Abstract
Congenital heart disease (CHD) affects up to 1 % of live births1. Although a genetic etiology is indicated by an increased recurrence risk2,3, sporadic occurrence suggests that CHD genetics is complex4. Here, we show that hypoplastic left heart syndrome (HLHS), a severe CHD, is multigenic and genetically heterogeneous. Using mouse forward genetics, we report what is, to our knowledge, the first isolation of HLHS mutant mice and identification of genes causing HLHS. Mutations from seven HLHS mouse lines showed multigenic enrichment in ten human chromosome regions linked to HLHS5–7. Mutations in Sap130 and Pcdha9, genes not previously associated with CHD, were validated by CRISPR–Cas9 genome editing in mice as being digenic causes of HLHS. We also identified one subject with HLHS with SAP130 and PCDHA13 mutations. Mouse and zebrafish modeling showed that Sap130 mediates left ventricular hypoplasia, whereas Pcdha9 increases penetrance of aortic valve abnormalities, both signature HLHS defects. These findings show that HLHS can arise genetically in a combinatorial fashion, thus providing a new paradigm for the complex genetics of CHD.
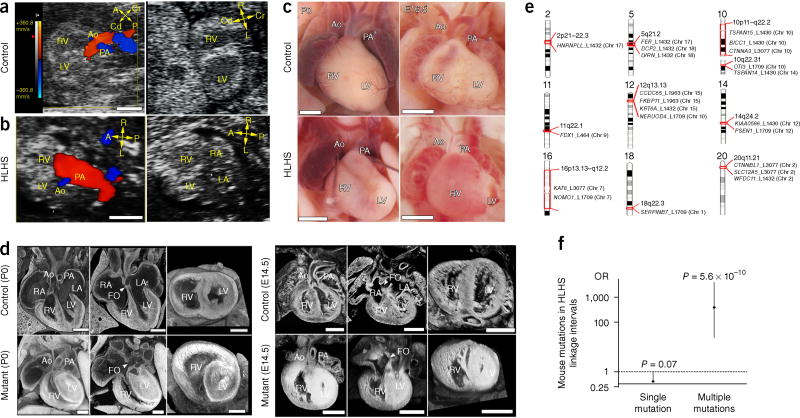
Mouse chemical mutagenesis coupled with ultrasound phenotyping of 100,000 fetal mice has yielded a wide spectrum of CHD mutants8,9, including the recovery of eight HLHS mutant lines (Supplementary Table 1 and Supplementary Figs. 1 and 2). Ultrasound detection of HLHS lines, relative to control (Fig. 1a), showed a narrowed aortic flow (Fig. 1b and Supplementary Video 1) and a small left ventricle (LV) with poor contractility (Fig. 1b and Supplementary Video 2). Hypoplasia of the LV, aorta, and mitral valve (MV)—essential features of HLHS—were confirmed by necropsy and histopathology (Fig. 1c,d and Supplementary Videos 3 and 4). As observed clinically, the small LV often appeared muscle bound with lumen obliteration (Fig. 1d). Unlike other CHD phenotypes, HLHS exhibited nonmendelian segregation, and only one HLHS mutant was recovered per line, except for the Ohia line, which had two mutants (Supplementary Table 1).
Figure 1.
Ohia HLHS phenotype and mouse HLHS mutations in human HLHS linkage intervals. (a–d) HLHS phenotype in Ohia mutants. (a,b) Ultrasound color-flow imaging of normal fetus (a), showing robust flow from the aorta (Ao) and pulmonary artery (PA). In the HLHS mutant (b), the aorta showed only a narrow flow stream, whereas the pulmonary artery showed robust flow. 2D imaging revealed hypoplastic LV (b), as compared with the normal-sized LV in the control (a). (c) Newborn (P0) or E16.5 hearts from wild-type and HLHS mutants. Hypoplastic aorta and LV are visible in the HLHS mutant. (d) Histopathology showing the cardiac anatomy of HLHS mutant and littermate control at birth (P0) and E14.5. Compared with controls, the HLHS mutant exhibited hypoplastic aorta and aortic valve atresia, hypertrophied LV with no lumen, and MV stenosis and patent foramen ovale (FO), arrowhead. LA, left atrium; RA, right atrium; A, anterior; P, posterior; L, left; R, right; Cr, cranial; Cd, caudal. Scale bars: a,b, 0.5 mm; c, 1 mm; d, 0.5 mm. (e) Mouse HLHS mutations in human chromosome regions linked to HLHS and left-ventricular outflow tract obstruction. Genes recovered from mouse HLHS lines found in human chromosome regions linked to HLHS and left-ventricular outflow tract obstructions are shown5–7. The mouse chromosome location of each gene is indicated in parentheses. Mouse mutant lines are identified with L and a numerical identifier. Chr, chromosome. (f) Enrichment of mouse HLHS mutations recovered from seven mutant lines in human chromosome linkage intervals. Genes with mouse HLHS mutations in groups of two or more showed significant enrichment in the HLHS linkage intervals previously identified5–7 (Pearson chi-squared test). This result was not observed for genes analyzed individually, thus indicating that HLHS involves multigenic interactions.
Exome sequencing of the eight HLHS lines recovered 330 coding or splicing mutations, none of which were shared, thus indicating that HLHS is genetically heterogeneous (Supplementary Data 1). Mutations in five Notch-related genes (Rbpj, Ep300, Nap1l1, Ncoa1, and Tspan12), a pathway implicated in HLHS10,11, were identified. Network analysis12 identified seven interconnected functional modules (0.64 modularity; 2.7 scaled modularity) significantly different from random networks (0.59 ± 0.019 modularity (n = 1,000, mean ± s.d.); P value = 0.003) (Supplementary Fig. 3). These results suggested that HLHS may involve disturbance of the cell cycle (KIFC3 and CDK1); muscle differentiation (TRIM63); mitochondrial and Notch signaling (STAT3, EGFR, and EP300); EGF, Wnt, and cadherin signaling (EGFR and CTNNB1); chromatin regulation (EP300); and cardiac hypertrophy (TRIM63, EGFR, and SLC4A1) (Supplementary Data 2). Five HLHS mouse lines had mutations in two or more genes in 10 of 14 human chromosome intervals associated (log of odds (lod) >2) with HLHS and LV outflow obstruction5–7 (Fig. 1e). Significant enrichment was observed when two or more mouse HLHS genes were interrogated across the intervals (odds ratio (OR), 322.5; confidence interval (CI), 24.9–4,177.2; P = 5.6 × 10−10), thus supporting multigenic etiology for HLHS (Fig. 1f).
The Ohia line with two founder HLHS mutants contained five homozygous mutations (Supplementary Data 1 and Supplementary Fig. 1), only two of which are required for HLHS (Table 1): a splice-site mutation in Sap130, which encodes a Sin3A-associated protein in the histone deacetylase complex13,14, and a missense mutation in Pcdha9, which encodes the cell-adhesion protein protocadherinA9 (Supplementary Fig. 4a,b). Both genes are expressed in the embryonic heart (Supplementary Fig. 4e,h–l). Ohia showed transmission of HLHS and related CHD phenotypes, including isolated hypoplastic LV or aorta, and outflow malalignment defects. Some mutants had a double-outlet right ventricle (RV) with hypoplastic LV and normal aorta, or hypoplastic aorta and normal LV (Table 1 and Supplementary Fig. 5a–c), thus indicating that, in contrast to a previous suggestion15, hemodynamic factors are not primary drivers of HLHS. In agreement with this conclusion, fetal echocardiography from embryonic day (E) 13.5–18.5 showed that only right-sided heart structures grew in HLHS fetuses, whereas right and left cardiac chambers and valves grew steadily in normal littermates (Supplementary Videos 1 and 2 and Supplementary Fig. 5d,e). HLHS mutants exhibited poor LV contractility (Supplementary Fig. 5f) with abnormal left-to-right flow through the foramen ovale (Supplementary Video 1). Premature foramen ovale closure and restriction, as previously hypothesized to cause LV hypoplasia, was not observed (Supplementary Fig. 5g).
Table 1.
Genotype–phenotype analysis of Ohia mutants
| CHD Phenotype (n = 142) |
Sap130m/m Pcdha9m/m (n = 88)g |
Sap130m/m Pcdha9m/+ (n = 27)h |
Sap130m/m Pcdha9+/+ (n = 59)i |
Sap130m/+ Pcdha9m/m (n = 12)j |
Sap130+/+ Pcdha9m/m (n = 27)k |
Sap130m/+ or +/+ Pcdha9m/+ or +/+ (n = 380)l |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLHSa (n = 28) | 23 | 0.26 | 3 | 0.11 | 2 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 |
| LV/MV hypoplasiab (n = 13) | 6 | 0.07 | 1 | 0.04 | 6 | 0.10 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hypo Aoc/ aortic stenosisd (n = 37) | 14 | 0.16 | 4 | 0.15 | 13 | 0.22 | 3 | 0.25 | 3 | 0.11 | 0 | 0 |
| Other CHDe (n = 64) | 30 | 0.34 | 14 | 0.52 | 18 | 0.31 | 2 | 0.17 | 0 | 0 | 0 | 0 |
| Totalf | 73 | 0.83 | 22 | 0.81 | 39 | 0.66 | 5 | 0.42 | 3 | 0.11 | 0 | 0 |
Five were DORV-subtype HLHS.
CHD associated with hypoplastic LV and hypoplastic MV, but with normal-sized aorta.
CHD associated with normal-sized LV but hypoplastic aorta (hypo Ao).
One mutant with Sap130+/+; Pcdha9m/m associated with aortic stenosis; two mutants associated with immature thickened aortic valves.
CHD with normal-sized aorta and normal-sized LV, but with an outflow tract malalignment defect, aortic arch anomalies, and hypoplastic RV with tricuspid hypoplasia.
Ohia offspring with CHD in different genotypes.
Left columns, number of mutants; right columns, percentage of mutants with CHD in the indicated genotype.
Comparisons by Fisher’s exact test. To control for multiple testing, Bonferroni correction was applied for six tests (five mouse groups plus an overall group), thus resulting in a P-value threshold of 0.0125 for significance. Genotype g versus i, P = 0.0003; genotype g versus h–j, P = 6.3 × 10−5; genotype g versus i + k, P = 6.0 × 10−7; genotype g versus h–k, P = 6.0 × 10−7, genotype g versus k, P = 0.00093. No significance for comparisons of g versus h, j, or h + j; h versus k; or i versus k, h + i, or j + k.
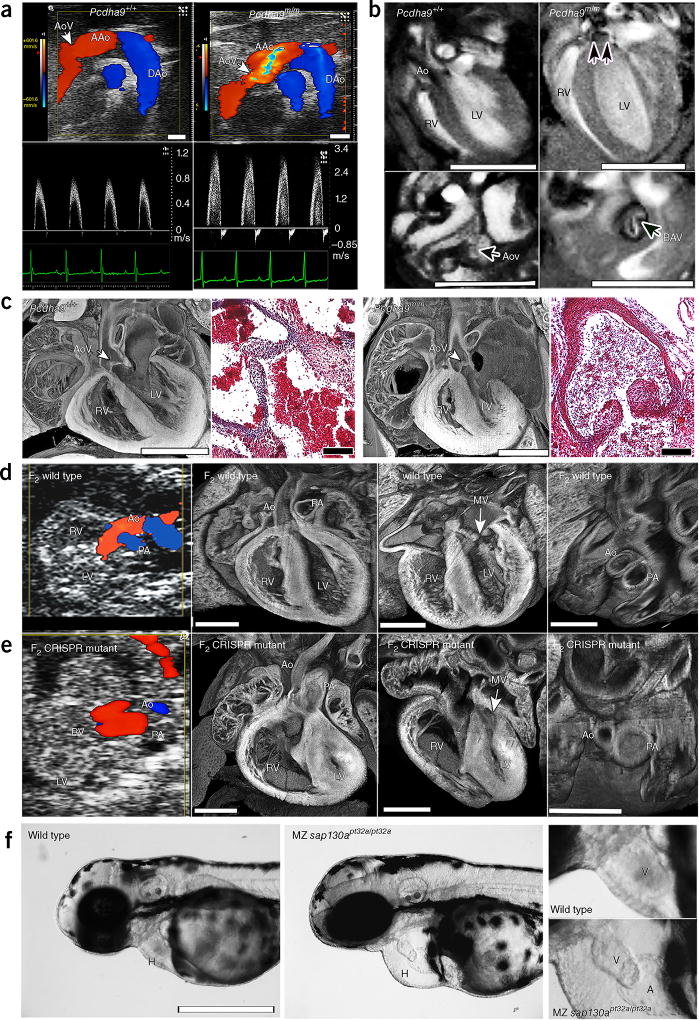
HLHS exhibited the highest penetrance in Sap130m/m; Pcdha9m/m mutants. Penetrance decreased by half in Sap130m/m; Pcdha9m/+ mutants and ninefold in Sap130m/m; Pcdha9+/+ mutants (Table 1). CHD comprising isolated hypoplastic LV was observed in only Sap130m/m mutants (Table 1). Pcdha9m/m mutants demonstrated normal-sized LV with isolated aortic hypoplasia and aortic stenosis and bicuspid aortic valves (BAV), phenotypes linked to HLHS5 (Fig. 2a–c and Table 1). These findings were accompanied by cardiac hypertrophy, which was probably secondary to the aortic stenosis (Supplementary Videos 5–7). This same phenotype was observed with increased penetrance in Pcdha9m/m; Sap130m/+ mutants (25% versus 11%; Table 1). In single mutants or double-heterozygous Sap130; Pcdha9 mutants, no CHD was observed. Overall, HLHS appeared to arise from synergistic interactions between the Sap130 and Pcdha9 mutations, and HLHS was significantly enriched in Sap130m/m; Pcdha9m/m mutants.
Figure 2.
Phenotypes of Pcdha9m/m mice and CRISPR–Cas9-generated Sap130; Pcdha9 mice and sap130a-mutant zebrafish. (a–c) Ohia Pcdha9m/m mutants exhibit aortic valve defects. (a) Echocardiography of adult Pcdha9m/m and wild-type mice. Color-flow (top) and spectral Doppler (bottom) imaging, showing a high-velocity jet (>3 m/s) across the aortic valve, thus indicating aortic stenosis (supplementary Video 5). (b) Functional magnetic resonance imaging (MRI) showing the bicuspid aortic valve. MRI of Pcdham/m mouse from a, showing BAV with doming (double arrows) during systole (supplementary Videos 6 and 7). LV hypertrophy, as determined by M-mode imaging (LV anterior wall/posterior wall: 0.45/0.64 mm in control versus 0.91/0.91 mm in mutant). (c) Histopathology of newborn Pcdha9m/m versus wild-type mice. Compared with wild type, the mutant mouse showed immature thickened aortic valves with increased trichrome staining, thus indicating abnormal matrix deposition. (d,e) CRISPR–Cas9-targeted Sap130; Pcdha9 mutants exhibit HLHS. The CRISPR-generated F2Sap130m/m; Pcdha9m/m mutant showed a small LV with a small flow stream from the ascending aorta. Aortic and pulmonary blood flow originated from the RV, thus indicating a double-outlet RV variant of HLHS (supplementary Video 8), as confirmed by histopathology showing a small LV with hypoplastic aorta and MV (supplementary Video 9). (f) CRISPR–Cas9-targeted sap130a zebrafish exhibiting a small ventricle. Maternal–zygotic (MZ) sap130apt32a/pt32a mutants, compared with wild type, exhibited hearts (H) with small ventricles and pericardial effusion. An enlarged view of the ventricle (V) is shown at right. AAo, ascending aorta; DAo, descending aorta; AoV, aortic valve; LV, left ventricle; RV, right ventricle; Ao, aorta; MV, mitral valve; PA, pulmonary artery; H, heart; V, ventricle; A, atrium. Scale bars: b, 5 mm; a,c (histopathology), 1 mm; c (trichrome stain), 0.1 mm; (d–f) 0.5 mm.
The role of Sap130 and Pcdha9 mutations in causing HLHS was confirmed with CRISPR–Cas9 gene editing16. Analysis of 90 E14.5 F0 CRISPR embryos showed that 52 had at least one gene-targeting event (Supplementary Fig. 6). Of these 52 embryos, 19 exhibited muscle-bound hypoplastic LV with MV stenosis and/or hypoplastic aorta (Supplementary Fig. 7). In one double-gene-targeted embryo, HLHS was observed with hypoplastic LV, aortic valve and MV, and bicuspid aortic stenosis (Supplementary Fig. 7, CC-74). Several Pcdha9-targeted F0 embryos had hypoplastic LV and MV, a result probably reflecting tissue mosaicism commonly observed in F0 CRISPR animals17,18. One CRISPR mouse line showed germline transmission of doubly targeted Pcdha9 and Sap130 alleles. This line expressed the same anomalously spliced Sap130 transcript encoding the 36–amino acid in-frame deletion seen in Ohia mutants (Supplementary Figs. 4d and 8). The Pcdha9 allele comprised a single in-frame amino acid insertion with a deletion of two adjacent amino acids near the site of the Ohia Pcdha9 missense mutation (Supplementary Fig. 8). F1 intercrosses yielded HLHS in double-homozygous F2 offspring, thus confirming the sufficiency of CRISPR-targeted Sap130 and Pcdha9 mutations in causing HLHS (Fig. 2d,e, Supplementary Figs. 1 and 8, and Supplementary Videos 8 and 9). These findings suggested that the Ohia and CRISPR Pcdha9 mutations are both likely to be loss-of-function mutations.
Using a Sap130-lacZ-knockout (KO) first allele (KOMP Repository; URLs), we found that Sap130 is broadly expressed in development (Supplementary Fig. 4f). Homozygous Sap130 KO (Sap130−/−) and compound heterozygous Sap130 KO; Ohia (Sap130−/m) mice exhibited peri-implantation lethality, thus suggesting that the Sap130Ohia allele is hypomorphic (Supplementary Table 2). In agreement with these findings, staining with antibody to Sap130 revealed strong nuclear localization in the Sap130m/m and wild-type heart tissue (Supplementary Fig. 4g–i). Zebrafish sap130a antisense-morpholino knockdown revealed a small-ventricle phenotype at 72 h postfertilization, when the heart comprises only first-heart-field derivatives equivalent to mouse LV progenitors (Supplementary Fig. 9a–c). Quantification showed a decrease in ventricular cardiomyocytes (Supplementary Fig. 9d). The same small-ventricle phenotype was observed with 36% penetrance in maternal–zygotic zebrafish embryos homozygous for a CRISPR-targeted sap130a loss-of-function allele (Fig. 2f and Supplementary Fig. 9e,f). Together with the isolated hypoplastic LV observed in Sap130Ohia mutants (Table 1), these findings indicated that Sap130 is essential for LV growth.
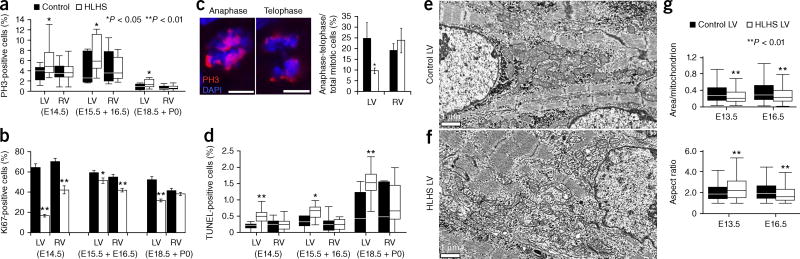
Ohia-mutant hearts exhibited increased phosphorylated histone 3 (PH3) and TUNEL labeling, but decreased Ki67-positive cardiomyocytes in HLHS LV (Fig. 3 and Supplementary Fig. 10). Mitotic cardiomyocytes in anaphase–telophase were decreased (Fig. 3c). Together, these results indicated cell-cycle arrest, in agreement with findings from a previous HLHS human study19. Electron microscopy indicated a cardiomyocyte differentiation defect with short myofilament bundles and more branching and poorly defined Z bands in the HLHS LV myocytes (Fig. 3e,f and Supplementary Fig. 11). The HLHS RV showed similar but milder changes (Supplementary Fig. 12). Mitochondria in the HLHS LV had a low-density matrix with fewer cristae (Fig. 3e,f), as well as shape changes and decreased size; together, these observations suggested a mitochondrial maturation defect (E13.5 and E16.5; Fig. 3g).
Figure 3.
Ohia HLHS mutants show defects in cardiomyocyte proliferation, differentiation, and mitochondria. (a,b) Cardiomyocyte proliferation defects in the HLHS heart. Cardiomyocytes (MF20 positive) in Ohia HLHS LV, compared with controls, showed increased PH3 but decreased Ki67 at E14.5−P0. For RV, a decrease in only Ki67 was observed at E14.5, E15.5–E16.5. Equal numbers of embryos were analyzed for HLHS and littermate controls, with n = 4 independent experiments for E14.5; n = 3 for E15.5 and E16.5; and n = 3 for E18.5 + P0. Wilcoxon rank-sum test in a yielded: E14.5, P = 0.0398 (LV) and P = 0.9248 (RV); E15.5 + E16.5, P = 0.032 (LV) and P = 0.9081 (RV); E18.5 + P0, P = 0.021 (LV) and P = 0.3771 (RV). Unpaired Student’s t-test in b yielded: E14.5, P = 1.6178 × 10−7 (t = 11.62, df = 11) (LV) and P = 0.000329, (t = 5.338, df = 10) (RV); E15.5 + E16.5, P = 0.032 (t = 2.374, df = 14) (LV) and P = 0.001 (t = 4.05, df = 14) (RV); E18.5 + P0, P = 0.000067 (t = 6.526, df = 10) (LV) and P = 0.278 (t = 1.148, df = 10) (RV). (c) Decrease in mitotic cells in anaphase or telophase in HLHS LV. PH3 and DAPI nuclear staining was used to examined anaphase and telophase in three E14.5 HLHS and littermate-control heart samples (n = 3 independent experiments). Total mitotic cells counted: n = 636 control LV, n = 1,347 HLHS LV, n = 635 control RV, and n = 567 HLHS RV. Unpaired Student’s t-test yielded P = 0.048 (t = 2.181, df = 13) (LV) and P = 0.48 (t = −0.726, df = 14) (RV). (d) TUNEL labeling shows increased apoptosis in HLHS LV but not RV. Analysis of same samples from a and b. Wilcoxson rank-sum test yielded: E14.5, P = 0.0001 (LV) and P = 0.9841 (RV); E15.5 + E16.5, P = 0.0122 (LV) and P = 0.6865 (RV); E18.5 + P0, P = 0.0042 (LV) and P = 0.4099 (RV). (e–g) Electron microscopy showing mitochondrial defects in HLHS LV. Control LV (e) and HLHS LV(f) hearts at E13.5 showed HLHS LV with shorter myofibrils with less distinct z-bands and fewer cristae (f). Quantification (g) of E13.5 and E16.5 HLHS LV and control LV samples showed that mitochondria in the HLHS LV were smaller and elongated but became more rounded at E16.5. (Number of mitochondria analyzed (n): E13.5, n = 611 (control) and n = 238 (HLHS); E16.5, n = 886 (control) and n = 1,241 (HLHS). Wilcoxon rank-sum test on mitochondrial area: E13.5, P = 0.000022; E16.5, P = 7.9448 × 10−10. Aspect ratio: E13.5, P = 0.00012; E16.5, P = 1.2282 × 10−13. Scale bars, 5 gm (c) and 1 µm (e,f).
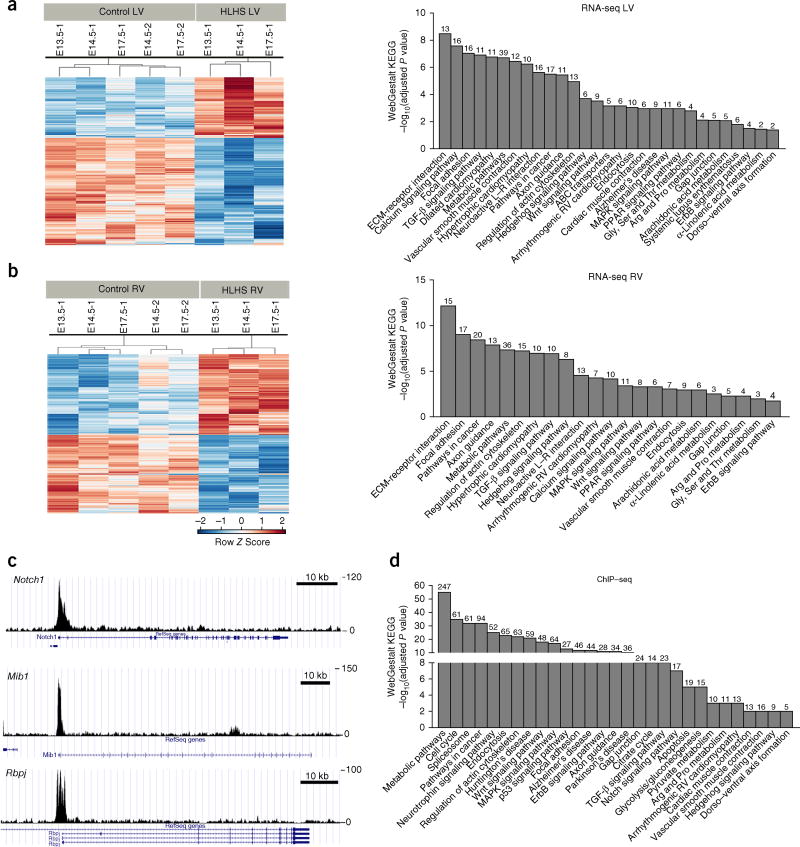
RNA-seq transcriptome profiling in the HLHS LV (Supplementary Data 3) showed changes in metabolic and mitochondria-related pathways, including TGFβ, Wnt, and ErbB signaling; cancer pathway; calcium signaling; cardiac muscle contraction; and dilated/hypertrophic cardiomyopathy (Fig. 4a and Supplementary Data 4). HLHS RV analysis yielded similar pathways with lower fold changes (Fig. 4b and Supplementary Data 4). Sap130 chromatin immunoprecipitation sequencing (ChIP–seq) to identify Sap130 targets yielded many of the same pathways, including TGFβ, Wnt, and ErbB signaling (Fig. 4c,d and Supplementary Data 5). Notable among the Sap130 targets was Meis1, a gene regulating mitochondrial metabolism20 and postnatal cardiomyocyte cell-cycle arrest21 (Supplementary Fig. 13). Also observed were Notch components known to regulate valve development and ventricular compaction22,23 (Rbpj, Mib1, Ep300, and Notch1; Fig. 4c,d and Supplementary Fig. 13). Together, the enriched pathways according to RNA-seq and ChIP–seq showed extensive overlap with the functional modules recovered from the network analysis of the mouse HLHS mutations (Supplementary Fig. 3). ToppGene ToppCluster network analysis24,25 of the RNA-seq and ChIP–seq data together identified pathways related to cardiac differentiation, including contractile fiber, mitochondria, cation transport and ion channels, and calcium binding and signaling from among the downregulated genes, but identified developmental pathways including mesenchyme determination, heart development, and Wnt, TGFβ, and growth-factor signaling from among the upregulated genes, thus indicating dysregulated heart developmental programs (Supplementary Fig. 14 and Supplementary Data 6).
Figure 4.
RNA-seq and Sap130 ChIP–seq analyses show similar pathway enrichment. (a,b) Left, heat maps of RNA-seq conducted with HLHS LV or RV, or littermate-control LV or RV tissues. Shown are log2 c.p.m. values with false discovery rate ≤0.05 and fold change ≥1.5. Right, WebGestalt KEGG pathway enrichment observed for the HLHS LV and HLHS RV RNA-seq. (c) Sap130 ChIP–seq of wild-type mouse whole heart at E12.5, showing Sap130 occupancy over the promoter regions of genes in the Notch signaling pathway (Notch1, Mib1, and Rbpj). Scale bar, 10 kb. (d) WebGestalt KEGG pathway enrichment analysis of Sap130-target genes recovered from Sap130 ChIP–seq. Numbers above bars indicate numbers of genes mapped to each term in the data set.
To interrogate the molecular mechanisms underlying the Sap130–Pcdha9 interactions, we generated a cardiomic expression atlas comprising a compendium of microarray expression data from 118 heart-related samples at different stages of development26–33 (Supplementary Fig. 15). This analysis showed that Pcdha9 expression was largely restricted to an E9.5 cardiac cushion comprising the valve anlage, whereas Sap130 was broadly expressed (Supplementary Fig. 15 and Supplementary Data 7). Pcdha9 and Sap130 expression in the cardiac cushions was confirmed by real-time PCR (Supplementary Fig. 15). Importantly, the Pcdha9-co-regulated genes included many associated with cardiac valve development, including Notch signaling, epithelial–mesenchymal transformation (EMT), and mesenchymal-cell differentiation (Supplementary Figs. 16 and 17). Also observed were genes related to cartilage and bone development, in agreement with known overlap between cartilage/bone and valve developmental pathways34. Recovery of neural crest–related genes reflected the known contribution of the cardiac neural crest in valve development35,36. In contrast, Sap130 co-regulated genes were associated with the mitotic cell cycle, mitotic checkpoint control, and chromatin-regulatory components (Eed, Ezh2, and Suz12) of the Polycomb repressive complex 2 (PRC2). Notably, previous studies have shown that the repressor SIN3A, histone deacetylase (HDAC), PRC2, and lysine-specific demethylase 1 (LSD1) play important roles in cadherin repression regulating EMT37–39. These findings suggest that Sap130 is essential for epigenetic regulation of EMT. This possibility is supported by the presence in the PCDHA gene cluster, of enhancers with SIN3A-binding sites and SIN3A-binding sites mapped by ChIP–seq (Supplementary Fig. 18a,b). Interestingly, the assembled Sap130 and Pcdha9 networks showed interconnections via chromatin and DNA- and histone-modification-related pathways (Supplementary Fig. 17). Also observed to interconnect the Sap130 and Pcdha9 networks was Med23, a transcriptional coactivator known as a cancer-metastasis-suppressor gene that represses EMT40. Together, these findings suggest that Sap130–Pcdha9 interactions may increase HLHS penetrance through epigenetic regulation of EMT mediating cardiac valve development.
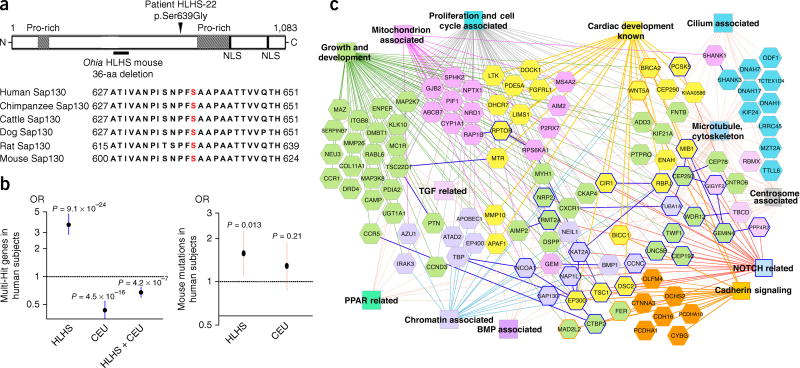
To further examine the genetic etiology of HLHS, we conducted exome sequencing of 68 subjects with HLHS and identified one SAP130 missense mutation in a highly conserved region of the SAP130 protein (Fig. 5a and Supplementary Fig. 19) and 17 PCDHA mutations in 15 subjects (Supplementary Table 3). The subject (HLHS-22) with the SAP130 mutation also had a novel PCDHA13 mutation, a co-occurrence that was unlikely to have been by chance (P = 0.0019; Online Methods). As compared against the ExAC database, our HLHS cohort exhibited greater than tenfold enrichment in PCDHA gene mutations. The PCDHA gene cluster is highly homologous, and the PCDHA13 mutation is situated in a region highly conserved across the gene family (Supplementary Fig. 20). Interestingly, a 16.7-kb deletion spanning PCDHA9 and PCDHA10 is found in multiple human populations41. Although this deletion is said to be present in unaffected individuals, BAV is typically not diagnosed until late adult life and would not affect reproductive fitness. Notably, African Americans have a lower frequency of the PCDHA deletion allele than do Europeans, and they also have a lower BAV incidence42 (P = 0.001). Mice with deletion of Pcdha2 through Pcdha11 (ref. 43), although viable, have not been examined for cardiac phenotype. We found that Pcdha9 mutants with BAV were adult viable. Further studies are warranted to determine the risk of BAV in human subjects with the PCDHA-deletion allele.
Figure 5.
Mutant genes recovered from HLHS mice and subjects with HLHS. (a) SAP130 mutation in subject HLHS-22 with HLHS: a missense mutation affects a highly conserved serine residue (p.Ser639Gly) (red). (b) Left, 68 subjects with HLHS showed enrichment for multihit genes with loss-of-function mutations (plotted as log2 values). This result was not observed for multihit genes shared among the CEU control subjects or between the subjects with HLHS and the CEU control subjects. Right, mouse HLHS-associated genes were in excess among loss-of-function mutations in human subjects with HLHS (plotted as log2 values), but this enrichment was not observed in these subjects compared with 95 CEU control subjects (Pearson chi-squared test). (c) ToppCluster-generated interactome network of multihit genes recovered from subjects with HLHS and HLHS mutant mice. Genes (hexagons) associated with heart development and various gene-ontology biological processes (color rectangles) are shown with corresponding color. Dark-blue-outlined hexagons are NOTCH-related genes. Protein–protein interactions are indicated as solid dark-blue lines between genes.
To assess the multigenic model of disease in HLHS, we interrogated subjects with HLHS for multihit genes, i.e., genes encompassing loss-of-function variants found in two or more subjects (Supplementary Fig. 21 and Supplementary Data 8). Ancestry-matched 1000 genomes CEU subjects served as controls44 (Supplementary Fig. 22 and Supplementary Data 9). Multihit genes were enriched among subjects with HLHS relative to CEU controls (P = 9.1 × 10−24; Fig. 5b and Supplementary Table 4). Mutations from the eight HLHS mouse lines were significantly enriched among multihit genes in the subjects with HLHS (P = 0.013; Fig. 5b and Supplementary Table 4) but not CEU controls (Fig. 5b). ToppGene ToppCluster network analysis of the HLHS mouse and human multihit genes together yielded pathways similar to those observed in the Ohia RNA-seq and ChIP–seq analyses (Fig. 5c). A notable observation was the recovery of the Notch pathway, given previously reported findings of Notch1 variants in subjects with HLHS10,11, with 12 NOTCH-related mutations identified in 8 genes from 12 subjects (Supplementary Table 5). Also recovered were genes in the TGFβ pathway, with eight mutations in eight subjects (Supplementary Table 6). Other pathways identified included mitochondrial related, proliferation and cell cycle, cadherin signaling, and chromatin associated, pathways consistent with the Ohia mouse-model findings and functional modules identified from mutations recovered from the eight HLHS mouse lines (Fig. 5c and Supplementary Fig. 3).
In conclusion, mouse forward genetics facilitated recovery of what are, to our knowledge, the first animal models of HLHS. The multigenic etiology is likely to account for the previous failure to obtain HLHS mutant mice. Sap130 and Pcdha9 mutations were identified and validated as genetic causes of HLHS. This digenic etiology is consistent with a two-locus model previously proposed for HLHS45. The animal modeling indicated that Sap130 drives LV hypoplasia by perturbing cardiomyocyte proliferation and differentiation, whereas protocadherin mutation drives the aortic valve defects. These findings suggest that HLHS is not merely a valve disease46, thereby indicating important therapeutic implications47. The incomplete penetrance observed in mice and zebrafish may reflect epigenetic effects, an expected result, given that Sap130 is a chromatin-modifying protein. Epigenetics, whose critical role in regulating EMT is well described48,49, may affect cardiac valve development not only in HLHS but probably also in other CHD. Although environmental effects also may contribute to incomplete penetrance, similar results were observed for the mouse and zebrafish models with vastly different settings for embryonic development. Together, our findings suggest a new paradigm in which modular phenotypes elicited genetically in a combinatorial fashion may contribute to the complex genetics of HLHS and perhaps to other CHD. These findings provide what is, to our knowledge, the first demonstration of the efficacy of mouse forward genetics in interrogating complex genetics. These findings should have broad relevance for future studies of not only CHD but also other human diseases.
URLs. KOMP Repository Knockout Mouse Project, http://www.komp.org/; 1000 Genomes Project, http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/; BioGRID, http://thebiogrid.org/; Human Protein Reference Database (HPRD), http://www.hprd.org/; ToppGene training gene set, https://toppgene.cchmc.org/enrichment.jsp.
ONLINE METHODS
Ultrasound and MRI assessment of cardiac structure and function
Mouse studies were conducted under an approved University of Pittsburgh Institutional Animal Care and Use Committee protocol. Breeding for the mutagenesis screen was conducted with a G2 female × G1 male backcross breeding scheme in C57BL/6J mice8. In utero ultrasound scanning was performed with a two-tier ultrasound imaging protocol with Acuson Sequoia and Vevo2100 biomicroscopes9. For LV contractility, the areas of the LV chamber in diastole (LVEDA) and systole (LVESA) were measured, and the fraction area change (FAC%) was calculated as (LVEDA – LVESA) × 100/LVEDA. The foramen ovale was imaged by color-flow Doppler in four-chamber view.
Pcdha9m/m adult female mutants and age-matched C57BL/6J female mice (2 months old) were ultrasound interrogated, then analyzed with in vivo cine MRI for further hemodynamic evaluation with a Bruker 7-Tesla MRI (BioSpec 70/30 USR) with a 35-mm quadrature transceiver coil. ECG and respiration-gated multislice cine MRI was acquired with FLASH sequence. The equivalent temporal resolution for the cine loops was ~12.8 ms, with 20–24 phases per cardiac cycle. Cine MRI was acquired with FO, 2.5 cm; slice thickness, 1 mm; in-plane resolution, 0.13 mm; TE, 2.26 ms; TR, 12.92 ms; and flip angle, 150.
Episcopic confocal microscopy histopathology
Fetal and neonatal mice with ultrasound findings indicating CHD were necropsied and embedded in paraffin for episcopic confocal microscopy (ECM)50 for detailed histopathological assessment. The 3D-reconstructed image stacks generated by ECM served as the gold standard for CHD diagnosis. Paraffin sections were collected for histochemical staining, including Masson’s trichrome staining for cardiac valve analysis.
Mouse whole-exome sequencing
Genomic DNA from HLHS mutant mice were sequence captured with an Agilent SureSelect Mouse All Exon kit V1 and sequenced with the Illumina HiSeq 2000 platform (BGI Americas) with a minimum average of 50× target-sequence coverage. Sequence reads were aligned to the C57BL/6J mouse reference genome (mm9), analyzed with GATK, and further processed as previously described8. Genotyping was performed by PCR amplification and Sanger sequencing with forward (F) and reverse (R) primers for point mutations in the five candidate genes (Supplementary Table 7).
Network functional module analysis
The human interactome was assembled with protein–protein interactions (PPIs) with data obtained from BioGRID (URLs)51 and human PPIs from HPRD52 (URLs). We then added PPIs predicted by the computational model High Precision Protein-Protein Interaction Prediction (HiPPIP)53. Next, we identified network modules among HLHS genes and their linker genes with Netbox12. Netbox reports modularity and a scaled modularity score, as compared with the modularity observed in 1,000 random permutations of the subnetwork. Scaled modularity refers to the s.d. difference between the observed subnetwork and the mean modularity of the random networks54. An empirical P value was computed by comparing the observed modularity and the modularity of the randomly permuted subnetworks.
CRISPR–Cas9 Sap130 and Pcdha9 gene editing in the mouse genome
Individual guides were selected on the basis of proximity to the desired base substitution. sgRNA DNA templates were generated by overlapping PCR with a forward primer containing a T7 promoter plus the guide sequence and a common reverse primer containing the stem loop. Forward guide primers (5′ 3′) are listed in Supplementary Table 7. PCR products were subsequently used for in vitro transcription (MegaShort Script; Ambion) to generate sgRNAs.
Single-stranded oligonucleotide (ssODN) donors for Sap130 and Pcdha9 were synthesized as Ultramers (IDT). The Sap130 (T>C) donor contains only a single-nucleotide substitution (underlined) (Supplementary Table 7). The Pcdha9 (C>T) donor carries the desired C>T substitution (underlined) and an additional four silent mutations (uppercase) to prevent recutting of the targeted allele (Supplementary Table 7). The injected embryos (C57BL/6J) were transferred to psuedopregnant mothers, and embryos were collected at E14.5 and genotyped from tail-tip DNA with gene-specific primers (Supplementary Table 7). PCR products were TA cloned and analyzed by Sanger sequencing. Some litters were allowed to deliver, and the resulting F0offspring were further bred to assess germline transmission of the CRISPR–Cas9-targeted Sap130 and Pcdha9 alleles.
Zebrafish antisense-morpholino gene knockdown
The zebrafish experiments were carried out with an approved University of Pittsburgh IACUC protocol with the AB* zebrafish strain. Antisense-morpholino oligonucleotides (MOs) were designed and purchased from Gene Tools. Sap130a (5 ng) translation blocking (Supplementary Table 7) or scrambled MO (5 ng) were injected into one-cell-stage embryos carrying the Tg(5.7myl7:nDsRed2)f2 transgene marker labeling cardiomyocyte nuclei with DsRed (gift from C.G. Burns)55. At 72 h postfertilization (hpf), injected embryos were anesthetized with Tricane, and the hearts were imaged with an Olympus Fluoview FV1000 MPE multiphoton confocal microscope with a 1.12-NA, 25× ELWD objective with a 750-nm laser line, and 3-µm optical slices were collected. 3D reconstructions of confocal stacks were made with Imaris 8.1.2 X64 software (BITPLANE, Andor) and quantification of red-labeled cardiomyocyte nuclei was performed interactively on 3D-reconstructed images.
Generation of zebrafish sap130a mutants
gRNAs were designed to target exon2 of the sap130a gene, according to the methods described by Gagnon et al.56. Briefly, gRNAs containing the target sequence (Supplementary Table 7) were synthesized in vitro and microinjected with codon-optimized Cas9 mRNA into one-cell-stage embryos of the AB* zebrafish strain57. F0 founders were identified with RFLP analysis (with primers in Supplementary Table 7). F1 mutants were confirmed by Sanger sequencing. In one line, pt32a, a 2-bp insertion and 46-bp deletion was detected in sap130a and was used to study heart development.
Sap130 immunostaining
IMCD3 cells (from ATCC) and paraffin sections of mouse embryos were immunostained with mouse anti-Sap130 antibody (ABMART, custom mouse monoclonal ID 14915-1-2/C508_130918; 1:500 for immunofluorescence; 1:100 for DAB immunohistochemistry) and AF555 donkey anti-mouse IgG (A31570, Life Technologies; 1:2,000). Horseradish peroxidase labeling was used for immunodetection of Sap130 in paraffin sections. Anti-Sap130 antibodies were verified by Sap130 siRNA knockdown analysis (described below).
Sap130 siRNA knockdown
The Sap130 siRNA (GS269003) was obtained from Qiagen. IMCD3 cells were transfected with 50 nM Sap130 siRNA with Lipofectamine RNAiMax reagent (Invitrogen), according to the manufacturer’s protocol, and incubated for 4 h at 37 °C. Transfected cells were cultured with fresh medium at 37 °C. After 24 h, cells were fixed and immunostained with the antibody to Sap130.
In situ hybridization
Embryos were fixed in 4% paraformaldehyde–DEPC-treated PBS at 4 °C overnight and processed as described previously43. Plasmid for Pcdha9 in situ probes was obtained from T. Yagi (Osaka University) and has been described previously43. After in situ hybridization, embryos were postfixed in 4% paraformaldehyde at 4 °C overnight, then embedded in O.C.T (Tissue-Tek) and cryosectioned at 20 µm.
Analysis of cardiomyocyte cell proliferation and apoptosis
Cell proliferation was assessed by immunostaining of paraffin sections with antibodies to PH3 (Millipore, Ser10, 2664259, 1:100) and Ki67 (Abcam, ab15580, 1:500). Cardiomyocytes were identified with monoclonal MF20 antibodies (Developmental Studies Hybridoma Bank, AB_2147781, 1:100). Secondary antibodies included AF555 goat anti-rabbit IgG (A21429, 1:1,000) and AF488 goat anti-mouse IgG2b (A21042, 1:1,000) from Life Technologies. DAPI staining allowed for visualization of cell nuclei and cells in anaphase or telophase versus prometaphase or metaphase. Apoptosis was analyzed with terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling (TUNEL) with a Roche TUNEL assay kit (12156792910). Sections were imaged with confocal microscopy and quantitatively analyzed with ImageJ. Antibody validation is provided on the manufacturers’ websites.
Production of Sap130-knockout mice
The Sap130 LacZ-knockout allele (Sap130tm1a(KOMP)Mbp) was obtained from the Knockout Mouse Project (KOMP) via the Jackson Laboratory. Sap130-knockout mice were examined for β-galactosidase expression through X-gal staining. Briefly, Sap130+/LacZ embryos were fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in PBS at 4 °C, rinsed in PBS, and X-Gal stained. Embryos were postfixed in 10% formalin.
Electron microscopy analysis
Heart samples (equal numbers of hearts were analyzed for controls and HLHS mutants, with n = 2 for E13.5 in 4 independent experiments; n = 2 for E16.5 in 4 independent experiments) were fixed in 2% glutaraldehyde and 4% paraformaldehyde in 0.1 M Millonig’s buffer, pH 7.4, and postfixed in 1% buffered osmium tetroxide, dehydrated in a graded ethanol series, and embedded in EPON/Araldite. Thin (70-nm) sections were stained with uranyl acetate/lead citrate and photographed with a Hitachi 7650 TEM, as previously described58. Mitochondrial area (in square micrometers), and aspect ratio (unitless, major axis/minor axis, a measure of length and elongation, 1 = a circle) were analyzed with 6,000/7,000× EM images with ImageJ, as previously described59,60.
Transcript and RNA-seq analysis
RNA was isolated from RV and LV tissue from embryos at E13.5, E14.5, and E17.5 (three total, with one at each stage), and littermate controls (five total, with one at E13.5 and two each at E14.5 and E17.5). cDNA was prepared with a high-capacity RNA-to-cDNA kit (Applied Biosystems). Real-time PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) and an ABI 7900HT Fast Real Time PCR System. The PCR primers used are listed in Supplementary Table 7. Expression data were normalized to β-actin. For splicing analysis, PCR was performed with PrimeSTAR HS DNA Polymerase with the primers listed in Supplementary Table 7. Each PCR fragment was gel purified and sequenced.
For RNA-seq, libraries were constructed with a TruSeq RNA Sample Preparation Kit v2 (Illumina) and sequenced with an Illumina HiSeq 2000 platform (BGI Americas) with 100-bp paired-end reads. The reads were aligned to mm9 (NCBI build 37) with TopHat2 (v2.0.9)61, and gene-level counts were calculated with HTSeq-count (v0.5.4p5)62. Differential expression analyses were performed with edgeR63. Factors of variation were assessed via multidimensional scaling, and dispersion parameters were estimated accounting for region (LV or RV), stage (E13.5, E14.5, or E17.5), and disease (HLHS or control), and their pairwise interactions. For each gene, the maximum of common, trended, and tag-wise dispersion estimates was used for subsequent analyses. Differential expression of HLHS LV versus control LV, and HLHS RV versus control RV were determined with likelihood-ratio tests63. To examine changes in gene expression shared across the three stages, we aggregated each sample type (HLHS LV, HLHS RV, control LV, and control RV), and differentially expressed genes were recovered for HLHS LV and HLHS RV with false discovery rate ≤0.05 (Benjamini–Hochberg) and ≥1.5-fold change. KEGG pathway enrichment was identified with WebGestalt64 (adjusted P ≤0.05).
ChlP–seq analysis
Sap130 chromatin immunoprecipitation was performed with rabbit anti-Sap130 antibody (A302-491A, Bethyl laboratories) and an iDeal ChIP–seq Kit for Transcription Factors (Diagenode). Briefly, E12.5 embryonic hearts were homogenized, pelleted, resuspended in PBS containing 1% formaldehyde, and then incubated for 20 min. Glycine was added to stop cross-linking, and samples were then washed with PBS, resuspended at 4 °C, and sonicated to 100- to 300-bp fragments with a Covaris S2 instrument. Sonicated lysates were cleared by pelleting of insoluble material and were then incubated with antibody-bound Protein A magnetic beads (2 µ;g antibody per 20 µ;l beads) in 300 µl ChIP buffer overnight at 4 °C. Immunoprecipitated material was subsequently un-cross-linked in elution buffer, extracted with magnetic beads, and resuspended in elution buffer. ChIP–seq libraries were generated with an NEBNext Ultra DNA Library Prep Kit Illumina (NEB), and sequencing was carried out on an Illumina HiSeq 4000 platform (BGI Americas). Reads were aligned to the mouse reference genome (mm10) with Bowtie1 (version 1.1.2)65, and 14,888 potential Sap130 peaks were identified with MACS1.4.2 software66. Enriched KEGG pathways were identified with WebGestalt (top 4,000 genes selected on the basis of MACS score).
Cardiomic gene expression atlas and Pcdha9–Sap130 interaction analysis
We used microarray data compiled from nine published microarray data sets from the GEO database (GSE IDs in Supplementary Data 7) encompassing 29 different sample types and 118 independent samples profiled on the Affymetrix mouse MOE430 plus 2.0 gene microarray platform26–33. CEL files were processed with the RMA algorithm, and gene sets were recovered on the basis of filtering for absolute RMA expression levels and Pearson correlation of global median-normalized values. Pcdha-related probes were identified on the basis of similarity to the expression of the 1420798_s_at probe and were probes contained within the 3′ UTR used by multiple members of the Pcdha cluster. Because Pcdha9 expression was strongest in the E9.5 atrioventricular canal (i.e., AV cushion tissue), we filtered for probe sets that exhibited RMA-normalized expression of 8.0 or greater in the E9.5 AV canal and identified a Pcdha9-related gene module of 105 probe sets (69 genes) with Pearson correlation >0.52. Similarly, a second gene module (150 probe sets, 138 genes) that exhibited Sap130-correlated expression in the same samples was identified, and the two modules were enriched and compared with ToppCluster.
Human whole-exome sequencing of subjects with HLHS
All human studies were approved by the Institutional Review Board of the University of Pittsburgh, Cincinnati Children’s Hospital Medical Center and University of California, San Diego, and appropriate informed consent was obtained. Whole-exome capture was performed on 22 subjects with HLHS recruited from Cincinnati, by using NimbleGen EZ human Exome V2 with triplex enrichment, and on 46 subjects from Pittsburgh (n = 26) and San Diego (n = 20), by using an Agilent V4 Exome Capture kit (23.5% females; dbGAP phs001256. v1.p1). Sequencing was performed on the Illumina HiSeq2000 platform with 100 paired-end reads at 100× coverage. Fastq files of all samples were processed with a pipeline as follows: alignment, BWA v0.5.9 with the bwa aln/sampe option; miscellaneous BAM file bookkeeping tasks, Picard v1.120.1579; and variant calling and filtering, GATK v3.3-0 with GATK′s HaplotypeCaller module. Variants were filtered with GATK′s Variant Quality Score Recalibration (VQSR) workflow. Variants that passed VQSR filtration were annotated with predicted transcript and protein changes in the UCSC known genes table (hg19, accessed 23 November 2015). Variants with predicted loss of function (LOF) comprising frameshift, nonsense, or splice site disruption were recovered. Mutations in subject HLHS-22 were validated by Sanger sequencing.
Multihit gene analysis in subjects with HLHS and HLHS mouse mutations
The number of genes used for human comparisons was 21,569. For the multihit gene analysis, we obtained exome sequencing data from 95 ancestry-matched human CEU control subjects from the 1000 Genomes Project (URLs). GATK Select Variants was used to extract only the CEU founders from each chromosome’s data set, which were then combined into a single VCF.
For this analysis, multihit genes are defined as genes with LOF variants seen in two or more subjects (i.e., variant unique to one subject). Subjects with HLHS had 1,736 unique LOF variants in 1,278 genes, whereas controls had 1,550 unique LOF variants in 1,372 genes. The number of multihit genes was compared with the expected number with 2 × 2 contingency tables and Pearson χ2 analysis. For sharing within the hHLHS cohort, the expected number was based on chance sharing ((frequency of variants per gene)2 × (number of possible combinations of 2 out of 68 cases) × (total number of genes)). For sharing within controls and within human cohorts (controls + hHLHS, n = 2,345 genes), the expected number was based on the proportion of sharing seen within the hHLHS cohort.
For comparison of gene sharing between mice and humans, we identified 16,939 genes present in both the mouse and human exome sequencing platforms. The 1,125 LOF variants in genes with mouse homologs recovered in the subjects with HLHS were compared with the 329 mutations recovered in the eight HLHS mouse mutants. For the control comparison, there were 1,149 LOF variants in genes with mouse homologs. Comparison of the overlap between the groups was performed with 2 × 2 contingency tables and Pearson χ2 analysis.
ToppFun and ToppCluster network analysis
With a list of known disease-related genes as a training set (‘seeds’), differentially expressed genes derived from RNA-seq data obtained from HLHS LV and HLHS RV, and target genes identified from Sap130 ChIP–seq were analyzed for functional enrichment with the ToppFun application within the ToppGene Suite24,25. Gene rankings of mutations recovered by exome sequencing analysis were evaluated with a training set of genes strongly associated with cardiac development67 (URLs).
For interactome analysis of genes with unique LOFs, ToppGene also ranked mutations recovered from the HLHS mice and subjects with HLHS on the basis of the connectivity of their protein–protein interactions, as compared with those in the training gene set. Genes that were hit twice in any combination of mouse, human, or mouse–human overlaps were subjected to a global multifeature network analysis with ToppGene with gene ontology, mouse phenotype, and pathway annotation options. All results were downloaded from ToppGene, clustered semantically on the basis of the abstracted annotation terms shown, and rendered with Cytoscape68.
Statistics
In animal experiments, sample sizes as small as three animals per group had 80% power to detect a 3.04-s.d. unit difference between groups at α = 0.05 with a two sample t-test. This effect size was reasonable, given the marked changes observed. The experiments were not randomized, though investigators were blinded to allocation during experiments and outcome assessments. In all studies, comparison of groups was conducted with unpaired Student’s t-test, nonparametric Wilcoxon rank-sum test, or Fisher’s exact test. Statistical analysis was performed with SPSS11.5 or SAS. A two-tailed P value <0.05 was considered statistically significant. For genotype–phenotype analysis (Table 1), Bonferroni correction was applied for six tests (five mouse groups plus an overall group), thus resulting in a P-value threshold of 0.0125 for significance. All values shown are mean ± s.e.m. except for qPCR analyses, which are shown as mean ± s.d.
Enrichment of mouse HLHS genes in human HLHS linkage regions
To determine whether mutations (missense, nonsense, splice, or frameshift) in HLHS mouse lines might play a role in human HLHS, we examined the overlap of the mouse genes with 14 human linkage peaks associated with HLHS5–7. The 1-lod-unit support interval of these linkage peaks encompassed 290,687,087 bp. Under the assumption that the exome is 1% of the genome, there were 2,906,871 potential variants within and 27,093,129 bp outside the linkage region. Variants were classified as being inside or outside the linkage region, and 2 × 2 contingency tables and Pearson χ2 analysis were used to test the significance of enrichment of single hits within the linkage region. To test whether animals were more likely than chance to have multiple hits, we calculated the probability of a specific base pair to have a functional variant (number of variants recovered/number of animals/exome size = 329/7 × 30,000,000 = 0.000001566). The probability of a mouse having two variants within the human linkage intervals equals (the probability of functional variant)2 × number of exomal base pairs in linkage interval × number of combinations for average number of variants (47) = (0.000001566)2 × 2,906,871 × 1,081 = 0.007705. To determine the probability of five of seven HLHS mutant mouse line having two or more putative functional variants in the linkage region, binomial distribution was used. To determine the odds ratio, the expected number was based on 1/probability of two variants present in the linkage interval. The eighth HLHS mutant line, Ohia, was excluded from this analysis, because SAP130 and the PCDHA gene cluster are not found in the human linkage intervals.
Co-occurence of SAP130 and PCDHA mutations
To determine the probability of co-occurrence of Sap130 and Pcdha double mutations in subjects with HLHS, the rate of protein-coding (missense/LOF) variants seen in a cohort of nearly 60,000 people was verified in the ExAC database. The frequency of Sap130 protein-coding variants in ExAC was 0.0012. Likewise, the frequency of Pcdha protein-coding variants in ExAC was 0.0234. Thus, the probability that both alleles would be present in a single individual would be 0.000028. The probability of seeing at least 1 person out of 68 individuals would be 0.0019.
Data availability
All eight HLHS mouse lines are curated in the Mouse Genome Informatics (MGI) database, and cryopreserved sperm is available from the Jackson Laboratory for reanimation. Whole mouse exome sequencing data have been deposited in the CvDC Datahub (https://b2b.hci.utah.edu/gnomex/gnomexGuestFlex.jsp?topicNumber567/), and ChIP–seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE77799. Human exome sequencing data are available from the Database of Genotypes and Phenotypes (dbGaP) under accession number phs001256.v1.p1.
Supplementary Material
Acknowledgments
We thank T. Yagi (Osaka University) for providing plasmid for Pcdha9 in situ probes and A. Handen (University of Pittsburgh) for assisting with programming. We also thank C.G Burns (Harvard Medical School) for providing the transgene marker, Tg (5.7myl7: nDsRed2). This work was supported by funding from the NIH (U01-HL098180 (C.W.L.), R01-HL132024 (C.W.L.), R01-GM104412 (C.W.L.), S10-OD010340 (C.W.L.), R01-MH094564 (M.K.G.), and OD011185 (S.A.M.)), the Children’s Heart Foundation (L.J.M. and D.W.B.), and the Junior Cooperative Society (D.W.B.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Study design, C.W.L.; fetal ultrasound imaging and mutant recovery, X.L.; mouse phenotype analysis, X.L., S.S., Z.C., W.D., L.L., G.C.G. Y.W., M.C.S., W.T.R., and B.G.; mouse breeding and genotyping, X.L., S.S., Z.C., H.Y., and B.C.; mouse exome sequencing analysis, Y.L. and N.T.K.; network analysis, B.J.A. and M.K.G.; cardiomyocyte proliferation and apoptosis, S.S., Z.C., and X.L.; in situ hybridization, Z.C., X.L., and H.Y.; mitochondria function analysis and electron microscopy, Z.C., X.L., G.A.P., and K.L.d.M.B.; Sap130 gene and protein expression analysis, H.Y. and S.S.; LV and RV cardiac-tissue harvesting, RNA extraction, and RNA-seq, X.L., B.G., C.W.L., Z.C., and H.Y.; RNA-seq bioinformatics and ToppGene analysis, A.S.B., D.K., and B.J.A.; Sap130 ChIP–seq, H.Y. and A.S.B.; zebrafish morpholino experiments, M.S., M.T., S.W., and B.G.; zebrafish CRISPR mutant generation and analysis, M.S. and M.T.; CRISPR Sap130 guide-RNA design, W.P.; CRISPR–Cas9 F0 mouse embryo production and analysis, K.A.P., S.A.M., Z.C., G.C.G., M.C.S., and X.L.; CRISPR–Cas9 F0 founder-mouse production and F1 offspring propagation, K.A.P., S.A.M., and X.L.; mouse microarray analysis, B.J.A., C.W.L., and H.Y.; recruitment of subjects with HLHS and sample collection, C.W.L., O.K., L.J.M., D.W.B., and P.G.; human exome sequencing analysis and multigene human and mouse comparisons, C.W.L., X.L., B.J.A., A.B., A.S.B., L.J.M., and P.D.; statistics, L.J.M., M.Z., and X.L.; manuscript preparation, C.W.L., X.L., L.J.M., D.W.B., G.A.P., K.A.P., M.T., B.J.A., D.K., A.S.B., P.D., and H.Y.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 2.Gill HK, Splitt M, Sharland GK, Simpson JM. Patterns of recurrence of congenital heart disease: an analysis of 6,640 consecutive pregnancies evaluated by detailed fetal echocardiography. J. Am. Coll. Cardiol. 2003;42:923–929. doi: 10.1016/s0735-1097(03)00853-2. [DOI] [PubMed] [Google Scholar]
- 3.Øyen N, et al. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- 4.Benson DW, Martin LJ, Lo CW. Genetics of hypoplastic left heart syndrome. J. Pediatr. 2016;173:25–31. doi: 10.1016/j.jpeds.2016.02.052. [DOI] [PubMed] [Google Scholar]
- 5.Hinton RB, et al. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J. Am. Coll. Cardiol. 2009;53:1065–1071. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride KL, et al. Linkage analysis of left ventricular outflow tract malformations (aortic valve stenosis, coarctation of the aorta, and hypoplastic left heart syndrome) Eur. J. Hum. Genet. 2009;17:811–819. doi: 10.1038/ejhg.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin LJ, et al. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum. Genet. 2007;121:275–284. doi: 10.1007/s00439-006-0316-9. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, et al. Interrogating congenital heart defects with noninvasive fetal echocardiography in a mouse forward genetic screen. Circ Cardiovasc Imaging. 2014;7:31–42. doi: 10.1161/CIRCIMAGING.113.000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iascone M, et al. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin. Genet. 2012;81:542–554. doi: 10.1111/j.1399-0004.2011.01674.x. [DOI] [PubMed] [Google Scholar]
- 11.Theis JL, et al. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum. Genet. 2015;134:1003–1011. doi: 10.1007/s00439-015-1582-1. [DOI] [PubMed] [Google Scholar]
- 12.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Taketazu M, Barrea C, Smallhorn JF, Wilson GJ, Hornberger LK. Intrauterine pulmonary venous flow and restrictive foramen ovale in fetal hypoplastic left heart syndrome. J. Am. Coll. Cardiol. 2004;43:1902–1907. doi: 10.1016/j.jacc.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen ST, et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver D, Yuan S, McSwiggin H, Yan W. Pervasive genotypic mosaicism in founder mice derived from genome editing through pronuclear injection. PLoS One. 2015;10:e0129457. doi: 10.1371/journal.pone.0129457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaber N, et al. Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. Am. J. Pathol. 2013;183:720–734. doi: 10.1016/j.ajpath.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 20.Tomoeda M, et al. Role of Meis1 in mitochondrial gene transcription of pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2011;410:798–802. doi: 10.1016/j.bbrc.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 21.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luxán G, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 23.MacGrogan D, Luna-Zurita L, de la Pompa JL. Notch signaling in cardiac valve development and disease. Birth Defects Res. A Clin. Mol. Teratol. 2011;91:449–459. doi: 10.1002/bdra.20815. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–W102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MP, Yutzey KE. Twist1 directly regulates genes that promote cell proliferation and migration in developing heart valves. PLoS One. 2011;6:e29758. doi: 10.1371/journal.pone.0029758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera-Feliciano J, et al. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development. 2006;133:3607–3618. doi: 10.1242/dev.02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano H, et al. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 2013;4:1564. doi: 10.1038/ncomms2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Loo PF, et al. Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol. Cell. Biol. 2007;27:8571–8582. doi: 10.1128/MCB.01350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manuylov NL, Tevosian SG. Cardiac expression of Tnnt1 requires the GATA4-FOG2 transcription complex. Scientific World Journal. 2009;9:575–587. doi: 10.1100/tsw.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijaya M, et al. Differential gene expression profiles during embryonic heart development in diabetic mice pregnancy. Gene. 2013;516:218–227. doi: 10.1016/j.gene.2012.12.071. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Fernandez A, Li X, Hartjes KA, Terzic A, Nelson TJ. Natural cardiogenesis-based template predicts cardiogenic potential of induced pluripotent stem cell lines. Circ Cardiovasc Genet. 2013;6:462–471. doi: 10.1161/CIRCGENETICS.113.000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magarin M, Schulz H, Thierfelder L, Drenckhahn JD. Transcriptional profiling of regenerating embryonic mouse hearts. Genom. Data. 2016;9:145–147. doi: 10.1016/j.gdata.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty S, Cheek J, Sakthivel B, Aronow BJ, Yutzey KE. Shared gene expression profiles in developing heart valves and osteoblast progenitor cells. Physiol. Genomics. 2008;35:75–85. doi: 10.1152/physiolgenomics.90212.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Lange FJ, et al. Lineage and morphogenetic analysis of the cardiac valves. Circ. Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ. Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- 37.Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 2004;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- 38.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardin WA, Haier J, Mees ST. Epigenetic regulation and role of metastasis suppressor genes in pancreatic ductal adenocarcinoma. BMC Cancer. 2013;13:264. doi: 10.1186/1471-2407-13-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noonan JP, et al. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am. J. Hum. Genet. 2003;72:621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra S, et al. Bicuspid aortic valve: inter-racial difference in frequency and aortic dimensions. JACC Cardiovasc. Imaging. 2012;5:981–989. doi: 10.1016/j.jcmg.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noguchi Y, et al. Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J. Biol. Chem. 2009;284:32002–32014. doi: 10.1074/jbc.M109.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride KL, et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am. J. Med. Genet. A. 2005;134A:180–186. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinton RB., Jr Hypoplastic left heart syndrome is heritable. J. Am. Coll. Cardiol. 2007;50:1590–1595. doi: 10.1016/j.jacc.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 47.Freud LR, et al. Fetal aortic valvuloplasty for evolving hypoplastic left heart syndrome: postnatal outcomes of the first 100 patients. Circulation. 2014;130:638–645. doi: 10.1161/CIRCULATIONAHA.114.009032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christofori G. Snail1 links transcriptional control with epigenetic regulation. EMBO J. 2010;29:1787–1789. doi: 10.1038/emboj.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Y, Dong C, Zhou BP. Epigenetic regulation of EMT: the Snail story. Curr. Pharm. Des. 2014;20:1698–1705. doi: 10.2174/13816128113199990512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Tobita K, Francis RJ, Lo CW. Imaging techniques for visualizing and phenotyping congenital heart defects in murine models. Birth Defects Res. C Embryo Today. 2013;99:93–105. doi: 10.1002/bdrc.21037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keshava Prasad TS, et al. Human Protein Reference Database: 2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganapathiraju MK, et al. Schizophrenia interactome with 504 novel protein-protein interactions. NPJ Schizophr. 2016;2:16012. doi: 10.1038/npjschz.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Zhang J. In search of the biological significance of modular structures in protein networks. PLOS Comput. Biol. 2007;3:e107. doi: 10.1371/journal.pcbi.0030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 56.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hom JR, et al. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev. Cell. 2011;21:469–478. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hom J, Yu T, Yoon Y, Porter G, Sheu SS. Regulation of mitochondrial fission by intracellular Ca2+ in rat ventricular myocytes. Biochim. Biophys. Acta. 2010;1797:913–921. doi: 10.1016/j.bbabio.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picard M, White K, Turnbull DM. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J. Appl. Physiol (1985) 2013;114:161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricci M, et al. Myocardial alternative RNA splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PLoS One. 2012;7:e29784. doi: 10.1371/journal.pone.0029784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu C, Wu C, Aronow BJ, Jegga AG. Computational approaches for human disease gene prediction and ranking. Adv. Exp. Med. Biol. 2014;799:69–84. doi: 10.1007/978-1-4614-8778-4_4. [DOI] [PubMed] [Google Scholar]
- 68.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All eight HLHS mouse lines are curated in the Mouse Genome Informatics (MGI) database, and cryopreserved sperm is available from the Jackson Laboratory for reanimation. Whole mouse exome sequencing data have been deposited in the CvDC Datahub (https://b2b.hci.utah.edu/gnomex/gnomexGuestFlex.jsp?topicNumber567/), and ChIP–seq and RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE77799. Human exome sequencing data are available from the Database of Genotypes and Phenotypes (dbGaP) under accession number phs001256.v1.p1.