Abstract
Long noncoding RNAs (lncRNAs) are an emerging class of regulators involved in a myriad of biological processes. Recent studies have revealed that many lncRNAs play pivotal roles in regulating adipocyte development. Due to the prevalence of obesity and the serious effects of adiposity on human health and society development, it is necessary to summarize functions and recent advances of lncRNAs in adipogenesis. In this review, we highlight functional lncRNAs contributed to the regulation of adipogenesis, discussing their potential use as therapeutic targets to combat human obesity.
Keywords: ncRNAs, Adipose tissue, Beige cells, SRA, HOTAIR, NEAT1, Blnc1
Introduction
Obesity refers to a condition of abnormal or excessive fat accumulation that is harmful to human health. Over the last several decades, the population of obese individuals has rapidly increased not only in developed countries but also in developing countries [1, 2]. According to data from the World Health Organization (WHO) in 2014, the prevalence of obesity worldwide more than doubled since 1980, and over 13 % of adults in the world were obese, with the body mass index (BMI) greater than or equal to 30.0. Although there is disagreement on the obesity definition and BMI classification method [3, 4], it is clear that obesity is a high risk factor for several serious human diseases, such as type 2 diabetes, cardiovascular disease, and certain types of cancers [5, 6]. Up to now, despite great progress in our understanding of adipose biology, an effective method for preventing/treating obesity remains to be developed. Further understanding molecular mechanisms controlling adipogenesis is critical to identify new targets for combating obesity.
In general, there are two types of adipose tissue in human and other mammals, white adipose tissue (WAT) and brown adipose tissue (BAT). WAT is mainly involved in the storage and mobilization of energy in the form of triglycerides and secretes adipokines that influence systemic energy homeostasis. Differing from WAT, BAT dissipates energy and generates heat through UCP1 (uncoupling protein 1) mediated uncoupling of respiration from ATP synthesis, defending against hypothermia and obesity [7]. Moreover, BAT- and WAT- derived fat cells possess both common features as well as individual properties regarding their origins, morphologies, functions, and gene expression patterns. For example, both PPARγ (peroxisome proliferator-activated receptor gamma) and CEBPs (CCAAT/enhancer binding proteins) are the master regulators driving BAT- and WAT- adipogenesis, while UCP1, PGC1α (peroxisome proliferator-activated receptor gamma coactivator 1 alpha), EBF2 (early B cell factor 2), and PRDM16 (PR-domain containing protein 16) only function in the transcriptional cascade of brown adipogenesis [8–10]. Of note, except for classical white or brown adipocytes, recent data suggest that inducible brown-like adipocytes, also referred to as beige or brite adipocytes, emerge in WAT in response to various stimuli (such as cold or β-adrenergic stimulation) [11, 12]. Distinct from white or brown adipocytes, the beige cells have high mitochondrial contents but express low levels of UCP1 in the basal (unstimulated) state; upon stimulation, beige cells elevate UCP1 expression and turn on robust programs of respiration and energy expenditure that are similar to those of classic brown fat cells, contributing to thermogenesis and obesity prevention [12].
Despite important progress toward revealing molecular regulations of adipogenesis, our knowledge on the regulatory mechanism of adipose development is rudimentary. Recent investigations on adipose development suggest a significant number of long noncoding RNAs (lncRNAs) participate in the regulatory networks of adipogenesis and play a key role in regulating adipogenic commitment and differentiation [13, 14]. This review summarizes recent advances on the most prominent lncRNAs that regulate adipogenesis (Table 1), providing novel insights into possible solutions to prevent or treat obesity.
Table 1.
Direct roles of lncRNAs in regulating adipogenesis
| lncRNA | Characteristics | Cell type/model | Functions in adipogenesis | References |
|---|---|---|---|---|
| Functional white adipocyte regulators | ||||
| SRA | Polyadenylated; intergenic | 3T3–L1; mouse marrow-derived ST2 cells | Promotes preadipocyte differentiation partly via coactivation of PPARγ | [14, 36] |
| lnc-RAP-n (n = 1–10) | Polyadenylated | Primary white adipocytes from mice | Promotes white preadipocyte differentiation; lnc-RAP-1 functions via binding to hnRNP-U | [13, 39] |
| slincRAD | Non-polyadenylated; intergenic; primarily in the nucleus | 3T3–L1 | Promotes preadipocyte differentiation with unknown mechanisms | [45] |
| PU.1 AS | Antisense | 3T3–L1; primary porcine preadipocytes | Promotes adipogenesis through preventing PU.1 mRNA translation via binding to PU.1 mRNA | [42, 43] |
| HOTAIR | Antisense/intergenic | Primary preadipocytes from human | Promotes preadipocyte differentiation with unknown mechanisms | [34] |
| ADINR | Polyadenylated; bidirectional; exclusively in the nucleus | Human MSCs | Promotes adipogenesis by activating CEBPα | [46] |
| NEAT1 | Exclusively in the nucleus | Primary mouse preadipocytes | Promotes adipogenesis with unknown mechanisms | [50] |
| Functional brown adipocyte regulators | ||||
| Blnc1 (AK038898) | Polyadenylated; intergenic; primarily in the nucleus | Primary brown and beige preadipocytes from mice | Promotes brown and beige adipocyte differentiation and function via forming the EBF2 ribonucleoprotein complex | [33] |
| lnc-BATE-1 | Polyadenylated; intergenic; similarly abundant in the cytoplasm and nucleus | Primary brown adipocytes from mice | Promotes brown adipogenesis possibly via forming a functional ribonucleoprotein complex with hnRNP-U | [51] |
lncRNAs
An overview of lncRNAs
Traditionally, messenger RNA is considered to deliver information from DNA to protein [15]. However, the rapid development of sequencing technology has revealed RNA also plays important regulatory functions in various biological processes of cells [16]. In mammals, most of the genome is transcribed in a developmentally regulated pattern, yielding a complex network of overlapping transcripts [15, 17, 18]. The sequencing of the human genome indicated that there are only around 20,000–25,000 protein-coding genes, representing less than 2 % of the total genomic sequence; the vast majority of transcripts are non-protein coding RNAs [19, 20]. Generally, noncoding RNAs (ncRNAs) can be sorted into two classes: housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and are usually expressed constitutively, while regulatory RNAs consists of piwi-interacting RNAs (piRNAs), microRNAs (miRNAs), and lncRNAs [21, 22]. In addition to the better known housekeeping ncRNAs, regulatory ncRNAs especially lncRNAs have been recently established as functional regulators in many differentiation and development processes and aberrant functions of lncRNAs may lead to a wide range of human diseases, such as cancer, cardiovascular disease, neurodegenerative disease, and metabolic syndrome, etc. [15, 23, 24]. Moreover, an increasing number of lncRNAs have been identified to regulate adipose development [13, 14].
The discovery of lncRNAs
Transcribed from the genome but rarely possessing protein-coding sequences, lncRNAs are a newly discovered class of ncRNAs with greater than 200 nucleotides in length. Instead of accumulating silence in cells, an increasing number of lncRNAs were found to be closely related with human health. Actually, the first recognized lncRNA, imprinted H19, was reported in 1990 [25], which is preferentially expressed from the maternal allele and participates in embryogenesis and human carcinogenesis [26]. Shortly after the discovery of H19, the silencing X-inactive-specific transcript (XIST) was also brought to light [27]. However, the discovery of the first miRNA lin-14 and subsequent breakthroughs in the miRNA field rapidly drew the foucus of ncRNA research from lncRNAs to miRNAs [28]. lncRNA related research grew dramatically with the advent of whole transcriptome sequencing techniques. New technologies combined with the novel functional annotation of a few lncRNAs greatly accelerated lncRNA discovery and its functional exploration.
Characterization and roles of lncRNAs
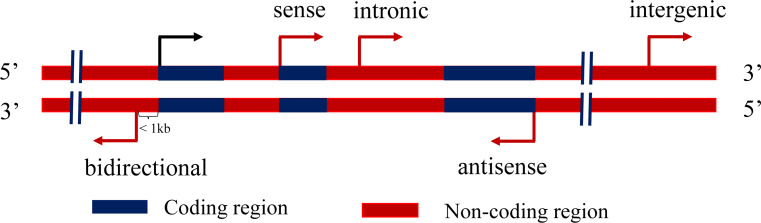
Generally, lncRNAs possess heterogeneous structures, may or may not be polyadenylated (polyA+ or polyA−; the polyA+ phenotype consists of multiple adenosine monophosphates at the 3′ end), and are located within the nuclear or cytosolic fractions. In addition, lncRNAs tend to be expressed at lower levels compared with mRNAs [29–31], and a high proportion of lncRNAs are preferentially expressed in specific tissues or cell lines [30, 32]. The expression of adipose lncRNAs is low and depot-specific [33, 34]. According to its position with respect to protein coding genes, a lncRNA can be classified into one or more of five categories [21]: sense, antisense, bidirectional, intronic, and intergenic (Fig. 1).
Fig. 1.
Categories of lncRNAs classified on the basis of their relatively locations to protein coding region. Black arrow represents the transcription initiation site and direction of a protein coding gene; red arrows represent positions and directions of five types of lncRNAs, including sense, antisense, bidirectional, intronic, and intergenic
Heterogeneity in both sequences and genomic location determine the diversity and complexity of lncRNA functions. Up to now, lncRNAs have been implicated in a wide range of biological processes, including epigenetic regulation, small RNA processing, pattern formation and development, transcriptional/post-transcriptional regulation, protein metabolism, and tumorigenesis [22, 35], as well as a recent discovery that lncRNA possess the ability to regulate adipogenesis [13]. Considering functions of lncRNAs in adipogenesis are poorly characterized, this review focused on the recent advances of lncRNA in adipogenesis, including their discovery, functions, and clinical value, in order to facilitate research in this emerging field.
lncRNAs in white adipogenesis
SRA
SRA (steroid receptor RNA activator) was initially identified as a lncRNA that functioned as a transcriptional coactivator of steroid receptors [36], and was subsequently found to interact directly with a large variety of proteins including non-steroid receptors [37]. A recent study revealed that adipose-enriched SRA was capable of binding to PPARγ and enhancing PPARγ transcriptional activity to promote 3T3–L1 preadipocyte differentiation [14], for the first time showing the involvement of lncRNAs in adipogenesis (Fig. 2). During 3T3–L1 preadipocyte differentiation, SRA expression was induced by around twofold; overexpression of SRA in ST2 mesenchymal precursor cells with differentiation induction media (DMI) promoted adipogenesis, with enhanced expressions of adipocyte master regulators PPARγ and CEBPα, as well as FABP4 (fatty acid binding protein 4) and AdipoQ (adiponectin) [14]. RNAi-mediated SRA loss-of-function showed opposite effects on 3T3–L1 cells, illustrating SRA as an essential pro-adipogenic regulator. Microarray analysis revealed that SRA regulated various cellular processes in adipocytes, including cell cycle and insulin-related signal transduction pathways, suggesting SRA might enhance adipogenesis and adipocyte function through multiple pathways [14].
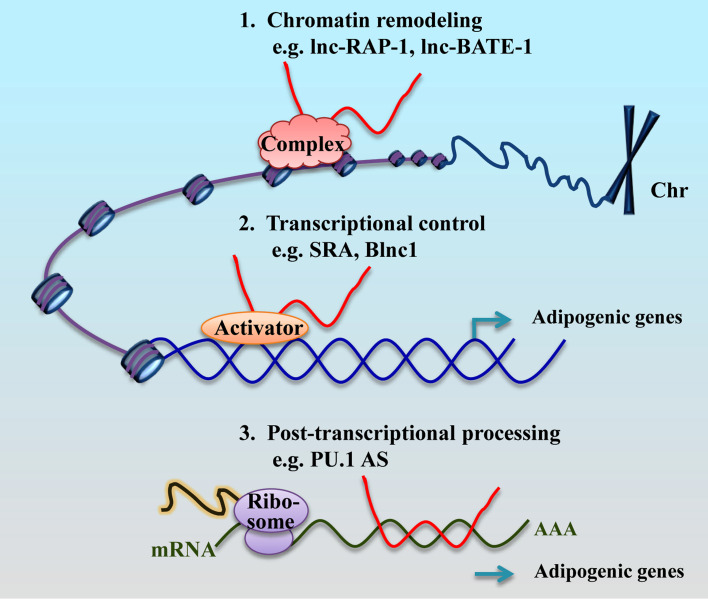
Fig. 2.
Functions of lncRNAs in adipogenesis. lncRNAs participate in chromatin remodeling, transcriptional control, and post-transcriptional processing in cells to regulate adipogenesis. For example, lnc-RAP-1 or lnc-BATE-1 participates in chromatin remodeling via binding hnRNP-U; SRA acts as a coactivator of adipogenic transcriptional factors to enhance PPARγ transcriptional activity; PU.1 AS inhibits PU.1 mRNA translation by forming an mRNA/AS lncRNA duplex to enhance expressions of adipogenic genes
Consistently, SRA expression was induced in WAT of HFD (high fat diet) induced obese mice [38]. To directly investigate SRA function in vivo, a whole SRA gene knockout mice was generated. Consistent with the previous data in vitro, SRA knockout mice were resistant to HFD-induced obesity, with reduced fat mass, decreased expression of a cluster of adipocyte marker genes and inflammation genes, lower plasma TNFα (tumour necrosis factor α) levels, and improved insulin sensitivity [38]. These data clearly indicate an important role of SRA in adipose tissue development, providing a potential target to control obesity and metabolic syndrome.
lnc-RAP-n
Research on lncRNAs as functional adipogenic regulators was largely promoted by large-scale genomic studies. Transcriptome profiling was used to examine the systematic implication of polyadenylated lncRNAs during adipogenesis, 175 poly-A tailed lncRNAs were significantly up- or down-regulated during differentiation of both white and brown mouse adipocytes; a substantial fraction of these lncRNAs were adipose-enriched and were tightly controlled by key transcription factors involved in adipogenesis (such as PPARγ and CEBPα) [13]. Several of these lncRNAs were strongly induced during differentiation, and siRNA-mediated loss of function demonstrated ten specific lncRNAs that could independently impair white preadipocyte differentiation, with reduced lipid accumulation as well as lower expressions of the majority of adipocyte markers (including PPARγ, CEBPα, FABP4, and AdipoQ), implicating that these lncRNAs were required for maturation of preadipocytes [13]. These 10 lncRNAs were therefore termed lnc-RAP-n (lncRNAs regulated in adipogenesis; n = 1–10). Among them, lnc-RAP-1, also known as FIRRE (functional intergenic repeating RNA element) or 6720401G13Rik, is an intergenic lncRNA that localized across a ~5 Mb domain on the X-chromosome which is in spatial proximity to five distinct trans-chromosomal loci within the nucleus, interfacing with and modulating nuclear architecture across chromosomes [39]. Furthermore, FIRRE binds hnRNP-U (heterogeneous nuclear ribonucleoprotein U, which is required for adipogenesis) to mediate the expression of adipogenic factors [39], providing a potential mechanism for the regulation of adipogenesis.
HOTAIR
HOTAIR, HOX antisense intergenic RNA, is located in the HOXC locus, and has been demonstrated to be involved in metastasis [40, 41]. Recently, a study has shown that HOTAIR participated in human subcutaneous preadipocyte differentiation [34]. HOTAIR was expressed in human gluteal fat but not in abdominal subcutaneous adipose tissue, and HOTAIR expression increased by twofold during gluteal preadipocyte differentiation in vitro [34]. Ectopic expression of HOTAIR in abdominal preadipocytes enhanced the percentage of differentiated cells and increased expressions of functional adipogenic markers including PPARγ, LPL (lipoprotein lipase), FABP4, and AdipoQ, with no effect on preadipocyte proliferation rate [34], suggesting an underlying transcriptional mechanism for HOTAIR to modulate preadipocyte differentiation. However, further study is required to decipher the molecular mechanism by which HOTAIR acts to regulate adipogenesis.
PU.1 AS
AS lncRNA (antisense long non-coding RNA) is a class of lncRNAs that are transcribed from the opposite DNA strand and overlap in part with its sense mRNA. Recent studies indicated that PU.1 AS lncRNA regulated adipogenesis through preventing PU.1 mRNA translation by forming an mRNA/AS lncRNA duplex [42, 43]. PU.1 is a transcription factor with inhibitory effects on preadipocyte differentiation [44]. Through attenuating PU.1 AS lncRNA binding to its mRNA, PU.1 AS lncRNA knockdown in 3T3–L1 preadipocytes promoted PU.1 protein expression and inhibited adipogenesis, as reflected by decreased lipid accumulation and reduced master gene expressions of PPARγ and CEBPα [42], indicating PU.1 AS lncRNA also plays a role in regulating adipogenesis.
slincRAD
A dynamic expressional profiling was performed in the induced differentiation of 3T3–L1 cells to examine potential functions of non-polyadenylated lncRNAs in adipogenesis; a super-long intergenic RNA transcript with a calculated size of 136 kb was shown to play a critical role in adipogenesis [45]. This gene was therefore named slincRAD (super-long intergenic non-coding RNA functioning in adipocyte differentiation) [45]. Quantitative analysis indicated that slincRAD inhibition in 3T3–L1 cell cultures reduced lipid accumulation and PPARγ expression, while the proliferation of the cells was not affected [45], demonstrating that slincRAD is a contributing factor in adipogenesis. Further investigation is needed to understand the underlying mechanism of slincRAD on fat cell development.
ADINR
Using mRNA-lncRNA-combined microarray technology, a recent study revealed 1423 differentially expressed lncRNAs on days 0, 3, and 6 of adipogenic differentiation of human mesenchymal stem cells (hMSCs) [46]. One of the highly induced lncRNAs, named ADINR (adipogenic differentiation induced noncoding RNA), was divergently transcribed from a position ~450 bp upstream of the CEBPα gene and was shown to be co-expressed with CEBPα during adipogenic differentiation [46]. Depletion of ADINR led to a dramatic adipogenic defect, as shown by decreased lipid accumulation and reduced adipogenic transcripts CEBPα, PPARγ, FABP4, and LPL. In addition, lentivirus-mediated ectopic expression of CEBPα enhanced expression of CEBPα as well as other adipogenic markers, and restored the severe adipogenic defects caused by the depletion of endogenous ADINR [46]. Mechanistically, ADINR RNA specifically binds to PA1 and recruits MLL3/4 histone methyl-transferase complexes to increase H3K4me3 and decrease H3K27me3 histone modification in the CEBPα locus, resulting in CEBPα activation as well as enhanced adipogenesis [46]. This data demonstrates that ADINR plays important roles in regulating the differentiation of hMSCs into adipocytes by modulating CEBPα in cis, demonstrating a mechanism of lncRNAs regulation of nearby gene expressions.
NEAT1
NEAT1 (nuclear enriched abundant transcript 1, also known as nuclear paraspeckle assembly transcript 1, MENε/β) is a nuclear lncRNA that is necessary for the formation of paraspeckles [47–49]. A recent study showed that miR-140 physically interacted with NEAT1 and increased NEAT1 expression [50]. In primary mouse preadipocytes, miR-140 knockout resulted in downregulation of NEAT1 as well as remarkable decreased lipid accumulation and the expression levels of PPARγ and CEBPα; re-expression of NEAT1 rescued the adipogenic phenotype, showing NEAT1 participates in miR-140 induced adipogenesis [50]. More functional essays are required to further understand NEAT1 roles in adipogenesis.
lncRNAs in brown/beige adipocyte development
Blnc1
It was recently illustrated that Blnc1 (brown fat lncRNA 1; AK038898), which was identified through global profiling of lncRNA expression during mouse thermogenic adipocyte formation, functioned as a driver of thermogenesis in brown and beige adipocytes, bringing a new layer of BAT developmental regulation. During adipogenesis, Blnc1 expression was highly induced by transcription factor EBF2; in addition, Blnc1 formed a ribonucleoprotein complex with EBF2 to enhance EBF2 expression, which promoted mouse brown and beige adipocyte differentiation and function by modulating the thermogenic gene program [33]. Overexpression of Blnc1 in brown preadipocytes significantly increased mRNA expressions of a subset of genes functioning in mitochondria that included UCP1 and PRDM1, with enhanced mitochondrial mass and DNA content and modest effect on lipid accumulation [33]. Further, mRNA expression of key thermogenic markers, including UCP1, was higher in Blnc1-expressing beige adipocytes [33]. In fat pads formed from transplanted Blnc1-transduced preadipocytes, UCP1 mRNA and protein levels reached approximately 30–40 % of that of endogenous brown fat [33], indicating Blnc1 as an effective activator of brown adipogenesis and brown fat formation. On the contrary, RNAi knockdown of Blnc1 severely impaired adipogenesis in cultured cells and in vivo [33].
lnc-BATE1
lnc-BATE1 is another BAT-specific lncRNA required for proper development and maintenance of mature, thermogenic brown adipocytes in mice. A recent study found that lnc-BATE1 was dramatically upregulated during brown adipogenesis or cold-induced beige adipocyte expansion [51]. lnc-BATE1 inhibition in brown preadipocytes resulted in decreased expression of brown fat markers (such as UCP1, PRDM16, and PGC1α), mitochondrial markers and, to a lesser extent, common adipogenic markers (PPARγ, CEBPα, FABP4, and AdipoQ), with limited effects on lipid accumulation and cell morphology; similar effects of lnc-BATE1 knockdown in beige adipocytes were also found [51]. Moreover, depletion of lnc-BATE1 in mouse mature brown adipocytes reduced BAT, mitochondrial, and common adipogenic markers, indicating lnc-BATE1 was essential for development and maintenance of mature brown adipocytes [51]. Indeed, lnc-BATE1 acts in trans to selectively sustain the core BAT gene program and repress WAT-selective genes through binding hnRNP-U, and both lnc-BATE1 and hnRNP-U were required for brown adipogenesis [51]. These data demonstrate that lnc-BATE1 is an essential factor during brown adipogenesis for induction of multiple mitochondrial proteins and for thermogenesis in brown adipocytes.
Potential lncRNAs that involved in adipogenesis
ecCEBPA
Recently, a functional lncRNA arising from the CEBPα locus, ecCEBPA (extra-coding CEBPα), was shown to bind directly to DNMT1 (DNA methyltransferase 1) and prevent CEBPα gene methylation, resulting in elevated expression of the CEBPα mRNA [52]. Loss and gain-of function experiments in HL-60, U937, and K562 cell lines suggested that CEBPα gene locus methylation levels were inversely correlated with ecCEBPA levels [52]. As CEBPα is a key driver of adipogenesis, epigenetic regulation of ecCEBPA on CEBPα expression might play an important role in adipogenesis. However, whether ecCEBPA is expressed in fat cells is unclear and further experiments are needed to evaluate ecCEBPA effects on adipogenesis.
AK142386 and AK133540
Via lncRNA microarray technology, Chen et al. evaluated differences in the lncRNA expression profiles of WAT and BAT [53]. In this study, hundreds of lncRNAs were identified to be differentially expressed between the two adipose depots [53]. GO (gene ontology) and pathway analyses of the differentially expressed lncRNAs showed that AK142386 and AK133540 might be involved in BAT and WAT development through their target genes Hoxa3 and Acad10 to regulate adipogenesis and metabolism [53]. Therefore, AK142386 and AK133540 might potentially serve as the required components for proper adipogenesis, but confirmation and elucidation of this assumption warrants further study.
Gm15051, Tmem189, and Cebpd
Additionally, differential expressions of lncRNAs on day 0 and day 8 during brown adipocyte differentiation were also profiled by microarray technology, providing a comprehensive analysis of lncRNA transcripts during classical brown adipocyte differentiation [54]. Among 1064 differentially expressed lncRNAs, lncRNA Gm15051, Tmem189, and Cebpd were identified to be important in the adipogenesis pathway via potential targets (Hoxa1, CEBPβ and CEBPδ) and might contribute to brown adipogenesis [54]. Further investigation of the molecular and biological functions of these candidate lncRNAs is required.
Clinical significance of lncRNAs for fighting against obesity
The worldwide prevalence of obesity has created heightened interest in understanding the detailed mechanisms regulating adipogenesis. Over the past several decades, great progress has been made in revealing master genes (such as PPARγ) and signal pathways in the complex transcriptional networks regulating fat development. Recent research on lncRNAs allow us further understanding factors that regulate adipogenesis and expand our traditional knowledge about fat development. Although lncRNAs are among the least well-understood transcripts, studies on adipose tissue have identified the lncRNAs as a novel class of adipogenic regulators, which play an essential role in regulating both white and brown/beige adipogenesis.
Clear evidences indicate that SRA knockout mice are resistant to HFD-induced obesity, with reduced expression of inflammation genes [14, 38], showing its potential clinical value since obesity is usually associated with a state of low-grade inflammation. Interestingly, improved insulin sensitivity is also detected in SRA−/− mice [38], which might partly be due to reduced inflammatory signaling observed in the experiment. Furthermore, as these mice have a global loss of SRA, other tissues, such as liver, in addition to fat tissue, may contribute to the whole body insulin sensitivity. It should be noted, however, that the primary SRA transcript can be alternatively spliced to generate a lncRNA (SRA) and a protein (SRAP) [55, 56]. Although the previous in vitro study indicates that SRA exerts the major adipogenic effects observed in SRA gene knockout mice [14], a possible role of SRAP in adipocytes cannot be completely excluded, as the knockout abolishes expressions of both SRA and SRAP, and the function of SRAP remains largely unclear; similar situation may exist in cell culture knockdown experiments of SRA. Nevertheless, the role of SRA on obesity and the metabolic syndrome deserves further attention. Followed to SRA, lnc-RAP-n, slincRAD, PU.1 AS, HOTAIR, ADINR, and NEAT1 are also found to be involved in white preadipocyte differentiation [13, 34, 42, 45, 46, 50]. Furthermore, Blnc1 and lnc-BATE-1 positively regulate brown or beige adipogenesis via forming a ribonucleoprotein complex with distinct transcriptional factors [33, 51].
Unlike microRNAs, a familiar class of ncRNAs modulating adipogenesis, diverse mechanisms for lncRNAs in regulating adipogenesis have been revealed, indicating their complexity and diversity. Although lncRNAs have been indicated to impact genetic output at almost every step of a gene’s life cycle [22, 35], the current functions of adipogenic lncRNAs focus on chromatin remodeling, transcriptional control (coactivator of transcriptional factors), and post-transcriptional processing (inhibition of translation) in cells to regulate adipogenesis (Fig. 2). In short, emerging lncRNAs function as important contributors to the intricate regulation network of fat cell development, and studies are further needed to elucidate their detailed mechanisms regulating fat accumulation.
As reviewed by Sun and Kraus, lncRNAs have been implicated in metabolism, endocrinology, reproduction, immunology, neurobiology, muscle biology, and cancer [57], and many of them possess tremendous clinical values [57, 58]. Recently, lncRNAs have been increasingly recognized as viable biomarkers for a number of diseases. For example, PCA3 lncRNA in urine samples has been developed into a clinical test for detecting human prostate cancer [59]. In addition to lncRNA-based diagnostics, lncRNA also serves as targets for lncRNA-based therapies, though many of which have focused on the treatment of cancers [57]. For combating human obesity, several possible lncRNA-based approaches might include developing pharmacological compounds that specifically activate/suppress lncRNA expressions to regulate white adipocyte differentiation. Notably, a new discovered drug ribocil is a synthetic small molecule selectively targeting riboswitches (the specific non-coding RNA structures) in mRNAs, indicating that non-coding RNA structural elements may successfully be targeted by synthetic small molecules [60]. As some adipogenic related lncRNAs have been uncovered functional structural elements (e.g. HOTAIR and NEAT1) or the complete secondary structure (e.g. SRA) with potential functions [37, 61], developing small molecule targeting specific lncRNA structures may be an effective therapeutic approach for combating metabolic related diseases. Additionally, overexpression or suppression of targeted lncRNAs regulating white adipogenesis through gene therapy provides a possibility to treat adiposity; one of the promising targets is SRA [38], though lncRNA-based gene therapies still have a long way to go before entering clinical trials in obese patients. Alternatively, lncRNA NEAT1 suggests that certain lncRNAs interact with miRNAs during fat cell differentiation [50], thus regulating lncRNA levels or activity via miRNAs may also prove useful. Another possible way for the development of possible approaches against obesity might involve the brown/beige adipogenesis, as some lncRNAs (such as Blnc1) can increase energy expenditure of brown fat to physiologically protect against obesity [33]. Strategies for stimulating brown/beige adipocyte abundance or function via lncRNAs have unique advantages as lncRNAs possess strong tissue specificity.
Collectively, similar with other modulators identified in adipogenesis, a few lncRNAs, not all, that regulating adipogenesis may have profound implications and clinical potential in obesity, although functions and mechanisms need to be explored in much more detail.
Conclusions
lncRNAs are a newly discovered class of regulatory RNAs that participate in a variety of cellular activities. Emerging evidence from both gain- and loss-of-function studies strongly indicates that lncRNAs are involved in the regulation of adipogenesis and play an important role in both white and brown/beige adipose tissue development and function. lncRNA functions in adipocytes are gradually being revealed, serving as a theoretical basis for understanding fat biology. Although this area warrants further investigation, the identification of lncRNA molecules with adipogenic activity may open up new possibilities of potential therapeutic targets and strategies for combating human obesity.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31501930), the Fundamental Research Funds for the Central Universities of China (KYZ201414, KJQN201606), the Natural Science Foundation of Jiangsu Province of China (BK20150656), and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2015BAD03B01).
Compliance with ethical standards
Conflict of interest
The authors have declared that no conflicts of interest exist.
Contributor Information
Lifan Zhang, Phone: +86-025-84399068, Email: lifanzhang@njau.edu.cn.
Michael V. Dodson, Phone: +1-509-335-9644, Email: dodson@wsu.edu
References
- 1.Low S, Chin MC, Deurenberg-Yap M. Review on epidemic of obesity. Ann Acad Med Singapore. 2009;38:57–59. [PubMed] [Google Scholar]
- 2.Prentice AM. The emerging epidemic of obesity in developing countries. Int J Epidemiol. 2006;35:93–99. doi: 10.1093/ije/dyi272. [DOI] [PubMed] [Google Scholar]
- 3.Katz DL. Perspective: obesity is not a disease. Nature. 2014;508:S57. doi: 10.1038/508S57a. [DOI] [PubMed] [Google Scholar]
- 4.Rothman KJ. BMI-related errors in the measurement of obesity. Int J Obes (Lond) 2008;32(Suppl 3):S56–S59. doi: 10.1038/ijo.2008.87. [DOI] [PubMed] [Google Scholar]
- 5.Dodson MV, Boudina S, Albrecht E, Bucci L, Culver MF, Wei S, Bergen WG, Amaral AJ, Moustaid-Moussa N, Poulos S, Hausman GJ. A long journey to effective obesity treatments: is there light at the end of the tunnel? Exp Biol Med (Maywood) 2013;238:491–501. doi: 10.1177/1535370213477603. [DOI] [PubMed] [Google Scholar]
- 6.Despres JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–887. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 7.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 9.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, Chen XW, Cawthorn WP, MacDougald OA, Koenig RJ. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5:e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Wong GK, Passey DA, Yu J. Most of the human genome is transcribed. Genome Res. 2001;11:1975–1977. doi: 10.1101/gr.202401. [DOI] [PubMed] [Google Scholar]
- 18.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein LD. Human genome: end of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 20.International Human Genome Sequencing C Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 21.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1:2–21. doi: 10.1002/wcs.2. [DOI] [PubMed] [Google Scholar]
- 23.Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13:183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- 24.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/MCB.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariel I, de Groot N, Hochberg A. Imprinted H19 gene expression in embryogenesis and human cancer: the oncofetal connection. Am J Med Genet. 2000;91:46–50. doi: 10.1002/(SICI)1096-8628(20000306)91:1<46::AID-AJMG8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 27.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 29.Bono H, Yagi K, Kasukawa T, Nikaido I, Tominaga N, Miki R, Mizuno Y, Tomaru Y, Goto H, Nitanda H, Shimizu D, Makino H, Morita T, Fujiyama J, Sakai T, Shimoji T, Hume DA, RIKEN GER Group, GSL Members. Hayashizaki Y, Okazaki Y. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res. 2003;13:1318–1323. doi: 10.1101/gr.1075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babak T, Blencowe BJ, Hughes TR. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genom. 2005;6:104–115. doi: 10.1186/1471-2164-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Divoux A, Karastergiou K, Xie H, Guo W, Perera RJ, Fried SK, Smith SR. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring) 2014;22:1781–1785. doi: 10.1002/oby.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 36.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 37.Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2012;2:189–199. doi: 10.4161/bioa.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Sheng L, Miao H, Saunders TL, MacDougald OA, Koenig RJ, Xu B. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem. 2014;289:13000–13009. doi: 10.1074/jbc.M114.564658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang WJ, Lin LG, Xiong Y, Wei N, Wang Y, Shen QW, Yang GS. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J Cell Biochem. 2013;114:2500–2512. doi: 10.1002/jcb.24595. [DOI] [PubMed] [Google Scholar]
- 43.Wei N, Wang Y, Xu RX, Wang GQ, Xiong Y, Yu TY, Yang GS, Pang WJ. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet. 2015;46:133–140. doi: 10.1111/age.12275. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Tong Q. Transcription factor PU.1 is expressed in white adipose and inhibits adipocyte differentiation. Am J Physiol Cell Physiol. 2008;295:C213–C220. doi: 10.1152/ajpcell.00422.2007. [DOI] [PubMed] [Google Scholar]
- 45.Yi F, Yang F, Liu X, Chen H, Ji T, Jiang L, Wang X, Yang Z, Zhang LH, Ding X, Liang Z, Du Q. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol. 2013;10:991–1001. doi: 10.4161/rna.24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, Wang S, Dalton S, Zhao RC, Chen R. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Rep. 2015;5:856–865. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gernapudi R, Wolfson B, Zhang Y, Yao Y, Yang P, Asahara H, Zhou Q. MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol Cell Biol. 2015;36:30–38. doi: 10.1128/MCB.00702-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, Lim YC, Knoll M, Slavov N, Chen S, Chen P, Lodish HF, Sun L. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Cui X, Shi C, Chen L, Yang L, Pang L, Zhang J, Guo X, Wang J, Ji C. Differential lncRNA expression profiles in brown and white adipose tissues. Mol Genet Genomics. 2015;290:699–707. doi: 10.1007/s00438-014-0954-x. [DOI] [PubMed] [Google Scholar]
- 54.You LH, Zhu LJ, Yang L, Shi CM, Pang LX, Zhang J, Cui XW, Ji CB, Guo XR. Transcriptome analysis reveals the potential contribution of long noncoding RNAs to brown adipocyte differentiation. Mol Genet Genomics. 2015;290:1659–1671. doi: 10.1007/s00438-015-1026-6. [DOI] [PubMed] [Google Scholar]
- 55.Emberley E, Huang GJ, Hamedani MK, Czosnek A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC, Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem Biophys Res Commun. 2003;301:509–515. doi: 10.1016/S0006-291X(02)03070-X. [DOI] [PubMed] [Google Scholar]
- 56.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–171. doi: 10.1042/bj20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol. 2011;8:123–124. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 60.Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, Murgolo N, Barbieri CM, Mann PA, Carr D, Xia E, Zuck P, Riley D, Painter RE, Walker SS, Sherborne B, de Jesus R, Pan W, Plotkin MA, Wu J, Rindgen D, Cummings J, Garlisi CG, Zhang R, Sheth PR, Gill CJ, Tang H, Roemer T. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 61.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]