Abstract
Since the discovery of microRNAs (miRNAs), circulating miRNAs have been proposed as biomarkers for disease. Consequently, many groups have tried to identify circulating miRNA biomarkers for various types of diseases including cardiovascular disease and cancer. However, the replicability of these experiments has been disappointingly low. In order to identify circulating miRNA candidate biomarkers, in general, first an unbiased high-throughput screen is performed in which a large number of miRNAs is detected and quantified in the circulation. Because these are costly experiments, many of such studies have been performed using a low number of study subjects (small sample size). Due to lack of power in small sample size experiments, true effects are often missed and many of the detected effects are wrong. Therefore, it is important to have a good estimate of the appropriate sample size for a miRNA high-throughput screen. In this review, we discuss the effects of small sample sizes in high-throughput screens for circulating miRNAs. Using data from a miRNA high-throughput experiment on isolated monocytes, we illustrate that the implementation of power calculations in a high-throughput miRNA discovery experiment will avoid unnecessarily large and expensive experiments, while still having enough power to be able to detect clinically important differences.
Keywords: MicroRNA, High-throughput screens, Biomarkers, Small sample size error, Array, Methodology
1. Introduction
Since their discovery as regulators of gene expression almost two decades ago, microRNAs (miRNAs) have been reported to be involved in crucial biological processes, such as cell differentiation, proliferation and apoptosis [1]. Following the first description of miRNA involvement in cancer [2], many studies have focused on the role of miRNAs in a wide variety of diseases. In 2007, the first study was published that revealed that miRNAs were also present in microvesicles and that these short RNA strands could be preserved in the extracellular space [3]. This important finding was quickly followed by the first detection of miRNAs in the circulation, opening up a whole new field of research [4].
Following these discoveries, circulating miRNAs became a burgeoning area of research, because of two reasons. First, miRNAs are protected from degradation in the extracellular environment by binding to Argonaute proteins and through encapsulation by high density lipoprotein particles, exosomes and microvesicles, which make them easy to detect in the circulation [[5], [6]]. Second, previous studies extensively reported the dysregulation of specific cell-based miRNAs in diseased states. It is thought that, along with the usual cargo, deregulated miRNAs are shed from diseased tissue into the circulation and that expression levels of specific miRNAs in the circulation could reflect the presence of disease [3]. Taken together, these characteristics led to the hypothesis that circulating miRNAs are suitable biomarker candidates.
In order to identify circulating miRNA candidate biomarkers, usually, first a high-throughput screen is performed in which a large number of miRNAs are quantified in the circulation. During this process several challenges are encountered. Here, we will discuss the challenges of performing such a high-throughput screen and highlight the impact of an important source of potential error. This is the low number of study subjects that is often used in these experiments which causes the so-called small sample size error.
2. Methods
2.1. Literature study methods
We searched Pubmed for recent reviews that summarized all circulating miRNAs associated to CAD. To avoid missing more recently published papers on promising miRNA candidates that had not appeared yet in reviews, all miRNAs that showed an up- or downregulation in CAD in the most recent reviews, were individually checked for their relation with CAD. The final search was performed on June 12th, 2017. Details on the search strategy can be found in the supplemental material.
2.2. Subsampling experiment methods
We performed a miRNA microarray experiment on isolated monocytes in a large cohort of 61 individuals to compare the expression levels of 461 miRNAs between 36 patients with premature coronary artery disease (CAD) and 25 controls. The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) in a MIAME compliant format and are accessible under GEO Series accession number GSE105449. MiRNA expression profiles were determined using the Agilent Human 8 × 15k miRNA microarray platforms based on Sanger miRBase release 19.0. A detailed description of the sample collection and data pre-processing can be found in de Ronde et al. [7]
All miRNAs were normalized and log2 transformed using a similar method as described in de Ronde et al. [7] To detect miRNAs that were differentially expressed between patients and controls, gene-wise linear models were fit with patient/control status as explanatory variable, corrected for Body Mass Index (BMI) and age, followed by a moderated t-test (limma R package) between patients and controls in each comparison. Resulting p-values were adjusted to correct for multiple hypothesis testing using the Benjamin-Hochberg false discovery rate.
To illustrate the increasing occurrence of false positive findings with small sample sizes, we performed a subsampling experiment on the above described dataset. From the 36 patients and 25 controls, random selections of 5 patients and 5 controls were chosen 10,000 times and each time differential expression between patients and controls was tested for all 461 miRNAs. The analysis was repeated using 10,000 randomly chosen subsamples of 10 versus 10, 15 versus 15, 20 versus 20 and 25 versus 25 individuals. In each individual subsample, the number of differentially expressed miRNAs (corrected p-value of <0.1) was counted. Also, to illustrate the occurrence of inflated effect sizes, in each subsample, the effect size of the most significant miRNA was recorded.
To illustrate that small sample sizes can also lead to false negative results, we modified the original dataset in such a way that 100 (out of 461) miRNAs were differentially expressed between patients and controls. Artificial differences in this perturbed dataset were created by choosing a random number between 0.3 and 0.5 and adding this number to the log2 expression level of each sample in 50 of the 461 measured miRNAs and subtracting this number from each sample in another 50 of the 461 measured miRNAs. Then, in order to analyze the effect of sample sizes in this perturbed dataset, we performed the above described subsampling experiment and recorded the number of miRNAs differentially expressed between patients and controls.
2.3. Power calculation methods
The power calculation was performed with the ssize.fdr R package [8]. Based on the above described experiment of miRNA expression levels between CAD patients and controls, a common standard deviation of 0.56, and an estimated proportion of non-differentially expressed miRNAs of 0.83 with a false discovery rate (FDR) controlled at 10% was used. The true difference between mean expressions in the two groups as well as the standard deviations of expressions were assumed to be identical for all miRNAs. The common value for the standard deviations was estimated from the data and set conservatively to the 90th percentile of the gene residual standard deviations. The proportion of non-differentially expressed genes π0 was estimated using the qvalue function (qvalue R package) on the vector of p-values obtained from the differential expression analysis.
3. Challenges in miRNA quantification
When quantifying miRNA expression using high-throughput screens, the miRNA profiling method has to deal with several challenges that result from the principal characteristics of miRNAs. These include the short length of the miRNAs, the low abundance of free miRNAs in plasma and serum, the high degree of sequence homology within miRNA families and the existence of isomiRs, in which sequences of miRNAs can differ by a single nucleotide from the reference miRNA [9]. The most commonly used techniques for miRNA profiling are miRNA microarrays, quantitative polymerase chain reaction (qPCR) and next generation sequencing (NGS) [[10], [11], [12]]. These techniques are all highly sensitive [13], but encounter several difficulties.
Although qPCR is the most sensitive of these methods, it is less appropriate for high-throughput experiments and much more expensive when profiling a large number of miRNAs [14]. For a high-throughput analysis, the costs of microarrays are lower, but so is their sensitivity. Even though an excellent intra-platform replicability that was reported, microarrays showed only limited concordance between platforms, suggesting a low sensitivity and specificity [15]. For NGS, a major difficulty lies in robust library preparation to obtain non-biased data. Although much progress has been made over the past years, most popular library preparation protocols being used today may still introduce serious variations in RNA sample composition, which might eventually result in misinterpretation of the data [16]. The majority of the published studies used microarrays for miRNA profiling in the circulation, probably due to higher costs of NGS experiments in the past and more challenging data analysis. However, nowadays the falling costs and high specificity of small RNA sequencing make NGS a more attractive tool for high-throughput screening.
4. Replicability issues
Over the past years, multiple groups have searched for circulating miRNAs as biomarker for various diseases. However, it has been suggested that circulating miRNAs that have been associated with disease are difficult to replicate. To quantify this lack of replicable results, we performed a literature study and summarized all coronary artery disease (CAD) associated circulating miRNAs.
4.1. Results literature study
In total, the literature search for reviews on CAD-associated circulating miRNAs yielded 309 hits. After screening titles and abstracts, 5 reviews remained [[17], [18], [19], [20], [21]]. In these 5 reviews, published between 2015 and 2016, 60 miRNAs were found to be up- or downregulated in CAD in a total of 29 different studies (Supplemental Table S1). Of these 60 miRNAs, 13 were found to be up- or downregulated in more than one study. However, more than half of these miRNAs (7 out of 13), showed a contradicting result between studies (e.g. for miR-21, 2 studies showed upregulation and 1 study showed downregulation). Concluding, similar studies investigating the same disease not only found many different miRNAs to be associated with this disease, also the miRNAs that have been associated with CAD in multiple studies, show inconsistent results between different studies.
4.2. Reasons for replicability issues
Reasons for these discordant results are multiple. First, the studies under investigation all had a different study design, with different in- and exclusion criteria, thereby creating different study cohorts which are difficult to compare. Second, a large part of the discrepancies encountered might be due to the use of different platforms, data pre-processing methods and/or statistical analysis methods [[22], [23], [24], [25]]. Furthermore, often incomplete names are used to refer to a miRNAs (e.g. miR-1 instead of miR-1-3p) and miRNA nomenclature also changes as a consequence of new miRBase versions, which could lead to confusion as to which miRNA was measured in a study. Another important factor driving the discordance in results between studies are the low number of study subjects (e.g. 5 healthy versus 5 diseased subjects) that are often used in these experiments. Independent of the method used for high throughput screening, this can lead erroneous study results of the experiment: the so-called small sample size error.
5. The influence of sample size on biomarker discovery
The quest for new miRNAs as biomarker for disease is often a search for a needle in a haystack. Since miRNA profiling experiments are costly, the initial step of the search is very often performed in only a small number of study subjects [26]. However, from a scientific perspective it is essential to include enough study subjects to have sufficient power to be able to detect clinically relevant differences and thus avoid small sample size error. In underpowered studies, three problems occur that contribute to the occurrence of unreliable findings. First, by definition, the chance of discovering true effects is low in studies with low power, leading to a high false-negative rate (type II error) [4]. This is a particularly undesired source of error since the expensive and laborious experiment would have been done in vain. Second, due to the relative large contribution of accidental outliers to the overall effect in a small sample, the number of false-positive findings (type I errors) will increase, decreasing the positive predictive value [[27], [28]]. Last, the magnitude of a true effect is often exaggerated in small sample studies since, due to lack of power, mainly large effects are significant. This leads to a specific type of publication bias, since the underpowered study that, by chance, discovers such a large effect is more likely to be published and to receive more attention and impact than competing, perhaps sufficiently powered studies that do not show any effect [[29], [30]]. This is supported by the observation that the effect sizes of associations reported by highly cited biomarker studies are often larger than the effect sizes of those same associations in subsequent meta-analyses [31]. Besides, when many unlikely hypotheses are tested, many of the detected effects may be wrong. This is the case for the large majority of circulating miRNA biomarker high-throughput studies, where only a few of the measured miRNAs are expected to be differentially expressed between disease and control [32].
To overcome these problems while still maintaining low experimental costs, several groups have pooled the blood samples of different subjects [[33], [34], [35], [36]]. Although pooling leads to a decrease in biological variation, which may help to detect significant differences between groups, these studies provide only limited due the loss of individual-specific information that is essential in a biomarker study [37]. Moreover, because compared to measuring all individuals, fewer measurements are carried out after pooling the samples, increasing random technical variation, which can lead to more false-positive and false-negative results. Therefore, we recommend not to pool blood samples.
6. Consequences of small sample sizes in a real microarray experiment
In order to illustrate the influence of small sample sizes on the number of false positive findings with inflated effect sizes and on false negative results, we used data from a circulating miRNA microarray experiment. First, analyses were performed on the complete original dataset. Then a subsampling experiment was performed on the original dataset. Finally, analyses were performed on the perturbed dataset, where 100 out of the 461 miRNAs were artificially up- or downregulated, as described in detail in the Methods section.
6.1. Original dataset
In the original dataset, the analysis of the miRNA expression profiles revealed that none of the detected miRNAs was differentially expressed between premature CAD patients and healthy controls with a multiple testing corrected p-value of less than 0.1. In fact, the smallest corrected p-value that was obtained in this study was 0.26 and the highest absolute fold change only reached 1.46 (Control/CAD).
6.2. Small sample sizes can lead to an increase in false positive results and exaggerated effect sizes
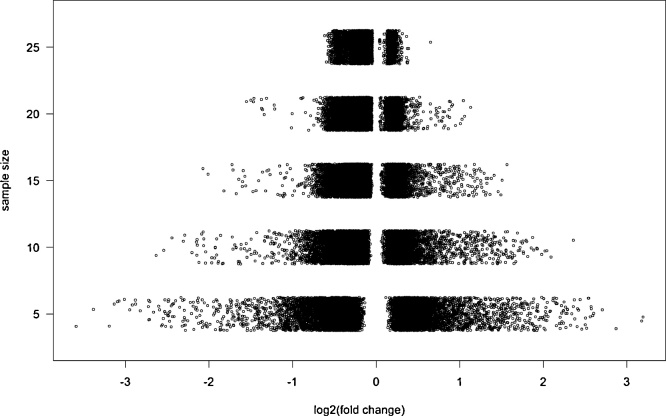
Analysis revealed that when choosing a slightly smaller sample of 25 patients versus 25 controls from the original dataset, 9 out of 10,000 subsamples showed differential expression for >10 different miRNAs (corrected p-value<0.1). Using a sample size of 5 versus 5 individuals, 145 out of 10,000 subsamples showed a differential expression of >10 miRNAs between patients and controls (Table 1A), of which one individual subsample even found 190 out of 461 miRNAs to be differentially expressed (Table 1B). This shows that the number of false positive findings rapidly increases with decreasing sample size. Besides, we found that in small subsamples, the observed fold changes were more heterogeneous and generally larger compared to larger subsamples (Fig. 1). In other words, enlarging the size of the subsamples resulted in more realistic effect sizes, an effect commonly referred to as regression to the mean. The above results underline that in small sample size studies, there is a large chance of false positive findings and exaggerated effect sizes as compared to studies with larger sample sizes.
Table 1.
Both numbers of false-negative and false-positive results increase with a decreasing sample size.
| 5 vs 5 | 10 vs 10 | 15 vs 15 | 20 vs 20 | 25 vs 25 | |
|---|---|---|---|---|---|
| Original dataset (no differences between patients and controls) | |||||
| A. # of subsamples with >10 miRNAs differentially expressed | 145/10,000 | 127/10,000 | 93/10,000 | 36/10,000 | 9/10,000 |
| B. Highest # of differentially expressed miRNAs (from 461) identified in one subsample | 190 | 176 | 201 | 105 | 13 |
| Perturbed dataset (100 miRNAs set to differentially expressed between patients and controls) | |||||
| C. Mean # of miRNAs differentially expressed between patient and control | 47/100 | 73/100 | 85/100 | 91/100 | 93/100 |
Results of the subsampling experiments. The column header indicates the number of patients and controls randomly sampled 10,000 times from the original dataset (A-B) and the perturbed dataset (C). A) Number of subsamples (out of 10,000) from the original dataset with at least 10 differentially expressed miRNAs between patients and controls. B) Maximal number of differentially expressed miRNAs in any of the 10,000 subsamples from the original dataset. C) Mean number of miRNAs (out of 100) that were differentially expressed in subsamples of the perturbed dataset. Differential expression corresponds to a Benjamin-Hochberg false discovery rate adjusted p-value <0.1. # = number.
Fig. 1.
Inflation of the effect size in small sample size studies.
Fold changes of the most significant miRNA in each of 10,000 subsamples from the original dataset for five different sample sizes (n = 5, 10, 15, 20, 25). In subsamples of 5 versus 5 individuals the heterogeneity in effect sizes is larger compared to subsamples of 25 versus 25 individuals, with larger observed fold changes.
6.3. Small sample sizes can lead to an increase in false negative results
The analysis of the perturbed dataset showed that, using a sample size of 25 versus 25 individuals, from 10,000 subsets, an average 93 from the 100 artificially up- or downregulated miRNAs were differential in expression between patients and controls. However, with the smallest sample size of 5 versus 5, an average of only 47 out of 100 miRNA were differentially expressed, showing that a small sample size led to more than 50% false negative results (Table 1C).
These analyses illustrate that using a cohort that is too small, the study may be underpowered. This not only leads to the identification of false positive candidate biomarkers and exaggerated effect sizes but also to non-identification of true biomarker candidates. The identification of false positive biomarkers may lead to putative biomarker sets that cannot be replicated in larger cohorts. Although, results of the replication/validation phase are largely dependent on the patient population, sample collection, measurement platform and statistical analysis, small sample size error is highly likely to lead to an inability to replicate candidate biomarkers. Indeed, in several published studies only few of the candidate biomarkers could be replicated in larger independent cohorts [[38], [39], [40], [41]]. But perhaps an even larger problem is the fact that true biomarkers may be missed in underpowered studies. In this case, a lot of money is spent on an experiment that is futile. Moreover, researchers may be discouraged to repeat the experiment because of the results of these previous underpowered studies that showed no effect, and some important miRNA biomarkers may never even be found.
7. Sample size calculation, the only solution to the problem of underpowered studies
The problem of small sample size error has raised the question as to what a sufficient sample size in the search of miRNAs as biomarkers for disease actually is. Because every experiment to detect circulating miRNAs differs from other experiments (e.g. differences in disease state, variance of the miRNA levels, fold changes and proportion of non-differentially expressed miRNAs), there is, unfortunately, no general answer on what the sample size should be. Therefore, despite the extra costs, the only way to design a sufficiently powered, valid experiment is to perform a pilot study before performing the actual discovery study.
7.1. Performing a power calculation
In a pilot experiment, the appropriate in- and exclusion criteria for patients and controls should be set. These criteria should be the same as for the intended final discovery experiment. Using these in- and exclusion criteria, a pilot study with a subset of the intended study population should be conducted (see Fig. 2 for flowchart). After conducting the pilot experiment, the variance and fraction of non-differentially expressed miRNAs estimated on the pilot study data can be used in a power calculator to calculate the required sample size [[8], [42], [43], [44], [45]]. Of special note is the required effect size that is used in the power calculation; meaning the minimal effect size that can still be reliably detected using the calculated sample size. What this minimal detectable effect size should be is entirely up to the researcher. Setting lower minimal effect sizes enables the detection of more subtle differences between patient and control. On the other hand, the required sample size rapidly increases when the researcher sets the minimally detectable effect size, or fold change, to a low value, increasing the costs of the discovery experiment. Therefore, before the initiation of a large and costly discovery experiment, the researcher has to determine which minimum effect size is deemed to be clinically relevant. Conversely, when the experiment has already been performed, one can use a power calculation to determine whether the underlying sample size provided enough power to find clinically relevant biomarker candidates.
Fig. 2.
Flowchart for setup of a miRNA biomarker experiment.
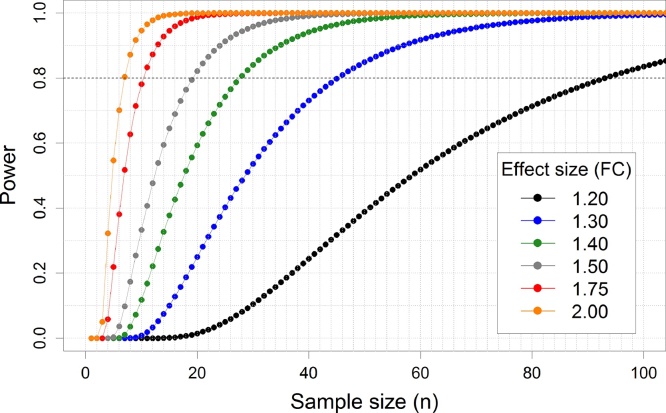
We can illustrate this again by using the above experiment that compared miRNA expression between premature CAD patients and healthy controls. In this experiment, we estimated that a minimum fold change of 1.5 would be biologically and clinically relevant. As described in the methods section, we performed a power calculation to test whether our sample size provided enough power to detect a fold change of more than 1.5. To detect a 1.5-fold change in expression levels, we calculated that a sample size of 19 subjects per group was needed to achieve more than 80% average power with a false discovery rate of 10% and an estimated proportion of non-differentially expressed miRNAs of 0.83. Because our experiment consisted of a sample size of 25 per group, this confirmed that our sample size had been large enough to reliably detect the required 1.5-fold expression differences. When the researcher, however, after execution of this experiment decides that (s)he wants to be able to reliably detect a fold change of 1.20, the power calculation shows that a sample size of 93 individuals per group is needed (Fig. 3).
Fig. 3.
Impact of sample size and the minimally detectable effect size on power.
The graph shows the statistical power for a given sample size (per group) in different scenarios of minimally detectable effect sizes ranging from 1.20 to 2. Commonly, a power of 80% is used in power calculation. Therefore, the sample size (X-axis) at which the dashed horizontal grey line crosses the line with the desired effect size is the sample size needed to achieve enough power to detect that effect size. Effect sizes are indicated as fold-change (FC). Smaller effect sizes require a larger sample size.
After having specified the sample size, the actual discovery experiment can be performed (Fig. 2). In this experiment, the exact same experimental protocols and measurement platform should be used as in the pilot [[46], [47]]. After selection of the most promising candidates from the discovery study, these candidates can then be validated in a subsequent validation study on an independent, larger, prospective cohort, preferably using a different quantification system like qPCR. Also this experiment should be performed using the same in- and exclusion criteria and a pre-specified and adequate number of subjects.
8. Conclusion
In the search for new biomarkers for diseases many miRNAs have been identified. However, in subsequent validation studies only few of these findings could be replicated. The use of small sample sizes and its consequences are still a large and underappreciated problem that may have contributed to many discordant results in circulating miRNA research. Here we illustrate that a small sample size has a major impact on the replicability of a study. Therefore, carefully designed small sample size pilot studies should be conducted and used in a power calculator to calculate a sufficient sample size to detect meaningful differences in the discovery study. All published research on biomarker identification should provide data about such pilot studies with its matching power calculation to support the choices of the sample size used. Neglecting this essential epidemiological element will lead to a high degree of results that cannot be replicated, wasting a large amount of money and effort.
Conflict of interest
Nothing to declare.
Handled by Jim Huggett
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.bdq.2017.11.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 3.Valadi H. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann L.M. Sample sizes of studies on diagnostic accuracy: literature survey. BMJ. 2006;332(7550):1127–1129. doi: 10.1136/bmj.38793.637789.2F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida-Aoki N., Ochiya T. Interactions between cancer cells and normal cells via miRNAs in extracellular vesicles. Cell. Mol. Life Sci. 2015;72(10):1849–1861. doi: 10.1007/s00018-014-1811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 7.de Ronde M.W.J. High miR -124-3p expression identifies smoking individuals susceptible to atherosclerosis. Atherosclerosis. 2017 doi: 10.1016/j.atherosclerosis.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 8.Orr M., Liu P. Sample size estimation while controlling false discovery rate for microarray experiments using the ssize.fdr package. R J. 2009;1(1):47–53. [Google Scholar]
- 9.Morin R.D. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker M. MicroRNA profiling: separating signal from noise. Nat. Methods. 2010;7(9):687–692. doi: 10.1038/nmeth0910-687. [DOI] [PubMed] [Google Scholar]
- 11.Git A. Systematic comparison of microarray profiling: real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16(5):991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zampetaki A., Mayr M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb. Haemost. 2012;108(4):592–598. doi: 10.1160/TH12-02-0097. [DOI] [PubMed] [Google Scholar]
- 13.Chugh P., Dittmer D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA. 2012;3(5):601–616. doi: 10.1002/wrna.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato F. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS One. 2009;4(5):e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijk E.L., Jaszczyszyn Y., Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 2014;322(1):12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Busch A., Eken S.M., Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann. Transl. Med. 2016;4(12):236. doi: 10.21037/atm.2016.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahlin F. MicroRNAs as circulating biomarkers in acute coronary syndromes: a review. Vascul. Pharmacol. 2016;81:15–21. doi: 10.1016/j.vph.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Schulte C., Zeller T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc. Diagn. Ther. 2015;5(1):17–36. doi: 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navickas R. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc. Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romaine S.P. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101(12):921–928. doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J.J. Reproducibility of microarray data: a further analysis of microarray quality control (MAQC) data. BMC Bioinf. 2007;8:412. doi: 10.1186/1471-2105-8-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannidis J.P. Repeatability of published microarray gene expression analyses. Nat. Genet. 2009;41(2):149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 24.Novianti P.W., Roes K.C., Eijkemans M.J. Evaluation of gene expression classification studies: factors associated with classification performance. PLoS One. 2014;9(4):e96063. doi: 10.1371/journal.pone.0096063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschmann D. Toward reliable biomarker signatures in the age of liquid biopsies − how to standardize the small RNA-Seq workflow. Nucleic Acids Res. 2016;44(13):5995–6018. doi: 10.1093/nar/gkw545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballman K.V. Genetics and genomics: gene expression microarrays. Circulation. 2008;118(15):1593–1597. doi: 10.1161/CIRCULATIONAHA.107.714600. [DOI] [PubMed] [Google Scholar]
- 27.Tong T., Zhao H. Practical guidelines for assessing power and false discovery rate for a fixed sample size in microarray experiments. Stat. Med. 2008;27(11):1960–1972. doi: 10.1002/sim.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J. Sources of variation in false discovery rate estimation include sample size, correlation, and inherent differences between groups. BMC Bioinf. 2012;13(Suppl. 13):S1. doi: 10.1186/1471-2105-13-S13-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ioannidis J.P. Excess significance bias in the literature on brain volume abnormalities. Arch. Gen. Psychiatry. 2011;68(8):773–780. doi: 10.1001/archgenpsychiatry.2011.28. [DOI] [PubMed] [Google Scholar]
- 30.van Enst W.A. Small-study effects and time trends in diagnostic test accuracy meta-analyses: a meta-epidemiological study. Syst. Rev. 2015;4:66. doi: 10.1186/s13643-015-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis J.P., Panagiotou O.A. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA. 2011;305(21):2200–2210. doi: 10.1001/jama.2011.713. [DOI] [PubMed] [Google Scholar]
- 32.Krzywinski M., Altman N. POINTS OF SIGNIFICANCE power and sample size. Nat. Methods. 2013;10(12):1139–1140. [Google Scholar]
- 33.Liu Y. Analysis of plasma microRNA expression profiles in a Chinese population occupationally exposed to benzene and in a population with chronic benzene poisoning. J. Thorac. Dis. 2016;8(3):403–414. doi: 10.21037/jtd.2016.02.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borgonio Cuadra V.M. Altered expression of circulating microRNA in plasma of patients with primary osteoarthritis and in silico analysis of their pathways. PLoS One. 2014;9(6):e97690. doi: 10.1371/journal.pone.0097690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci. Rep. 2015;5:11251. doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T. Catheter ablation restores decreased plasma miR-409-3p and miR-432 in atrial fibrillation patients. Europace. 2016;18(1):92–99. doi: 10.1093/europace/euu366. [DOI] [PubMed] [Google Scholar]
- 37.Kendziorski C. On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. U. S. A. 2005;102(12):4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzel E. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int. J. Cancer. 2015;136(4):875–879. doi: 10.1002/ijc.29054. [DOI] [PubMed] [Google Scholar]
- 39.Qu X. Circulating microRNA 483-5p as a novel biomarker for diagnosis survival prediction in multiple myeloma. Med. Oncol. 2014;31(10):219. doi: 10.1007/s12032-014-0219-x. [DOI] [PubMed] [Google Scholar]
- 40.Ren J. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS One. 2013;8(12):e80738. doi: 10.1371/journal.pone.0080738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sondermeijer B.M. Platelets in patients with premature coronary artery disease exhibit upregulation of miRNA340* and miRNA624*. PLoS One. 2011;6(10):e25946. doi: 10.1371/journal.pone.0025946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim K.Y., Chung H.C., Rha S.Y. A weighted sample size for microarray datasets that considers the variability of variance and multiplicity. J. Biosci. Bioeng. 2009;108(3):252–258. doi: 10.1016/j.jbiosc.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 43.Liu P., Hwang J.T. Quick calculation for sample size while controlling false discovery rate with application to microarray analysis. Bioinformatics. 2007;23(6):739–746. doi: 10.1093/bioinformatics/btl664. [DOI] [PubMed] [Google Scholar]
- 44.Oura T., Matsui S., Kawakami K. Sample size calculations for controlling the distribution of false discovery proportion in microarray experiments. Biostatistics. 2009;10(4):694–705. doi: 10.1093/biostatistics/kxp024. [DOI] [PubMed] [Google Scholar]
- 45.Pang H., Jung S.H. Sample size considerations of prediction-validation methods in high-dimensional data for survival outcomes. Genet. Epidemiol. 2013;37(3):276–282. doi: 10.1002/gepi.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira J.A., Zwinderman A. Approximate sample size calculations with microarray data: an illustration. Stat. Appl. Genet. Mol. Biol. 2006;5 doi: 10.2202/1544-6115.1227. [DOI] [PubMed] [Google Scholar]
- 47.van Iterson M. Relative power and sample size analysis on gene expression profiling data. BMC Genomics. 2009;10:439. doi: 10.1186/1471-2164-10-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.