/– cells, at a similarly significant level (14.3% versus 16.8%; Figure 2D and inset). This appears to suggest that H2AX deficiency does not change the level of SSA or MMEJ in this context.
To validate the effect of H2AX deficiency on mNHEJ-mediated CRISPR/Cas9 editing of a natural site in the genome, we designed three gRNAs (gR26–1, gR26–2 and gR26–3) targeting the ROSA26 locus of the murine genome (Figure 2E). By transiently transfecting mouse H2AX+/+ and H2AX–/– ES cells with the plasmids expressing these gRNAs and Cas9, we induced site-specific CRISPR/Cas9 genome editing. Using Illumina deep sequencing to analyze the edited sites, we found that the editing efficiencies (i.e. the efficiencies of mNHEJ) normalized with transfection efficiencies are 22.7%, 39.9% and 46.3% respectively with gR26–1, gR26–2 and gR26–3 in H2AX–/– cells, lower than those in H2AX+/+ cells (versus 56.4%, 68.0% and 68.2%) (Figure 2E). This indicates that H2AX is required for efficient CRISPR/Cas9 genome editing by mNHEJ.
By further examination of the length of deletions induced by Cas9/gR26–3, we found 1–2bp deletion incurs less in H2AX+/+ cells (26.9%) and more in H2AX–/– cells (32.2%) (χ2 test: P< 0.0001), while deletion exceeding 2 bp was detected in 73.1% of H2AX+/+ cells and only 67.8% of H2AX–/– cells (χ2 test: P< 0.0001; Figure 2F and inset). This suggests that H2AX deficiency also causes a shift of bias towards modestly shorter deletions in mNHEJ-mediated CRISPR/Cas9 genome editing. Notably, 6bp deletions are prominent in NHEJ products. Analysis of the junctions revealed that most of these events are MMEJ with deletion of ‘CAAAGG’ or ‘GAAAGG’ due to the presence of the ‘AAAGG’ repeat at the junctions (Figure 2F and inset).
H2AX deficiency reduces mNHEJ with a bias towards shorter deletion
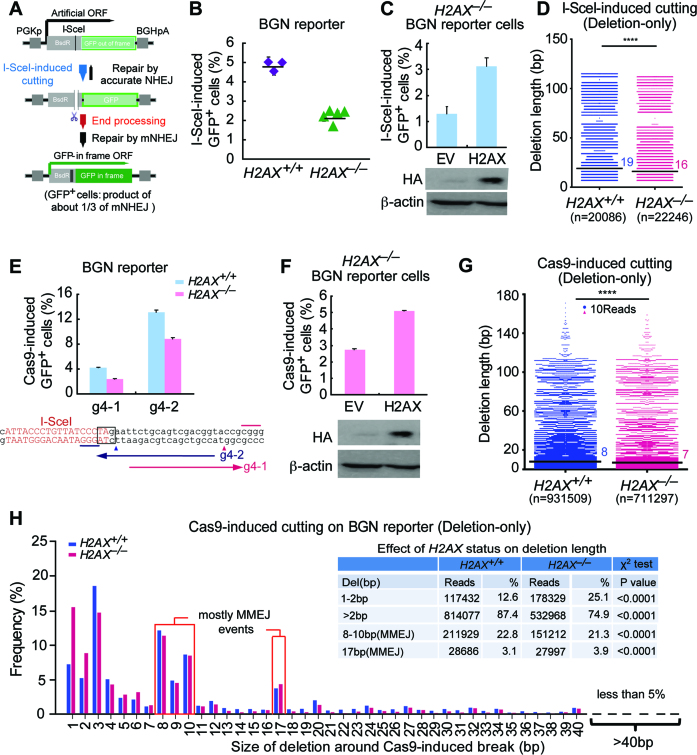
Although we were able to detect and quantitate mNHEJ using the sGEJ reporter above, it could not be done rapidly and readily. We either had to distinguish I-SceI-induced mNHEJ from accurate NHEJ by DNA sequencing or to elicit CRISPR/Cas9-induced mNHEJ, which is interfered by SSA or MMEJ between tandem I-SceI sites of the sGEJ reporter. Moreover, the CRISPR/Cas9-induced DSB in the sGEJ reporter is too close to the coding region of GFP and the promoter region, restricting the length of deletion in both directions in mNHEJ. Hence, we developed a new reporter for rapid quantification of mNHEJ and for epistasis analysis of H2AX’s NHEJ function. This NHEJ reporter contains the fusion gene of partial Blasticidin S deaminase (Bsd) gene and out-of-frame GFP gene (named ‘BGN’ reporter), with an I-SceI site embedded in the linker region between Bsd and GFP (Figure 3A). Upon I-SceI expression, a site-specific DSB is induced at the I-SceI site. Repair of this DSB by mNHEJ induces additional frame-shifts, which in theory have one third of probability (‘3n’, ‘3n+1’ or ‘3n+2’-bp frame-shift) to correct the reading frame of the downstream GFP gene, generating GFP+ cells, whereas products of accurate NHEJ cannot be differentiated from the uncut reporter. Thus, the frequency of I-SceI-induced GFP+ cells represents the relative level of mNHEJ.
Figure 3.
H2AX deficiency reduces mNHEJ with a bias towards shorter deletions. (A) Schematic of the BGN reporter for quantitatively measuring mNHEJ. An I-SceI site was placed in a linker region between BsdR and GFP of the BsdR-GFP fusion gene for induction of a site-specific DSB. With no I-SceI expression, the BsdR-GFP fusion gene generates no GFP+ cells because GFP is out-of-frame. With I-SceI expression, a site-specific DSB could be induced at the I-SceI site, and subsequent repair of the DSB by mNHEJ has 1/3 probability (i.e. 3n+1 frame-shift) to correct out-of-frame GFP, generating GFP+ cells. Frequency of I-SceI-induced GFP+ cells reflects relative efficiency of mNHEJ. (B) Percentage of I-SceI-induced GFP+ cells from H2AX+/+ and H2AX–/– BGN reporter cell clones. Values are the mean ± S.D. of three independent experiments, each performed in triplicates. One-way Anova between ‘H2AX+/+’ and ‘H2AX–/–’: P < 0.0001. (C) Percentage of I-SceI-induced GFP+ cells from H2AX–/– BGN reporter cells transiently transfected with mouse H2AX expression plasmids. Values are the mean ± S.D. of three independent experiments, each performed in triplicates. Student's paired t-test between ‘EV’ and ‘H2AX’: P = 0.036. Exogenous HA-tagged H2AX was detected by anti-HA antibody as indicated with β-actin as the loading control. (D) Deletion distributions of ‘Del’ events in I-SceI-induced mNHEJ from H2AX+/+ and H2AX–/– BGN reporter mouse ES cells. The median deletion length is indicated. ****P <0.0001 (two-tailed Mann–Whitney test). (E) Percentage of Cas9-induced GFP+ cells from H2AX+/+ and H2AX–/– BGN reporter cells. Two gRNAs targeting the region surrounding the I-SceI site are shown under the bar chart. Bars represent the mean ± S.D. of three independent experiments, each performed in triplicates. Student's paired t-test between ‘H2AX+/+’ and ‘H2AX–/–’: P = 0.036 for g4–1; P = 0.046 for g4–2. (F) Percentage of Cas9/g4–1-induced GFP+ cells from H2AX–/– BGN reporter cells transiently transfected with mouse H2AX expression plasmids. Bars represent the mean ± S.D. of three independent experiments, each performed in triplicates. Student's paired t-test between ‘EV’ and ‘H2AX’: P = 0.022. Exogenous HA-tagged H2AX was detected by anti-HA antibody as indicated with β-actin as the loading control. (G) Deletion distributions of ‘Del’ events in Cas9/g4–2-induced mNHEJ from H2AX+/+ and H2AX–/– BGN reporter mouse ES cells. The median deletion length is indicated. Each blue or red dot represents 10 reads. ****P <0.0001 (two-tailed Mann–Whitney test). (H) Frequency of deletions with different deletion length in ‘Del’ events of Cas9/g4–2-induced mNHEJ at the BGN reporter from H2AX+/+ and H2AX–/– cells. The mNHEJ events were grouped into 1–2 bp, >2 bp, 8–10 bp (MMEJ) and 17 bp (MMEJ), and their combined reads and frequencies summarized in inset with P values from a χ2 test indicated. The sequence surrounding the I-SceI site (red) in the BGN reporter is shown with the ‘TAG’ stop codon highlighted in black box. The distance between this ‘TAG’ and the GFP start codon ‘ATG’ is 45bp as indicated.
To validate this BGN reporter system, we established XRCC4flox/flox mouse ES cells containing a single copy of the BGN reporter integrated into the genome (Supplementary Figure S2A–C). In these cells, without I-SceI-induced DSB, the reporter generates few GFP+ cells with a frequency of 0.01–0.1% indicating a low background, whereas I-SceI expression induces a high frequency of GFP+ cells, representing the relative level of mNHEJ (Supplementary Figure S2D). We derived five isogenic XRCC4+/+, 4 XRCC4+/– and five XRCC4–/– reporter clones after infection of adenovirus encoding the Cre recombinase (Ad-Cre), and expression of I-SceI produced GFP+ cells at ∼2% in XRCC4+/+ and XRCC4+/– reporter clones and 4% in XRCC4–/– reporter clones (Supplementary Figure S2E). Given that XRCC4 promotes accurate NHEJ (33), it is no surprise that loss of XRCC4 leads to a 2-fold increase of the mNHEJ efficiency. Furthermore, transient expression of wild-type mouse XRCC4 reduces mNHEJ in XRCC4–/– cells using the BGN reporter (P = 0.004; Supplementary Figure S2F). These results demonstrate that the BGN reporter is suitable for analysis of mNHEJ in mammalian cells.
To determine the role of H2AX in mNHEJ using the BGN reporter, we generated an H2AXflox/flox mouse ES clone carrying a single-copy BGN reporter targeted at the ROSA26 locus in the genome (Supplementary Figure S2C), and derived three H2AX+/+ and five H2AX–/– isogenic BGN reporter clones by Ad-Cre infection (Figure 3B). Deletion of H2AX reduces the efficiency of I-SceI-induced mNHEJ by ∼50% (P = 0.017; Figure 3B), and stable expression of wtH2AX restores I-SceI-induced mNHEJ in H2AX–/– cells to a level similar to that in H2AX+/+ cells (P = 0.036; Figure 3C), suggesting that H2AX is required for efficient mNHEJ. However, despite being reduced significantly, the activity of mNHEJ remains robust in H2AX–/– cells, indicating the existence of H2AX-independent mNHEJ.
To determine whether H2AX deficiency is also biased towards shorter deletions during mNHEJ using the BGN reporter, we sorted I-SceI-induced GFP+ cells (i.e. I-SceI-induced mNHEJ products) by FACS and analyzed repair junctions of mNHEJ in which ‘Del’ is ∼65–72% of total mNHEJ events (Supplementary Table S3). Compared to H2AX+/+ cells, H2AX deficiency increases short-deletion events (7–10 bp deletion; 42.4% versus 34.1% in H2AX+/+ cells) and reduces long-deletion events (over 10 bp deletion; 57.6% versus 65.9% in H2AX+/+ cells) (χ2 test: P< 0.0001; Supplementary Figure S3). The median deletion length is shorter in H2AX–/– cells (16 bp versus 19bp in H2AX+/+ cells) (Mann–Whitney test: P< 0.0001; Figure 3D). This indicates a bias towards shorter deletions in H2AX-deficient cells, consistent with those using the sGEJ reporter and a nature site of the ROSA26 locus of the murine genome. Due to a stop codon ‘TAG’ localized downstream of the I-SceI site and within the same reading frame of GFP, at least 7bp deletion downstream is needed to destroy the ‘TAG’ stop codon and produce GFP+ cells (Supplementary Figure S3). Indeed, 7bp is the minimum length of deletion detected in repair junctions (Figure 3D, Supplementary Figure S3).
Because the distance from the I-SceI site to the downstream start codon ‘ATG’ of GFP is 51 bp, deletion longer than 51 bp downstream during I-SceI-induced mNHEJ would start to destroy the GFP gene, generating no GFP+ cells. As a result, deletions with such length downstream of the I-SceI site are not detected in directional analysis of repair junctions of mNHEJ (Supplementary Figure S4). In contrast, deletions upstream of the I-SceI site can reach to 411 bp in theory with the GFP gene intact. However, we did not find any deletions longer than 51 bp upstream, indicating such events are rare; any effect of H2AX status on generation of these events is not apparent (Supplementary Figure S4).
To exclude potential interference by the ‘TAG’ stop codon downstream of the I-SceI site in I-SceI-induced mNHEJ, and also to test whether H2AX is required for efficient CRISPR/Cas9-induced mNHEJ in the BGN reporter, we generated two gRNAs (g4–1 and g4–2) directing a double-strand DNA breakage respectively within and downstream of the ‘TAG’ stop codon in the BGN reporter (Figure 3E). CRISPR/Cas9-induced mNHEJ in H2AX–/– cells is about 50% of that in H2AX+/+ cells irrespective of the position of a DSB induced in the reporter (Figure 3E). Stable expression of wtH2AX increases Cas9/g4–1-induced mNHEJ in H2AX–/– cells by 2-fold (Figure 3F). We also examined repair junctions of Cas9/g4–2-induced mNHEJ products (unsorted) by deep sequencing to evaluate the effect of H2AX on deletion length of CRISPR/Cas9-induced mNHEJ in the BGN reporter. In these mNHEJ products, 69.2 ± 1.9% is ‘Del’, 21.8 ± 2.0% ‘Ins’ and 9.0 ± 3.7% ‘InDel’; H2AX deficiency decreases the percentage of ‘Del’ events to 51.9 ±3.4% and InDel to 8.0 ± 4.7%, but elevates ‘Ins’ events to 40.1 ± 2.9% (Supplementary Table S4). Consistent with I-SceI-induced mNHEJ and mNHEJ-mediated CRISPR/Cas9 genome editing, H2AX deficiency tends to drive deletions in ‘Del’ NHEJ events to shorter length, with the median length of deletions from 8 bp in H2AX+/+ cells to 7 bp in H2AX–/– cells (Mann–Whitney test: P< 0.0001; Figure 3G). Compared to H2AX+/+ cells, H2AX–/– cells have a larger proportion of mNHEJ events with 1–2 bp short deletions (25.1% versus 12.6% in H2AX+/+ cells; χ2 test: P< 0.0001), and smaller with long deletions over 2 bp (74.9% versus 87.4%; χ2 test: P< 0.0001) (Figure 3H and inset). Taken together, these results suggest that H2AX assists a subtype of NHEJ that requires end processing with a bias towards modestly longer deletion in mammalian cells. It is notable that 8–10 bp deletions and 17 bp deletions frequently occur and majority of these deletions are generated by MMEJ with 1–3nt microhomology (Figure 3H and inset).
MDC1 transduces H2AX-dependent NHEJ
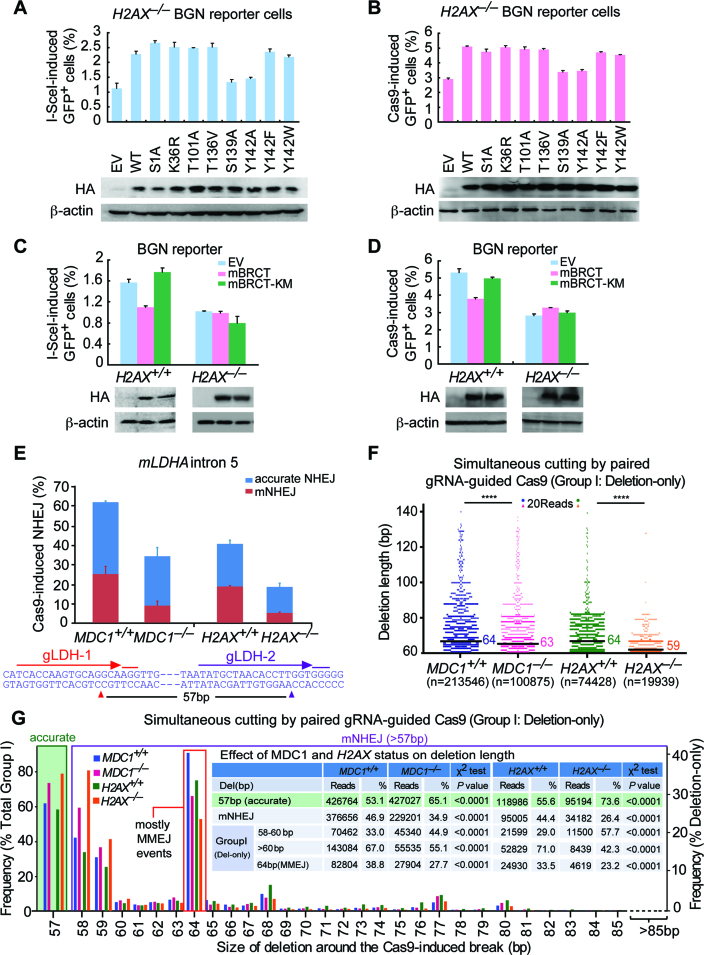
In addition to DNA damage-induced S139 phosphorylation, H2AX has many damage-dependent or independent post-translational modifications (34), which could regulate H2AX-mediated NHEJ. To determine which modification(s) has effect on H2AX’s NHEJ function, we tested various H2AX mutants for their ability to rescue mNHEJ in H2AX–/– BGN reporter cells. When transiently expressed, most of H2AX mutants, like wtH2AX, elevate I-SceI- and Cas9-induced mNHEJ in H2AX–/– cells efficiently, but neither S139A nor Y142A does (Figure 4A and B). Substitution of Y142 for another aromatic residue W or F does not abrogate the ability of H2AX to restore the efficiency of mNHEJ in H2AX–/– cells (Figure 4A and B). Compared to empty vector control (EV), stable expression of wtH2AX, not S139A or Y142A, increases the efficiency of mNHEJ (Supplementary Figure S5A and B).
Figure 4.
MDC1 transduces H2AX-dependent NHEJ. (A and B) Percentage of I-SceI (A) or Cas9 (B)-induced GFP+ cells from H2AX–/– BGN reporter cells transiently transfected with mouse H2AX expression plasmids. Bars represent the mean ± S.D. of three independent experiments, each performed in triplicates. In I-SceI-induced NHEJ assays (A), Student's paired t-test: P = 0.037 between ‘EV’ and ‘WT’; P = 0.015 between ‘WT’ and ‘S139A’; and P = 0.01 between ‘WT’ and ‘Y142A’. In Cas9-induced NHEJ assays (B), Student's paired t-test: P = 0.015 between ‘EV’ and ‘WT’; P = 0.044 between ‘WT’ and ‘S139A’; and P = 0.002 between ‘WT’ and ‘Y142A’. Exogenous HA-tagged H2AX and its variants were detected by anti-HA antibody with β-actin as the loading control. (C and D) Percentage of I-SceI (C) or Cas9 (D)-induced GFP+ cells from H2AX+/+ and H2AX–/– BGN reporter cells transiently transfected with EV, HA-tagged mBRCT or mBRCT-KM expression plasmids. Bars represent the mean ± S.D. of three independent experiments, each performed in triplicates. Student's paired t-test between ‘EV’ and ‘mBRCT’ in H2AX+/+ BGN reporter cells: P = 0.004 in I-SceI-induced NHEJ assays (C) and P = 0.042 in Cas9-induced NHEJ assays (D); between ‘EV’ and ‘mBRCT’ in H2AX–/– BGN reporter cells and between ‘EV’ and ‘mBRCT KM’ in both H2AX+/+ and H2AX–/– BGN reporter cells: NS. Exogenous HA-tagged mBRCT proteins detected by anti-HA antibody is indicated below with β-actin as the loading control. (E) Paired gRNA-guided Cas9-induced NHEJ at the LDHA locus (LDHA intron 5) in mouse ES cells lacking MDC1, H2AX or not. The NHEJ efficiency was calculated as ratios of total NHEJ reads to total reads from Illumina sequencing and normalized by transfection efficiency. Distributions of accurate NHEJ and mNHEJ indicated were calculated by proportionating their respective reads to total NHEJ reads. Pop-out of specific 57 bp DNA region caused by accurate NHEJ is indicated. The LDHA target sequence for paired gRNAs is shown under the bar chart. (F) Deletion distributions of ‘Del’ events in NHEJ of paired gRNAs-guided simultaneous cuts (Group I) from mouse ES cells lacking MDC1, H2AX or not. The median deletion length is indicated, and deletion distributions demonstrate a shift towards shorter deletions in cells lacking MDC1 or H2AX. Each blue, red, green or orange dot represents 20 reads. ****P < 0.0001 (two-tailed Mann–Whitney test). (G) Frequency of accurate NHEJ (57bp pop-out) in NHEJ of paired gRNAs-guided simultaneous cuts (Group I; left), and frequency of deletions with different deletion length in ‘Del’ events of Group I NHEJ (right) from mouse ES cells lacking MDC1, H2AX or not. ‘Del’ NHEJ events were grouped into 58–60 bp, >60 bp and 64 bp (MMEJ), and their respective reads and frequencies were summarized in inset and compared by a χ2 test with P values indicated.
As both S139 phosphorylation and aromatic group of Y142 are important for the interaction of γH2AX with MDC1, which orchestrates the assembly of repair factors on the damaged chromatin (46,47), we speculated that MDC1 might mediate the function of H2AX in NHEJ. To test this hypothesis, we used tandem BRCT domain of mouse MDC1 (mBRCT) as a dominant negative to block the interaction of γH2AX with endogenous MDC1 as done previously (38), with BRCT K1554M (mBRCT-KM) as a negative control that does not bind to γH2AX, and analyzed their effect on H2AX’s NHEJ function. Transient and stable expression of mBRCT reduces the efficiency of I-SceI- and Cas9-induced mNHEJ in H2AX+/+ cells, but mBRCT-KM does not (Figure 4C and D, Supplementary Figure S5C and D). However, neither mBRCT nor mBRCT-KM expressed has effect on mNHEJ in H2AX–/– cells (Figure 4C and D, Supplementary Figure S5C and D), implying the inhibitory effect of mBRCT on mNHEJ is dependent upon H2AX and, by extension, the interaction of γH2AX with MDC1.
To directly determine whether MDC1 plays a role similar to that of H2AX in NHEJ, we generated isogenic murine MDC1+/+ ES clones and MDC1–/– ES clones by CRISPR/Cas9 (Supplementary Figure S6). By using paired gRNA-guided Cas9 to simultaneously induce two DSBs that are 57bp apart on the intron 5 of the LDHA locus in both MDC1+/+ and MDC1–/– ES clones as well as in both H2AX+/+ and H2AX–/– ES cells, in combination with deep sequencing of repair junctions amplified by PCR, we were able to analyze joining of one end of one DSB with one end of the other DSB simultaneously induced (Figure 4E). DNA breakage from simultaneous cutting is termed ‘Group I’ as defined before and accurate NHEJ is detectable only for this group. Due to its major contribution to total repair of DSBs (Supplementary Tables S5 and S6), our analysis is mainly focused on NHEJ-mediated repair of ‘Group I’ DSBs. Loss of either MDC1 or H2AX slightly reduces the overall efficiency of ‘Group I’ end joining (χ2 test: P< 0.0001; Figure 4E, Supplementary Tables S5 and S6). This is different from the previous observation with the sGEJ reporter that H2AX deficiency does not reduce the efficiency of I-SceI-induced NHEJ (Figure 1B). This discrepancy is likely due to more mNHEJ-mediated repair of Cas9-induced DSBs (e.g. 70.2% in H2AX+/+ cells; Supplementary Table S6) than that of I-SceI-induced DSBs (38.9% in H2AX+/+ cells; Supplementary Table S1).
Also, consistent with the previous results, while accurate end joining slightly increases, mNHEJ is reduced nearly by half in MDC1–/– or H2AX–/– cells (χ2 test: P< 0.0001; Figure 4E, Supplementary Tables S5 and S6). In other words, MDC1 deficiency, like H2AX deficiency, is biased towards accurate NHEJ (from 62.0%±3.6% in MDC1+/+ cells to 74.0 ± 1.5%; χ2 test: P< 0.0001) and away from mNHEJ (from 38.0 ± 3.6% MDC1+/+ cells to 26.0 ± 1.5%; χ2 test: P< 0.0001) (Figure 4E, Supplementary Tables S5 and S6). Furthermore, the median length of mNHEJ deletions is 64 bp in MDC1+/+ cells and 63 bp in MDC1–/– cells (Mann–Whitney test: P< 0.0001), and 64 bp in H2AX+/+ cells and 59 bp in H2AX–/– cells (Mann–Whitney test: P< 0.0001; Figure 4F), suggesting that MDC1 deficiency, similar to H2AX deficiency, shift the bias of deletions towards shorter length. In addition, compared with MDC1+/+ cells and H2AX+/+ cells, the deficiencies of MDC1 and H2AX cause more frequent mNHEJ events respectively with deletion of 58–60 bp (i.e. 1–3 bp + 57 bp pop-out) (44.9% versus 33.0% between MDC1–/– and MDC1+/+ cells, and 57.7% versus 29.0% between H2AX–/– and H2AX+/+ cells; χ2 test: P< 0.0001), and less with deletion of over 60 bp (i.e. 3 bp + 57 bp pop-out) (55.1% versus 67.0%, and 42.3% versus 71.0%; χ2 test: P< 0.0001) (Figure 4G and inset). These data demonstrate that the effect of MDC1 deficiency on NHEJ perfectly mimics that of H2AX deficiency and suggest that MDC1 mediates H2AX-dependent NHEJ. Of note, the frequencies of 64bp deletions are surprisingly high and we found that most of these deletions are products of MMEJ with the microhomology ‘GTG’ at the junctions (Figure 4G and inset).
The NHEJ core factors XRCC4 and DNA-PKcs mediate H2AX-dependent NHEJ
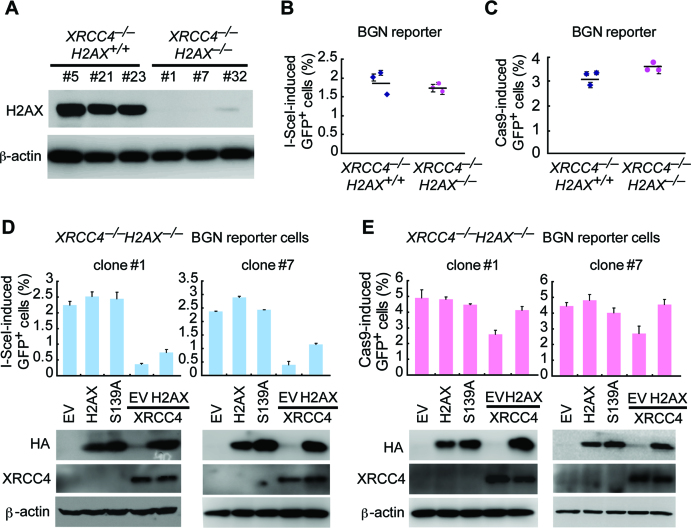
A-NHEJ acts independently of either of core NHEJ factors such as XRCC4 and DNA-PKcs and is characterized by excessive deletions/insertions and frequent use of microhomology (3,4). Although the use of microhomology in mNHEJ is not altered by H2AX deficiency (data not shown), the observation that H2AX is required for efficient mNHEJ associated with deletions and insertions with a bias towards modestly longer deletions prompted us to ask whether H2AX-dependent NHEJ is part of A-NHEJ. To address this question, we first analyzed the role of XRCC4 in this NHEJ pathway. We deleted H2AX in XRCC4–/– BGN reporter cells by CRISPR/Cas9 gene editing and generated 3 isogenic XRCC4–/–H2AX–/– clones and 3 XRCC4–/–H2AX+/+ clones (Figure 5A). Surprisingly, little change is observed in I-SceI- and Cas9-induced mNHEJ between XRCC4–/–H2AX–/– clones and XRCC4–/–H2AX+/+ clones, suggesting that H2AX-mediated NHEJ is inactive without XRCC4 (Figure 5B and C). Moreover, with EV as a negative control, neither wtH2AX nor S139A mutant ectopically expressed in XRCC4–/–H2AX–/– clones affects I-SceI- and Cas9-induced mNHEJ (Figure 5D and E). Complementation of XRCC4 reduces both I-SceI- and Cas9-induced mNHEJ in XRCC4–/–H2AX–/– clones, suggesting that XRCC4 suppresses H2AX-independent mNHEJ. Nevertheless, in XRCC4-complemented H2AX–/– cells, additional complementation of H2AX elevates I-SceI- and Cas9-induced mNHEJ (Figure 5D and E). This further confirms that H2AX-mediated NHEJ requires XRCC4.
Figure 5.
XRCC4 mediates H2AX-dependent NHEJ. (A) Deletion of H2AX in XRCC4–/– BGN reporter cells by CRISPR/Cas9. XRCC4–/– BGN reporter mouse ES cells were transfected twice with Cas9 expression plasmids and H2AX gRNA expression plasmids and then plated on MEF. In about 2 weeks, individual clones were picked, and deletion of H2AX was identified by Western blot (upper). (B and C) Percentage of I-SceI (B) and Cas9 (C)-induced GFP+ cells from XRCC4–/–H2AX+/+ and XRCC4–/–H2AX–/– BGN reporter cells. Bars represent the mean ± S.D. of three independent experiments, each performed in triplicates. One-way Anova between ‘XRCC4–/–H2AX+/+’ and ‘XRCC4–/–H2AX–/–’: NS. (D and E) Percentage of I-SceI (D) and Cas9 (E)-induced GFP+ cells from two XRCC4–/–H2AX–/– BGN reporter cell clones transiently transfected with mouse H2AX and/or XRCC4 expression plasmids. Bras represent the mean ± S.D. of three independent experiments, each in triplicates. In I-SceI-induced NHEJ assays (D), paired t-test in clone#1 (left): NS between ‘EV’ and ‘H2AX’; P = 0.005 between ‘EV’ and ‘EV+XRCC4’; and P = 0.024 between ‘EV+XRCC4’ and ‘H2AX+XRCC4’. Paired t-test in clone#7 (right): NS between ‘EV’ and ‘H2AX’; P = 0.001 between ‘EV’ and ‘EV+XRCC4’; and P = 0.007 ‘EV+XRCC4’ and ‘H2AX+XRCC4’. In Cas9-induced test (E), paired t-test in clone#1 (left): NS between ‘EV’ and ‘H2AX’; P = 0.047 between ‘EV’ and ‘EV+XRCC4’; and P = 0.002 between ‘EV+XRCC4’ and ‘H2AX+XRCC4’. Paired t-test in clone#7 (right): NS between ‘EV’ and ‘H2AX’; P = 0.034 between ‘EV’ and ‘EV+XRCC4’; and P = 0.035 between ‘EV+XRCC4’ and ‘H2AX+XRCC4’. Expression of exogenous H2AX (HA-tagged) and XRCC4 was detected by western blot as indicated.
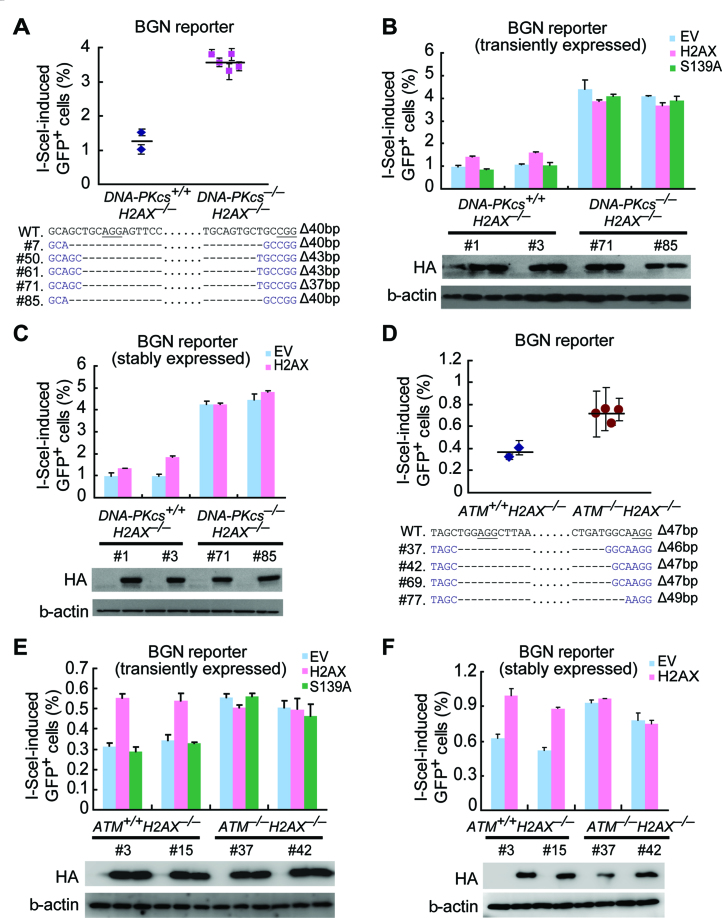
We then asked whether H2AX-mediated mNHEJ requires another core NHEJ factor DNA-PKcs. By deleting DNA-PKcs in H2AX–/– BGN reporter cells through CRISPR/Cas9 gene editing, we established 2 isogenic DNA-PKcs+/+H2AX–/– clones and 5 DNA-PKcs–/–H2AX–/– clones (Figure 6A). Like XRCC4, DNA-PKcs is suppressive to H2AX-independent mNHEJ, as deletion of DNA-PKcs increases I-SceI-induced mNHEJ in H2AX–/– BGN reporter cells (one-way Anova: P <0.0001; Figure 6A). As expected, transient expression of H2AX, not the S139A mutant, as compared with EV, increases I-SceI- and Cas9-induced mNHEJ in DNA-PKcs+/+H2AX–/– clones by nearly 2-fold (Figure 6B). In contrast, transient expression of wtH2AX does not alter the level of mNHEJ in DNA-PKcs–/–H2AX–/– clones (Figure 6B), indicating H2AX-mediated NHEJ is inactive in the absence of DNA-PKcs. Consistently, stable complementation of H2AX stimulates I-SceI-induced mNHEJ in DNA-PKcs+/+H2AX–/– clones, but has no effect in DNA-PKcs–/–H2AX–/– clones (Figure 6C), suggesting that H2AX-mediated NHEJ requires DNA-PKcs. Therefore, despite its association with deletions and insertions, H2AX-mediated NHEJ is likely part of C-NHEJ that requires XRCC4 and DNA-PKcs. Of note, PARP1 is a key regulator of A-NHEJ (48,49), and its inhibition by Olaparib reduces mNHEJ in both XRCC4–/– cells and DNA-PKcs–/– cells, not in WT cells (Supplementary Figure S7). This further confirms that XRCC4-independent or DNA-PKcs-independent mNHEJ, as part of A-NHEJ, is promoted by PARP1.
Figure 6.
DNA-PKcs and ATM are essential for H2AX-dependent NHEJ. (A) Percentage of I-SceI-induced GFP+ cells from DNA-PKcs+/+H2AX–/– and DNA-PKcs–/–H2AX–/– BGN reporter cell clones. Bars represent the mean ± S.D. of three independent experiments, each in triplicates. One-way Anova: P< 0.0001 between ‘DNA-PKcs+/+H2AX–/–’ and ‘DNA-PKcs–/–H2AX–/–’. To generate DNA-PKcs–/–H2AX–/– clones using CRISPR/Cas9, H2AX–/– BGN reporter mouse ES cells were transfected twice with Cas9 and gRNAs targeting DNA-PKcs and then plated on MEF. In ∼2 weeks, individual clones were picked, and DNA-PKcs–/–H2AX–/– clones with identical deletions in two DNA-PKcs alleles were verified by Sanger sequencing of the edited site. DNA-PKcs+/+H2AX–/– clones were also obtained from the same treatment. (B and C) Percentage of I-SceI-induced GFP+ cells from two DNA-PKcs+/+H2AX–/– BGN reporter clones (clone#1 and #3) and two DNA-PKcs–/–H2AX–/– BGN reporter clones (clone#71 and #85) transiently (B) or stably (C) transfected with EV, wtH2AX and/or S139A expression plasmids. Bars represent the mean ± S.D. of three independent experiments, each in triplicates. Student's paired t-test between ‘EV’ and ‘H2AX’ in DNA-PKcs+/+H2AX–/– clones: P = 0.044 in clone #1 and P = 0.026 in clone #3 in transient transfection experiments; P = 0.019 in clone #1 and P = 0.04 in clone #3 in stable transfection experiments; in DNA-PKcs–/–H2AX–/– clones #71 and #85: NS in either transient transfection or stable transfection experiments. Expression of exogenous HA-tagged H2AX and/or its mutant S139A was detected by Western blot as indicated. (D) Percentage of I-SceI-induced GFP+ cells from ATM+/+H2AX–/– and ATM–/–H2AX–/– BGN reporter cell clones. Bars represent the mean ± S.D. of three independent experiments, each in triplicates. One-way Anova: P = 0.014 between ‘ATM+/+H2AX–/–’ and ‘ATM–/–H2AX–/–’. ATM–/–H2AX–/– clones and ATM+/+H2AX–/– were similarly generated by CRISPR/Cas9 gene editing as in (A). ATM–/–H2AX–/– clones with identical deletions in two ATM alleles were verified by Sanger sequencing of the edited site. (E and F) Percentage of I-SceI-induced GFP+ cells from two ATM+/+H2AX–/– BGN reporter clones (clone #3 and #15) and two ATM–/–H2AX–/– BGN reporter clones (clone#37 and #42) transiently (E) or stably (F) transfected with EV, wtH2AX and/or S139A expression plasmids. Bars represent the mean ± S.D. of three independent experiments, each in triplicates. Student's paired t-test between ‘EV’ and ‘H2AX’ in ATM+/+H2AX–/– clones: P = 0.024 in clone #3 and P = 0.037 in clone #15 in transient transfection experiments; P = 0.011 in clone #1 and P = 0.001 in clone #3 in stable transfection experiments; in ATM–/–H2AX–/– clones #37 and #42: NS in either transient transfection or stable transfection experiments. Expression of exogenous HA-tagged H2AX and/or its mutant S139A was detected by Western blot as indicated.
The ATM kinase is essential for H2AX-dependent NHEJ
DNA-PKcs may mediate H2AX-dependent NHEJ as a core NHEJ factor. However, DNA-PKcs is among the kinases responsible for DSB-induced S139 phosphorylation of H2AX (9,12), which is essential for H2AX’s function in NHEJ. Therefore, DNA-PKcs may alternatively promote H2AX-dependent NHEJ by phosphorylating H2AX. If this is the case, as the primary kinase for H2AX phosphorylation, ATM may be dispensable for H2AX-dependent NHEJ. To test this hypothesis, we thus deleted ATM in H2AX–/– BGN reporter cells by CRISPR/Cas9 gene editing and generated two isogenic ATM+/+H2AX–/– clones and four ATM–/–H2AX–/– clones (Figure 6D). Deletion of ATM stimulates mNHEJ in H2AX–/– cells (one-way Anova: P = 0.014; Figure 6D). This demonstrates that like DNA-PKcs, ATM suppresses H2AX-independent mNHEJ. Compared with EV and S139A, transient expression of wtH2AX, as expected, increases I-SceI-induced mNHEJ in ATM+/+H2AX–/– clones by nearly 2-fold (Figure 6E), indicating reinstatement of H2AX-mediated mNHEJ. However, transient expression of wtH2AX has little effect on the efficiency of mNHEJ in ATM–/–H2AX–/– clones (Figure 6E), suggesting that ATM is required for H2AX-mediated NHEJ. Similarly, stable expression of H2AX, compared with S139A, stimulates mNHEJ in ATM+/+H2AX–/– clones, but not in ATM–/–H2AX–/– clones (Figure 6F). This shows that H2AX-dependent NHEJ requires ATM.
In line with the role of ATM in suppressing H2AX-independent mNHEJ, the ATM inhibitor KU60019 stimulates I-SceI-induced mNHEJ by similar extent in both DNA-PKcs–/–H2AX–/–H2AX cells and DNA-PKcs–/–H2AX–/–EV cells, in both of which H2AX-dependent NHEJ is inactivated (Supplementary Figure S8A). Similarly, the DNA-PKcs inhibitor NU7441 also increases I-SceI-induced mNHEJ to the same level in both ATM–/–H2AX–/–H2AX cells and ATM–/–H2AX–/–EV cells, in both of which H2AX-dependent NHEJ is inactivated, substantiating the previous observation that DNA-PKcs suppresses H2AX-independent mNHEJ (Supplementary Figure S8B). Taken together, the present data indicate that both ATM and DNA-PKcs have two different roles in mNHEJ: one promoting or mediating H2AX-dependent NHEJ and the other suppressing H2AX-independent mNHEJ. These opposing roles could be tightly controlled and are yet to be further elucidated. Of note, NU7441 does not have any effect on mNHEJ in DNA-PKcs–/–H2AX–/–H2AX cells and DNA-PKcs–/–H2AX–/–EV cells, indicating this inhibitor is quite specific to DNA-PKcs (Supplementary Figure S8A). On the other hand, KU60019 appears to have some off-target effect on mNHEJ, as it causes reduction in mNHEJ in both ATM–/–H2AX–/–H2AX cells and ATM–/–H2AX–/–EV cells (Supplementary Figure S8B).
The chromatin remodeler Tip60–TRRAP–P400 is required for H2AX-dependent NHEJ
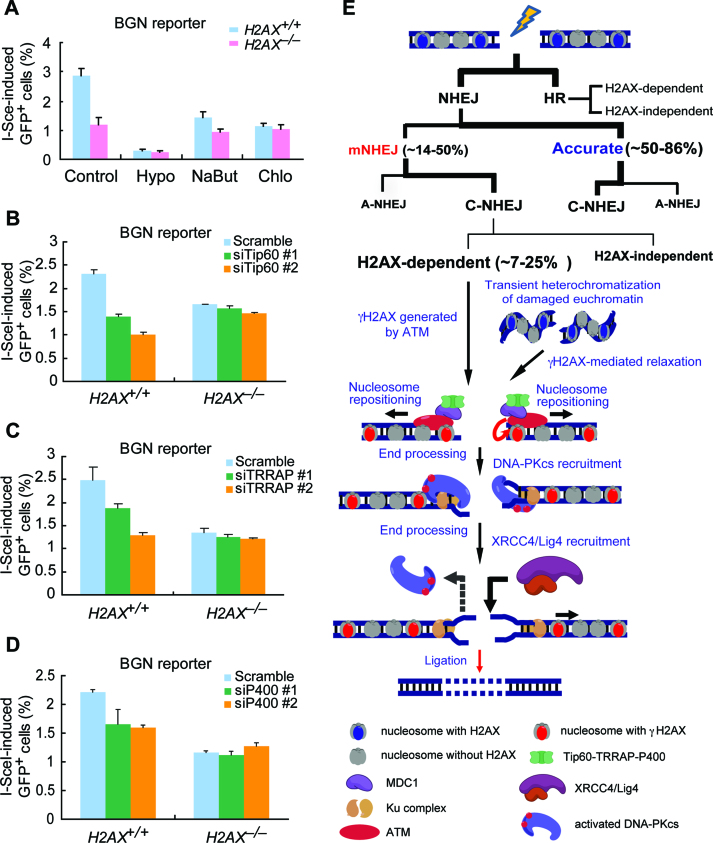
The length of deletion during NHEJ is determined mostly by the extent to which two DNA ends of a DSB are processed before ligation. As shown thus far, H2AX deficiency reduces mNHEJ and also has a bias towards shorter deletions, suggesting that end processing in NHEJ is more restricted in H2AX–/– cells than in H2AX+/+ cells. Given that the chromatin architecture near a DSB may present a barrier to both end processing and access to the NHEJ machinery, it is possible that formation of the γH2AX chromatin domain is at least in part to increase the mobility of nucleosomes adjacent to DSBs, preparing nucleosome-free DNA ends for NHEJ activities (31). Initiation of such mobilization of nucleosomes may be disabled by the absence of H2AX, thus restricting end processing for efficient NHEJ. Therefore, we predicted that by forcing chromatin relaxation independently of H2AX surrounding I-SceI-induced DSBs, H2AX would not be needed in I-SceI-induced mNHEJ. Indeed, when mouse H2AX+/+ and H2AX–/– BGN reporter cells were treated with hypotonic medium, sodium butyrate (2mM) and chloroquine (5μg/ml), all of which causes chromatin decondensation (50), the difference in the efficiency of mNHEJ that is observed with control treatment between H2AX+/+ and H2AX–/– cells is reduced or even abolished (Figure 7A). It is also noted that overall mNHEJ is reduced in both H2AX+/+ and H2AX–/– cells by these treatments, possibly through global influence.
Figure 7.
The chromatin remodeler Tip60–TRRAP–P400 is required for H2AX-dependent NHEJ. (A) Effect of hypotonic condition (Hypo), sodium butyrate (NaBut, 2 mM) and chloroquine (Chlo, 5 μg/ml) on I-SceI-induced mNHEJ in H2AX+/+ and H2AX–/– BGN reporter cells. Bars here and below represent the mean ± S.D. of three independent experiments, each in triplicates. Student's paired t-test between H2AX+/+ and H2AX–/– BGN reporter cells: P = 0.024 in Control; P = 0.122 in Hypo; P = 0.124 in NaBut; P = 0.076 in Chlo. (B–D) Percentage of I-SceI-induced GFP+ cells from BGN reporter cells transfected with RNAi against Tip60 (B), TRRAP (C) and P400 (D). Student's paired t-test in H2AX+/+ BGN reporter cells: P = 0.032 between ‘siTip60 #1’ and ‘Scramble’ and P = 0.039 between ‘siTip60 #2’ and ‘Scramble’ in (B); P = 0.008 between ‘siTRRAP #1’ and ‘Scramble’ and P = 0.003 between ‘siTRRAP #2’ and ‘Scramble’ in (C); P = 0.032 between ‘siP400 #1’ and ‘Scramble’ and P = 0.039 between ‘siP400 #2’ and ‘Scramble’ in (D). Student's paired t-test in H2AX–/– BGN reporter cells: NS between ‘Scramble’ and any other siRNA. (E) Model for the function of H2AX in NHEJ. H2AX-dependent NHEJ is responsible for about a half of classical but mutagenic NHEJ, contributing to ∼7–25% of total NHEJ. Upon DSBs, ATM can phosphorylate H2AX and help initiate the formation of the γH2AX/MDC1 chromatin domain. This domain may recruit the remodeler Tip60–TRRAP–P400. Alternatively, if DSBs induce transient heterochromatization of the damaged chromatin, the γH2AX/MDC1 chromatin domain may help relax the heterochromatized region via Tip60–TRRAP–P400 recruited. Tip60–TRRAP–P400 thus repositions nucleosomes locally around the breaks and allow sufficient processing of DNA ends for efficient DSB repair by C-NHEJ at the expense of losing a few extra nucleotides at NHEJ junctions. See details in the Discussion.
Previous studies have shown that γH2AX/MDC1 can recruit the remodeler complex Tip60–TRRAP–P400 to the damaged chromatin, promoting local chromatin relaxation for DSB repair (50–53). We thus asked whether Tip60–TRRAP–P400 is involved in H2AX-mediated mNHEJ. Since loss of any component of this complex is lethal to cells, we used small interference RNA (siRNA), two for each, to deplete these three components individually in H2AX+/+ and H2AX–/– BGN reporter cells (Supplementary Figure S9), and determined its effect on H2AX-mediated mNHEJ. The efficiency of I-SceI-induced mNHEJ is significantly reduced by transient depletion of Tip60, TRRAP or P400 in H2AX+/+ BGN reporter cells, but not in H2AX–/– BGN reporter cells (Figure 7B–D). This suggests that the Tip60–TRRAP–P400 complex may mediate H2AX-dependent NHEJ. Taken together, we posited that the Tip60–TRRAP–P400 complex recruited by γH2AX/MDC1 increases the mobility of nucleosomes adjacent to DSBs and, when needed, allows exposure of certain length of nucleosome-free DNA ends for sufficient end processing and access to NHEJ machinery.
DISCUSSION
H2AX deficiency causes genomic instability characterized with more frequent chromosomal and chromatid breaks as well as translocations in the murine genome (14–18). Early studies have revealed that H2AX is required for efficient HR (14,19,20), indicating that inactivation of H2AX-dependent HR is among the mechanisms for increased genomic instability in H2AX-deficient cells. During V(D)J recombination and CSR in developing lymphocytes, H2AX also appears to either stabilize broken DNA strands for proper NHEJ, suppress aberrant hairpin opening and subsequent end resection of RAG-initiated DSBs, or promote synapsis of two distantly located DSBs for efficient NHEJ (22,23,29,30). In general settings, however, the molecular functions of H2AX in NHEJ are poorly understood.
In this study, we demonstrated that H2AX deficiency causes significant reduction of mNHEJ. This reduction is accompanied with a slight increase in accurate NHEJ or naNHEJ (when DSBs are induced by I-SceI) and a bias towards shorter deletions in H2AX-deficient cells. At first glance, this seemingly suggests that H2AX-dependent NHEJ is a threat to genome integrity, contradicting the role of H2AX in suppressing genomic instability. However, it is possible that a subset of DSBs in cells can only be efficiently repaired by H2AX-dependent NHEJ. As H2AX deficiency can inactivate both H2AX-dependent NHEJ and H2AX-dependent HR (Figure 7E) (14,19), combined inactivation of these two pathways is expected to leave a number of DSBs in H2AX-deficient cells to be either unrepaired or repaired alternatively, but with delay and lower efficiency, by H2AX-independent repair mechanisms. Unrepaired DSBs and improper DSB repair may lead to an environment of genomic instability for oncogenic transformation (29,54).
H2AX is also required for efficient rejoining of CRISPR/Cas9-induced DSBs, in addition to I-SceI-induced DSBs. As a powerful tool in genome editing, CRISPR genome editing is achieved through induction of site-specific DSBs and subsequent DSB repair by several repair mechanisms including the HR and NHEJ pathways in mammalian cells (36). While current knowledge of DSB repair is important for us to understand and control CRISPR genome editing, DSB repair that mediates CRISPR genome editing may be unique in some aspects due to timing of DSB exposure from the Cas9-sgRNA-DNA ternary complex, residence duration of Cas9-sgRNA on target DNA, or distinct end configurations (e.g. blunt end in Cas9-induced DSBs and 5′-overhang in Cpf1-induced DSBs vs. 3′-overhang in I-SceI-induced DSBs) (35,55–57). Therefore, it is of interest to compare repair of CRISPR/Cas9-induced DSBs with that of I-SceI-induced DSBs. In fact, when accurate end-joining is included, H2AX deficiency has little effect on the overall efficiency of I-SceI-induced NHEJ (33,34), but reduces that of CRISPR/Cas9-induced NHEJ (Figure 4E, Supplementary Table S6). Nevertheless, both I-SceI-induced mNHEJ and CRISPR/Cas9-induced mNHEJ are less efficient and generate deletions biased towards shorter length in H2AX- or MDC1-deficient cells. In addition, XRCC4 suppresses I-SceI-induced mNHEJ more strongly than Cas9-induced mNHEJ (Figure 5D versus E). As target gene knockout is one of major applications in CRISPR genome editing and can be mediated by CRISPR/Cas9-induced mNHEJ (36,37,39,58), our results indicate that H2AX-dependent NHEJ is important for this type of CRISPR/Cas9 genome editing. The molecular details of H2AX-dependent NHEJ should thus provide insight into the mechanistic basis of NHEJ-mediated CRISPR/Cas9 genome editing and help better understand the mechanisms and safety of this technology. This could yield a potential strategy to improve the efficiency of CRISPR/Cas9-mediated gene knockout.
H2AX-dependent NHEJ is inactivated by deletion of XRCC4 or DNA-PKcs, two genes required for C-NHEJ, pointing to the notion that H2AX-dependent NHEJ is part of C-NHEJ, rather than a subtype of A-NHEJ. This is also in line with the deep sequencing results showing the use of microhomology is rather infrequent in products of H2AX-dependent NHEJ, as compared to XRCC4-independent NHEJ (data not shown). This raises a question: what proportion of NHEJ and C-NHEJ does H2AX control? Earlier work has indicated that H2AX, along with ATM and DNA-PKcs, repairs ∼10–15% of IR-induced DSBs through NHEJ (32). As shown previously (33) and here, ∼14–30% of I-SceI-induced NHEJ (see Group I in Figure 1B and C and Supplementary Table S1) and 40–50% of Cas9-induced NHEJ (see Group I in Figure 4G and Supplementary Tables S5 and S6) are mNHEJ in wild-type mouse ES cells. Reduction of mNHEJ by about a half due to H2AX deficiency indicates that H2AX-dependent NHEJ accounts for ∼7–15% of total I-SceI-induced NHEJ or 20–25% of Cas9-induced NHEJ. However, due to partial suppression of XRCC4-independent A-NHEJ in XRCC4+/+ cells and the unknown extent of this suppression, it is difficult to determine the contribution of H2AX-dependent NHEJ to C-NHEJ.
If H2AX-dependent NHEJ is responsible for repairing a subset of DSBs as part of C-NHEJ, is this subset of DSBs randomly selected or predetermined by certain configuration of these DSBs in the context of chromatin for H2AX-dependent NHEJ? Considering the difference between H2AX-dependent NHEJ and H2AX-independent NHEJ and the role of Tip60/TRRAP/P400 in H2AX-dependent NHEJ, random selection is unlikely. Instead, the context of the γH2AX chromatin surrounding a DSB may be an important determinant in this regard. Previous studies have implied that H2AX may promote synapsis between two distal DSBs for NHEJ during CSR, suppress end resection of RAG-initiated DSBs, or stabilize broken ends for efficient NHEJ in V(D)J recombination (22,23,29,30). If these were the case in general NHEJ between two ends of a DSB, H2AX deficiency would reduce both accurate NHEJ and mNHEJ and shift deletions in NHEJ products towards longer length; but this is opposite to what this study has revealed.
Hence, we propose a different model explaining how H2AX acts in C-NHEJ in general settings. In this model (Figure 7E), the position of the nucleosomes adjacent to a DSB determines whether rejoining of this DSB requires H2AX. While the majority of DSBs repaired by C-NHEJ do not involve the γH2AX chromatin domain, it is conceivable that the DNA ends of a subset of DSBs remain occupied by nucleosomes due to the timing and positioning of breakage induction. The free ends of these DSBs may not be exposed from the nucleosomes, or the length of the free ends exposed may not be sufficient for C-NHEJ (59–61). In order for DNA ends to be efficiently processed or rejoined by core NHEJ factors such as DNA-PKcs/Ku70/Ku80 and XRCC4/DNA ligase 4, nucleosomes adjacent to the DSB must be removed or repositioned away from the DNA ends. This requires the activity of the Tip60/TRRAP/P400 remodeling complex that can be recruited to the damaged chromatin in a γH2AX/MDC1-dependent manner (50–52). This is indeed supported by the present data indicating that H2AX-dependent NHEJ requires ATM, γH2AX, MDC1, DNA-PKcs, XRCC4 and Tip60/TRRAP/P400 (Figure 7E). However, direct testing of this model must await the development of an approach that can purposefully place nucleosomes at selected positions on DNA ends for studying rejoining of these ends.
Alternatively, γH2AX may help overcome the barrier of DSB-induced transient heterochromatization around the breaks in H2AX-dependent NHEJ (Figure 7E). H2AX was previously found to be responsible for repairing about 10–15% of IR-induced DSBs in mammalian cells by NHEJ (32). This repair not only requires short-range end processing (27), but is also affected by heterochromatin-associated factors (62). As double-strand DNA breakage in euchromatin can induce a transient compaction of chromatin around the breaks, adding a layer of control over DSB repair (63,64), this ‘heterochromatized region’ may require the γH2AX chromatin domain for subsequent chromatin relaxation prior to DSB repair by NHEJ (62). It is possible that γH2AX and its associated factors are needed for decondensation of the ‘heterochromatized region’ surrounding I-SceI- and CRISPR/Cas9-induced DSBs, allowing exposure and short-range processing of DNA ends for efficiently engaging the C-NHEJ core factors (e.g. DNA-PKcs/Ku70/Ku80 and XRCC4/DNA ligase 4) for repair (Figure 7E).
H2AX-dependent NHEJ exhibits loss of a few nucleotides at the repair junctions, indicating the involvement of H2AX-dependent short-range end resection. One would wonder: is this resection essential for H2AX-dependent NHEJ? Several studies have demonstrated that C-NHEJ of two distant ends that is 3.2 kb apart is a slow repair process and requires orchestrated end resection (27,65,66), whereas C-NHEJ of two close ends that is 34 bp apart could be rapid and is less dependent on resection (33,65). As γH2AX-mediated nucleosome repositioning requires space and time to prepare DNA ends of a subset of DSBs for efficient C-NHEJ, this may provide opportunities for end resection (65,67). Yet, this resection may occur passively in H2AX-dependent NHEJ of close ends, increasing the frequency and length of deletions without affecting the NHEJ efficiency. On the other hand, the longer the distance between two ends, the less efficient NHEJ and the more important end resection (27,30,65). Thus, the γH2AX chromatin domain may need to coordinate with end resection or even directly recruit resection factors such as 53BP1 and BRCA1 to promote C-NHEJ of distant ends (27,65,68). However, in the case of 53BP1 and BRCA1 that have an antagonistic relationship in end resection, loss of H2AX causes concomitant defects in recruiting either factor onto the damaged chromatin (13). As a result, it is unlikely that H2AX deficiency would alter the resection balance maintained by 53BP1 and BRCA1 associated with the γH2AX chromatin. In addition, the synapsis between two distant ends was previously proposed as a critical step in their C-NHEJ (22,30,65). It is possible that the γH2AX chromatin domain assist this synapsis in general NHEJ as in CSR (22,30); but this has yet to be tested.
ATM, DNA-PKcs and ATR can phosphorylate H2AX to generate γH2AX upon DSBs (9–12, 69, 70). Both ATM and DNA-PKcs are required for H2AX-dependent NHEJ; but it remains unclear which one acts in H2AX-dependent NHEJ by phosphorylating H2AX. As the primary kinase for IR-induced phosphorylation of H2AX (9,10,12), we speculate that ATM may promote H2AX-dependent NHEJ by phosphorylating H2AX to form the γH2AX chromatin domain while DNA-PKcs functions as a core NHEJ factor to mediate H2AX-dependent NHEJ.
The formation of the γH2AX chromatin domain helps recruit Tip60/TRRAP/P400 and other chromatin remodelers to damaged chromatin (21,31,51). It has been shown that the Tip60/TRRAP/P400 complex promotes end resection, HR and NHEJ (21,50,53,71–73). While this complex can function to decondense the expansive regions of damaged chromatin upon DSBs, promoting long-range end resection for HR (21,50,71,73), one of the effects from the chromatin decondensation might just move nucleosomes a few base pairs away locally from the ends, exposing sufficient free DNA ends for efficient rejoining (53,72). It is conceivable that in H2AX-deficient cells, the Tip60/TRRAP/P400 complex is not recruited, thus restricting mobilization of nucleosomes along the DNA ends and preventing end processing and the binding of core NHEJ factors for efficient NHEJ. Consequently, DSBs supposedly repaired by H2AX-dependent NHEJ are not repaired timely, giving opportunities for rejoining with distal DSBs (i.e. translocations) by H2AX-independent repair mechanisms, or not repaired at all, thus progressing into chromosomal and chromatid breaks (29,54). Therefore, H2AX-dependent NHEJ, as part of C-NHEJ, is an important mNHEJ pathway to actively promote repair of a subset of DSBs and prevent non-repair and cancer-predisposing translocations at the expense of limited deletions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Xie laboratory for helpful discussions. We thank Penny Jeggo (University of Sussex, UK), Markus Loebrich (Darmstadt University of Technology, Germany) and William Folk (University of Missouri at Columbia, USA) for helpful discussions and critically reading the manuscript. We apologize to authors that are not quoted in references, due to space limitations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81472755, 31671385, 81661128008 to A-Y.X.,. 31401189 to Y.-L.F.]; Natural Science Foundation of Zhejiang Province [LZ17C060001 to A.-Y.X.]; Department of Science and Technology of Zhejiang Province [2015C03047 to A.-Y.X.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pâques F., Haber J.E.. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999; 63:349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boboila C., Alt F.W., Schwer B.. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 2012; 116:1–49. [DOI] [PubMed] [Google Scholar]
- 5. Iliakis G., Murmann T., Soni A.. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015; 793:166–175. [DOI] [PubMed] [Google Scholar]
- 6. Frit P., Barboule N., Yuan Y., Gomez D., Calsou P.. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst.). 2014; 17:81–97. [DOI] [PubMed] [Google Scholar]
- 7. Smeenk G., van Attikum H.. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013; 82:55–80. [DOI] [PubMed] [Google Scholar]
- 8. Rogakou E.P., Boon C., Redon C., Bonner W.M.. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Savic V., Yin B., Maas N.L., Bredemeyer A.L., Carpenter A.C., Helmink B.A., Yang-Iott K.S., Sleckman B.P., Bassing C.H.. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 2009; 34:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J.. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001; 276:42462–42467. [DOI] [PubMed] [Google Scholar]
- 11. Ward I.M., Chen J.. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 2001; 276:47759–47762. [DOI] [PubMed] [Google Scholar]
- 12. Stiff T., O’Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P.A.. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004; 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- 13. Scully R., Xie A.. Double strand break repair functions of histone H2AX. Mutat. Res. 2013; 750:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bassing C.H., Chua K.F., Sekiguchi J., Suh H., Whitlow S.R., Fleming J.C., Monroe B.C., Ciccone D.N., Yan C., Vlasakova K. et al. . Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bassing C.H., Suh H., Ferguson D.O., Chua K.F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F.W.. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003; 114:359–370. [DOI] [PubMed] [Google Scholar]
- 16. Celeste A., Difilippantonio S., Difilippantonio M.J., Fernandez-Capetillo O., Pilch D.R., Sedelnikova O.A., Eckhaus M., Ried T., Bonner W.M., Nussenzweig A.. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003; 114:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J. et al. . Genomic instability in mice lacking histone H2AX. Science. 2002; 296:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franco S., Gostissa M., Zha S., Lombard D.B., Murphy M.M., Zarrin A.A., Yan C., Tepsuporn S., Morales J.C., Adams M.M. et al. . H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006; 21:201–214. [DOI] [PubMed] [Google Scholar]
- 19. Xie A., Puget N., Shim I., Odate S., Jarzyna I., Bassing C.H., Alt F.W., Scully R.. Control of sister chromatid recombination by histone H2AX. Mol. Cell. 2004; 16:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J.E., Koshland D.. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004; 16:991–1002. [DOI] [PubMed] [Google Scholar]
- 21. van Attikum H., Gasser S.M.. The histone code at DNA breaks: a guide to repair. Nat. Rev. Mol. Cell Biol. 2005; 6:757–765. [DOI] [PubMed] [Google Scholar]
- 22. Bassing C.H., Alt F.W.. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004; 3:149–153. [DOI] [PubMed] [Google Scholar]
- 23. Helmink B.A., Tubbs A.T., Dorsett Y., Bednarski J.J., Walker L.M., Feng Z., Sharma G.G., McKinnon P.J., Zhang J., Bassing C.H. et al. . H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011; 469:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin B., Savic V., Juntilla M.M., Bredemeyer A.L., Yang-Iott K.S., Helmink B.A., Koretzky G.A., Sleckman B.P., Bassing C.H.. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J. Exp. Med. 2009; 206:2625–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zha S., Guo C., Boboila C., Oksenych V., Cheng H.-L., Zhang Y., Wesemann D.R., Yuen G., Patel H., Goff P.H. et al. . ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011; 469:250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C.. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 2003; 197:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biehs R., Steinlage M., Barton O., Juhász S., Künzel J., Spies J., Shibata A., Jeggo P.A., Löbrich M.. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell. 2017; 65:671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petersen S., Casellas R., Reina-San-Martin B., Chen H.T., Difilippantonio M.J., Wilson P.C., Hanitsch L., Celeste A., Muramatsu M., Pilch D.R. et al. . AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001; 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alt F.W., Zhang Y., Meng F.-L., Guo C., Schwer B.. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013; 152:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Daniel J.A., Nussenzweig A.. The AID-induced DNA damage response in chromatin. Mol. Cell. 2013; 50:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Downs J.A., Lowndes N.F., Jackson S.P.. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000; 408:1001–1004. [DOI] [PubMed] [Google Scholar]
- 32. Riballo E., Kühne M., Rief N., Doherty A., Smith G.C.M., Recio M.-J., Reis C., Dahm K., Fricke A., Krempler A. et al. . A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004; 16:715–724. [DOI] [PubMed] [Google Scholar]
- 33. Xie A., Kwok A., Scully R.. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009; 16:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie A., Odate S., Chandramouly G., Scully R.. H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell Cycle. 2010; 9:3602–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Komor A.C., Badran A.H., Liu D.R.. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017; 168:20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doudna J.A., Charpentier E.. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 38. Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J. et al. . Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007; 28:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B.S.. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004; 14:611–623. [DOI] [PubMed] [Google Scholar]
- 41. Ghezraoui H., Piganeau M., Renouf B., Renaud J.-B., Sallmyr A., Ruis B., Oh S., Tomkinson A.E., Hendrickson E.A., Giovannangeli C. et al. . Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell. 2014; 55:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang J., Kobert K., Flouri T., Stamatakis A.. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014; 30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith T.F., Waterman M.S.. Identification of common molecular subsequences. J. Mol. Biol. 1981; 147:195–197. [DOI] [PubMed] [Google Scholar]
- 44. Prlić A., Yates A., Bliven S.E., Rose P.W., Jacobsen J., Troshin P.V., Chapman M., Gao J., Koh C.H., Foisy S. et al. . BioJava: an open-source framework for bioinformatics in 2012. Bioinformatics. 2012; 28:2693–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rouet P., Smih F., Jasin M.. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994; 14:8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M.A., Celeste A., Manis J.P., van Deursen J., Nussenzweig A., Paull T.T. et al. . MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006; 21:187–200. [DOI] [PubMed] [Google Scholar]
- 47. Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P.. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005; 123:1213–1226. [DOI] [PubMed] [Google Scholar]
- 48. Audebert M., Salles B., Calsou P.. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004; 279:55117–55126. [DOI] [PubMed] [Google Scholar]
- 49. Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G.. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006; 34:6170–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murr R., Loizou J.I., Yang Y.-G., Cuenin C., Li H., Wang Z.-Q., Herceg Z.. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006; 8:91–99. [DOI] [PubMed] [Google Scholar]
- 51. Xu Y., Sun Y., Jiang X., Ayrapetov M.K., Moskwa P., Yang S., Weinstock D.M., Price B.D.. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol. 2010; 191:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Downs J.A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., Bouchard N., Kron S.J., Jackson S.P., Côté J.. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell. 2004; 16:979–990. [DOI] [PubMed] [Google Scholar]
- 53. Taty-Taty G.-C., Chailleux C., Quaranta M., So A., Guirouilh-Barbat J., Lopez B.S., Bertrand P., Trouche D., Canitrot Y.. Control of alternative end joining by the chromatin remodeler p400 ATPase. Nucleic Acids Res. 2016; 44:1657–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bunting S.F., Nussenzweig A.. End-joining, translocations and cancer. Nat. Rev. Cancer. 2013; 13:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al. . In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al. . Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ochi T., Wu Q., Chirgadze D.Y., Grossmann J.G., Bolanos-Garcia V.M., Blundell T.L.. Structural insights into the role of domain flexibility in human DNA ligase IV. Structure. 2012; 20:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walker J.R., Corpina R.A., Goldberg J.. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001; 412:607–614. [DOI] [PubMed] [Google Scholar]
- 61. West R.B., Yaneva M., Lieber M.R.. Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol. Cell. Biol. 1998; 18:5908–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 63. Feng Y.-L., Xiang J.-F., Kong N., Cai X.-J., Xie A.-Y.. Buried territories: heterochromatic response to DNA double-strand breaks. Acta Biochim. Biophys. Sin. (Shanghai). 2016; 48:594–602. [DOI] [PubMed] [Google Scholar]
- 64. Gursoy-Yuzugullu O., House N., Price B.D.. Patching broken DNA: nucleosome dynamics and the repair of DNA breaks. J. Mol. Biol. 2016; 428:1846–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guirouilh-Barbat J., Gelot C., Xie A., Dardillac E., Scully R., Lopez B.S.. 53BP1 protects against CtIP-dependent capture of ectopic chromosomal sequences at the junction of distant double-strand breaks. PLoS Genet. 2016; 12:e1006230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S.. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009; 16:819–824. [DOI] [PubMed] [Google Scholar]
- 67. Bétermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process?. PLoS Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Panier S., Boulton S.J.. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014; 15:7–18. [DOI] [PubMed] [Google Scholar]
- 69. Falck J., Coates J., Jackson S.P.. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005; 434:605–611. [DOI] [PubMed] [Google Scholar]
- 70. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. . ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 71. Courilleau C., Chailleux C., Jauneau A., Grimal F., Briois S., Boutet-Robinet E., Boudsocq F., Trouche D., Canitrot Y.. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J. Cell Biol. 2012; 199:1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J.-P., Roques C., Pandita R.K., Paquet E., Herst P., Gingras A.-C., Pandita T.K. et al. . The TIP60 complex regulates bivalent chromatin recognition by 53BP1 through direct H4K20me binding and H2AK15 acetylation. Mol. Cell. 2016; 62:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Robert F., Hardy S., Nagy Z., Baldeyron C., Murr R., Déry U., Masson J.-Y., Papadopoulo D., Herceg Z., Tora L.. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell. Biol. 2006; 26:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.