Abstract
The glucocorticoid receptor (GR) is a ligand-regulated transcription factor that controls the expression of extensive gene networks, driving both up- and down-regulation. GR utilizes multiple DNA-binding-dependent and -independent mechanisms to achieve context-specific transcriptional outcomes. The DNA-binding-independent mechanism involves tethering of GR to the pro-inflammatory transcription factor activator protein-1 (AP-1) through protein-protein interactions. This mechanism has served as the predominant model of GR-mediated transrepression of inflammatory genes. However, ChIP-seq data have consistently shown GR to occupy AP-1 response elements (TREs), even in the absence of AP-1. Therefore, the current model is insufficient to explain GR action at these sites. Here, we show that GR regulates a subset of inflammatory genes in a DNA-binding-dependent manner. Using structural biology and biochemical approaches, we show that GR binds directly to TREs via sequence-specific contacts to a GR-binding sequence (GBS) half-site found embedded within the TRE motif. Furthermore, we show that GR-mediated transrepression observed at TRE sites to be DNA-binding-dependent. This represents a paradigm shift in the field, showing that GR uses multiple mechanisms to suppress inflammatory gene expression. This work further expands our understanding of this complex multifaceted transcription factor.
INTRODUCTION
Glucocorticoid receptor (GR) is a ligand-regulated transcription factor in the nuclear receptor superfamily, which activates and represses the expression of thousands of genes (1). GR contains a modular domain architecture common to the superfamily: an unstructured N-terminal domain (NTD) which contains the activation function-1 (AF-1) region that interacts with coregulator proteins; a zinc finger DNA binding domain (DBD); a hinge region; and a ligand-binding domain (LBD) which contains the ligand-sensitive AF-2 surface that also enables interaction with coregulators (2,3). In humans, GR activity is regulated by the steroid hormone cortisol, or by exogenous glucocorticoids (GCs) such as dexamethasone, or DEX (1). Most unliganded GR resides in the cytoplasm bound to chaperone proteins. Upon binding ligand, GR undergoes a conformational change exposing nuclear localization signals and subsequently translocates to the nucleus (4,5). In the nucleus, GR interacts with genomic response elements via multiple DNA-binding-dependent and -independent mechanisms (6–8).
GR binds directly to DNA at canonical GR binding sequence (GBS) composed of two pseudo-palindromic hexameric AGAACA repeats separated by a three-base pair spacer (9,10) (Figure 1Ai). GR binding to GBS sequences occurs in a head-to-head fashion through a protein-protein interaction between two GR DBD proteins (11). GR also binds to a newly characterized inverted repeat-GBS (IR-GBS/nGREs), which shows monomeric GR binding to a CTCC(N)0–2GGAGA motif (12,13) (Figure 1Aii). Unlike binding to a canonical GBS, GR binding to IR-GBS sequences occurs in a tail-to-tail fashion where GR DBD proteins do not interact (13,14). Lastly, GR binds degenerate GBSs that are found in conjunction with other transcription factor (TF) binding sites or composite elements (15,16) (Figure 1Aiii). However, in contrast to these DNA-binding-dependent mechanisms, GR also represses transcription without direct interaction with DNA (17,18).
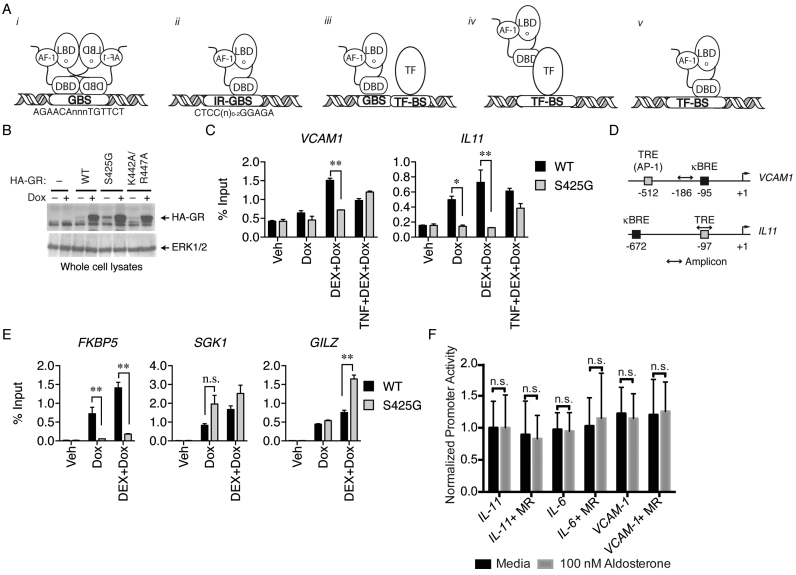
Figure 1.
WT GR, but not GR Ser425Gly, is recruited to TREs in the absence of tethering factors. (A) Cartoon representation of GR-DNA interactions. (B–E) MCF-7 cells were transfected and steroid-deprived with or without 1 μg/ml Dox as described in methods, stimulated for 1 h with 100 nM dexamethasone (DEX) alone or in combination with 10 ng/ml TNFα, and analyzed by ChIP assay using anti-HA antibody. HA-GR occupancies at the IL11 and VCAM1 promoters were determined by ChIP-PCR and shown as percent input (mean ± S.E.M.) relative to un-stimulated transfectants. (*Padj < 0.005, **Padj < 0.0001). (B) MCF7 whole cell lysates analyzed by western blot using anti-HA and anti-ERK1/2 antibodies. (C) WT GR occupies TRE target genes where GR Ser425Gly has reduced occupancy at these sites (D) Schematic of IL11 and VCAM1 promoters. Both promoters contain a TRE and NF-κB (κBRE) recognition element. (E) WT GR and GR Ser425Gly are similarly recruited to canonical GBS containing promoters from GILZ and SGK1, but reduced at FKBP5 (F) MR is unable to transrepress a constitutively active luciferase reporter containing a portion of the IL11, IL6, and VCAM1 promoters upon treatment with 100 nM aldosterone, a MR agonist.
GR-mediated transrepression involves binding of GR to other TFs, such as activator protein-1 (AP-1) and nuclear factor-kappa-light-chain-enhancer of activated B cells (NF-κB), through protein-protein interactions (18,19) (Figure 1Aiv). This mechanism, referred to as tethering, has been the accepted model for GR-mediated transrepression of inflammatory genes (20,21). GR is a potent repressor of AP-1 activity, driving its popularity as a sought-after drug target for anti-inflammatory therapies (22,23). GR-targeted therapeutics, collectively known as glucocorticoids (GCs), are the predominant treatment for chronic inflammatory diseases such as arthritis and asthma (24). However, continued administration of GCs results in numerous side effects, including diabetes, muscle wasting, and Cushing's syndrome, which diminish the effectiveness of treatment (25,26). These side effects have driven extensive efforts to develop dissociative GCs that separate the beneficial anti-inflammatory properties from side effects (23). Unfortunately, efforts to make these selective modulators have been unsuccessful, encouraging a reexamination into the mechanisms of GR-mediated transrepression of inflammation.
With advances in genome-wide sequencing, studies using chromatin immunoprecipitation-sequencing (ChIP-seq) have consistently found GR to occupy AP-1 response elements (TREs) in the absence of AP-1 as a tethering factor, suggesting the current tethering model is insufficient to explain GR occupancy of TRE motifs (27–31). Additionally, the discovery of DNA-binding-dependent repression at IR-GBSs revealed GR can interact directly with genomic DNA to repress gene transcription (12,13). Therefore, we asked whether DNA binding was required for transrepression at TRE-containing inflammatory genes. Using a combination of structural and biochemical techniques, we show GR binds to a GBS half-site located within TRE motifs in a sequence-specific fashion. We found that direct interaction of GR with the GBS DNA half-site within TREs is vital for transcriptional repression and monomeric GR is preferred at these sites, a mechanism similar to repressive GR-IR:GBS interactions (13,14). Taken together, our findings suggest that in addition to tethering, GR is able to regulate a subset of AP-1 driven inflammatory genes through a DNA-binding-dependent mechanism (Figure 1Av).
MATERIALS AND METHODS
Reporter gene assays
The U-2 OS human osteosarcoma cells were obtained from the American Type Culture Collection (ATCC) and cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM), 10% stripped fetal bovine serum (FBS), and 5% penicillin/streptomycin without phenol red. A 150 bp region of the IL11, IL6 and VCAM1 TRE-containing inflammatory gene promoters was cloned in the pGL3 plasmid in between the SV40 enhancer and promoter with a Firefly luciferase reporter gene downstream, as described previously (12,13). The sequences cloned into the pGL3 vectors are as follows: IL11, –17 to –167 upstream of the transcription start site, VCAM1, –385 to –585 upstream of the transcription start site, and IL6, –938 to –1131 upstream of the transcription start site. All numbers correspond to the human GRCh38.p7 genome, accessed through Ensembl (32). U-2 OS cells were transfected with 50 ng of the indicated reporter; 10 ng of wild-type (WT) or mutant full-length GR, or full-length MR in a pcDNA3 vector; and 5 ng of constitutively active Renilla luciferase under the control of pRL-TK promoter with FuGene HD in OptiMEM (Invitrogen) according to the manufacturer's protocol. Twenty-four hours after transfection, cells were treated with media alone, 100 nM dexamethasone (DEX), or 100 nM aldosterone for MR experiments. Twenty-four hours post-treatment, firefly and Renilla luciferase activity was detected using the Duel-Glo Luciferase Assay system (Promega) and read on a Biotek Synergy 4 plate-reader. Data were plotted as firefly luciferase activity divided by Renilla luciferase activity, normalized to control for each well and plotted using GraphPad Prism (v7). Data are representative of three independent biological replicates, and normalized values were compared using Tukey's multiple comparison test.
ChIP-PCR
Wild type and mutant GR were expressed in HEK293T cells using the Tet-On® Advanced inducible gene expression system (Takara Bio USA, Inc., Mountain View, CA, USA). Cells in six-well plates were co-transfected with the TransIT®-LT1 transfection reagent (Mirus Bio LLC, Madison, WI, USA) and 1.25 μg/well of pTet-On Advanced reverse tetracycline-controlled transactivator (rtTA), and 1.25 μg/well of pTight-FRT-Hygro2-HA-GR-WT, -Ser425Gly or -Lys442Ala/Arg447Ala expression plasmids. Control cells were transfected with empty pTight vector instead of the GR expression plasmid. After 24 h, the media were replaced with phenol red-free DMEM + 10% csFBS, with or without 1 ug/ml Doxycycline (Dox). The next day, the cells were treated with 100 nM Dex alone or in combination with 10 ng/ml TNFα for 1 h.
Quantitative ChIP assay was performed as previously described with some modification (33). Cells were fixed in 11% formaldehyde for 15 min, quenched with 0.125 M glycine for 10 min, and rinsed with cold 1× PBS. The cells were disrupted in lysis buffer (33), incubated at 4°C for 1 h and then sonicated. The lysates were then incubated with 100 μl Dynabeads protein G (Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, USA) and 10 μl pre-immune rabbit IgG for 1 h at 4°C, and centrifuged at 12 000 rpm for 15 min at 4°C. 100 μl of the pre-cleared supernatant was mixed with an HA antibody (Y-11, Santa Cruz Biotechnology Inc., Dallas, TX, USA), 25 μl Dynabeads protein G (Invitrogen, Thermo Fisher Scientific Inc.), and lysis buffer to make 200 μl IP mixtures that were rotated overnight at 4°C. The precipitates were sequentially washed in low salt, high salt, and LiCl buffers, and twice in 1× TE buffer. The crosslinks were then reversed at 65°C for 3 h. DNA fragments were isolated using QIAquick PCR purification kit (QIAGEN, Hilden, Germany), and analyzed by qPCR using TaqMan® 2× master mix and the following custom TaqMan® real-time PCR assays (Applied Biosystems, Thermo Fisher Scientific Inc.):
| IL11 | |
| Fwd | 5’ -AACTTTTCCTTCCGTGCCCT-3’ |
| Rev | 5’-TGACACATCCTGACTCACCC-3’ |
| Hyb | 5’-TGAATGGAAAAGGCAGGCAG-3’ |
| VCAM1 | |
| Fwd | 5’-CCAGGACAGAGAGAGGAGCT-3’ |
| Rev | 5’-AGTTTAACAGACACCCAGCCA-3’ |
| Hyb | 5’-TCAGCAGTGAGAGCAACTGA-3’ |
| FKBP5 | |
| Fwd | 5’-AGGGTGTTCTGTGCTCTTCAA-3’ |
| Rev | 5’-CGAGCTGCAAAACATCACTT-3’ |
| Hyb | 5’-CTGCCCTAGAGCAATTTTGTT-3’ |
| GILZ | |
| Fwd | 5’-CCGTTGCTGCTCTGCTATTG-3’ |
| Rev | 5’-TTCCCTGTCAGAGCAAGCAC-3’ |
| Hyb | 5’-GCTGTTGCCAGACATCCAAT-3’ |
| SGK1 | |
| Fwd | 5’-TGTCAGCGTCCATCCAAATG-3’ |
| Rev | 5’-ACAGCATGATTGATCCTCAGC-3’ |
| Hyb | 5’-TGGGCACAGTGAGATGACTC-3’ |
Data are representative of three independent biological replicates and were compared two-way ANOVA and multiple comparisons test for each site/gene. Sidak's multiple comparison test gave adjusted p values (*Padj < 0.005, **Padj < 0.0001).
Protein expression and purification
GR DBD (residues 417–506) was expressed and purified as described previously (13) as a 6X-Histidine tag fusion protein using the pMCSG7 vector. GR DBD was expressed in BL21 (DE3)pLysS Escherichia coli and induced with 0.3 mM IPTG and grown for 4 h at 32°C. Cells were lysed in 20 mM Tris–HCl (pH 7.4), 1 M NaCl, 25 mM imidazole, and 5% glycerol via sonication. Protein was purified using affinity chromatography (His-Trap) followed by gel filtration chromatography. Protein was then concentrated to 3–4 mg/ml in 20 mM Tris–HCl (pH 7.4), 150 mM NaCl, and 5% glycerol, flash frozen in liquid N2, and stored at –80°C. Full-length GR was a gift from Prof. David Bain. Briefly, full-length GR was expressed in baculovirus-infected SF9 cells treated with 1 μM triamcinolone acetonide (TA) for 24 h and purified as described previously (34). 15N-GR DBD was expressed in E. coli BL21(DE3) pLysS cells using a standard minimal media protocol with 15NH4Cl as the sole nitrogen source and purified as described above. The 6X-His tag was cleaved with TEV protease overnight at 4°C, passed through an Ni-NTA column, and the flow through containing purified 15N-GR DBD was collected and verified to be >99% pure by SDS-PAGE.
Nucleic acid binding assays
Synthesized FAM-labeled nucleic acid duplexes (Integrated DNA Technologies) of various TREs were annealing by heating to 90°C followed by slow cooling to room temperature. Fluorescence polarization assays were performed by adding increasing concentrations of WT or mutant GR DBD (1 nM–50 μM) and 10 nM of the FAM-labeled DNA. Reactions were performed in 20 mM Tris–HCl (pH 7.4), 150 mM NaCl and 5% glycerol. Polarization was monitored on a Biotek Synery 4 plate-reader at an excitation/emission wavelength of 485/528 nm. Three technical replicates and three biological replicates were conducted. Data plotted using GraphPad Prism (v7) are a compilation of all data collected and error bars represent standard error of measurement (SEM). Binding data was analyzed with an F-test to compare a two-site binding event to a one-site binding event with Hill slope; this test generated an F-statistic and P-value for a two-site binding model, which are represented in Figure 3 along with dissociation values (Kd) and coefficient of determination (r2).
Figure 3.
GR binds a variety of TRE sequences. (A) Fluorescence polarization binding assays monitored the ability of full-length GR to bind to the IL11 TRE and a canonical GBS from the SGK promoter. (B) GR DBD binding to TREs from the IL11, IL6, VCAM1, CSF1, IL1a and MMP13 promoters. (C) Values of curve fits of the graphs shown in (B).
Sequences of DNA constructs used for fluorescence polarization assays were: IL6: 5’-(FAM) CAAGTGCTGAGTCACTAATAA-3’, 5’-TTATTAAGACTCAGCACGTTG-3’; IL11 5’-(FAM) GGGTGAGTCAGGATGTGTCAGG-3’, 5’-CCTGACACATCCTGACTCACCC-3’; VCAM1 5’-(FAM) TTCCGGCTGACTCATCAAGCG-3’, 5’-CGCTTGATGAGTCAGCCGGAA-3’; IL1(F5’-FAM) GAAGAAGACTGACTCTCAGGCTTAAGC-3’, 5’-GCTTAAGCCTGAGAGTCAGTCTTCTTC-3’; MMP13 5’-(FAM) ATAAGTGATGACTCACCATTGCA-3’, 5’-TGCAATGGTGAGTCATCACTTAT-3’, In all cases, (FAM) indicates the position of 6-FAM (fluorescein).
NMR analysis
NMR data were collected on a Bruker 700 MHz NMR instrument equipped with a QCI cryoprobe. For NMR studies to monitor binding to DNA, the 19-nt IL11 TRE DNA duplex was reconstituted in 20 mM phosphate (pH 6.7), 100 mM NaCl, 1 mM TCEP, 10% D2O buffer to a final concentration of 437 μM, subsequently annealed by denaturing at 95°C for 3 min and then equilibrated to room temperature (20–23°C) overnight. Two-dimensional [1H,15N]-HSQC spectra were collected at 25°C for free 15N-GR DBD in the absence or presence of 0.44:1 or 2.3:1 of IL11 TRE DNA duplex; or 1.5× GBS consensus DNA sequence. Data were processed using Bruker Topspin (v3.2) and analyzed using NMRViewJ (OneMoon Scientific, Inc.). Chemical shift perturbations were calculated based on previously published GR DBD NMR chemical shifts (35) using the minimum chemical shift perturbation procedure (34).
Structure determination of GR DBD-TRE complexes
Crystals of the GR DBD:IL11 complex were grown by hanging drop vapor diffusion in 0.2 M lithium nitratie, 15% PEG 3350, 1% glycerol with a 2:1 protein:DNA molar ratio. Crystals were cryo-protected with 30% PEG 3350 and 15% glycerol and flash cooled in liquid N2. Crystals of the GR DBD:VCAM1 complex were grown by hanging drop vapor diffusion in 0.05 M sodium cacodylate (pH 7), 5 mM magnesium chloride, 1 mM spermine, and 2% t-butanol with a 2:1 protein:DNA molar ratio. Crystals were cryo-protected with 50% glycerol and flash cooled in liquid N2. Data were collected at 1.00 Å wavelength at the 22-ID beamline (Advanced Photon Source, Argonne, IL) and processed using the HKL-2000 software (36). The structures were phased using a previously solved structure of GR DBD:IR-GBS complex (PDB 4hn5) in Phenix (13,37). Structure refinement and validation was performed using PHENIX refine software and model building was performed in COOT (37,38). PDB Redo was used iteratively to optimize refinement parameters and geometry (39). PyMOL v1.8.2 was used to visualize structures and generate figures (Schrödinger, LLC).
Generation of GR Lys442Ala/Arg447Ala ‘DNA Dead’ and GR Ala458Thr/Ile634Ala (GRmon) mutants
The QuikChange Site-directed Mutagenesis kit (Stratagene) was used to generate the GR Lys442Ala/Arg447Ala and GR Ala458Thr/Ile634Ala mutants. The ‘DNA Dead’ mutation was made in both the GR DBD only for bacterial expression and in the full-length GR PCDNA3 vector for reporter gene assays. The GRmon mutant was also made in the full-length GR PCDNA3 vector for reporter gene assays.
TR-FRET competition assays
Lyophilized Lumi4-Tb Cryptate anti-HIS6 antibody (Cisbio) was reconstituted in 250 μL of distilled water to create a working stock per the manufacturer instructions (40) and subsequently diluted 1:100 in reaction buffer containing 20 mM Tris–HCl (pH 7.4), 150 mM NaCl, and 5% glycerol. The Lanthascreen TR-FRET-based assay was performed by adding increasing concentrations of HIS-tagged AP-1 (1.5–50 nM), 10 nM of the FAM-labeled IL11 TRE DNA, and 5 μl of diluted antibody. Competition experiments were conducted by adding increasing concentrations of GR DBD (50 nM–1 μM) lacking the 6xHIS tag. TR-FRET was measured on a Biotek Neo (Winooski, VT) plate-reader at an excitation and emission wavelength of 340 and 520 nm, respectively. Two technical replicates and three biological replicates were conducted. Plots generated using GraphPad Prism (v7) are a compilation of all data collected; data are shown as a ratio of acceptor to donor values; and error bars represent SEM.
RESULTS
GR is recruited to AP-1 target genes in a DNA-binding-dependent manner
ChIP-seq data have found GR to be highly enriched at TRE motifs (27–31). AP-1 subunits are largely cytoplasmic until treatment with an activator, such as lipopolysaccharide (LPS) or tumor necrosis factor alpha (TNF-α); therefore, AP-1 would not be present in the nucleus or at these GR occupied sites in the absence of an AP-1 activator. However, ChIP-seq for GR in DEX-treated mouse primary macrophages without an AP-1 activator showed the TRE motif was the second most enriched site compared to a canonical GBS (27). Furthermore, even with AP-1 activation the ChIP-seq profiles revealed that 50.3% of the GR occupancy is without a tethering factor (27).
Based on these findings that indicate GR can occupy TREs in the absence of a tethering factor, we hypothesized that GR may bind to some TRE sequences directly. To test GR recruitment to TREs, we performed ChIP using a tetracycline-inducible system in HEK293T cells. The exogenous receptors were detected using their N-terminal hemagglutinin (HA) epitope tag (Figure 1B). We tested GR occupancy on two TRE-containing promoters; interleukin-11 (IL11) and vascular cell adhesion molecule-1 (VCAM1). In the absence of an AP-1 activator, HA-tagged WT GR occupies the TRE motif from both promoters in the presence of a GR agonist, DEX (Figure 1C). GR occupancy is maintained with cellular pretreatment of an AP-1 activator, TNF-α. As both promoters contain a TRE and a NF-κB response element (κBRE), it is likely that DNA fragments generated for qPCR could encompass both response element motifs (Figure 1D). As a control, WT GR occupies several canonical GBS motifs that show comparable levels of GR recruitment (Figure 1E). We have observed a DNA-binding-dependent mechanism is also possible at κBREs (Ortlund Lab, unpublished). These data, when considered with genome-wide ChIP analysis (27,31), suggests that tethering alone is inadequate to explain GR recruitment to these sites.
GR Ser425Gly mutation cannot be used to distinguish DNA-binding-dependent from -independent mechanisms
Compared to other steroid receptors, GR is unique in its ability to counter AP-1 activity (41,42). Despite sharing 90% sequence identity in the DBD and complete conservation of DNA-contacting residues, the mineralocorticoid (MR), which is able to transactivate from a canonical GBS, is unable to repress a TRE-containing reporter (35,41,43) (Figure 1F). Swapping the GR DBD into the full-length MR protein partially restores transrepression, suggesting that the GR DBD is necessary to repress inflammatory genes (41).
To determine the specific residues within GR that allow for this transrepressive function, the GR and MR DBDs were aligned, which revealed seven amino acid differences—including Ser425 in GR, which is a Gly in MR. When this GR residue is mutated to the equivalent site in MR (GR Ser425Gly), the mutation renders GR incapable of repressing select inflammatory genes but maintains the ability to bind to a canonical GBS (41). This mutation did not affect binding and transactivation from canonical sites, and it was therefore concluded that GR did not need to directly bind DNA in order to repress, solidifying support for a tethering hypothesis. However, this conclusion was made before DNA-binding-dependent repression at IR-GBSs was identified (12–14). At IR-GBS sites, the GR Ser425Gly mutation reduced the affinity for IR-GBS DNA sequences and was deficient in transrepression of the TSLP IR-GBS in cells (13,14). Based on these findings, the GR Ser425Gly mutation cannot be used to discriminate DNA-binding-dependent from independent mechanisms.
Despite conflicting reports on the effect this mutation has on GR-mediated transrepression at TREs, we tested whether GR Ser425Gly had an effect on GR recruitment to TREs in cells (44). Compared to WT GR, the Ser425Gly mutant showed a reduced occupancy on TREs (Figure 1C). This reduced occupancy is in line with the effect observed at IR-GBSs (13,14), suggesting mechanisms of transrepression might be similar. Conversely, both WT GR and the Ser425Gly mutant are similarly recruited to canonical GBS sites in the promoters of, GILZ, and SGK1, but reduced occupancy at FKBP5 (Figure 1D).
GR represses AP-1 target genes in the absence of tethering factors
Since GR can occupy genomic TREs without tethering factors, we hypothesized that GR might also repress transcription without tethering factors. Canonical AP-1 target genes are expressed at low levels in the absence of pro-inflammatory signaling, making the study of DNA-dependent GR-mediated transrepression difficult (19,45,46). Stimulation of AP-1 and its target genes would confound our aims of delineating DNA-binding-dependent from -independent transrepression. To circumvent this issue, we constructed reporter plasmids that measure transcriptional repression without prior activation of AP-1. The reporters contain a strong SV40 enhancer and promoter that constitutively expresses luciferase; and in between the enhancer and promoter regions we inserted ∼150 bases of the promoters from IL11, VCAM1, and interleukin-6 (IL6), all of which contain TREs and are known to be upregulated by AP-1 (47–50). With co-transfection of full-length GR or mutants, a loss of luciferase signal is a readout for gene repression (12). When these reporter plasmids were transfected with WT GR into U-2 OS cells, which do not express endogenous GR, only in the presence of GR and upon DEX treatment is GR able to repress transcription of the reporter, indicating that GR alone is sufficient to repress these TRE-containing inflammatory genes (Figure 2A).
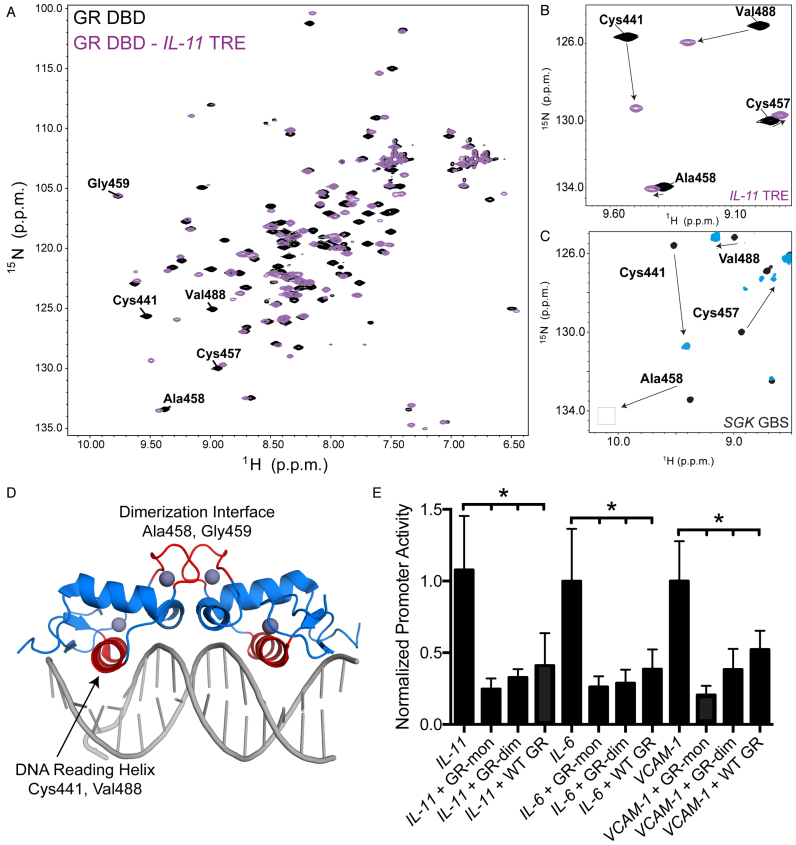
Figure 2.
WT GR, but not GR Lys442Ala/Arg447Ala, is able to transrepress inflammatory genes. (A) WT GR transrepresses constitutively active luciferase reporters containing portions of the IL11, IL6, and VCAM1 promoters upon treatment with 100 nM DEX but not media alone; GR Lys442Ala/Arg447Ala (GR DNA Dead ‘DD’) is unable to repress inflammatory gene reporters. (B) GR uses K442 and R447 to make base specific interactions on a canonical GBS (PDB: 3FYL), these side chains were mutated to Ala to disrupt sequence-specific DNA interactions. (C) Compared to WT GR, GR Lys442Ala/Arg447Ala does not occupy TREs or to canonical GBS from FKBP5, GILZ, and SGK1 (*Padj < 0.005, **Padj < 0.0001).
To test the requirement of DNA-binding for the observed repression, we generated a new mutant GR Lys442Ala/Arg447Ala, termed ‘DNA-Dead’ GR, which prevents binding of GR to multiple DNA response elements due to a loss of sequence-specific contacts (9,13) (Figure 2B). ChIP experiments revealed that GR Lys442Ala/Arg447Ala is not recruited to TREs or to canonical GBS, and this mutant is also unable to repress transcription (Figure 2B and C), indicating the importance of DNA binding.
GR binds directly to TREs
Since ChIP and transrepression analysis suggested DNA-dependent interactions were critical for GR action at inflammatory genes, we hypothesized that GR could bind directly to TREs. To test this, we used fluorescence polarization assays to monitor full-length GR binding to IL11 TRE and SGK GBS DNA sequences (51). Surprisingly, GR bound to the IL11 TRE with similar affinity (42 nM) to the canonical SGK GBS (34 nM) (Figure 3A). Additionally, we found that the GR DNA binding domain (DBD) bound to TREs from the IL6, IL11, VCAM1, macrophage colony-stimulating factor 1 (CSF1), interleukin 1 alpha (IL1α), and collagenase 3 precursor (MMP13) promoters (Figure 3b,c) with similar affinities as IR-GBS (13). Full-length GR has a higher affinity for TREs, suggesting that domains other than the DBD contribute to DNA binding, potentially by increased non-sequence specific interactions, pre-ordering the DBD, or by making additional contacts with DNA (41,52,53).
The crystal structure of the GR DBD:IR-GBS complex revealed two monomers of GR bound to opposite sides of the DNA in an everted fashion (13,14). NMR analysis of the GR DBD:IR-GBS complex confirmed that residues critical for dimerization on GBS are not perturbed, suggesting monomeric GR is likely sufficient at these elements (14). To test whether the GR DBD adopts a canonical head-to-head dimeric or IR-GBS-like monomeric conformation on TREs, we performed 2D [1H,15N]-HSQC NMR on the GR DBD:IL11 TRE complex. Binding of IL11 TRE to 15N-labeled GR DBD causes large NMR chemical shift perturbations for residues within the DNA reading helix, such as Cys441 and Val488 (Figure 4A). However, in contrast to the dimeric GR-GBS complex, residues within the GR DBD dimerization loop, such as Ala458 and Gly459, showed no change upon binding IL11 TRE (Figure 4B–D) (9,11), indicating GR binds as a monomer to the IL11 TRE (14).
Figure 4.
Monomeric GR is preferred to repress inflammatory genes. (A) 2D [1H,15N]-HSQC NMR analysis of 15N-GR DBD binding to IL11 TRE DNA; data for GR DBD alone is shown in black and the GR DBD:IL11 complex shown in purple. (B) Zoom-in view of the 2D NMR data show that residues contacting DNA in the complex, including Cys441 and Val488, show significant chemical shift perturbations upon binding DNA. Residues in the dimerization loop (D loop) do not show perturbations, suggesting GR binds as a monomer to IL11 TRE. (C) D loop residues show perturbations when GR binds as a dimer on a canonical GBS. (D) GR DBD-GBS crystal structure (PDB: 3FYL) with the DNA reading helix and D loop highlighted in red. Residues highlighted in panels a-c are located within these two regions. (E) Dimerization deficient mutants GRdim (Ala458Thr) and GRmon (Ala458Thr/Ile634Ala) cause more repression of the constitutively active TRE luciferase reporters compared to WT GR.
To determine whether dimerization is required for transrepression, we performed transcriptional reporter assays using two well-characterized full-length GR dimerization mutants, GRdim (Ala458Thr) and GRmon (Ala458Thr/Ile634Ala) (20,54). Both mutants repressed the constitutively active SV40 TRE luciferase reporters in the presence of GR agonist, suggesting monomeric GR preferred at these sites (Figure 4e). It is not surprising that GRdim maintained transrepressive function as this mutant represses inflammation in vivo (20). However, because GRdim can form dimers on canonical GBSs (11), we also tested GRmon, which was shown to be predominantly monomeric in cells (54). Based on our NMR analysis and transrepression assays with GR monomeric mutants in cells, indicates that monomeric GR is responsible for GR-mediated repression of this subset of genes.
GR recognizes TREs in a sequence specific manner
To determine how GR recognizes TRE sequences, we determined crystal structures of GR DBD bound to the IL11 and VCAM1 TREs (Figure 5). The GR DBD: IL11 TRE complex crystallized in the P212121 space group and data were collected to 2.15 Å (Figure 5A, Table 1). The GR DBD: VCAM1 TRE complex crystallized in the P212121 space group and data were collected to 2.29 Å (Figure 5B, Table 1). Both structures show two GR monomers bound to opposite sides of the TRE DNA sequences in an everted fashion, similar to the GR DBD:IR-GBS structure (13). However, in the TRE structures, one of the GR monomers straddles the end-stacking junction where the DNA makes a pseudo-continuous helix; this GR DBD does not make base-specific interactions and only contacts the DNA backbone. Based on our cellular transrepression and NMR footprinting analysis above, it is likely this GR DBD is only important for efficient crystal packing and may not be biologically relevant in vivo.
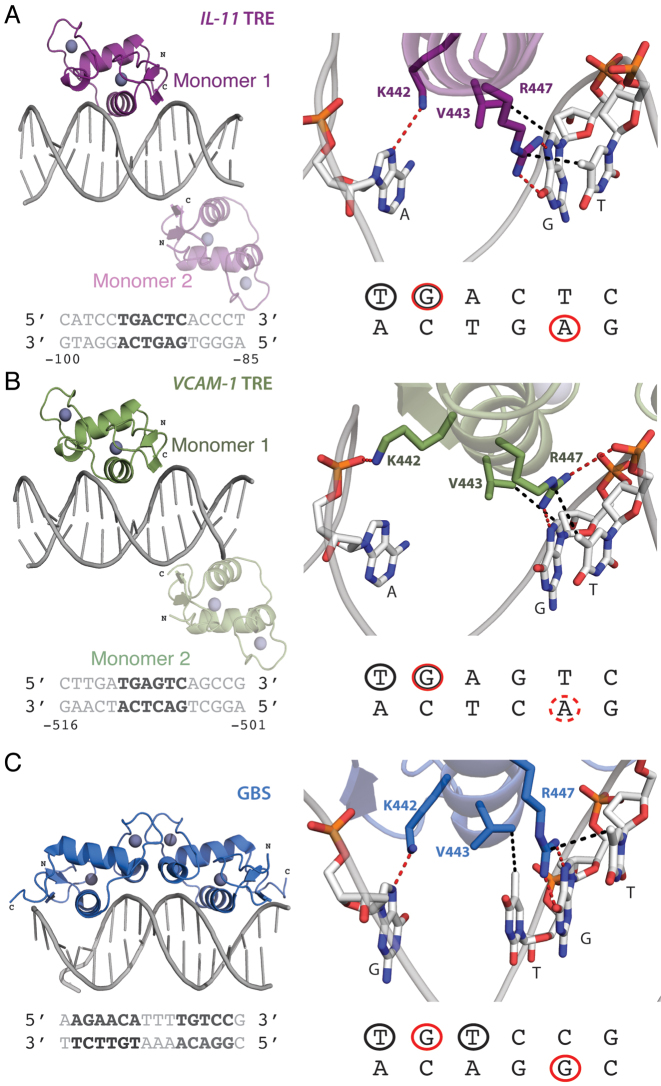
Figure 5.
Crystal structures of GR DBD bound to the IL11 TRE, VCAM1 TRE, and a GBS. (A, B) Structures of the GR DBD bound to the (A) IL11 (purple) and (B) VCAM1 (green) TRE sequences. The sequence contacted by GR is colored dark grey. In both structures, two GR monomers were bound to opposite sides of the DNA sequence. The darker colored monomer (#1) sits over the GR binding footprint, highlighted in red, and the faded monomer (#2) sits over an end-stacking junction without making direct DNA contacts. (C) In contrast, the structure of GR DBD bound to a canonical GBS shows GR binds as a homodimer. For all structures, GR base-specific contacts are shown to the right. GR contacts the DNA through hydrogen bonds (red) and Van der Waals contacts (black) made between Arg447, Lys442 and Val443. These contacts are highlighted on the sequence below, where red circles represent contacts made through hydrogen bonds and black circles are bases contacted by Van der Waals interactions. The dotted red circle in the VCAM1 footprint indicates contacts being made by GR with the backbone of the adenine base but not a base-specific interaction. This data shows that GR makes similar contacts on all sites.
Table 1. Summary of crystal data collection and refinement statistics.
| GR DBD – IL11 | GR DBD – VCAM1 | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimension | a = 39.1, b = 96.9, c =104.6 | a = 39.4, b = 96.3, c = 105.2 |
| Resolution (Å) | 2.15 (2.23–2.15)a | 2.29 (2.37–2.29)a |
| R sym | 7.7 (47.5) | 11.9 (58.2) |
| I/σ | 8.9 (1.7) | 9.7 (2.1) |
| Completeness | 98.3 (82.9) | 94.4 (98.1) |
| Redundancy | 5.3 (3.5) | 7.0 (6.6) |
| Refinement | ||
| Resolution | 2.15 | 2.29 |
| No. of reflections | 21901 | 17582 |
| R work/Rfree | 20.1/22.7 | 19.4/22.8 |
| No. of atoms: | ||
| Protein | 1082 | 1094 |
| DNA | 650 | 650 |
| Water | 47 | 42 |
| B-factors: | ||
| Protein | 47.3 | 42.3 |
| DNA | 64.7 | 50.9 |
| Water | 48.2 | 40.5 |
| R.m.s. deviations: | ||
| Bond lengths (Å) | 0.009 | 0.008 |
| Bond angles (°) | 1.06 | 1.88 |
| PDB code | 5VA7 | 5VA0 |
aData collected from a single crystal; values in parentheses are for the highest-resolution shell.
In both structures, GR recognizes a hexameric TGA(G/C)TC sequence; though the third base differs, our structures show that GR does not directly contact this base. Analysis of the GR-DNA structural interfaces using PISA (55) revealed a favorable free energy gained with the GR DBD (Monomer 1)–DNA interaction. The free energy gain upon complex formation is –9.7 kcal/mol for IL11 TRE and –8.9 kcal/mol for VCAM1 TRE, values similar to GR DBD:IR-GBS complex formation (13). Monomer 1 of the GR DBD:IL11 TRE complex positions the DNA reading helix in the major groove (Figure 5A). Three side chains, Arg447, Lys442 and Val443, participate in base-specific interactions with the DNA and are consistently used by the GR DBD to recognize DNA sequences (Figure 5C) (9,13). Arg447 makes hydrogen bonds to the N7 position and the terminal oxygen on guanine 107, and van der Waals contacts to the methyl group on thymine 106. Lys442 makes hydrogen bonds to the N7 position on adenine 91, and Val443 makes van der Waals contacts to guanine 107 (Figure 5A). Monomer 1 of the GR DBD:VCAM1 TRE complex only uses two side chains to make base-specific contacts with the DNA (Figure 5B). Arg447 makes hydrogen bonds to the N7 position on guanine 523, and Val443 makes van der Waals contacts to the same base. Arg447 also makes van der Waals contacts to the methyl group on thymine 522. In both structures, additional side chains participate in backbone interactions, marking the boundaries of the GR binding footprint.
In all structures, GR recognizes similar DNA bases with consistent spacing. In the IL11 structure, Arg447 hydrogen bonds to a guanine but also makes side-on hydrophobic contacts with the methyl group on a neighboring thymine base. Additionally, Lys442 makes hydrogen bonds to a pyrimidine base. We do not observe a direct interaction between GR and this base in the VCAM1 structure; instead, Lys442 interacts with the DNA backbone at the same position. In the GBS structure, Val443 has an additional base contact though van der Waals forces with a thymine; TREs have a pyrimidine base in this position, and therefore the Val residue is shifted to make hydrophobic contacts with the guanine base instead. Our structures show that GR recognizes a GBS half-site sequence embedded within the TRE.
GR and AP-1 likely compete for the same binding site
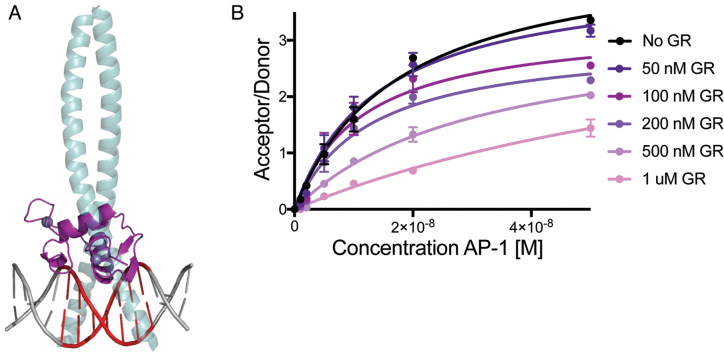
Structural alignment of the GR DBD:IL11 TRE structure with the AP-1:TRE structure reveals that GR and AP-1 would likely compete for the same DNA binding site (Figure 6A). To test if GR competes with AP-1 for binding to the TRE, we performed a time resolved-fluorescence resonance energy transfer (TR-FRET) competition assay by monitoring the concentration-dependent effect of GR DBD on the interaction between AP-1 and IL11 TRE DNA. Titration of AP-1 resulted in an increase in TR-FRET signal, showing binding of AP-1 to IL11 TRE. Notably, when increasing amounts of GR DBD is added, TR-FRET signal is reduced indicating that GR competes with AP-1 binding to the IL11 TRE (Figure 6B).
Figure 6.
GR and AP-1 Compete for the same DNA binding site. (A) Overlay of GR DBD-IL11 TRE complex and AP-1 TRE complex (GR, purple; AP-1 green; TRE, red) (PDB code: 1FOS). (B) TR-FRET assay showing GR competes with AP-1 for binding to FAM-labeled IL11 TRE DNA via decreased acceptor/donor signal.
DISCUSSION
GR is a ligand-regulated transcription factor that controls distinct gene networks across numerous biological processes including development, metabolism, and inflammation (1). Current models suggest that GR uses distinct mechanisms to recognize promoters with different DNA sequences (Figure 1A). GR can cooperatively homodimerize at GBSs or bind as a monomer to IR-GBSs and composite elements (9,13). Conversely, GR can tether onto other DNA-bound TFs, such as AP-1, but not make direct contact with DNA (19). However, new genome-wide methodologies have revealed a new complexity of GR-genome interactions and suggest there may be alternative mechanisms beyond the current view of GR signalling (8,27,56). Here, we propose a new mechanism by which GR binds directly to a GBS-like half-site located within a canonical AP-1 TRE, such as sequences found in the promoter regions of IL11 and VCAM1 (Figure 1Av).
As detailed above, genome-wide ChIP-seq studies find that GR occupies the TRE motif across numerous cell types (27–31) (Figure 1B). These experiments are conducted with GR agonist alone, which does not alter the subcellular localization of AP-1 subunits, and in the absence of AP-1 activation—suggesting GR occupies these sites without AP-1 (27). However, loss of AP-1 results in a significant reduction of GR DNA occupancy (29). This could suggest that GR requires tethering with AP-1 in order to bind DNA or could also be explained by increased chromatin accessibility gained by AP-1 activation (30,57). GR, like most nuclear receptors, predominantly binds to accessible chromatin (58). Therefore, we propose that AP-1 would be recruited to inflammatory gene TREs to drive transcription and act as a pioneering factor for subsequent GR recruitment (29,30). While GR and AP-1 have been shown to interact directly (52), other studies were unable to validate this interaction (53,59). Instead, we show GR can directly bind TREs and the GR’s DNA-binding function is required for transrepression at these elements. Structural overlays of GR and AP-1 bound to a TRE sequence suggests they likely compete for the same DNA binding site on some elements (Figure 6A), and we confirmed that GR and AP-1 indeed compete for binding to the TRE sequence (Figure 6B), consistent with previous studies (52,53).
Further support for a GR DNA-binding-dependent repressive mechanism comes from single molecule tracking experiments that revealed only 3% of cellular GR is likely to be tethered to other TFs, indicating that DNA-binding-dependent mechanisms represent the majority of GR-chromatin interactions (27,60,61). We therefore hypothesized that GR might be able to bind directly to TREs. We show here that full-length GR is able to bind the IL11 TRE with an affinity nearly identical to canonical GBS (Figure 3A). Furthermore, we demonstrate that GR alone is sufficient to transrepress. Ablation of GR DNA binding results in an attenuation of GR occupancy at TREs as well as GR-mediated transrepression (Figures 1B and 2B). The latter data is in line with other reports that show the importance of the GR DNA binding domain for repression of inflammatory genes (41,44,52,53,62). Another striking example is that GR has already been shown to bind directly to a TRE-like sequence in the rat tyrosine hydroxylase (TH) gene promoter (63). Though the promoter contains a TRE and a canonical GBS, GR was shown to regulate transcription through the TRE sequence by directly binding to the TRE-like site (TGACTAA). This sequence is almost identical to the GR footprint identified by our structural analysis (Figure 5). Binding at this site is conserved in humans, suggesting GR DNA-binding-dependent mechanisms at TREs may represent an evolutionary conserved model (64).
Initial support for the tethering hypothesis stemmed from studies of dimerization deficient GR mutants, which generated complex and often conflicting interpretations (65). The GRdim mutant, designed with the aim of breaking intramolecular protein-protein interactions at the GR homodimerization interface (9), was the main driver for a DNA-binding-independent mechanism at inflammatory genes. GRdim was shown to not bind DNA and displays a reduced ability to transactivate genes, whereas transrepression was unaffected (41,66). Furthermore, whereas complete GR knock-out mice die quickly after birth, GRdim knock-in mice lived and maintained the ability to combat inflammatory challenge (20,67). These results drove the conclusions that dimerization of GR is required for DNA binding and that dimerization is not necessary for GR to repress inflammatory gene expression (21). However, it was later shown that GRdim does not affect GR stoichiometry on DNA and GRdim can still forms dimers in vitro and in cells. Instead, GRdim affects cooperative binding to DNA (11,54). These results suggest that the GRdim mutant cannot be used to rule out a DNA-binding-dependent mechanism at TREs.
The GR Ser425Gly mutant was previously used to show that DNA binding was dispensable at TREs; this mutant binds to canonical GBSs but is unable to repress inflammatory genes (41). However, these interpretations were made before the DNA-binding-dependent GR mechanisms at IR-GBS were identified. It has since been shown that the GR Ser425Gly mutant not only has diminished binding to an IR-GBS but also affects GR transrepression from IR-GBS sites (14). In this work, we further show that GR Ser425Gly mutant is poorly recruited to TREs (Figure 1B). Taken together, our findings suggest that GR repression of inflammatory genes likely occurs using a similar mechanism to repression of IR-GBSs through a preference for monomeric GR.
Our findings represent a shift in our understanding of GR-mediated repression of inflammation. Our data support a DNA-binding-dependent mechanism for GR repression at TREs, however a variety of mechanisms are likely involved (17,23). What drives the selection between DNA-binding-dependent repression and tethering/transrepression remains unclear, but this work adds yet another layer of complexity to the role of GR in regulating transcription. This work will be paramount as the field continues to seek selective gene modulators for the treatment of chronic inflammatory diseases. We not only propose a new mechanism of repression but also show that monomeric GR is preferred at these and other repressive DNA sites (14). This distinction of oligomeric state on different DNA sequences could provide an avenue for future therapeutic design.
ACCESSION NUMBERS
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5VA7 and 5VA0.
ACKNOWLEDGEMENTS
We thank David Bain, PhD for providing full-length GR used for binding studies. We thank Sam Hong, PhD for providing AP-1 used for competition assays. We thank Oskar Laur of the Emory Cloning Core for assistance with generating reporter constructs. X-ray data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members/html. Use of the Advanced Photon source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Present addresses:
Ian Mitchelle S. de Vera, Department of Pharmacology and Physiology, Saint Louis University School of Medicine, MO 63104, USA.
William H. Hudson, Department of Microbiology and Immunology, Emory University School of Medicine, Atlanta, GA 30322, USA.
FUNDING
National Institutes of Health General Medical Sciences [1F31GM113397-01A1 to E.R.W.]; National Institutes of Health [R01DK095750 to E.A.O.]; AHA [14GRNT20460124 to E.A.O.]; W.M. Keck Foundation Medical Research Grant (to E.A.O.); National Institutes of Health [R01DK101871 to D.J.K.]; National Institutes of Health [R01GM114420 to D.J.K.]. J.C.N is supported by the BallenIsles Men's Golf Association. Funding for open access charge: W.M. Keck Foundation Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kadmiel M., Cidlowski J.A.. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013; 34:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carson-Jurica M.A., Schrader W.T., O’Malley B.W.. Steroid receptor family: structure and functions. Endocr. Rev. 1990; 11:201–220. [DOI] [PubMed] [Google Scholar]
- 3. Kumar R., Thompson E.B.. The structure of the nuclear hormone receptors. Steroids. 1999; 64:310–319. [DOI] [PubMed] [Google Scholar]
- 4. Pratt W.B., Toft D.O.. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997; 18:306–360. [DOI] [PubMed] [Google Scholar]
- 5. Picard D., Khursheed B., Garabedian M.J., Fortin M.G., Lindquist S., Yamamoto K.R.. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990; 348:166–168. [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto K.R. Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 1985; 19:209–252. [DOI] [PubMed] [Google Scholar]
- 7. Meijsing S.H. Mechanisms of Glucocorticoid-Regulated Gene Transcription. Adv. Exp. Med. Biol. 2015; 872:59–81. [DOI] [PubMed] [Google Scholar]
- 8. Weikum E.R., Knuesel M.T., Ortlund E.A., Yamamoto K.R.. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 2017; 18:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luisi B.F., Xu W.X., Otwinowski Z., Freedman L.P., Yamamoto K.R., Sigler P.B.. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991; 352:497–505. [DOI] [PubMed] [Google Scholar]
- 10. Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R.. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009; 324:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watson L.C., Kuchenbecker K.M., Schiller B.J., Gross J.D., Pufall M.A., Yamamoto K.R.. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat. Struct. Mol. Biol. 2013; 20:876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Surjit M., Ganti K.P., Mukherji A., Ye T., Hua G., Metzger D., Li M., Chambon P.. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011; 145:224–241. [DOI] [PubMed] [Google Scholar]
- 13. Hudson W.H., Youn C., Ortlund E.A.. The structural basis of direct glucocorticoid-mediated transrepression. Nat. Struct. Mol. Biol. 2013; 20:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hudson W.H., Kossmann B.R., de Vera I.M., Chuo S.W., Weikum E.R., Eick G.N., Thornton J.W., Ivanov I.N., Kojetin D.J., Ortlund E.A.. Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miner J.N., Yamamoto K.R.. The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev. 1992; 6:2491–2501. [DOI] [PubMed] [Google Scholar]
- 16. Schiller B.J., Chodankar R., Watson L.C., Stallcup M.R., Yamamoto K.R.. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 2014; 15:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ratman D., Vanden Berghe W., Dejager L., Libert C., Tavernier J., Beck I.M., De Bosscher K.. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol. Cell. Endocrinol. 2013; 380:41–54. [DOI] [PubMed] [Google Scholar]
- 18. De Bosscher K., Vanden Berghe W., Haegeman G.. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev. 2003; 24:488–522. [DOI] [PubMed] [Google Scholar]
- 19. Herrlich P. Cross-talk between glucocorticoid receptor and AP-1. Oncogene. 2001; 20:2465–2475. [DOI] [PubMed] [Google Scholar]
- 20. Reichardt H.M., Kaestner K.H., Tuckermann J., Kretz O., Wessely O., Bock R., Gass P., Schmid W., Herrlich P., Angel P. et al. . DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998; 93:531–541. [DOI] [PubMed] [Google Scholar]
- 21. Karin M. New twists in gene regulation by glucocorticoid receptor: is DNA binding dispensable. Cell. 1998; 93:487–490. [DOI] [PubMed] [Google Scholar]
- 22. De Bosscher K., Vanden Berghe W., Haegeman G.. Mechanisms of anti-inflammatory action and of immunosuppression by glucocorticoids: negative interference of activated glucocorticoid receptor with transcription factors. J. Neuroimmunol. 2000; 109:16–22. [DOI] [PubMed] [Google Scholar]
- 23. Clark A.R., Belvisi M.G.. Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol. Ther. 2012; 134:54–67. [DOI] [PubMed] [Google Scholar]
- 24. Cato A.C., Wade E.. Molecular mechanisms of anti-inflammatory action of glucocorticoids. Bioessays. 1996; 18:371–378. [DOI] [PubMed] [Google Scholar]
- 25. Kleiman A., Tuckermann J.P.. Glucocorticoid receptor action in beneficial and side effects of steroid therapy: lessons from conditional knockout mice. Mol. Cell. Endocrinol. 2007; 275:98–108. [DOI] [PubMed] [Google Scholar]
- 26. Schacke H., Docke W.D., Asadullah K.. Mechanisms involved in the side effects of glucocorticoids. Pharmacol. Ther. 2002; 96:23–43. [DOI] [PubMed] [Google Scholar]
- 27. Uhlenhaut N.H., Barish G.D., Yu R.T., Downes M., Karunasiri M., Liddle C., Schwalie P., Hubner N., Evans R.M.. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell. 2013; 49:158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim H.W., Uhlenhaut N.H., Rauch A., Weiner J., Hubner S., Hubner N., Won K.J., Lazar M.A., Tuckermann J., Steger D.J.. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 2015; 25:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.H., Trump S., Lightman S.L. et al. . Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011; 43:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A.. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011; 43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reddy T.E., Pauli F., Sprouse R.O., Neff N.F., Newberry K.M., Garabedian M.J., Myers R.M.. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009; 19:2163–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. et al. . Ensembl 2013. Nucleic Acids Res. 2013; 41:D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nwachukwu J.C., Srinivasan S., Bruno N.E., Parent A.A., Hughes T.S., Pollock J.A., Gjyshi O., Cavett V., Nowak J., Garcia-Ordonez R.D. et al. . Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. Elife. 2014; 3:e02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robblee J.P., Miura M.T., Bain D.L.. Glucocorticoid receptor-promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry. 2012; 51:4463–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hudson W.H., Youn C., Ortlund E.A.. Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA. PLoS One. 2014; 9:e107000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 37. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Joosten R.P., Salzemann J., Bloch V., Stockinger H., Berglund A.C., Blanchet C., Bongcam-Rudloff E., Combet C., Da Costa A.L., Deleage G. et al. . PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009; 42:376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mathis G. Probing molecular interactions with homogeneous techniques based on rare earth cryptates and fluorescence energy transfer. Clin. Chem. 1995; 41:1391–1397. [PubMed] [Google Scholar]
- 41. Heck S., Kullmann M., Gast A., Ponta H., Rahmsdorf H.J., Herrlich P., Cato A.C.. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994; 13:4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cato A.C., Konig H., Ponta H., Herrlich P.. Steroids and growth promoting factors in the regulation of expression of genes and gene networks. J. Steroid Biochem. Mol. Biol. 1992; 43:63–68. [DOI] [PubMed] [Google Scholar]
- 43. Pearce D., Yamamoto K.R.. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993; 259:1161–1165. [DOI] [PubMed] [Google Scholar]
- 44. Wei P., Inamdar N., Vedeckis W.V.. Transrepression of c-jun gene expression by the glucocorticoid receptor requires both AP-1 sites in the c-jun promoter. Mol. Endocrinol. 1998; 12:1322–1333. [DOI] [PubMed] [Google Scholar]
- 45. Angel P., Imagawa M., Chiu R., Stein B., Imbra R.J., Rahmsdorf H.J., Jonat C., Herrlich P., Karin M.. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987; 49:729–739. [DOI] [PubMed] [Google Scholar]
- 46. Karin M., Liu Z., Zandi E.. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997; 9:240–246. [DOI] [PubMed] [Google Scholar]
- 47. Ahmad M., Theofanidis P., Medford R.M.. Role of activating protein-1 in the regulation of the vascular cell adhesion molecule-1 gene expression by tumor necrosis factor-alpha. J. Biol. Chem. 1998; 273:4616–4621. [DOI] [PubMed] [Google Scholar]
- 48. Sawa Y., Ueki T., Hata M., Iwasawa K., Tsuruga E., Kojima H., Ishikawa H., Yoshida S.. LPS-induced IL-6, IL-8, VCAM-1, and ICAM-1 expression in human lymphatic endothelium. J. Histochem. Cytochem. 2008; 56:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tohjima E., Inoue D., Yamamoto N., Kido S., Ito Y., Kato S., Takeuchi Y., Fukumoto S., Matsumoto T.. Decreased AP-1 activity and interleukin-11 expression by bone marrow stromal cells may be associated with impaired bone formation in aged mice. J. Bone Miner. Res. 2003; 18:1461–1470. [DOI] [PubMed] [Google Scholar]
- 50. Tang W., Yang L., Yang Y.C., Leng S.X., Elias J.A.. Transforming growth factor-beta stimulates interleukin-11 transcription via complex activating protein-1-dependent pathways. J. Biol. Chem. 1998; 273:5506–5513. [DOI] [PubMed] [Google Scholar]
- 51. Bain D.L., Yang Q., Connaghan K.D., Robblee J.P., Miura M.T., Degala G.D., Lambert J.R., Maluf N.K.. Glucocorticoid receptor-DNA interactions: binding energetics are the primary determinant of sequence-specific transcriptional activity. J. Mol. Biol. 2012; 422:18–32. [DOI] [PubMed] [Google Scholar]
- 52. Yang-Yen H.F., Chambard J.C., Sun Y.L., Smeal T., Schmidt T.J., Drouin J., Karin M.. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990; 62:1205–1215. [DOI] [PubMed] [Google Scholar]
- 53. Schule R., Rangarajan P., Kliewer S., Ransone L.J., Bolado J., Yang N., Verma I.M., Evans R.M.. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990; 62:1217–1226. [DOI] [PubMed] [Google Scholar]
- 54. Presman D.M., Ogara M.F., Stortz M., Alvarez L.D., Pooley J.R., Schiltz R.L., Grontved L., Johnson T.A., Mittelstadt P.R., Ashwell J.D. et al. . Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 2014; 12:e1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 56. Sacta M.A., Chinenov Y., Rogatsky I.. Glucocorticoid signaling: an update from a genomic perspective. Annu. Rev. Physiol. 2016; 78:155–180. [DOI] [PubMed] [Google Scholar]
- 57. John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., Voss T.C., Davis S.R., Meltzer P.S., Stamatoyannopoulos J.A. et al. . Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008; 29:611–624. [DOI] [PubMed] [Google Scholar]
- 58. Ray A., Prefontaine K.E.. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:752–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lucibello F.C., Slater E.P., Jooss K.U., Beato M., Muller R.. Mutual transrepression of Fos and the glucocorticoid receptor: involvement of a functional domain in Fos which is absent in FosB. EMBO J. 1990; 9:2827–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baschant U., Culemann S., Tuckermann J.. Molecular determinants of glucocorticoid actions in inflammatory joint diseases. Mol. Cell Endocrinol. 2013; 380:108–118. [DOI] [PubMed] [Google Scholar]
- 61. Gebhardt J.C., Suter D.M., Roy R., Zhao Z.W., Chapman A.R., Basu S., Maniatis T., Xie X.S.. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013; 10:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oro A.E., Hollenberg S.M., Evans R.M.. Transcriptional inhibition by a glucocorticoid receptor-beta-galactosidase fusion protein. Cell. 1988; 55:1109–1114. [DOI] [PubMed] [Google Scholar]
- 63. Rani C.S., Elango N., Wang S.S., Kobayashi K., Strong R.. Identification of an activator protein-1-like sequence as the glucocorticoid response element in the rat tyrosine hydroxylase gene. Mol. Pharmacol. 2009; 75:589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sheela Rani C.S., Soto-Pina A., Iacovitti L., Strong R.. Evolutionary conservation of an atypical glucocorticoid-responsive element in the human tyrosine hydroxylase gene. J. Neurochem. 2013; 126:19–28. [DOI] [PubMed] [Google Scholar]
- 65. Beck I.M., De Bosscher K., Haegeman G.. Glucocorticoid receptor mutants: man-made tools for functional research. Trends Endocrinol. Metab. 2011; 22:295–310. [DOI] [PubMed] [Google Scholar]
- 66. Tuckermann J.P., Reichardt H.M., Arribas R., Richter K.H., Schutz G., Angel P.. The DNA binding-independent function of the glucocorticoid receptor mediates repression of AP-1-dependent genes in skin. J. Cell Biol. 1999; 147:1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cole T.J., Blendy J.A., Monaghan A.P., Krieglstein K., Schmid W., Aguzzi A., Fantuzzi G., Hummler E., Unsicker K., Schutz G.. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995; 9:1608–1621. [DOI] [PubMed] [Google Scholar]