Abstract
The current study examined quantitative measures of psychosis proneness in a nonpsychotic population, in order to elucidate their underlying genetic architecture and to observe if there is any commonality to that already detected in the studies of individuals with overt psychotic conditions, such as schizophrenia and bipolar disorder. Heritability, univariate and multivariate genome-wide association (GWAs) tests, including a series of comprehensive gene-based association analyses, were developed in 4269 nonpsychotic persons participating in the Northern Finland Birth Cohort 1966 study with information on the following psychometric measures: Hypomanic Personality, Perceptual Aberration, Physical and Social Anhedonia (also known as Chapman’s Schizotypia scales), and Schizoidia scale. Genome-wide genetic data was available for ~9.84 million SNPs. Heritability estimates ranged from 16% to 27%. Phenotypic, genetic and environmental correlations ranged from 0.04–0.43, 0.25–0.73, and 0.12–0.43, respectively. Univariate GWAs tests revealed an intronic SNP (rs12449097) at the TMC7 gene (16p12.3) that significantly associated (P = 3.485 × 10–8) with the hypomanic scale. Bivariate GWAs tests including the hypomanic and physical anhedonia scales suggested a further borderline significant SNP (rs188320715; P-value = 5.261 × 10–8, ~572 kb downstream the ARID1B gene at 6q25.3). Gene-based tests highlighted 20 additional genes of which 5 had previously been associated to schizophrenia and/or bipolar disorder: CSMD1, CCDC141, SLC1A2, CACNA1C, and SNAP25. Altogether the findings explained from 3.7% to 14.1% of the corresponding trait heritability. In conclusion, this study provides preliminary genomic evidence suggesting that qualitatively similar biological factors may underlie different psychosis proneness measures, some of which could further predispose to schizophrenia and bipolar disorder.
Keywords: genome-wide association study, heritability, Finnish population, psychoses proneness, schizophrenia, bipolar disorder
Introduction
Psychosis is a serious mental condition by which affected persons experience delusions, hallucinations and severe behavioral abnormalities usually leading to losing contact with reality. While acute forms of psychosis characterize disorders such as schizophrenia, schizoaffective disorder or psychotic bipolar disorder, subclinical types are also present in the general population in the form of odd behaviors, social withdrawal/anxiety, lack of feelings, perceptual abnormalities and magical ideation.1 These psychotic-like experiences, also termed “schizotypy,” are regarded as signs of an underlying predisposition (psychosis proneness) to undergo a clinically meaningful psychotic episode. Evidence from population data on this regard is steadily accumulating,2,3 and as a consequence there is a current consensus amongst clinicians and researchers on the idea that psychosis proneness is a continuum ranging from a normal dissociative to full-blown diagnosable primary psychotic disorders.4
This continuum understanding of psychosis implies that individual differences exist in people’s vulnerability due to the combined effect of their personal genetic background and certain environmental stressors, and only those most susceptible would be pushed over a disease threshold.5,6 It is then possible that the genetic bases of these psychosis-like feelings and experiences (the lower and central portion of the continuum) are to a certain extent common to those for schizophrenia and other disorders with psychotic components (the upper end of the continuum, beyond the disease threshold).6 Consequently, unveiling the genes mediating psychosis proneness in nonpsychotic individuals could help to understand part of the genetic architecture of disorders with psychotic components, and in future to accurately identify individuals at putative risk prior to any clinical manifestation.
To date, large meta-analyses of multinational case-control samples on psychotic disorders have successfully identified numerous common polymorphisms with moderately robust effects, mainly on individuals diagnosed with schizophrenia.7,8 However significant, many of these common polymorphisms showed very limited predictive power. This implies the need for complementary strategies capable of unveiling less common and population specific genetic polymorphisms of importance.9 Studying psychosis proneness in the general population could make this feasible due to 2 main reasons. First, it allows incorporating standard psychiatric epidemiological data that provides information on carefully defined, quantifiable phenotypes and symptoms well below the disease threshold. Second, these data are normally gathered from well-characterized nation-wide registers and cohorts, which can take better advantage of population specific haplotype blocks and subtle linkage disequilibrium patterns to isolate less common, population specific variants with unanticipated roles that may well pass undetected in meta-analyses of multinational case-control samples.10–12
Current understanding of the genetics underlying psychosis proneness in the general population derives principally from traditional quantitative genetic methods, applied to phenotypic data alone, and obtained from close relatives such as twins or parents-offspring. These have offered heritability estimates ranging from 15% to 65% depending upon symptom evaluated or population age.13–18 The most complete study thus far investigated phenotypic data on self-rated paranoia, hallucinations, cognitive disorganization, grandiosity, and anhedonia, as well as parent-rated negative symptoms, from 5059 adolescent twin pairs from England and Wales.19 Consistent with findings from smaller studies, their results showed genetic influences ranging between 15% and 59%, and genetic correlations between 0.27 and 0.63. A relatively similar study in 3685 Australian young-adult twins focusing on Perceptual Aberration, Magical Ideation, Hypomania, Impulsivity, and Physical and Social Anhedonia, provided comparable genetic influences ranging from 24% to 50%.20
Importantly, recent methodological and experimental studies have suggested that heritability estimates obtained through traditional quantitative genetic methods would represent an upper bound estimate of the overall genetic effects on the trait, and that applying current association methods to the available genomic data may well never reveal a significant part of those overall effects.21 This in turn imposes the need of finding distinctive, more conservative heritability estimates that can provide a sensible description of the extent to which psychoses proneness is affected by polymorphisms susceptible to be detected through the analyses of population genomic data. This type of lower level estimate can nowadays be obtained by applying modern statistical methods that rely on the joint analysis of phenotypic and genomic information from large samples of genetically unrelated individuals. At present, only the study by Sieradzka et al22 has considered this methodological approach, which resulted in heritability estimates of 20% for anhedonia, 19% for cognitive disorganization, 17% for grandiosity and 14% for paranoia, and an estimate of 0 for hallucinations.
Given the relevance of the genetic effects on psychosis proneness in the normal population, and the current availability of genome-wide association (GWAs) data in psychiatric research, looking for specific genes and markers affecting these traits is a fundamental and sensible action. However, to date only a few studies have provided suggestive evidence for genetic loci of potential interest, such as the TCF4, COMT and DISC1 genes.23–26 Therefore, we aim to elucidate the genetic architecture of psychosis proneness within a Finnish population-based cohort, maximizing the phenotypic and genomic information available in order to: compute an estimate of the heritability of psychoses proneness traits; disentangle whether shared genetic influences may account for the observed relation between different psychoses proneness scales; and examine genomic regions potentially harboring genes affecting 1 or more of the traits.
Methods
Participants
This study used data collected as part of the larger “Northern Finland Birth Cohort 1966” (NFBC66), a population based longitudinal cohort study comprising 12 058 persons (~96.3% of all possible) with an expected date of birth in 1966 in the northern Finnish provinces of Oulu and Lapland. Data from the participants and their mothers were recorded during pregnancy and at birth with additional follow-up data being collected when they were 1, 14, and 31 years of age, by means of postal questionnaires. Supplementary data was obtained from hospital records, national registers and a physical examination at age 31 years. This investigation analyzed the data gathered at the 31-year follow-up. A detailed explanation of the study protocol for this follow-up may be found elsewhere.27 In brief, 8394 of the cohort members alive and living in Northern Finland or the Helsinki metropolitan area were initially contacted and invited to participate. A total of 6033 persons participated, of which 5122 provided answers to a series of standard psychometric instruments. Here we focus on the study of the summary scores from the following scales: Perceptual Aberration Scale (PAS),28 Hypomanic Personality Scale (HPS),29 Revised Social Anhedonia Scale (SAS),30 Revised Physical Anhedonia Scale (PHAS)30 (also known as Chapman’s Schizotypia Scales), and Schizoidia Scale (SCHS).31 PAS consists of 35 true/false items evaluating psychotic-like experiences including uncommon bodily discomforts, discontinuities and experiences (eg, “My hearing is sometimes so sensitive that ordinary sounds become uncomfortable”). HPS includes 48 true/false items designed to identify hyperactive or exhibitionistic behaviors, feelings of euphoria, impulsivity, irritability, or mood swings (eg, “I have often been so excited about an involving project that I didn’t care about eating or sleeping”). PHAS comprises 61 true/false items assessing difficulties experiencing pleasure from physical stimuli that are usually pleasurable such as food, sex, visual or acoustic settings, etc. (eg, “One food tastes as good as another to me”). SAS consists of 40 true/false items evaluating problems experiencing pleasure from nonphysical stimuli and social interactions such as people’s company, talking, etc. (eg, “I prefer watching television to going out with other people.”). Finally, SCHS includes 7 true/false items revealing key characteristics reckoned to be associated with schizotypal personality (eg, “I am more sensitive than most other people”).
These scales are well described and characterized in clinical and epidemiological psychiatric literature and their reliability and validity have been extensively examined previously,32–35 including in the current sample,36 and deemed acceptable (eg, test–retest reliability values for the Chapman scales ranging between 0.75 to 0.85), which facilitates subsequent interpretation and understanding of the genetic results obtained. In addition a 12-item version of the Infrequency Scale was included to identify individuals offering random answers.37
Exclusion procedures were applied to maximize the quality of the phenotypic data. First, when a participant left 1 or more items of a psychometric scale blank, the rest of the items for that scale were rejected, as full information was required to accurately build the corresponding score. Further, data from participants endorsing >2 items of the Infrequency Scale were disregarded as it was considered evidence of careless response. As we were interested in understanding the genetic predisposition to undergo psychosis-like experiences in the general population without overt psychotic disorders, individuals diagnosed between 1982 (when the cohort members turned 18 years old, the legal age of majority in Finland) to 1997 (when this last 31-years follow-up was completed) with schizophrenia or other psychotic disorders (eg, schizophreniform disorder, schizoaffective disorder, delusional disorder, major depression with psychotic features, bipolar disorder with psychotic features, and psychotic disorders not otherwise specified) were excluded. This information was obtained from the Finnish Hospital Discharge Register. Finally, we disregarded participants who did not provide DNA or who showed any of the confounding characteristics for rejection of genomic information (see below). Supplementary table 1 summarizes the numbers excluded at every step and the final sample sizes available for each trait.
Genomic Data
The NFBC66 members participating at the 31-year follow-up were genotyped using the Illumina HumanHap CNV 370k array (Illumina Inc). Quality control analyses were performed on raw genotype data: genotypes missing in >5% of the samples or samples missing >5% of genotypes, as well as samples showing signs of excessive genome-wide heterozygosity, gender discrepancies, or close consanguinity were excluded.
These cleaned genotypes formed the basis for imputing additional genomic variants, using the IMPUTE2 software,38 considering the reference genomes from the 1000 Genomes Project (Phase I integrated variant set release from March 2012, Includes 93 Finnish individuals), and outlined as in the National Center for Biotechnology Information (NCBI) human genome assembly 37 (GRCh37). Quality control filters were subsequently applied to reduce the number of SNPs to a highly informative set of ~9.8 million SNPs. The exclusion criteria were: poorly imputed (an imputation informativeness INFO statistic from IMPUTE software < 0.4), undefined genotypes (an allele posterior probability < .95, genotypes set to “missing”), too rare (a minor allele frequency < .001), poorly characterized (missing in >10% of the samples), or genetically unbalanced (Hardy-Weinberg Equilibrium test P-value < 1 × 10–6).
Statistical Methods (Extended in Supplementary Information)
Descriptive statistics of the psychoses proneness traits were initially computed (supplementary table 2). Phenotypes were then corrected for sex and population substructure (10 principal components), and transformed to a Gaussian distribution by applying inverse normal transformation methods.
Estimates of the proportion of the phenotypic variances explained by the SNPs in autosomal chromosomes were obtained by genome-wide restricted maximum likelihood (GREML) models in the Genome-wide Complex Trait Analysis software (GCTA).39 The patterns of association between pairs of psychoses proneness traits were studied through examining their pairwise Pearson correlation coefficients. To understand the influences underlying such patterns observed at the phenotypic level, we further computed estimates of their genetic and environmental correlation through GCTA methods.
Phenotypes showing a significant heritability were examined in univariate association analyses using conventional linear regression modeling, while correlated phenotypes also showing significant heritability were entered into a series of bivariate association tests using canonical correlation analyses.40 Both univariate and multivariate phenotype-SNP association tests were implemented through the corresponding extensions of the PLINK software.40,41
Additional analysis of statistical power indicated that the sample size available here (N = 4269, or the maximum number of individuals with phenotypic and genetic data) conferred 86.4% power to detect SNPs explaining ≥1% of the total variance in the phenotypes analyzed (supplementary figure 1), assuming a type-I error rate of 5 × 10–8, complete LD between genotyped and causal markers, and a MAF ≥0.001.42,43
Genomic regions harboring SNPs with a suggestive phenotype-SNP association P-value < 5 × 10–6 were followed-up for gene-based association tests, using Versatile Gene-Based Test for Genome-wide Association methods within the VEGAS2 software.44 Because we are aware that this technique may have difficulties to pinpoint genes including one or very few significant SNPs (and depend somewhat on the LD structure of the region), we performed the analyses also taking into account only the top 10% most significant SNPs in each gene to partly correct for this limitation. Gene-based tests were consequently regarded in our study as an enhancement, rather than a replacement, to the single marker association tests.45 Genes offering P-values < .05 after Bonferroni correction taking into account the number of genes tested for each phenotype, were taken as significant.
Results
Genetic influences, as estimated from the ~9.6 million autosomal SNPs accounted for a significant part of the total variance for most of the phenotypes. The specific heritabilities (and SE) were as follows: = 27.4% (8.1); = 16.6% (6.9); = 26.6% (7.9); = 20.4% (7.9); and < 0.1% (6.9%). Since the heritability for SCHS was extremely low, we disregarded the phenotype from further tests.
Phenotypic correlations between traits were low to moderate (rp = .04–.43), while genetic (rg) and environmental (re) correlations were moderate to high (rg = .25–.73), and low to moderate (re = .12–.43), respectively (table 1). The strongest of the pairwise phenotypic associations were rp,PAS–HPS = .43 and rp,PHAS–SAS = .41. However, similar in magnitude, different patterns were detected in their underlying architecture, and genetic factors tend to account for a higher proportion of the association between PHAS-SAS than between PAS-HPS (rg,PHAS–SAS = .73 [SE rg,PHAS–SAS = .20] vs rg,PAS–HPS = .36 [SE rg,PAS–HPS = .22]). Yet, given the wide standard error of these estimates this differential pattern would not be statistically significant.
Table 1.
Phenotypic, Genetic, and Environmental Correlations Between Psychoses Proneness Traits
| Trait 1 | Trait 2 | N (pairwise) | r p (SE) | P-value | r g (SE) | P-value | r e (SE) |
|---|---|---|---|---|---|---|---|
| HPS | PAS | 3824 | .432 (0.015) | <1.00E-16 | .358 (0.217) | .084 | .429 (0.056) |
| HPS | PHAS | 3787 | −.204 (0.016) | <1.00E-16 | −.435 (0.197) | .023 | −.119 (0.076) |
| HPS | SAS | 3837 | −.041 (0.016) | 1.11E-02 | −.297 (0.262) | .122 | .140 (0.074) |
| PAS | PHAS | 3812 | .040 (0.016) | 1.14E-02 | .256 (0.261) | .148 | −.122 (0.069) |
| PAS | SAS | 3868 | .229 (0.016) | <1.00E-16 | .337 (0.263) | .121 | .205 (0.062) |
| PHAS | SAS | 3820 | .410 (0.015) | <1.00E-16 | .730 (0.203) | .005 | .352 (0.062) |
Note: PAS, Perceptual Aberration Scale; HPS, Hypomanic Personality Scale; PHAS, Revised Physical Anhedonia Scale. rp, rg, and re refers to estimates of phenotypic, genetic and environmental correlations (pair-wise); SE refers to standard errors of estimates; P-value obtained from tests on correlation estimates equal to 0 as null hypothesis.
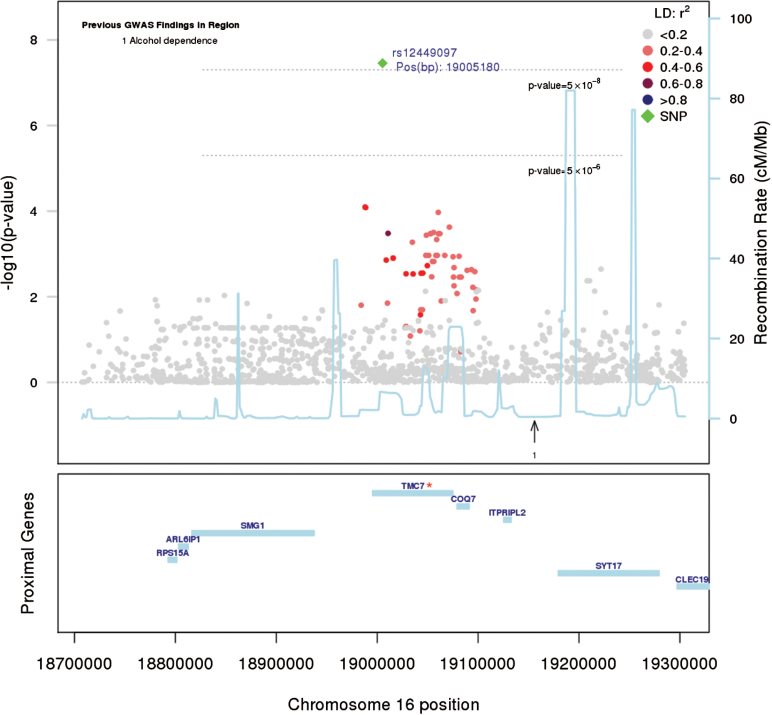
The univariate GWAs analysis highlighted an intronic variant (rs12449097; P-value = 3.49 × 10–8) at the TMC7 gene (16p12.3) associated to HPS (table 2, figure 1). Interestingly, this marker alone explained a meaningful 2.9% of the trait heritability. As this marker was imputed in our study sample, and was relatively uncommon, its imputation accuracy was verified by contrasting its minor allele frequency with respect to that observed in the Sequencing Initiative Suomi (SISu, www.sisuproject.fi46) population reference exome set, which is based upon exome sequence data from 1918 Finnish individuals. Given that the marker was present in both populations at similar frequencies (MAF = 0.003 here vs MAF = 0.004 in SISu), there are grounds to imply that rs12449097 was statistically imputed with reasonable precision in our study sample.
Table 2.
Relevant Loci Detected in Univariate and Bivariate Marker-Based Association Analyses
| Trait | SNP | Chr | Position | Gene | Allele | INFO | MAF | Missing | HWE | Test Statistic | P-value | Beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPS | rs12449097 | 16 | 19005180 | TMC7 | A/G | 0.606 | 0.003 | 0.045 | 1 | −5.527 | 3.485e-08 | −1.13 |
| PAS–PHAS | rs188320715 | 6 | 156526681 | ARID1B | C/T | 0.690 | 0.005 | 0.065 | 0.117 | 16.830 | 5.261e-08 | −.616/.609 |
Note: PAS, Perceptual Aberration Scale; HPS, Hypomanic Personality Scale; PHAS, Revised Physical Anhedonia Scale. Chr refers to chromosome; Position refers to marker position (NCBI human genome assembly 37, GRCh37); Gene refers to suggested candidate gene in the SNP region; INFO refers to imputation informativeness INFO statistic from IMPUTE2 software; MAF refers to minor allele frequency; missing refers to percentage of individuals with missing genotype; HWE refers to P-values from Hardy-Weinberg Equilibrium tests; Test statistic obtained the corresponding univariate (t-statistic) or bivariate (F-statistic) tests; P-value from the corresponding univariate or bivariate test; Beta from independent univariate association tests.
Fig. 1.
Regional Manhattan plot (16p12.3) based on P-values from a univariate model including HPS. The candidate gene in the locus is marked with a star (*). The leading SNP within the locus is highlighted in green; other SNPs colored according their linkage disequilibrium (LD) with it. Genomic position as in the National Center for Biotechnology Information (NCBI) human genome assembly 37, GRCh37. Information on previous genome-wide association (GWAs) findings (with P-value < 5e-8) retrieved from the National Human Genome Research Institute (NHGRI): http://www.genome.gov.
In total, 211 SNPs with suggestive P-value < 5 × 10–6, in any of the 4 univariate traits tested, were mapped in or near to 108 different genes (HPS: 43 genes; PAS: 29 genes; PHAS: 23 genes; and SAS: 13 genes). The subsequent gene-based analyses pinpointed a series of loci harboring 6 significant genes for HPS, 4 genes for PAS, 1 gene for PHAS, and 3 genes for SAS (table 3, and supplementary figures 2, 4, and 5). Altogether, these loci accounted for a meaningful proportion of the heritability of the corresponding psychosis proneness phenotypes: 13.9% of , 10.4% of , 3.7% of and 7.1% of . When taking into account the genetic variance explained by the SNPs pinpointed in marker-based GWAs tests, the genomic findings explained as much as 14.1% of .
Table 3.
Results From Gene-Based Association Analyses, Including the Most Likely Candidate Genes for Each Region
| Phenotype | Location | Gene | Start–End | N SNPs | SimGENE | VEGASGENE | P-valueGENE | SimTop10% | VEGASTop10% | P-valueTop10% | Best SNP | Position BP | P-valueSNP |
|
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPS | 1q32.1 | KISS1 | 204159468–204165619 | 301 | 1.00E+05 | 884.125 | 2.740E-03 | 1.00E+07 | 569.956 | 1.140E-05 | rs7513165 | 204147186 | 3.37E-07 | 0.965% | 0.642% | ||
| 8q24.13 | FBXO32 | 124510126–124553493 | 464 | 1.00E+07 | 1527.036 | 9.220E-05 | 1.00E+07 | 632.226 | 9.120E-05 | rs61510724 | 124568658 | 2.67E-07 | 0.928% | 0.628% | |||
| 9p21.3 | CDKN2B-AS1 | 21994789–22121093 | 404 | 1.00E+07 | 2512.052 | 5.400E-06 | 1.00E+07 | 820.222 | 6.300E-06 | rs3217992 | 22003223 | 6.57E-07 | 0.605% | 0.604% | |||
| 9p21.3 | CDKN2B | 22002901–22009312 | 194 | 1.00E+07 | 1400.944 | 2.280E-05 | 1.00E+07 | 383.383 | 1.290E-05 | rs3217992 | 22003223 | 6.57E-07 | 0.632% | 0.604% | |||
| 12p13.33 | CACNA1C | 2162415–2807115 | 1814 | 1.00E+05 | 3722.052 | 2.530E-03 | 1.00E+07 | 2624.117 | 2.040E-05 | rs34382810 | 2340798 | 9.98E-07 | 0.756% | 0.612% | |||
| 13q32.1 | CLDN10 | 96085852–96232010 | 816 | 1.00E+04 | 1819.670 | 1.050E-02 | 1.00E+07 | 940.510 | 7.477E-04 | rs17235257 | 96071345 | 1.20E-06 | 0.895% | 0.600% | |||
| HPS–PAS | 1q32.1 | KISS1 | 204159468–204165619 | 301 | 1.00E+05 | 783.959 | 6.290E-03 | 1.00E+07 | 485.473 | 5.890E-05 | rs7513165 | 204147186 | 1.64E-06 | 0.965%/>0.001% | 0.642%/0.020% | ||
| 8q24.13 | FBXO32 | 124510126–124553493 | 464 | 1.00E+07 | 1291.855 | 7.074E-04 | 1.00E+07 | 539.489 | 3.641E-04 | rs61510724 | 124568658 | 1.41E-06 | 0.928%/>0.001% | 0.628%/0.019% | |||
| 9p21.3 | CDKN2B-AS1 | 21994789–22121093 | 404 | 1.00E+07 | 2196.735 | 2.580E-05 | 1.00E+07 | 715.374 | 2.790E-05 | rs3217992 | 22003223 | 3.98E-06 | 0.605%/0.211% | 0.604%/0.117% | |||
| 9p21.3 | CDKN2B | 22002901–22009312 | 194 | 1.00E+07 | 1219.809 | 9.330E-05 | 1.00E+07 | 340.837 | 4.630E-05 | rs3217992 | 22003223 | 3.98E-06 | 0.632%/0.173% | 0.604%/0.117% | |||
| 13q12.2 | MTIF3 | 28009775–28024739 | 447 | 1.00E+05 | 1231.000 | 7.810E-03 | 1.00E+05 | 510.123 | 1.110E-03 | rs9581857 | 28027714 | 7.49E-07 | >0.001%/0.188% | 0.000%/0.535% | |||
| 17q23.2 | MRC2 | 60704761–60770962 | 250 | 1.00E+05 | 694.994 | 6.170E-03 | 1.00E+07 | 437.334 | 5.190E-05 | chr17:60762523:D | 60762523 | 3.07E-06 | 0.302%/0.314% | 0.130%/0.643% | |||
| HPS–PHAS | 1q32.1 | KISS1 | 204159468–204165619 | 301 | 1.00E+05 | 778.313 | 6.470E-03 | 1.00E+07 | 483.014 | 7.110E-05 | rs7513165 | 204147186 | 1.89E-06 | 0.965%/0.012% | 0.642%/>0.001% | ||
| 2q31.2 | CCDC141 | 179694483–179914786 | 1008 | 1.00E+05 | 2765.664 | 1.340E-03 | 1.00E+07 | 1187.733 | 4.911E-04 | rs16866587 | 179863363 | 4.40E-06 | 0.019%/1.002% | 0.103%/0.559% | |||
| 6q22.32 | TRMT11 | 126307575–126360420 | 128 | 1.00E+03 | 305.742 | 7.193E-02 | 1.00E+07 | 137.469 | 8.470E-04 | rs141259875 | 126387618 | 4.30E-06 | 0.157%/1.002% | 0.045%/0.449% | |||
| 8q24.13 | FBXO32 | 124510126–124553493 | 464 | 1.00E+07 | 1367.177 | 3.721E-04 | 1.00E+07 | 514.114 | 5.397E-04 | rs61510724 | 124568658 | 1.55E-06 | 0.928%/0.187% | 0.628%/0.034% | |||
| 9p21.3 | CDKN2B-AS1 | 21994789–22121093 | 404 | 1.00E+07 | 2185.916 | 2.860E-05 | 1.00E+07 | 706.810 | 3.150E-05 | rs3217992 | 22003223 | 3.66E-06 | 0.605%/0.178% | 0.604%/0.040% | |||
| 9p21.3 | CDKN2B | 22002901–22009312 | 194 | 1.00E+07 | 1263.299 | 6.590E-05 | 1.00E+07 | 343.480 | 4.220E-05 | rs3217992 | 22003223 | 3.66E-06 | 0.632%/0.174% | 0.604%/0.040% | |||
| 12p13.33 | CACNA1C | 2162415–2807115 | 1814 | 1.00E+05 | 3478.389 | 4.930E-03 | 1.00E+07 | 2267.963 | 8.250E-05 | rs34382810 | 2340798 | 2.44E-06 | 0.756%/0.001% | 0.612%/>0.001% | |||
| HPS–SAS | 1q32.1 | KISS1 | 204159468–204165619 | 301 | 1.00E+05 | 767.543 | 7.320E-03 | 1.00E+07 | 478.031 | 7.680E-05 | rs7513165 | 204147186 | 2.01E-06 | 0.965%/0.044% | 0.642%/>0.001% | ||
| 3p22.3 | CCR4 | 32993065–32996403 | 343 | 1.00E+04 | 1031.097 | 1.130E-02 | 1.00E+07 | 588.986 | 3.880E-05 | rs11919880 | 32962051 | 2.07E-06 | >0.001%/0.455% | 0.036%/0.587% | |||
| 8q24.13 | FBXO32 | 124510126–124553493 | 464 | 1.00E+07 | 1469.779 | 1.457E-04 | 1.00E+07 | 587.502 | 1.748E-04 | rs61510724 | 124568658 | 1.96E-07 | 0.928%/0.119% | 0.628%/0.063% | |||
| 9p21.3 | CDKN2B-AS1 | 21994789–22121093 | 404 | 1.00E+07 | 2032.304 | 6.370E-05 | 1.00E+07 | 684.368 | 4.280E-05 | rs3217992 | 22003223 | 4.39E-06 | 0.605%/>0.001% | 0.604%/>0.001% | |||
| 9p21.3 | CDKN2B | 22002901–22009312 | 194 | 1.00E+07 | 1134.339 | 1.831E-04 | 1.00E+07 | 319.903 | 8.850E-05 | rs3217992 | 22003223 | 4.39E-06 | 0.632%/>0.001% | 0.604%/>0.001% | |||
| 20p12.2 | SNAP25 | 10199476–10288066 | 438 | 1.00E+05 | 1073.322 | 2.990E-03 | 1.00E+07 | 531.011 | 3.029E-04 | rs6108491 | 10324253 | 2.81E-06 | 0.729%/0.386% | 0.420%/0.148% | |||
| PAS | 1p36.12 | VWA5B1 | 20617411–20681387 | 552 | 1.00E+04 | 1101.382 | 2.810E-02 | 1.00E+05 | 618.233 | 1.120E-03 | rs10916772 | 20677197 | 4.10E-06 | 0.364% | 0.553% | ||
| 12q24.33 | MMP17 | 132312940–132336316 | 299 | 1.00E+04 | 560.701 | 2.790E-02 | 1.00E+05 | 334.614 | 1.080E-03 | rs11246807 | 132275765 | 3.01E-06 | 0.806% | 0.538% | |||
| 16p13.11 | ABCC1 | 16043433–16236930 | 742 | 1.00E+04 | 1407.935 | 2.010E-02 | 1.00E+07 | 806.508 | 3.965E-04 | rs9931487 | 16204822 | 3.72E-06 | 0.448% | 0.546% | |||
| 17q23.2 | MRC2 | 60704761–60770962 | 250 | 1.00E+05 | 731.145 | 4.560E-03 | 1.00E+07 | 494.472 | 1.340E-05 | chr17:60762523:D | 60762523 | 6.71E-07 | 0.314% | 0.643% | |||
| PAS–PHAS | 8p11.22 | ADAM18 | 39442086–39587583 | 524 | 1.00E+05 | 1508.380 | 5.840E-03 | 1.00E+07 | 673.844 | 4.824E-04 | chr8:39394311:D | 39394311 | 3.45E-06 | 0.111%/0.335% | 0.205%/0.421% | ||
| 17q23.2 | MRC2 | 60704761–60770962 | 250 | 1.00E+04 | 657.311 | 1.080E-02 | 1.00E+07 | 443.403 | 4.150E-05 | chr17:60762523:D | 60762523 | 2.52E-06 | 0.314%/0.104% | 0.643%/>0.001% | |||
| PAS–SAS | 2q31.3 | CERKL | 182401400–182521834 | 533 | 1.00E+05 | 1673.159 | 4.770E-03 | 1.00E+05 | 580.911 | 1.950E-03 | rs13392054 | 182502438 | 3.76E-06 | 0.301%/0.330% | 0.347%/0.379% | ||
| 3p22.3 | CCR4 | 32993065–32996403 | 343 | 1.00E+05 | 1109.915 | 7.740E-03 | 1.00E+07 | 659.673 | 1.220E-05 | rs11919880 | 32962051 | 6.12E-07 | 0.085%/0.455% | 0.211%/0.587% | |||
| 17q23.2 | MRC2 | 60704761–60770962 | 250 | 1.00E+04 | 638.890 | 9.399E-03 | 1.00E+07 | 423.914 | 7.080E-05 | chr17:60762523:D | 60762523 | 2.93E-06 | 0.314%/>0.001% | 0.643%/>0.001% | |||
| PHAS | 2q31.2 | CCDC141 | 179694483–179914786 | 1008 | 1.00E+07 | 3050.673 | 4.887E-04 | 1.00E+07 | 1429.163 | 1.130E-04 | rs16866587 | 179863363 | 1.62E-06 | 1.002% | 0.559% | ||
| PHAS–SAS | 3p22.3 | CCR4 | 32993065–32996403 | 343 | 1.00E+05 | 1070.718 | 9.190E-03 | 1.00E+07 | 542.552 | 8.330E-05 | rs11919880 | 32962051 | 4.18E-06 | 0.074%/0.455% | 0.025%/0.587% | ||
| 11p13 | SLC1A2 | 35272751–35441610 | 772 | 1.00E+07 | 2926.337 | 2.665E-04 | 1.00E+07 | 1261.179 | 4.410E-05 | rs12804343 | 35244457 | 4.56E-06 | 0.148%/0.255% | 0.291%/0.050% | |||
| SAS | 3p22.3 | CCR4 | 32993065–32996403 | 343 | 1.00E+05 | 1217.351 | 4.960E-03 | 1.00E+07 | 653.640 | 1.520E-05 | rs11919880 | 32962051 | 6.90E-07 | 0.455% | 0.587% | ||
| 8p23.2 | CSMD1 | 2792874–4852328 | 14 485 | 1.00E+03 | 15907.856 | 2.478E-01 | 1.00E+05 | 7713.277 | 3.580E-03 | rs4518695 | 3568201 | 3.72E-06 | 0.490% | 0.521% | |||
| 12q14.3 | CAND1 | 67663060–67708388 | 485 | 1.00E+07 | 2644.596 | 1.838E-04 | 1.00E+07 | 736.263 | 1.900E-04 | chr12:67741435:I | 67741435 | 4.89E-06 | 0.289% | 0.522% |
Note: PAS, Perceptual Aberration Scale; HPS, Hypomanic Personality Scale; SAS, Revised Social Anhedonia Scale; PHAS, Revised Physical Anhedonia Scale. Start-End BP refers to base-pair position for the gene start and end (NCBI human genome assembly 37, GRCh37); NSNP refers to the number of SNPs mapped to the gene region and used in gene-based association tests; SimGENE indicates the number of simulations reached during the Vegas test for all markers mapped within the gene; SimTop10% indicates the number of simulations reached during the Vegas test for the top 10% most relevant markers; VEGASGENE indicates the VEGAS statistic for the test all SNPs mapped within the gene; VEGASTop10% indicate the VEGAS statistic for the test the top 10% most relevant SNPs within the gene; P-valueGENE refers to the P-value obtained in VEGAS tests all SNPs mapped within the gene; P-valueTop10% refers to the P-value obtained in VEGAS tests including the top 10% most relevant SNPs within the gene (significant P-values after Bonferroni correction are in bold); Position BP refers to the chromosomal base-pair position of the most relevant marker within the gene tested; P-valueSNP refers to the P-value obtained in the corresponding univariate or bivariate SNP-based test; refers to the heritability (genetic variance) explained by the gene region evaluated in VEGAS tests, using GCTA methods; refers to the heritability (genetic variance) explained by the best SNP within the gene evaluated in VEGAS tests.
The bivariate GWAs analyses suggested a further SNP with a borderline genome-wide significant P-value (rs188320715; P-value = 5.261 × 10–8) to HPS–PHAS. This SNP is located within an intergenic region at 6q25.3 with the nearest gene being ARID1B, ~572 kb upstream (table 2 and supplementary figure 2). A total of 263 SNPs showed a suggestive P-value < 5 × 10–6 and they were mapped in or near to 125 different genes (HPS–PAS: 32 genes; HPS–PHAS: 57 genes; HPS–SAS: 40 genes; PAS–PHAS: 33 genes; PAS–SAS: 18 genes; PHAS–SAS: 38 genes). No signs of genomic inflation were detected (inflation factor λ = 1.01–1.03, supplementary figure 3).
The subsequent gene-based analyses showed that genes at 1q32.1, 8q24.13, and 9p21.3 primarily associated to HPS also had residual effects on PAS, PHAS and SAS. Similarly, the gene CACNA1C (12p13.33) primarily associated to HPS also had residual effects on PHAS, MRC2 (17q23.2) associated with PAS also had effects on HPS, PHAS, and SAS; while CCR4 (3p22.3) associated to SAS also had effects on PHAS. These analyses revealed additional new loci harboring significant genes that passed undetected to all previous tests: SNAP25 (20p12.2) and ADAM18 (8p11.22) associated to both HPS and SAS; CERKL (2q31.3) to both PAS and SAS; and SLC1A2 (11p13) to both PHAS and SAS (table 3, and supplementary figures 2, 4, and 5).
Discussion
The initial part of this study provides heritability estimates for traits that relate to psychosis proneness, based on the analysis of ~9.6 million autosomal SNPs available from a large sample of unrelated individuals without overt psychotic disorders. It was earlier hypothesized that estimates obtained through this method would be lower than those detected in phenotypic analyses of twin samples.21 Our results provide support to this hypothesis as the estimates here are either similar or lower than those from previous twin studies with matching psychometric instruments (current vs earlier): HPS: 27.4% vs 28%; PAS: 16.6% vs 33%–49%; PHAS: 26.6% vs 36%–57%; and SAS: 20.4% vs 45%–67%.13,14,20 Accordingly, they constitute an upper estimate of genetic influences possible to identify by current genome wide association methods.21
Other results in this study provide further relevant conclusions. First, by employing multiple continuous measures of psychosis proneness and utilizing statistical methods that allowed scrutinizing genomic regions of interest, we were able to identify a series of genetic loci using a much smaller sample size than has been required to pinpoint most loci in earlier psychiatric case-control GWAs settings. These loci support some earlier findings from case based GWAs studies, but also provide novel insights on the molecular pathways potentially contributing to the development of clinical psychosis, such as schizophrenia or bipolar disorder, as well as other behavioral problems usually associated with psychoses proneness (eg, substance abuse,47 post-traumatic stress disorder,48 or some personality disorders49). For example, the locus at 1q32.1 showed multiple genes with significant P-values in gene-based association tests involving HPS. A detailed examination of the LD structure at this locus in relation to the leading SNP suggested that KISS1, a gene highly expressed in different brain regions is likely to be the candidate gene for the locus and explains a meaningful 3.5% of the trait heritability (2.3% accounted for by the leading SNP in the locus alone). The neuropeptide coded by this gene (kisspeptin) modulates neuronal calcium homeostasis in hypothalamic neurons involved in the production of gonadotropin-release hormone and initiates the hypothalamic–pituitary–gonadal axis.50–52 Importantly, studies have recognized that gonadotropin alterations may prompt mood and behavioral changes.53,54 Hypothalamic-pituitary dysfunctions have been earlier reported in relation to nonclinical psychotic feelings and experiences,55 and are also characteristic of schizophrenia patients.56–58 This finding thus provides preliminary molecular understanding on observations detected in earlier clinical studies among both psychiatrically healthy and ill populations.
Second, it is noticeable that our analyses also detected a series of genes previously linked to some extent either to schizophrenia alone or to both schizophrenia and bipolar disorder (CSMD1, CCDC141, SLC1A2, CACNA1C, and SNAP25).7,8,59–61 These genes accounted here for 2.8% of the heritability of HPS, 3.8% of PHAS and 1.8% of SAS. Particularly noteworthy is the effect on PHAS of CCDC141 alone (3.8% of the heritability), a gene that interacts directly with the schizophrenia candidate gene DISC1,61 and had been identified previously in this cohort in a GWAs conditioned on DISC1 genotypes.24 Altogether, these findings have important implications as they provide suggestive evidence of a genomic link between psychosis proneness in healthy adults and schizophrenia/bipolar disorder.
Previous positive evidence on this genomic link is limited. The study by Sieradzka et al investigated a large community sample of 2152 16-year-olds, utilizing the Specific Psychotic Experiences Questionnaire in relation to polygenic risk scores for schizophrenia and bipolar disorder, as well as 28 individual SNPs previously associated with schizophrenia, and reported 2 SNPs at the TCF4 gene significantly associated with the paranoia subscale.25 Smaller-sized candidate gene studies have also reported positive findings for some markers at the COMT gene associated with psychosis proneness scores in healthy samples of European and Chinese origin.26,62,63
Our results here provide preliminary support to the idea that a biological susceptibility to disorders such as schizophrenia or bipolar disorder may still be partly found even in individuals without a psychosis diagnosis. Still, observational studies have shown that very few of the persons who score high on psychosis proneness scales go on to develop psychotic disorders,64 with subclinical negative psychotic symptoms (eg, physical and social anhedonia) carrying stronger predictive power than others.65,66 Altogether this would imply that only those individuals most susceptible, who were further affected by key genetic and/or environmental influences (eg, an increased burden of large and rare chromosomal abnormalities67), would finally develop those diseases. Further evidence on this respect is nevertheless necessary.
A series of methodological considerations need to be acknowledged here. This study ensues from earlier recommendations on psychiatric genetics encouraging new investigations to explore approaches in which traits, rather than being classified as dichotomized entities, are deconstructed into a spectrum of lower-order, highly characterized set of traits with clinical relevance.68 This would initiate a dynamic process, in which refined phenotypes would allow pinpointing of new genetic signals and in turn generating further phenotype refinements leading to even lower-order traits.69 Considering the findings in this study, in which the identified loci explained up to 14.1% of the estimated trait heritability, we believe that our methodological approach was indeed an advantage. The results reported here from an array of inter-related and carefully characterized traits represent only the first step in the genetic dissection of lower-order, high-resolution phenotypes regarding psychosis proneness, and new studies considering further phenotypic refinements are warranted to discover additional genomic regions of importance.
Due to the genomic link that our data suggested between psychosis proneness and schizophrenia/bipolar disorder, we believe that evaluating psychosis proneness even in nonpsychotic individuals is an advisable strategy to understand part of the genetic basis of severe mental disorders with psychotic components. This conclusion however is in contrast to that of Zammit et al70 from an earlier GWAs study evaluating psychosis proneness in 3483 nonpsychotic adolescents. That previous study did not pinpoint any markers associated with psychotic-like experiences, and variants known to be associated with schizophrenia were also far from significant. The authors concluded that psychosis proneness in population-based samples may not share a comparable genetic architecture to schizophrenia, and thus utilizing a broader more common phenotype of psychotic experiences may not be an efficient approach to increase understanding of the genetic etiology of schizophrenia. This discrepancy with our conclusion is likely to be accounted for by 2 key methodological differences between the studies. Firstly our investigation evaluated a significantly larger pool of genetic markers (~9.8 million here vs ~2.5 million there). This in turn may explain the greater capability of our investigation to detect important loci, as variants that are in high LD with causal ones would be better captured. Secondly, and most importantly, we investigated here psychosis proneness utilizing summary (quantitative) scales built upon an ample number of answers provided to an array of questionnaires dissecting psychosis proneness into specific psychotic features. However, Zammit et al carried out a pre hoc categorization on participants as “cases and controls” depending on whether they displayed any type of psychotic-like experience during adolescence. Given that this categorization resulted in 424 “cases” (or 12.2% of the participants) and 3057 “controls,” their statistical power to detect relevant loci was considerably lower compared to ours. To reach a reasonable statistical power similar to the one in our study (eg, ≥80%) with their strategy of categorizing individuals as “cases and controls,” at least 7307 cases and as many as 52 682 controls would be required to detect variants significant genome-wide (P < 5 × 10−8) with a moderate effect (relative risk ≥ 1.5), not rare in the population (risk allele frequency ≥ 0.01), assuming a 12.2% prevalence, and 7.21 cases-to-controls ratio (as provided by Zammit et al). This illustrates further the high implications of utilizing quantitative endophenotypes in psychiatric genetic studies of persons not reaching a positive clinical diagnosis of schizophrenia or bipolar disorder.
Finally, it should be acknowledged that our study investigated a sample of adult individuals, while the 2 already mentioned genetic studies on the topic to date (by Sieradzka et al, and Zammit et al) investigated adolescents. Despite psychosis proneness features being more common among adolescents, it has been suggested that when observed at that period it may constitute a developmental phenomenon, carrying a somewhat different connotation and weight than when observed in adulthood.71 It is very likely however, that most genetic influences involved in adolescence psychotic-like experiences may still be present during adulthood, although a considerable proportion of them may well be age-specific, as genetic studies on many other complex diseases seem to indicate. Further longitudinal studies to clearly determine transitory and persistent genetic and environmental causes are thus warranted. In addition, despite that our descriptive statistics showed significant sex differences within the psychoses proneness scores, our genomic tests were limited to sex-corrected data, as sex-stratified analyses to detect potential sex-heterogeneity of the genetic effects would have resulted in a substantial reduction of the statistical power of detection.
In summary, this study highlights the value of using population-based data without overt psychiatric disorders for genetic association analyses concerning predisposition to psychoses and their related psychiatric conditions. Our results confirm that the genetic predisposition to psychosis proneness is moderate for most traits evaluated. We provide further evidence for overlap in genetic risk between psychiatric endophenotypes related to both schizophrenia and bipolar disorder. We also found association of several of these traits with 5 loci previously reported in connection with schizophrenia and/or bipolar disorder, and provide initial evidence for 15 additional loci that had not previously been connected to any psychiatric trait.
Supplementary Material
Supplementary material is available at Schizophrenia Bulletin online.
Funding
The Academy of Finland supported this work (grant numbers 265097 and 259589). NFBC1966 received financial support also from the Academy of Finland (project grants [104781, 120315, 129269, 268336, 1114194], Center of Excellence in Complex Disease Genetics and SALVE); University Hospital Oulu, Biocenter, University of Oulu, Finland (75617); the European Commission (EURO-BLCS, Framework 5 award [QLG1-CT-2000-01643]); National Heart, Lung and Blood Institute (NHLBI) grant (5R01HL087679-02) through the STAMPEED program (1RL1MH083268-01); National Institute of Health/ National Institute of Mental Health (5R01MH63706:02); ENGAGE project and grant agreement (HEALTH-F4-2007-201413); and the Medical Research Council, UK (G0500539, G0600705, PrevMetSyn/SALVE); EU Framework Programme 7 small-scale focused research collaborative project (EurHEALTHAgeing 277849).
Supplementary Material
Acknowledgments
Dr Nelson Freimer, Professor of Psychiatry at University of California, Los Angeles (United States), is gratefully acknowledged for his contribution to this study. The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Raine A. Schizotypal personality: neurodevelopmental and psychosocial trajectories. Annu Rev Clin Psychol. 2006;2:291–326. [DOI] [PubMed] [Google Scholar]
- 2. Binbay T, Drukker M, Elbi H, et al. Testing the psychosis continuum: differential impact of genetic and nongenetic risk factors and comorbid psychopathology across the entire spectrum of psychosis. Schizophr Bull. 2012;38:992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janssens M, Boyette LL, Heering HD, Bartels-Velthuis AA, Lataster T; Genetic Risk and Outcome of Psychosis Investigators Developmental course of subclinical positive and negative psychotic symptoms and their associations with genetic risk status and impairment. Schizophr Res. 2016;174:177–182. [DOI] [PubMed] [Google Scholar]
- 4. van Os J, Linscott RJ, Myin-Germeys I, Delespaul P, Krabbendam L. A systematic review and meta-analysis of the psychosis continuum: evidence for a psychosis proneness-persistence-impairment model of psychotic disorder. Psychol Med. 2009;39:179–195. [DOI] [PubMed] [Google Scholar]
- 5. Ingram RE, Luxton DD. Vulnerability-stress models. In: Hankin BL, Abela JRZ, eds. Development of Psychopathology: A Vulnerability-Stress Perspective. Thousand Oaks, CA: Sage Publications; 2005:510. [Google Scholar]
- 6. Tienari P, Wynne LC, Sorri A, et al. Genotype-environment interaction in schizophrenia-spectrum disorder. Long-term follow-up study of Finnish adoptees. Br J Psychiatry. 2004;184:216–222. [DOI] [PubMed] [Google Scholar]
- 7. Ripke S, O’Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MacRae CA, Vasan RS. Next-generation genome-wide association studies: time to focus on phenotype? Circ Cardiovasc Genet. 2011;4:334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shifman S, Kuypers J, Kokoris M, Yakir B, Darvasi A. Linkage disequilibrium patterns of the human genome across populations. Hum Mol Genet. 2003;12:771–776. [DOI] [PubMed] [Google Scholar]
- 11. Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28:309–310. [DOI] [PubMed] [Google Scholar]
- 12. Sarig O, Bercovici S, Zoller L, et al. Population-specific association between a polymorphic variant in ST18, encoding a pro-apoptotic molecule, and pemphigus vulgaris. J Invest Dermatol. 2012;132:1798–1805. [DOI] [PubMed] [Google Scholar]
- 13. Kendler KS, Hewitt J. The structure of self-report schizotypy in twins. J Pers Disord. 1992;6:1–17. [Google Scholar]
- 14. Lin CC, Su CH, Kuo PH, Hsiao CK, Soong WT, Chen WJ. Genetic and environmental influences on schizotypy among adolescents in Taiwan: a multivariate twin/sibling analysis. Behav Genet. 2007;37:334–344. [DOI] [PubMed] [Google Scholar]
- 15. Linney YM, Murray RM, Peters ER, MacDonald AM, Rijsdijk F, Sham PC. A quantitative genetic analysis of schizotypal personality traits. Psychol Med. 2003;33:803–816. [DOI] [PubMed] [Google Scholar]
- 16. Ericson M, Tuvblad C, Raine A, Young-Wolff K, Baker LA. Heritability and longitudinal stability of schizotypal traits during adolescence. Behav Genet. 2011;41:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bogdan R, Pizzagalli DA. The heritability of hedonic capacity and perceived stress: a twin study evaluation of candidate depressive phenotypes. Psychol Med. 2009;39:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Grootheest DS, Boomsma DI, Hettema JM, Kendler KS. Heritability of obsessive-compulsive symptom dimensions. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:473–478. [DOI] [PubMed] [Google Scholar]
- 19. Zavos HM, Freeman D, Haworth CM, et al. Consistent etiology of severe, frequent psychotic experiences and milder, less frequent manifestations: a twin study of specific psychotic experiences in adolescence. JAMA Psychiatry. 2014;71:1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hay DA, Martin NG, Foley D, Treloar SA, Kirk KM, Heath AC. Phenotypic and genetic analyses of a short measure of psychosis-proneness in a large-scale Australian twin study. Twin Res. 2001;4:30–40. [DOI] [PubMed] [Google Scholar]
- 21. Visscher PM, Yang J, Goddard ME. A commentary on “common SNPs explain a large proportion of the heritability for human height” by Yang et al. (2010). Twin Res Hum Genet. 2010;13:517–524. [DOI] [PubMed] [Google Scholar]
- 22. Sieradzka D, Power RA, Freeman D, Cardno AG, Dudbridge F, Ronald A. Heritability of individual psychotic experiences captured by common genetic variants in a community sample of adolescents. Behav Genet. 2015;45:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomppo L, Hennah W, Miettunen J, et al. Association of variants in DISC1 with psychosis-related traits in a large population cohort. Arch Gen Psychiatry. 2009;66:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomppo L, Ekelund J, Lichtermann D, Veijola J, Järvelin MR, Hennah W. DISC1 conditioned GWAS for psychosis proneness in a large Finnish birth cohort. PLoS One. 2012;7:e30643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sieradzka D, Power RA, Freeman D, et al. Are genetic risk factors for psychosis also associated with dimension-specific psychotic experiences in adolescence? PLoS One. 2014;9:e94398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma X, Sun J, Yao J, et al. A quantitative association study between schizotypal traits and COMT, PRODH and BDNF genes in a healthy Chinese population. Psychiatry Res. 2007;153:7–15. [DOI] [PubMed] [Google Scholar]
- 27. Haapea M, Miettunen J, Läärä E, et al. Non-participation in a field survey with respect to psychiatric disorders. Scand J Public Health. 2008;36:728–736. [DOI] [PubMed] [Google Scholar]
- 28. Chapman LJ, Chapman JP, Raulin ML. Body-image aberration in Schizophrenia. J Abnorm Psychol. 1978;87:399–407. [DOI] [PubMed] [Google Scholar]
- 29. Eckblad M, Chapman LJ. Development and validation of a scale for hypomanic personality. J Abnorm Psychol. 1986;95:214–222. [DOI] [PubMed] [Google Scholar]
- 30. Chapman LJ, Chapman JP, Raulin ML. Scales for physical and social anhedonia. J Abnorm Psychol. 1976;85:374–382. [DOI] [PubMed] [Google Scholar]
- 31. Golden RR, Meehl PE. Detection of the schizoid taxon with MMPI indicators. J Abnorm Psychol. 1979;88:217–233. [DOI] [PubMed] [Google Scholar]
- 32. Bailey B, West KY, Widiger TA, Freiman K. The convergent and discriminant validity of the Chapman Scales. J Pers Assess. 1993;61:121–135. [DOI] [PubMed] [Google Scholar]
- 33. Chapman LJ, Chapman JP, Kwapil TR, Eckblad M, Zinser MC. Putatively psychosis-prone subjects 10 years later. J Abnorm Psychol. 1994;103:171–183. [DOI] [PubMed] [Google Scholar]
- 34. Winterstein BP, Willse JT, Kwapil TR, Silvia PJ. Assessment of score dependability of the Wisconsin Schizotypy Scales Using generalizability analysis. J Psychopathol Behav. 2010;32:575–585. [Google Scholar]
- 35. Chan RC, Shi HS, Geng FL, et al. The Chapman psychosis-proneness scales: consistency across culture and time. Psychiatry Res. 2015;228:143–149. [DOI] [PubMed] [Google Scholar]
- 36. Miettunen J, Veijola J, Isohanni M, et al. Identifying schizophrenia and other psychoses with psychological scales in the general population. J Nerv Ment Dis. 2011;199:230–238. [DOI] [PubMed] [Google Scholar]
- 37. Roivainen E, Veijola J, Miettunen J. Careless responses in survey data and the validity of a screening instrument. Nordic Psychology. 2016;68:114–123. [Google Scholar]
- 38. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. [DOI] [PubMed] [Google Scholar]
- 39. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferreira MA, Purcell SM. A multivariate test of association. Bioinformatics. 2009;25:132–133. [DOI] [PubMed] [Google Scholar]
- 41. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. [DOI] [PubMed] [Google Scholar]
- 43. Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. Am J Hum Genet. 2000;66:1616–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18:86–91. [DOI] [PubMed] [Google Scholar]
- 45. Liu JZ, McRae AF, Nyholt DR, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Institute for Molecular Medicine Finland (FIMM); University of Helsinki (Finland). Sequencing Initiative Suomi project (SISu) http://sisuproject.fi. Accessed December 2015.
- 47. Kwapil TR. A longitudinal study of drug and alcohol use by psychosis-prone and impulsive-nonconforming individuals. J Abnorm Psychol. 1996;105:114–123. [DOI] [PubMed] [Google Scholar]
- 48. Spauwen J, Krabbendam L, Lieb R, Wittchen HU, van Os J. Impact of psychological trauma on the development of psychotic symptoms: relationship with psychosis proneness. Br J Psychiatry. 2006;188:527–533. [DOI] [PubMed] [Google Scholar]
- 49. Meyer TD, Hautzinger M. Prediction of personality disorder traits by psychosis proneness scales in a German sample of young adults. J Clin Psychol. 2002;58:1091–1101. [DOI] [PubMed] [Google Scholar]
- 50. Liu PY, Baker HW, Jayadev V, Zacharin M, Conway AJ, Handelsman DJ. Induction of spermatogenesis and fertility during gonadotropin treatment of gonadotropin-deficient infertile men: predictors of fertility outcome. J Clin Endocrinol Metab. 2009;94:801–808. [DOI] [PubMed] [Google Scholar]
- 51. Liu X, Lee K, Herbison AE. Kisspeptin excites gonadotropin-releasing hormone neurons through a phospholipase C/calcium-dependent pathway regulating multiple ion channels. Endocrinology. 2008;149:4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dhillo WS, Murphy KG, Bloom SR. The neuroendocrine physiology of kisspeptin in the human. Rev Endocr Metab Disord. 2007;8:41–46. [DOI] [PubMed] [Google Scholar]
- 53. Loosen PT, Purdon SE, Pavlou SN. Effects on behavior of modulation of gonadal function in men with gonadotropin-releasing hormone antagonists. Am J Psychiatry. 1994;151:271–273. [DOI] [PubMed] [Google Scholar]
- 54. McAdoo BC, Doering CH, Kraemer HC, Dessert N, Brodie HK, Hamburg DA. A study of the effects of gonadotropin-releasing hormone on human mood and behavior. Psychosom Med. 1978;40:199–209. [DOI] [PubMed] [Google Scholar]
- 55. Mittal VA, Orr JM, Pelletier A, Dean DJ, Smith A, Lunsford-Avery J. Hypothalamic-pituitary-adrenal axis dysfunction in non-clinical psychosis. Psychiatry Res. 2013;206:315–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Akhondzadeh S, Rezaei F, Larijani B, Nejatisafa AA, Kashani L, Abbasi SH. Correlation between testosterone, gonadotropins and prolactin and severity of negative symptoms in male patients with chronic schizophrenia. Schizophr Res. 2006;84:405–410. [DOI] [PubMed] [Google Scholar]
- 57. Ferrier IN, Cotes PM, Crow TJ, Johnstone EC. Gonadotropin secretion abnormalities in chronic schizophrenia. Psychol Med. 1982;12:263–273. [DOI] [PubMed] [Google Scholar]
- 58. Bradley AJ, Dinan TG. A systematic review of hypothalamic-pituitary-adrenal axis function in schizophrenia: implications for mortality. J Psychopharmacol. 2010;24:91–118. [DOI] [PubMed] [Google Scholar]
- 59. Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deng X, Shibata H, Ninomiya H, et al. Association study of polymorphisms in the excitatory amino acid transporter 2 gene (SLC1A2) with schizophrenia. BMC Psychiatry. 2004;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fukuda T, Sugita S, Inatome R, Yanagi S. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J Biol Chem. 2010;285:40554–40561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de Castro-Catala M, Barrantes-Vidal N, Sheinbaum T, Moreno-Fortuny A, Kwapil TR, Rosa A. COMT-by-sex interaction effect on psychosis proneness. Biomed Res Int. 2015;2015:829237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Avramopoulos D, Stefanis NC, Hantoumi I, Smyrnis N, Evdokimidis I, Stefanis CN. Higher scores of self reported schizotypy in healthy young males carrying the COMT high activity allele. Mol Psychiatry. 2002;7:706–711. [DOI] [PubMed] [Google Scholar]
- 64. Chapman JP, Chapman LJ, Kwapil TR. Does the Eysenck Psychoticism Scale predict psychosis - a 10-year longitudinal-study. Pers Indiv Differ. 1994;17:369–375. [Google Scholar]
- 65. Valmaggia LR, Stahl D, Yung AR, et al. Negative psychotic symptoms and impaired role functioning predict transition outcomes in the at-risk mental state: a latent class cluster analysis study. Psychol Med. 2013;43:2311–2325. [DOI] [PubMed] [Google Scholar]
- 66. Demjaha A, Valmaggia L, Stahl D, Byrne M, McGuire P. Disorganization/cognitive and negative symptom dimensions in the at-risk mental state predict subsequent transition to psychosis. Schizophr Bull. 2012;38:351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. International Schizophrenia C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Craddock N, Owen MJ. The beginning of the end for the Kraepelinian dichotomy. Br J Psychiatry. 2005;186:364–366. [DOI] [PubMed] [Google Scholar]
- 69. Owen MJ, Craddock N, Jablensky A. The genetic deconstruction of psychosis. Schizophr Bull. 2007;33:905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zammit S, Hamshere M, Dwyer S, et al. A population-based study of genetic variation and psychotic experiences in adolescents. Schizophr Bull. 2014;40:1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Verdoux H, van Os J, Maurice-Tison S, Gay B, Salamon R, Bourgeois M. Is early adulthood a critical developmental stage for psychosis proneness? A survey of delusional ideation in normal subjects. Schizophr Res. 1998;29:247–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.