Abstract
The development of efficacious and safe post transcriptional gene silencing (PTGS) agents is a challenging scientific endeavor that embraces “biocomplexity” at many levels. The target mRNA exhibits a level of structural complexity that profoundly limits annealing of PTGS agents. PTGS agents are macromolecular RNAs that must be designed to fold into catalytically active structures able to cleave the target mRNA. Pushing into and beyond the biological complexity requires new technologies for high throughput screening (HTS) to efficiently and rapidly assess a set of biological and experimental variables engaged in RNA Drug Discovery (RDD).
Keywords: Retinal degenerations, Macular degenerations, Gene therapy, Post-transcriptional gene silencing, Ribozyme, Hammerhead ribozyme, shRNA, siRNA, high throughput screening, breakthrough technology development
20.1 Introduction
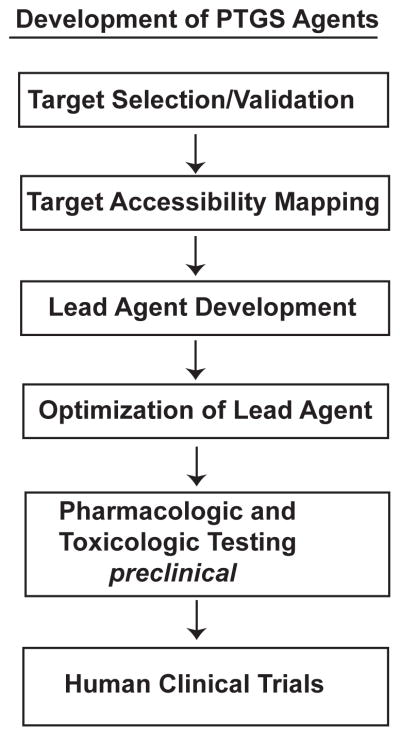
Currently, only one nucleic acid drug is approved for human disease by the Food and Drug Administration (Vitravene™, fomiversen, ISIS 2922). This singular success is testament to the difficulties that exist in development of PTGS agents over the last three decades. We encountered the biocomplexity intrinsic to development of PTGS agents in RDD for retina-directed therapeutics. The steps in the process of RDD are similar to the development of small molecule drugs except that in RDD the drug is a delivered or expressed macromolecular RNA molecule (Fig. 20.1). We critically detailed the bottlenecks at each step in RDD that limit the efficiency and success of PTGS agent development (Sullivan et al. 2008). Here, we present an overview of established and emerging technologies to address these bottlenecks, some of which were established in this lab.
Fig. 20.1.
Process of RNA Drug Discovery. Steps in PTGS development.
20.2 Materials & Methods
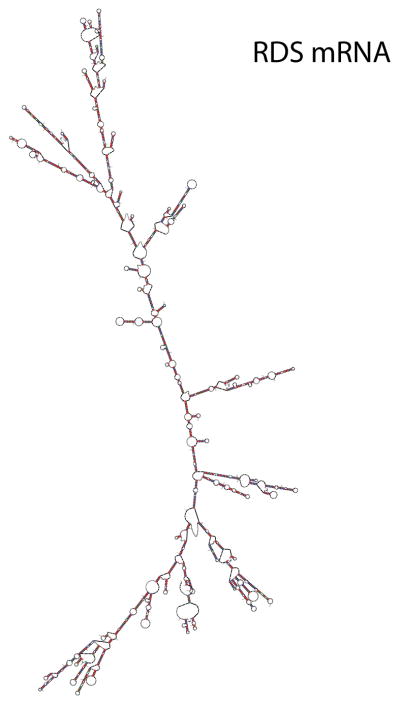
To demonstrate the complexity of mRNA structure we folded human peripherin mRNA (RDS, PRPH2) into secondary structures with Mfold, which first finds the minimal free energy structure (Zuker 2003). RDS is a disease target mRNA in autosomal dominant retinitis pigmentosa and macular dystrophies and is expressed in rod and cone photoreceptors (Boon et al. 2008).
20.3 Variables in RDD
20.3.1 Agents of PTGS
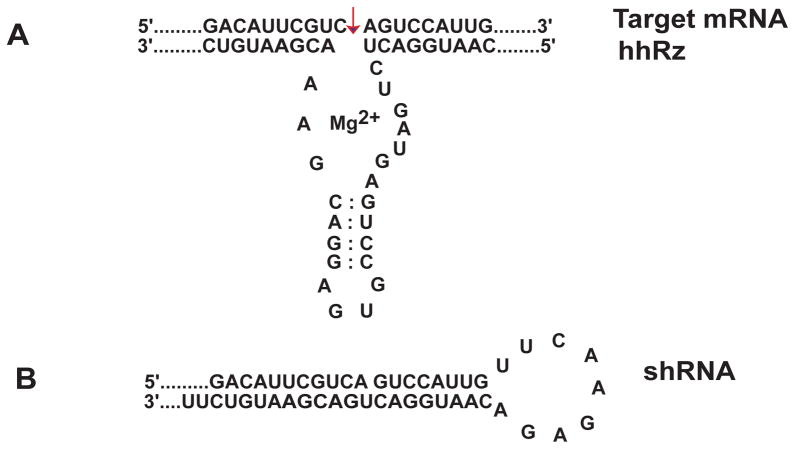
PTGS agents interact with target mRNAs on the basis of complementary Watson–Crick base pairing. We focus on ribozymes and shRNAs agents that could constitute gene-based therapeutics. PTGS agents contain an engineered region that is antisense to an accessible region in the target mRNA (Fig. 20.2). The hammerhead ribozyme (hhRz) has its catalytic core situated between two antisense flanks that anneal to the target mRNA and position the enzyme to cleave after an NUH↓ triplet (N= G, A, U, C; U=U, H= C, A, U). The hhRz catalyzes a transesterification reaction based on RNA chemistry. After cleavage the release of the target fragments is critical to allow enzymatic turnover and avoid product inhibition. The total antisense span of the hhRz should be at least 12 nt which is sufficient for good specificity with respect to the human transcriptome. The hhRz can be expressed within the context of a structured (chimeric) RNA that provides stability, appropriate cellular trafficking (to colocalize with target mRNA), and cellular stability (resistance to nucleases). The short hairpin RNA (shRNA) is a double-stranded stem loop element that contains a guide or antisense strand (complementary to the target mRNA region) and its complement separated by a short engineered loop. The cytoplasmic RNase Dicer cleaves off the loop to generate a short interfering RNA (siRNA). Cellular proteins interact with the siRNA, then select and orient the guide strand to form the RNA-induced silencing complex (RISC). RISC binds to the target mRNA and the protein components cleave the target RNA. RISC uses a seed sequence of 7–8 nt to interact with the target mRNA and within this seed the process is mismatch-tolerant. It is not surprising that there are typically many off-target effects with shRNA as compared to the hhRz which, relatively, is expected to have higher specificity for cleaving the target mRNA and potentially lower toxicity.
Fig. 20.2.
Ribozyme and shRNA Agents. (A) Hammerhead ribozyme structure. (B) shRNA structure.
20.3.2 Validating Appropriate Disease Target mRNAs
The target mRNA expresses a protein which is expected to contribute to the disease process. In an autosomal dominant hereditary retinal degeneration the mutant mRNA encodes a protein that could have gain-of-function toxic properties or dominant negative properties that promote cellular compromise and ultimate cell death and vision loss. Also, the loss of the wild type (WT) mRNA and protein may create a state of haploinsufficiency that may contribute to cellular demise. To solve the simultaneous problem of gain-of-function toxicity of mutant gene products and WT haploinsufficiency it may be necessary to express both a PTGS agent to knockdown the mutant (and WT) proteins and a variant WT allele to reconstitute WT protein expression with an mRNA that cannot be cleaved by the PTGS agent. Albeit complex, a knockdown-reconstitute strategy could allow use of a single therapeutic PTGS agent for all or most mutant alleles of a dominant disease gene.
PTGS agents have therapeutic potential for retinal degenerations where the targets are human WT mRNAs and proteins. For example, age-related macular degeneration is a multifactorial disease process with pathophysiological contributions originating from oxidative stress, accumulation of toxic retinoids (e.g. A2E), and local inflammation. Investigation of cellular disease pathways can validate WT mRNA targets and proteins that, if reduced by PTGS agents, could ameliorate disease states.
20.3.3 Target mRNA Structure and determinations of Accessibility
Annealing of a PTGS agent to a target mRNA is the rate limiting step in PTGS reaction kinetics. Annealing cannot occur if the targeted region is in a preexisting state of stable secondary or tertiary structure, or is protein coated. The PTGS agent must colocalize to the cellular compartment with the target mRNA to allow collision-mediated annealing, and the PTGS agent must be in sufficient local concentration to drive the second-order annealing reaction forward. The capacity of the PTGS to anneal will depend upon the local accessibility of the target mRNA. mRNA and viral RNA targets have profoundly limiting secondary structures that constrain the number of large and kinetically-stable single stranded platforms able to support PTGS annealing. Accessibility is rare in any target mRNA. An example of extensive secondary structures present in a disease target (human RDS) mRNA is shown (Fig. 20.3). There are few, if any, large single stranded regions. The primary and critical challenge in RDD is to identify those rare regions in the target mRNA which are accessible to rapid PTGS ligand annealing. Any perturbing intrinsic RNA structures or extrinsic structures (e.g. protein coating) that occlude the annealing platform will prevent or delay annealing events at physiological temperatures. The conformational folding space of the target mRNA can be sampled by combinatorial HTS procedures. Computational HTS approaches are used to assess RNA target secondary structures. There are number of algorithms available such as MFold (Zuker 2003), SFold (Ding and Lawrence 2004), and OligoWalk (Mathews et al. 1999). We combine the outputs of these algorithms to analyze target mRNA structure in a bioinformatics approach that we call multiparameter prediction of RNA accessibility (Abdelmaksoud et al. 2009).
Fig. 20.3.
Target mRNA Structure and Accessibility. The most stable PRPH2 mRNA structure predicted by Mfold (−993 kCal/mol).
It is also critical to experimentally probe the accessibility of a target mRNA to which PTGS agents will be designed. Randomized short oligodeoxynucleotides (ODN) can be presented to the folded target mRNA and then RNaseH added to promote cleavage within rare regions of annealing. RNA primer extension analysis can then be used to identify the region of the probe binding and cleavage (Ho et al. 1998). Similar approaches that use reverse transcriptase to initiate cDNA synthesis from regions of ODN binding can also be used to identify accessible platforms (Allawi et al. 2001). We recently developed a novel approach based on reverse transcription and ODN probes to rapidly identify stable single stranded accessible regions that contain hhRz cleavage sites in target mRNAs in their native folding state(s) (Sullivan and Taggart 2007).
20.3.4 Screening large numbers of PTGS agents to identify a Lead Candidate
In any accessible platform(s) there are typically multiple potential hhRz or shRNA cleavage sites. Once rare regions of accessibility are identified it may be necessary to construct a substantial set of PTGS agents to test efficacy and identify a lead candidate that exerts the greatest knockdown of target mRNA/protein. The design of a PTGS expression plasmid should be such that the small RNA is expressed to abundance in cultured cells (e.g. an RNA polymerase III promoter). The PTGS agent may be inserted within an RNA chimera to provide support, cellular stability, and cell compartment colocalization with target mRNA. The vector should be modified such that ligation of adaptors containing the PTGS cDNA is efficient. The cell line chosen should have high transfection efficiency with commercially available reagents (e.g. HEK293S cells, Lipofectamine). Conventional molecular biological tools such as RT/PCR and western analysis are slow, low throughput approaches with substantial variability and do not permit rapid, efficient and reliable identification of the lead PTGS candidate. A more efficient approach is to identify a means of directly or indirectly coupling the expression of the target gene to the expression of a convenient reporter molecule that can be rapidly and efficiently analyzed under HTS assays (Yau and Sullivan 2007; Yau and Sullivan submitted). Examples of such reporter genes could be secreted alkaline phosphatase or rapid decay forms of EGFP. The critical factor is that reporter expression represents the steady-state accumulation of the target mRNA. The PTGS agent and the target-reporter plasmid can be cotransfected into naïve cultured cells or the PTGS expression plasmid can be delivered into a stable cell line engineered to express the target-reporter construct. Output reads from the HTS reporter screen (e.g. SEAP enzyme assay, EGFP fluorescence) are then rank-ordered to identify the lead candidate PTGS plasmid that promotes the greatest target knockdown.
20.3.5 Optimizing the Lead Candidate
If the PTGS agent is a hhRz, the lead candidate may be subject to several rational modifications to obtain a more effective knockdown agent. The antisense flanks of the hhRz govern both target mRNA molecular recognition and specificity as well as the product leaving rates and enzymatic turnover efficiency (Stage-Zimmermann and Uhlenbeck 1998). The enzymatic core of the ribozyme is evolutionarily optimized and changes are typically deleterious, but stem II-loop elements that support the core can be altered with potential to enhance knockdown. Addition of upstream (5′) tertiary accessory elements can potentially enhance efficacy by driving the enzyme structure into a catalytically active state (Khvorova et al. 2003). The expression of the PTGS agent in a protective RNA chimera (e.g. a tRNA, adenoviral VAI RNA) with defined structure and cellular trafficking streams can facilitate colocalization with target mRNA and increase the half-life of the PTGS agent in the cell (Lieber and Strauss 1995; Koseki et al. 1999).
20.3.6 The Vector, PTGS Expression Construct, and Animal Testing
For a gene-based therapy there must be a vector to deliver the PTGS expression construct to appropriate cells. A variety of engineered viruses (e.g. rAAV) (Flannery et al. 1997) and nanoparticles, which have properties of synthetic viruses (Farjo et al. 2006), offer potential for high transduction efficiency to promote PTGS expression in as many diseased cells as possible. An optimized surgical approach is needed to maximize therapeutic vector delivery and target cell transduction. Visualization of surgical delivery of vector insures correct compartmental access in the eye (e.g. subretinal space) and allows quantitation of the areal extent of delivery (e.g. fraction of retina coverage by delivered volume). Quantitative measurement of target cell transduction (e.g. expression of a non-toxic fluorescent protein) should be used to normalize measured rescue (e.g. ERG). In many mouse-based preclinical studies such measures are limited by the imaging technology. The small mouse eye has a constrained pupillary aperture (~1 mm dilated) that limits both the input of light and the collection of the output specular image. As a facilitating technology we developed a stereo bright-field and fluorescence microscope that allows real time visualization within the neonatal mouse eye during and after subretinal or intraocular injections (Butler and Sullivan, 2010).
20.3.7 Appropriate targets (human)
Many animal models exist for retinal degenerations. In the context of PTGS agents under development for potential clinical translation, it is critical to understand that an animal mRNA is unlikely to represent the same conformational landscape as a PTGS receptor as would a human cognate mRNA target. This results due to sequence divergence at the 5′UT and 3′UT regions, codon differences, and third position bias. RNA folding into secondary structures is a local process, and even when the primary sequences of an animal and human target are identical over the region of PTGS annealing, the secondary structures can indeed be different and this can impact efficacy. Our view for PTGS studies intended toward human clinical translation is that the mRNA targets expressed in preclinical animal models be true full length human mRNAs.
20.4 Conclusions
PTGS agents have great therapeutic potential for retinal degenerations. The overview presented here will hopefully provide both direction and incentive for other motivated groups to share the challenges of RDD. Viral and nanoparticle vector technology for retinal or ocular gene therapy is advanced. Gene therapy clinical trail for Leber congenital amaurosis due to autosomal recessive (null) mutations in the RPE65 gene are demonstrating early safety and efficacy profiles when the delivered genetic construct expresses a WT protein (den Hollander et al. 2010). If fully successful, this initial therapeutic trial could establish the eye as an organ for vector based gene therapy for other recessive conditions, and ultimately for expressed PTGS agents that act to knockdown expression of specific validated disease targets as a path to cell preservation and sustenance of vision.
Acknowledgments
We thank the National Eye Institute (R01 EY13433, PI: Sullivan) (R24 EY016662, PI: M Slaughter), the Veterans Administration (Merit Grant 1I01BX000669-01), an Unrestricted grant from Research to Prevent Blindness, and a grant from the Oishei Foundation (Buffalo, NY).
References
- Abdelmaksoud H, Yau EH, Zuker M, et al. Development of lead hammerhead ribozyme candidates against human rod opsin for retinal degeneration therapy. Exp Eye Res. 2009;88:859–879. doi: 10.1016/j.exer.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allawi HT, Dong F, Ip HS, et al. Mapping of RNA accessible sites by extension of random oligonucleotide libraries with reverse transcriptase. RNA. 2001;7:314–327. doi: 10.1017/s1355838201001698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon CJF, den Hallander AI, Hoyng CB, et al. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Ret Eye Res. 2008;27:213–225. doi: 10.1016/j.preteyeres.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Butler MC, Sullivan JM. A novel real-time in vivo mouse retinal imaging system. Invest Ophthalmol Vis Sci. 2010;51:3103. doi: 10.1167/iovs.14-16370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hollander AI, Black A, Bennet J, et al. Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. J Clin Invest. 2010;120:3042–3053. doi: 10.1172/JCI42258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32(supp):W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farjo R, Skaggs J, Quiambao AB, et al. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS One. 2006;1:e38. doi: 10.1371/journal.pone.0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery JG, Zolotukhin S, Vaquero MI, et al. Efficient photoreceptor-targeted gene expression in vivo by recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:6916–6921. doi: 10.1073/pnas.94.13.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SP, Bao Y, Lesher T, Malhotra R, et al. Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat Biotechnol. 1998;16:56–63. doi: 10.1038/nbt0198-59. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, et al. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Koseki S, Tanabe T, Tani K, et al. Factors governing the activity in vivo of ribozymes transcribed by RNA polymerase III. J Virol. 1999;73:1868–1877. doi: 10.1128/jvi.73.3.1868-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber A, Strauss M. Selection of efficient cleavage sites in target mRNAs by using a ribozyme expression library. Mol Cell Biol. 1995;15:540–551. doi: 10.1128/mcb.15.1.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Burkard ME, Freier SM, et al. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Taggart RT. Novel and enhanced approaches to determined local mRNA accessibility. Invest Ophthalmol Vis Sci. 2007;48:4605. [Google Scholar]
- Yau EH, Sullivan JM. High throughput cellular screening for ribozyme development against arbitrary mRNA targets. Invest Ophthalmol Vis Sci. 2007;48:1681. [Google Scholar]
- Yau EH, Kolniak TA, Taggart RT, et al. Optimization of lead ribozyme agents for human rod opsin therapeutics. Invest Ophthalmol Vis Sci. 2008;49:5344. [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]