Abstract
Introduction
Expression of breast cancer metastasis suppressor 1 (BRMS1) is decreased in non–small cell lung cancer cells and tumors. We hypothesized that intratumoral BRMS1 expression is associated with lung adenocarcinoma (LUAD) histologic subtypes and overall survival (OS) and disease-free survival (DFS) in patients undergoing resection for early-stage LUAD.
Methods
Patients (n=1030) who underwent complete resection for LUAD with tissue available for histologic evaluation were identified. Tissue microarrays were constructed, and immunostaining was performed and scored for intensity of BRMS1 expression. OS and DFS were estimated (Kaplan-Meier method) and compared between groups (log-rank test), stratified by stage. Hazard ratios (HRs) for hazard of death and recurrence were estimated using univariable and multivariable Cox proportional hazards models. OS and DFS nomograms were created, and model performance was examined.
Results
Intratumoral BRMS1 expression was high in 632 (61%) and low in 398 (39%) patients. Low BRMS1 expression was associated with higher pathologic T stage (P=0.001), larger tumor size (P≤0.0001), greater lymphatic (P=0.032) and vascular (P=0.001) invasion, LUAD histologic subtypes (P=0.001), and intermediate and high architectural tumor grade (P=0.003). Low BRMS1 expression was an independent predictor of worse OS (HR, 1.35 [95% CI, 1.10–1.65]; P=0.004) and DFS (HR, 1.27 [95% CI, 1.05–1.54]; P=0.012). OS and DFS nomograms showed excellent predictive performance based on discrimination and calibration.
Conclusions
Among patients with surgically resected LUAD, OS and DFS were significantly worse in low intratumoral BRMS1 expression. Our findings suggest BRMS1 is an independent biomarker with prognostic significance in surgically resected LUAD.
Keywords: breast cancer metastasis suppressor 1, lung adenocarcinoma, histologic subtype, biomarker, metastasis
Introduction1
Risk factors to identify patients with surgically resectable lung adenocarcinoma (LUAD) who have a high risk of distant recurrence are poorly defined.1 Currently, the most important prognostic factor for LUAD is tumor-nodal-metastasis (TNM) stage.2 In addition to TNM stage, several clinicopathologic prognostic factors have been investigated, albeit in heterogeneous study populations. These include lymphovascular invasion,3–5 visceral pleural invasion,4,5 and tumor size.6,7 Our group and others have identified specific LUAD histologic subtypes that are associated with poor prognosis.8–10 This suggests that specific LUAD histologic subtypes have an underlying biology that is associated with an increased proclivity for developing metastases. More recently, genomic and immunologic profiling of LUAD has identified several genomic perturbations associated with poor prognosis, including mutations in KRAS, p53, and PI3K and tumor PD-L1 immunoreactivity.11–14 Whereas considerable effort has been focused on understanding the mechanisms of metastases in advanced-stage lung cancer, little work has been done to identify biomarkers of metastases in early-stage, surgically resected LUAD.
Breast cancer metastasis suppressor 1 (BRMS1), is expressed in all normal human tissues, maps to chromosome 11q13.1-13.2 and contains a helix-turn-helix DNA binding domain and coiled-coiled domains, which suggests it may be part of a transcription complex.15,16 BRMS1 is one of >25 known metastasis suppressor genes that, together with their encoded proteins, inhibit metastasis formation in vivo without altering primary tumor formation.17,18 Previous studies have shown that BRMS1 mRNA and protein are decreased in non-small cell lung cancer (NSCLC) cells and patient tumor samples, compared with normal epithelial lung cells and adjacent noncancerous lung tissues.19,16 These observations suggest that lower intratumoral BRMS1 expression may be a putative biomarker of an increased risk of developing metastases.
We hypothesized that intratumoral BRMS1 expression is an independent prognostic marker of overall survival (OS) and disease-free survival (DFS) and that it contributes to a prediction model of increased metastatic potential in a large cohort of patients with surgically resected LUAD. To experimentally address this hypothesis, we used the reporting recommendations for tumor marker prognostic study (REMARK) guidelines,20,21 as well as the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) prediction modeling criteria.22,23
Materials and Methods
Patients
This study was approved by the Institutional Review Board (WA0269-08) at Memorial Sloan Kettering Cancer Center (MSK). We reviewed 1030 patients who underwent an R0 surgical resection for LUAD at MSK between 1995 and 2009 and had tumor blocks available for construction of tissue microarrays (see CONSORT diagram, Supplemental Figure 1). Median follow-up was 5 years (range, 0–14 years). Clinicopathologic data were collected from a prospectively maintained database. Disease stage was based on the seventh edition of the American Joint Committee on Cancer Staging Manual.24
Tissue Microarrays
As previously reported by our group,25,26 histologic subtyping of tumors was based on review of H&E slides from the surgical resection specimens. We then used the formalin-fixed, paraffin-embedded tumor specimens to construction our tissue microarrays. In brief, four to nine representative tumor areas from the most predominant histologic pattern or the second most predominant pattern were marked on hematoxylin and eosin (H&E)–stained slides, and cylindrical 0.6-mm tissue cores were arrayed from the corresponding paraffin blocks into a recipient block by an automated tissue arrayer (ATA-27; Beecher Instruments, Sun Prairie, WI).
Immunohistochemical Analysis and Scoring of BRMS1
In brief, 4-μm-thick sections from the microarray blocks were deparaffinized. Antigen retrieval was performed using citrate buffer (pH 6.0). The standard avidin-biotin-peroxidase complex was used for immunostaining of anti-BRMS1 antibody (EPR7202/ab134968 [Abcam, Cambridge, MA], diluted at 1:250). Sections were stained using a Ventana Discovery XT automated immunohistochemical stainer (Ventana Medical Systems, Tucson, AZ). Noncancerous adjacent lung tissues were stained as positive controls in parallel with the study tissues.
BRMS1 expression was initially evaluated using two components: distribution and intensity. Nuclear BRMS1 expression was diffusely expressed in at least 50% of the tumor area in each core distribution and was therefore not a good discriminator. Other groups have used distribution as a scoring criterion, and this may be related to antibody type, dilution, or tissue type undergoing immunohistochemical analysis—all of which were different in our study. Accordingly, BRMS1 expression was evaluated on the basis of intensity of nuclear immunostaining. BRMS1 nuclear staining was scored on the basis of intensity, as follows: 0+ = no staining, 1+ = weak, 2+ = moderate, and 3+ = strong. Staining of only the tumor cells was used when determining BRMS1 staining intensity. One score per core was given. When a core exhibited heterogeneous tumor cell staining, the maximal score was used to give one score per core. For example, if 60% of the tumor cells showed moderate nuclear staining (2+) and 40% showed strong staining (3+), the core would be scored as 3+. Cores lacking tumor cells were disregarded. Four to 9 cores were used per patient; 395 patients had 4 cores each, 106 patients had 5 cores each, and 656 patients had 9 cores each. The median number of tumor cores per patient available for analysis was 5 (25th–75th percentile, 4–7 cores). The mean of all the patients’ cores was used as the total score. Consistent with other BRMS1 immunostaining scoring, low BRMS1 expression was defined as an intensity score of 0–2 and high expression as a score of 3.36,38 A pathologist blinded to patient outcomes and demographic characteristics independently analyzed the tissue microarrays. To determine concordance and agreement rates between pathologists, a second pathologist blinded to the analysis of the first pathologist examined 100 randomly selected patient samples (10% of cohort, > 500 cores).
Histologic Subtype Evaluation
H&E-stained tumor slides were reviewed by two pathologists, blinded to clinical outcomes. Discrepancies in assignment of predominant histologic subtype between pathologists were resolved by consensus. Invasive adenocarcinomas were classified according to the 2015 WHO classification27 and the 2011 IASLC/ATS/ERS classification. Invasive adenocarcinoma was subdivided into lepidic-, acinar-, papillary-, micropapillary-, and solid-predominant subtypes.28 Minimally invasive adenocarcinoma (n=29) was grouped with the lepidic histologic subtype. Tumors were also grouped by architectural grade, as low (lepidic predominant), intermediate (papillary or acinar predominant), or high (micropapillary or solid predominant).8,29 Nuclear features, lymphatic invasion, and vascular invasion were also assessed.
Statistical Analysis
Patient demographic and clinical characteristics were summarized using descriptive statistics. The association between clinicopathologic factors and BRMS1 expression (low vs. high) was analyzed using Fisher’s exact test for categorical variables and the Wilcoxon rank sum test for continuous variables.
The outcomes of interest were OS and DFS. Both OS and DFS were estimated by the Kaplan-Meier method: OS was defined from the time of surgery to the time of death from any cause; DFS was measured from the time of surgery to the time of any recurrence or death from any cause. Patients were censored at the date of last follow-up. Associations between factors and survival were analyzed using the log-rank test and univariable Cox proportional hazards regression model. Univariable analyses, including those with BRMS1 as the variable of interest, were stratified by pathologic stage. Key interactions between factors of interest were examined using Cox proportional hazards models for OS and DFS, including both main effects and the interaction term, and were reported as hazard ratios (HRs). The proportional hazards assumption for the Cox models of both outcomes was met on the basis of assessments of the scaled Schoenfeld residuals.
For multivariable models of OS and DFS, variable selection was guided by the all subset method using Akaike Information Criterion,30 including main and interaction terms with P<0.1 from the univariable analyses. On the basis of published literature identifying factors associated with OS and DFS in this population,1,8–10,31 some variables were chosen to remain in the multivariable models regardless of significance: IASLC predominant subtype, extent of resection (pneumonectomy, lobectomy, bilobectomy, and wedge), and pathologic stage. Continuous variables were tested for nonlinearity using restricted cubic splines, whereas categorical covariates were included as dummy variables. Overall P values of categorical factors were calculated on the basis of Wald tests of linear hypotheses of the dummy variables.
The performance of the nomogram models was evaluated by discrimination (Harrell’s C-index),32 calibration (calibration plots), and overall accuracy integrated up to 5 years after surgery (Integrated Brier Score).33 The C-index describes the proportion of pairs in which the responder has a higher predicted probability than the nonresponder; a C-index value closer to 1 implies that the nomogram has good discriminatory ability. Calibration plots visually compare the nomogram-predicted survival probabilities with the observed survival probabilities in groups with approximately equal sample sizes; the calibration curve would lie on the diagonal 45-degree line in an ideal nomogram. To overcome the issue of optimism from validating the multivariable models using the development cohort, we used bootstrapping to obtain bias-corrected (overfitting-corrected) estimates of the performance measures. We report results of the performance measures from both apparent performance (without bootstrap adjustments) and optimism-corrected performance (bootstrap corrected).
The OS and DFS nomograms were developed using the multivariable Cox models, which allowed for calculations of survival probability estimates. The predictions of both OS and DFS nomograms were calculated for 5 years after surgery, as 5 years was the median follow-up time distribution for the cohort.
To assess the interrater reliability (IRR) of the scoring system, we used two chance-corrected agreement measures: Cohen’s kappa and Gwet’s AC1. Cohen’s kappa is a conventional summary measure of the IRR given two raters. We also present the IRR measured by Gwet’s AC1, which takes into account prevalence bias and potential “prevalence paradox,” which occurs when one response is extremely common.34 Both measures range from −1 to 1. Following the guidelines from Landis et al., a kappa statistic of 0.00–0.20 is considered to have slight agreement, 0.21–0.40 has fair agreement, 0.41–0.60 has moderate agreement, 0.61–0.80 has substantial agreement, and 0.81–1.00 has almost perfect agreement.35 In addition, percent raw agreement was used to calculate the percentage of time that the raters agreed. Statistics were calculated with R 3.3.1., using the IRR package, and Gwet’s user-written R functions.36
All other statistical tests were 2-sided, and P<0.05 was considered statistically significant. Statistical analyses were performed using R 3.3.1 (R Development Core Team, Vienna, Austria) with the “glmulti,” “pec,” and “rms” packages (downloaded in January 2017) and Stata 13 (StataCorp, 2013, College Station, TX).
Results
Association between BRMS1 Expression and Clinicopathologic Factors
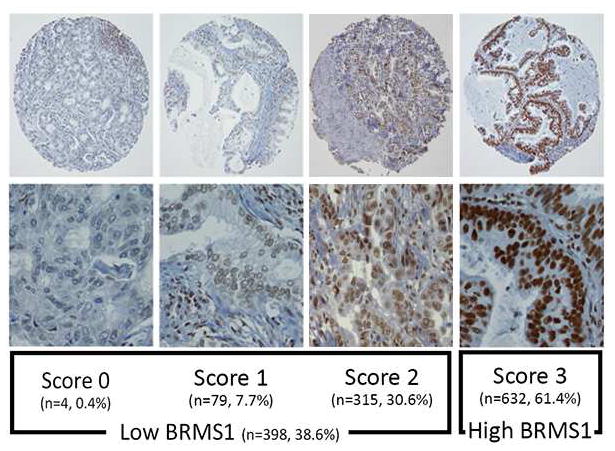
Representative photomicrographs of BRMS1 immunostaining are shown in Figure 1. Analysis of BRMS1 expression for the entire cohort (n=1030) revealed scores of 0 (n=4 [0.4%]), 1 (n=79 [8%]), 2 (n=315 [31%]), and 3 (n=632 [61%]). Thus, 398 LUAD specimens (39%) had low BRMS1 expression, and 632 (61%) had high expression (Figure 1).
Figure 1.
Immunohistochemical analysis of BRMS1. Representative photomicrographs show different expression scores of BRMS1 immunostaining in lung adenocarcinoma. Magnification: upper row, 10X; lower row, 40X.
BRMS1 expression levels relative to specific clinicopathologic variables were then examined (Table 1). Of the 1030 patients with clinical stage I disease, 880 (85%) had pathologic stage I disease. There were slightly more women (65% vs. 58%; P=0.021) in the high- than the low-expression group. More high-expression tumors were T1a (47% vs. 34%; P=0.001), and high-expression tumors were smaller (2.0 vs. 2.4 cm; P<0.0001). We next examined common histopathologic markers associated with tumor aggressiveness, invasion, and metastases. Lymphatic invasion (P=0.032), vascular invasion (P=0.001), and combined lymphovascular invasion (P=0.015) were all more common in the low-expression group.
Table 1.
Association between BRMS1 Expression and Clinicopathologic Factors
| Variable | High BRMS1 (N=632; 61%) | Low BRMS1 (N=398; 39%) | P |
|---|---|---|---|
| Age, years, median (range) | 69.0 (62.0–75.0) | 69.0 (62.0–76.0) | 1.0 |
| Sex | |||
| Female | 411 (65) | 230 (58) | 0.021 |
| Male | 221 (35) | 168 (42) | |
| Smoking status (n=1026) | |||
| Never | 107 (17) | 63 (16) | 0.9 |
| Former | 445 (71) | 287 (72) | |
| Current | 76 (12) | 48 (12) | |
| Extent of resection | |||
| Pneumonectomy | 7 (1) | 6 (2) | 0.022 |
| Lobectomy/bilobectomy | 486 (77) | 327 (82) | |
| Segmentectomy | 47 (7) | 32 (8) | |
| Wedge | 92 (15) | 33 (8) | |
| Pathologic T category | |||
| 1a | 296 (47) | 135 (34) | 0.001 |
| 1b | 131 (21) | 106 (27) | |
| 2a | 189 (30) | 142 (36) | |
| 2b | 9 (1) | 7 (2) | |
| 3 | 6 (1) | 8 (2) | |
| 4 | 1 (<1) | 0 (0) | |
| Pathologic N category (n=1025) | |||
| 0 | 549 (87) | 348 (88) | 1 |
| 1 | 39 (6) | 25 (6) | |
| 2 | 40 (6) | 23 (6) | |
| x | 1 (<1) | 0 (0) | |
| Pathologic M category | |||
| 0 | 632 (100) | 398 (100) | NA |
| Pathologic stage | |||
| IA | 384 (61) | 227 (57) | 0.6 |
| IB | 154 (24) | 115 (29) | |
| IIA | 40 (6) | 24 (6) | |
| IIB | 8 (1) | 6 (2) | |
| IIIA | 46 (7) | 26 (6) | |
| Gross tumor size, cm, median (range) | 2.0 (1.5–2.8) | 2.4 (1.7–3.1) | <0.0001 |
| Lymphatic invasion (n=1029) | |||
| Absent | 386 (61) | 215 (54) | 0.032 |
| Present | 246 (39) | 182 (46) | |
| Vascular invasion (n=1029) | |||
| Absent | 448 (71) | 243 (61) | 0.001 |
| Present | 184 (29) | 154 (39) | |
| Lymphovascular invasion (n=1029) | |||
| Absent | 325 (51) | 173 (44) | 0.015 |
| Present | 307 (49) | 224 (56) | |
| IASLC/ATS/ERSa subtype (n=1028) | |||
| Lepidic | 84 (14) | 28 (7) | 0.001 |
| Acinar | 253 (40) | 164 (41) | |
| Papillary | 144 (23) | 97 (24) | |
| Micropapillary | 37 (6) | 43 (11) | |
| Solid | 112 (18) | 66 (17) | |
| Architectural grade | |||
| Low | 86 (14) | 28 (7) | 0.003 |
| Intermediate | 397 (63) | 261 (66) | |
| High | 149 (24) | 109 (27) | |
Data are no. (%), unless otherwise noted.
IASLC/ATS/ERS classification of lung adenocarcinoma.28
Lepidic-predominant tumors (P=0.001) were more common in the high-expression group. Additionally, intermediate and high histologic architectural tumor grade were more common in the low-expression group (P=0.003; Supplemental Figure 2). Interestingly, micropapillary-predominant tumors (n=80 [8% of cohort]) had the highest percentage of cells with low BRMS1 expression (54%); alternatively only 25% of lepidic-predominant tumors had low BRMS1 expression.
Relationship between BRMS1 Expression and OS
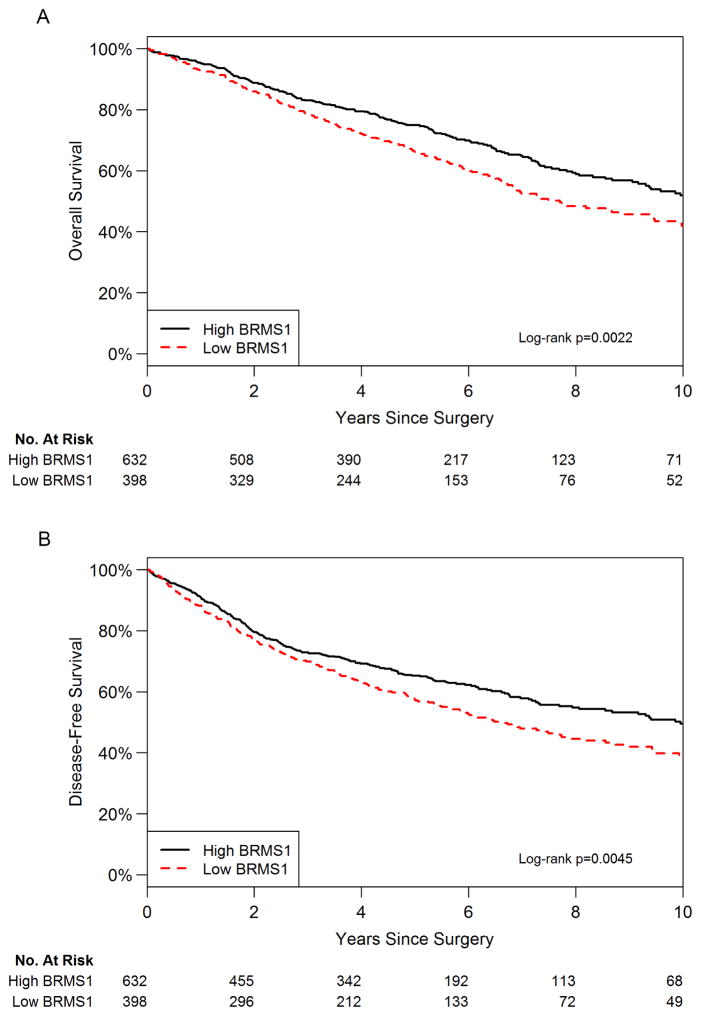
In the high-expression group, 209 (33%) patients died from any cause, with 423 (67%) alive at the end of the study. Comparatively, in the low-expression group, 215 (54%) patients died from any cause, with 183 (46%) alive at the end of the study. Five-year OS was 75% (95% confidence interval [CI], 71%–78%) for the high-expression group and 66% (95% CI, 61%–71%) for the low-expression group. Corresponding median OS were 10.7 (95% CI, 9.3–12.1) and 7.7 (95% CI, 6.7–9.5) years. Stratified log-rank test indicated significant difference in OS between the two groups (P=0.002; Figure 2A). In univariable analysis, low BRMS1 expression was significantly associated with worse OS (HR, 1.37 [95% CI, 1.12–1.67]; P=0.002; Table 2). As a continuous variable in reverse order, lower BRMS1 expression was associated with worse OS (HR, 1.21 [95% CI, 1.05–1.39]; P=0.007).
Figure 2.
Low intratumoral BRMS1 expression is associated with reduced (A) overall survival and (B) disease-free survival.
Table 2.
Univariable and Multivariable Cox Models for Overall Survival
| Variable | Univariable | Multivariable | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| HR | 95% CI | Pa | Overall Pa | HR | 95% CI | P | Overall P | |
| Age | 1.03 | 1.02–1.05 | <0.0001 | 1.03 | 1.02–1.04 | <0.0001 | ||
| Sex: male vs female | 1.47 | 1.20–1.79 | 0.0002 | 1.36 | 1.11–1.66 | 0.003 | ||
| Smoking status | ||||||||
| Never | 1.00 | — | — | 0.089 | ||||
| Former | 1.16 | 0.88–1.54 | 0.3 | |||||
| Current | 1.51 | 1.04–2.18 | 0.030 | |||||
| Extent of resection | ||||||||
| Lobectomy/bilobectomy | 1.00 | — | — | <0.0001 | 1.00 | — | — | <0.0001 |
| Pneumonectomy | 1.77 | 0.89–3.53 | 0.106 | 1.60 | 0.79–3.23 | 0.2 | ||
| Segmentectomy | 1.61 | 1.12–2.33 | 0.011 | 1.65 | 1.14–2.39 | 0.008 | ||
| Wedge | 2.05 | 1.54–2.72 | <0.0001 | 2.09 | 1.55–2.81 | <0.0001 | ||
| Pathologic stage | ||||||||
| IA | 1.00 | — | — | <0.0001 | 1.00 | — | — | <0.0001 |
| IB | 1.86 | 1.48–2.34 | <0.0001 | 1.50 | 1.16–1.93 | 0.002 | ||
| IIA | 2.67 | 1.81–3.94 | <0.0001 | 2.24 | 1.47–3.39 | 0.0002 | ||
| IIB/IIIA | 3.96 | 2.92–5.38 | <0.0001 | 3.15 | 2.15–4.63 | <0.0001 | ||
| Gross tumor size, cm | 1.08 | 0.99–1.17 | 0.073 | 1.06 | 0.98–1.16 | 0.2 | ||
| Lymphovascular invasion | 1.52 | 1.22–1.90 | 0.0002 | 1.38 | 1.09–1.74 | 0.007 | ||
| IASLC predominant subtype | ||||||||
| Lepidic | 1.00 | — | — | 0.016 | 1.00 | — | — | 0.3 |
| Acinar | 1.57 | 0.99–2.49 | 0.055 | 1.35 | 0.84–2.17 | 0.2 | ||
| Papillary | 1.74 | 1.08–2.81 | 0.023 | 1.45 | 0.90–2.36 | 0.13 | ||
| Micropapillary | 2.41 | 1.41–4.11 | 0.001 | 1.78 | 1.02–3.11 | 0.044 | ||
| Solid | 1.89 | 1.15–3.10 | 0.012 | 1.51 | 0.90–2.52 | 0.12 | ||
| Architectural grade | ||||||||
| Low | 1.00 | — | — | 0.008 | ||||
| Intermediate | 1.63 | 1.04–2.56 | 0.033 | |||||
| High | 2.05 | 1.27–3.30 | 0.003 | |||||
| BRMS1 score: low vs high | 1.37 | 1.12–1.67 | 0.002 | 1.36 | 1.11–1.67 | 0.003 | ||
CI, confidence interval; HR, hazard ratio.
P is from a model stratified by pathologic stage.
Although it was not the primary objective of our study, to assess the relationship between BRMS1 mRNA expression and OS in early-stage LUAD, we examined two independent cohorts. Both the stage I LUAD Nagoya cohort (N=79) and the University of Michigan cohort (N=128) of LUAD patients without nodal metastasis had a robust decrease in OS with low BRMS1 mRNA transcript levels (Supplemental Figure 3).37,38
In a multivariable model that adjusted for age, sex, surgery type, pathologic stage, tumor size, lymphovascular invasion, and histologic subtype, the hazard of death for patients with low BRMS1 expression was 1.36-fold higher than that for patients with high expression (95% CI, 1.10–1.65; P=0.003; Table 2).
The OS nomogram is used to predict the probability of death from any cause at 5 years (Supplemental Figure 4). Calibration plots for internal validation of OS at 5 years are shown in Supplemental Figure 5. The Harrell C-index for the OS nomogram was 0.708 (95% CI, 0.679–0.734), with an internal validation C-index of 0.693 (95% CI, 0.669–0.725; Supplemental Table 1).
Relationship between BRMS1 Expression and DFS
In the high-expression group, 249 (39%) patients died of disease or had locoregional or distant recurrence, with 383 (61%) alive without disease at the end of the study. Comparatively, in the low-expression group, 206 (52%) patients died of disease or had recurrence, with 192 (48%) alive without disease at the end of the study. The 5-year DFS was 65% (95% CI, 61%–69%) for the high-expression group and 57% (95% CI, 52%–62%) for the low-expression group. Corresponding median DFS were 10.0 (95% CI, 7.9–11.4) and 6.6 (95% CI, 5.5–8.2) years. Stratified log-rank test indicated significant difference in DFS between the two groups (P=0.0045; Figure 2B). In univariable analysis, low BRMS1 expression was significantly associated with worse DFS (HR, 1.30 [95% CI, 1.08–1.57]; P=0.005; Table 3). As a continuous variable in reverse order, lower BRMS1 expression was associated with worse DFS (HR, 1.19 [95% CI, 1.04–1.36]; P=0.009).
Table 3.
Univariable and Multivariable Cox Models for Disease-Free Survival
| Variable | Univariable | Multivariable | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| HR | 95% CI | Pa | Overall Pa | HR | 95% CI | P | Overall P | |
| Age | 1.02 | 1.01–1.03 | <0.0001 | 1.02 | 1.01–1.03 | 0.0005 | ||
| Sex: male vs female | 1.44 | 1.19–1.73 | 0.0001 | 1.34 | 1.11–1.62 | 0.003 | ||
| Smoking status | ||||||||
| Never | 1.00 | — | — | 0.3 | ||||
| Former | 1.11 | 0.86–1.44 | 0.4 | |||||
| Current | 1.32 | 0.93–1.86 | 0.12 | |||||
| Extent of resection | ||||||||
| Lobectomy/bilobectomy | 1.00 | — | — | <0.0001 | 1.00 | — | — | <0.0001 |
| Pneumonectomy | 1.36 | 0.68–2.72 | 0.4 | 1.11 | 0.55–2.21 | 0.8 | ||
| Segmentectomy | 1.52 | 1.08–2.13 | 0.016 | 1.66 | 1.18–2.35 | 0.004 | ||
| Wedge | 1.96 | 1.50–2.56 | <0.0001 | 2.18 | 1.65–2.88 | <0.0001 | ||
| Pathologic stage | ||||||||
| IA | 1.00 | — | — | <0.0001 | 1.00 | — | — | <0.0001 |
| IB | 2.00 | 1.62–2.47 | <0.0001 | 1.49 | 1.17–1.89 | 0.001 | ||
| IIA | 2.88 | 2.03–4.10 | <0.0001 | 2.22 | 1.52–3.24 | <0.0001 | ||
| IIB/IIIA | 4.19 | 3.15–5.57 | <0.0001 | 2.98 | 2.08–4.26 | <0.0001 | ||
| Gross tumor size, cm | 1.15 | 1.06–1.24 | 0.001 | 1.15 | 1.06–1.24 | 0.001 | ||
| Lymphovascular invasion | 1.52 | 1.23–1.87 | <0.0001 | 1.33 | 1.07–1.65 | 0.011 | ||
| IASLC predominant subtype | ||||||||
| Lepidic | 1.00 | — | — | 0.0001 | 1.00 | — | — | 0.027 |
| Acinar | 1.72 | 1.11–2.67 | 0.015 | 1.57 | 1.00–2.46 | 0.049 | ||
| Papillary | 2.09 | 1.33–3.29 | 0.001 | 1.83 | 1.16–2.89 | 0.010 | ||
| Micropapillary | 2.97 | 1.79–4.91 | <0.0001 | 2.23 | 1.32–3.78 | 0.003 | ||
| Solid | 2.26 | 1.41–3.60 | 0.001 | 1.81 | 1.11–2.94 | 0.017 | ||
| Architectural grade | ||||||||
| Low | 1.00 | — | — | 0.0001 | ||||
| Intermediate | 1.85 | 1.21–2.85 | 0.005 | |||||
| High | 2.48 | 1.58–3.89 | <0.0001 | |||||
| BRMS1 score: low vs high | 1.30 | 1.08–1.57 | 0.005 | 1.25 | 1.04–1.52 | 0.020 | ||
CI, confidence interval; HR, hazard ratio.
P is from a model stratified by pathologic stage.
We then performed multivariable analysis that adjusted for age, sex, surgery type, pathologic stage, tumor size, lymphovascular invasion, and histologic subtype. Low intratumoral BRMS1 expression was an independent predictor of worse DFS, compared with high BRMS1 expression (adjusted HR=1.25 [95% CI, 1.04–1.52]; P =0.02; Table 3).
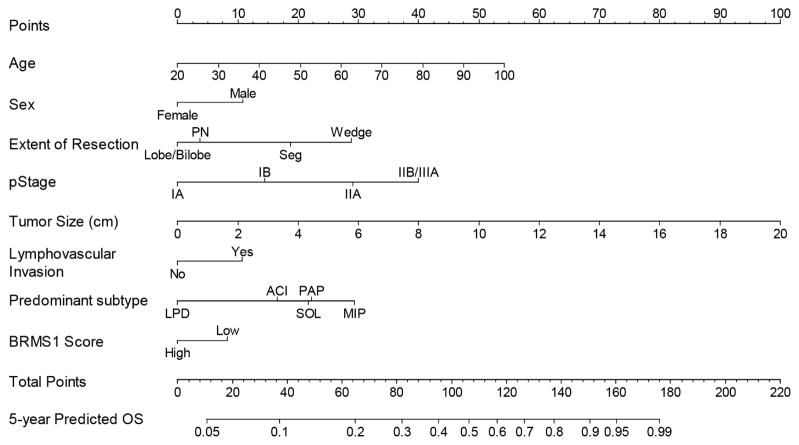
The DFS nomogram is used to predict the probability of death from recurrent LUAD at 5 years (Figure 3). Calibration plots for internal validation of DFS at 5 years are shown (Supplemental Figure 5). The Harrell C-index for the DFS nomogram was 0.707 (95% CI, 0.682–0.730), with an internal validation C-index of 0.692 (95% CI, 0.667–0.720; Supplemental Table 1).
Figure 3.
The disease-free survival (DFS) nomogram provides a graphical approach to calculate 5-year DFS after resection based on a patient’s combination of clinicopathologic covariates. First, locate the patient’s age, draw a line straight up to the points axis to derive the score associated with an age. Repeat for the other covariates on the nomogram. Add the scores for each covariate to determine the total score. Draw a vertical line from the total points axis to the 5-year DFS axis to obtain the predicted probability.
Discussion
We have shown that low intratumoral BRMS1 expression in surgically resected LUAD is associated with increased T stage, larger tumors, and more lymphatic and vascular invasion, compared with high BRMS1 expression. Moreover, we found that BRMS1 expression is an independent predictor of OS and DFS in our patient cohort. Patients with surgically resected LUAD and low intratumoral BRMS1 expression had worse OS and DFS than patients with high BRMS1 expression. Specifically, the hazard of death and hazard of disease recurrence or death among patients with low BRMS1 expression were 1.36- and 1.25-fold, respectively, compared with patients with high BRMS1 expression, after adjustment for relevant factors. Collectively, these findings show that BRMS1 expression has prognostic and functional significance in surgically resected LUAD.
Selected metastasis suppressor genes have been implicated in the metastatic cascade associated with NSCLC. Decreased levels of KAI1 and KISS1 have been associated with worse prognosis in a small series of mixed histologic types, treatment strategies, and variable stages.39,40 In contrast, the roles that other well known metastasis suppressor genes, such as nm23, MKK4, and RHOGDI2, play in the biology of lung cancer metastases are less well-characterized.41–44 We first reported that BRMS1 expression was reduced in NSCLC cells and human tumor tissues and was associated with worse OS, suggesting possible prognostic relevance.45 In that small (n=80), exploratory study of mixed NSCLC histologic types and stages, BRMS1 expression did not correlate with histologic grade or lymphatic invasion. However, in this larger study of surgically resected LUAD only, lymphatic invasion and tumor size correlated with intratumoral BRMS1 expression.
This study extends observations from other groups on the prognostic relevance of BRMS1 expression for solid tumors. Low BRMS1 expression, as measured by immunohistochemical analysis, has been identified as an independent predictive factor for poor prognosis in nasopharyngeal carcinoma,46 gallbladder adenocarcinoma,47 and melanoma.48 Collectively, this and other studies illustrate more broadly the consequences of BRMS1 loss for cancer progression and its impact on survival in patients with solid tumors of different histologic profiles and organ types.
Loss of BRMS1 expression in NSCLC and other solid tumors occurs at the chromatin level, where promoter methylation results in transcriptional repression.45,49 Specifically, we have demonstrated that RelA/p65-DNMT-1–mediated BRMS1 promoter methylation results in transcriptional repression of BRMS1 in lung cancer.49 Methylation of the BRMS1 promoter has also been shown to correlate with smoking history and poor survival.45,50 Although it was not the primary objective of this study, we found that decreased BRMS1 transcript levels were associated with decreased OS in two independent study cohorts of early stage, node-negative LUAD. In addition, we recently showed that BRMS1 expression is posttranslationally regulated via phosphorylation on serine 30 by CK2α′. This results in 14-3-3–mediated nuclear exportation of BRMS1 and its subsequent proteasome-mediated ubiquitination and degradation.51 Both of these mechanisms have potential clinical implications, as serum cfDNA BRMS1 promoter methylation has been shown to be an independent predictor of OS and DFS in early-stage NSCLC,52 and small-molecule inhibition of CK2α′ with CX4945, a drug currently in clinical trials, resulted in a 60-fold reduction of extrathoracic metastases in our orthotopic lung cancer model.51 Collectively, these observations suggest that, with appropriate validation, intratumoral BRMS1 expression may be pharmacologically modified; this is a testable hypothesis for future clinical trials.
The strengths of this study include the large number of patients (n=1030), the focus on a single tumor histology (LUAD), and the use of a homogeneous patient cohort. A further strength is the reproducibility of the scoring system: the raw observed agreement of the tested 100 cases was 96%, and only 4 cases were discordant. Cohen’s kappa indicates that the IRR was substantial (0.84 [95% CI, 0.68–0.99]), and Gwet’s AC1 indicates that the IRR was excellent (0.95 [95% CI, 0.90–1.00]). Additional strengths include the completeness of the clinical and pathologic annotation and the long follow-up, the internal validation of the data, and the development of nomograms to predict OS and DFS using strict REMARK and TRIPOD criteria. This is the first study to examine BRMS1 expression as a prognostic biomarker in surgically resected lung cancer independent of other well-described clinicopathologic variables. Finally, we make a hypothesis-generating observation that loss of intratumoral BRMS1 expression is associated with intermediate and high histologic architectural tumor grades in LUAD. This suggests that previously described epigenetic, posttranslational, and other unknown mechanisms that govern BRMS1 expression may be particularly relevant in specific LUAD histologic subtypes.
Limitations of this study include the examination of an isolated biomarker by the use of immunohistochemical analysis. However, an integrated approach that combines gene/protein expression with associated clinicopathologic information may be best for biomarker classification and discovery.37 A second limitation is the lack of inclusion of other tumor genomic mutations or translocations in our multivariable models. This was intentional, as we wanted to specifically examine the role of BRMS1, a metastasis suppressor gene, and not oncogenes, tumor suppressor genes, or oncogenic kinases. Moreover, it has become evident that discrete metastatic processes can be regulated independently of oncogene-driven tumor growth.17 A third limitation is that the contribution of BRMS1 to the predictive abilities of the nomogram is small. This is likely secondary to the a priori selection of clinicopathologic criteria that result in a very robust model without BRMS1, as well as a focus on early-stage LUAD—and not on node-positive, advanced-stage disease, for which the contribution of BRMS1 may be greater. Finally, a fourth limitation is the need for external validation of findings on the relevance of BRMS1 as a prognostic biomarker in LUAD.
In conclusion, loss of intratumoral BRMS1 expression is an independent predictor of decreased DFS and OS in a large cohort of patients with surgically resected LUAD. Targeting metastases is an increasingly attractive option to prevent initial metastases in high-risk patients, to shrink established lesions, and to prevent additional metastases in patients with limited disease.53 This study opens the door to future clinical trials enriched for specific high-grade LUAD histologic subtypes that will test therapies and their abilities to increase BRMS1 expression, decrease metastases, and improve DFS.
Supplementary Material
CONSORT diagram.
Expressional profiles of BRMS1 in histologic subtypes of lung adenocarcinoma. The expression of BRMS1 protein in different histologic subtypes of lung adenocarcinoma was assessed using immunohistochemistry.
Kaplan-Meier curves of overall survival based on the expression of BRMS1 mRNA in A) patients with stage I LUAD (Nagoya cohort, N=79) and B) LUAD patients without nodal metastasis (University of Michigan cohort, N=128).
The overall survival (OS) nomogram provides a graphical approach to calculate 5-year OS after resection based on a patient’s combination of clinicopathologic covariates. First, locate the patient’s age draw a line straight up to the points axis to derive the score associated with an age. Repeat for the other covariates on the nomogram. Add the scores for each covariate to determine the total score. Draw a vertical line from the total points axis to the 5-year OS axis to obtain the predicted probability.
Calibration plots for (A) overall survival nomogram and (B) disease-free survival nomogram. Values are optimism-corrected based on 500 bootstrap resamples. The predicted probabilities are calculated based on the respective nomogram, stratified in equally sized subgroups. In each subgroup, the average predicted probability (Predicted, x-axis) was plotted against the Kaplan-Meier estimate (Observed, y-axis). Calibration from an ideal nomogram lies along the 45-degree reference line. Vertical lines represent 95% confidence intervals of the Kaplan-Meier estimates. OS, overall survival; DFS, disease-free survival.
Acknowledgments
Funding: This work was supported by the National Institutes of Health/National Cancer Institute (R01 CA136705 to D.R.J., U54 CA137788 to P.S.A., and 5 T32 CA 9501-27 to P.R.B.). This work was also supported, in part, by National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748.
Footnotes
Abbreviations: BRMS1, breast cancer metastasis suppressor 1; CI, confidence interval; DFS, disease-free survival; H&E, hematoxylin and eosin; HR, hazard ratio; LUAD, lung adenocarcinoma; MSK, Memorial Sloan Kettering Cancer Center; NSCLC, non-small cell lung cancer; OS, overall survival; REMARK, reporting recommendations for tumor marker prognostic study; TNM, tumor-node-metastasis; TRIPOD, transparent reporting of a multivariable prediction model for individual prognosis or diagnosis.
Conflicts of interest: All authors report no conflicts of interest.
The funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koo HK, Jin SM, Lee CH, et al. Factors associated with recurrence in patients with curatively resected stage I–II lung cancer. Lung Cancer. 2011;73:222–229. doi: 10.1016/j.lungcan.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy CM, Klassen AF, Cano SJ, et al. Patient satisfaction with postmastectomy breast reconstruction: a comparison of saline and silicone implants. Cancer. 2010;116:5584–5591. doi: 10.1002/cncr.25552. [DOI] [PubMed] [Google Scholar]
- 3.Fujimoto T, Cassivi SD, Yang P, et al. Completely resected N1 non-small cell lung cancer: Factors affecting recurrence and long-term survival. Journal of Thoracic and Cardiovascular Surgery. 2006;132:499–506. doi: 10.1016/j.jtcvs.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Varlotto JM, Recht A, Flickinger JC, et al. Factors associated with local and distant recurrence and survival in patients with resected nonsmall cell lung cancer. Cancer. 2009;115:1059–1069. doi: 10.1002/cncr.24133. [DOI] [PubMed] [Google Scholar]
- 5.Cho S, Sung SW, Jheon S, et al. Risk of recurrence in surgically resected stage I adenocarcinoma of the lung: Histopathologic and immunohistochemical analysis. Lung. 2008;186:411–419. doi: 10.1007/s00408-008-9116-4. [DOI] [PubMed] [Google Scholar]
- 6.Tsutani Y, Miyata Y, Nakayama H, et al. Prognostic significance of using solid versus whole tumor size on high-resolution computed tomography for predicting pathologic malignant grade of tumors in clinical stage IA lung adenocarcinoma: a multicenter study. J Thorac Cardiovasc Surg. 2012;143:607–612. doi: 10.1016/j.jtcvs.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 7.Tsutani Y, Miyata Y, Nakayama H, et al. Solid tumor size on high-resolution computed tomography and maximum standardized uptake on positron emission tomography for new clinical T descriptors with T1 lung adenocarcinoma. Ann Oncol. 2013;24:2376–2381. doi: 10.1093/annonc/mdt230. [DOI] [PubMed] [Google Scholar]
- 8.Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol. 2010;34:1155–1162. doi: 10.1097/PAS.0b013e3181e4ee32. [DOI] [PubMed] [Google Scholar]
- 9.Ujiie H, Kadota K, Chaft JE, et al. Solid Predominant Histologic Subtype in Resected Stage I Lung Adenocarcinoma Is an Independent Predictor of Early, Extrathoracic, Multisite Recurrence and of Poor Postrecurrence Survival. J Clin Oncol. 2015;33:2877–2884. doi: 10.1200/JCO.2015.60.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin WA, Aisner DL, Varella-Garcia M. Prognostic patterns in the histopathology of pulmonary adenocarcinoma. J Clin Oncol. 2012;30:1401–1403. doi: 10.1200/JCO.2011.40.3964. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadal E, Chen G, Prensner JR, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2014;9:1513–1522. doi: 10.1097/JTO.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Shi L, Zhao X, et al. PIK3CA gene mutation associated with poor prognosis of lung adenocarcinoma. Onco Targets Ther. 2013;6:497–502. doi: 10.2147/OTT.S41643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou ZJ, Zhan P, Song Y. PD-L1 over-expression and survival in patients with non-small cell lung cancer: a meta-analysis. Transl Lung Cancer Res. 2015;4:203–208. doi: 10.3978/j.issn.2218-6751.2015.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seraj MJ, Samant RS, Verderame MF, et al. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 16.Samant RS, Seraj MJ, Saunders MM, et al. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2000;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- 17.Shoushtari AN, Szmulewitz RZ, Rinker-Schaeffer CW. Metastasis-suppressor genes in clinical practice: lost in translation? Nat Rev Clin Oncol. 2011;8:333–342. doi: 10.1038/nrclinonc.2011.65. [DOI] [PubMed] [Google Scholar]
- 18.Yan J, Yang Q, Huang Q. Metastasis suppressor genes. Histol Histopathol. 2013;28:285–292. doi: 10.14670/hh-28.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–8696. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman DG, McShane LM, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 2012;10:51. doi: 10.1186/1741-7015-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 22.Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 23.Moons KG, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 24.Edge SBBD, Compton CC, et al., editors. American Joint Committee on Cancer Cancer Staging Manual. New York, NY: Springer-Verlag New York; 2010. [Google Scholar]
- 25.Kadota K, Nitadori J, Sarkaria IS, et al. Thyroid transcription factor-1 expression is an independent predictor of recurrence and correlates with the IASLC/ATS/ERS histologic classification in patients with stage I lung adenocarcinoma. Cancer. 2013;119:931–938. doi: 10.1002/cncr.27863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eguchi T, Kadota K, Chaft J, et al. Cell cycle progression score is a marker for five-year lung cancer-specific mortality risk in patients with resected stage I lung adenocarcinoma. Oncotarget. 2016;7:35241–35256. doi: 10.18632/oncotarget.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 28.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 30.Burnham KPAD. Model Selection and Multimodel Inference. New York: SpringerVerlag; 2002. [Google Scholar]
- 31.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Brier G. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78:1–3. [Google Scholar]
- 34.Gwet KL. Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol. 2008;61:29–48. doi: 10.1348/000711006X126600. [DOI] [PubMed] [Google Scholar]
- 35.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 36.Gwet K. [Accessed January 31, 2017];R functions for calculating agreement coefficients. Available at http://www.agreestat.com/r_functions.html.
- 37.Director’s Challenge Consortium for the Molecular Classification of Lung A. Shedden K, Taylor JM, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomida S, Takeuchi T, Shimada Y, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 39.Sun YB, Xu S. Expression of KISS1 and KISS1R (GPR54) may be used as favorable prognostic markers for patients with non-small cell lung cancer. Int J Oncol. 2013;43:521–530. doi: 10.3892/ijo.2013.1967. [DOI] [PubMed] [Google Scholar]
- 40.Zhou L, Yu L, Wu S, et al. Clinicopathological significance of KAI1 expression and epithelial-mesenchymal transition in non-small cell lung cancer. World J Surg Oncol. 2015;13:234. doi: 10.1186/s12957-015-0657-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Chen D, Yang L, et al. A functional variant (-1304T>G) in the MKK4 promoter contributes to a decreased risk of lung cancer by increasing the promoter activity. Carcinogenesis. 2010;31:1405–1411. doi: 10.1093/carcin/bgq126. [DOI] [PubMed] [Google Scholar]
- 42.Niu H, Li H, Xu C, et al. Expression profile of RhoGDI2 in lung cancers and role of RhoGDI2 in lung cancer metastasis. Oncol Rep. 2010;24:465–471. doi: 10.3892/or_00000880. [DOI] [PubMed] [Google Scholar]
- 43.Smith SC, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer. 2009;9:253–264. doi: 10.1038/nrc2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomita M, Ayabe T, Matsuzaki Y, et al. Immunohistochemical analysis of nm23-H1 gene product in node-positive lung cancer and lymph nodes. Lung Cancer. 1999;24:11–16. doi: 10.1016/s0169-5002(99)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Smith PW, Liu Y, Siefert SA, et al. Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 2009;276:196–203. doi: 10.1016/j.canlet.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui RX, Liu N, He QM, et al. Low BRMS1 expression promotes nasopharyngeal carcinoma metastasis in vitro and in vivo and is associated with poor patient survival. BMC Cancer. 2012;12:376. doi: 10.1186/1471-2407-12-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Liu F, Yang ZL. BRMS1 and HPA as Progression, Clinical Biological Behaviors, and Poor Prognosis-related Biomarkers for Gallbladder Adenocarcinoma. Appl Immunohistochem Mol Morphol. 2016;24:275–282. doi: 10.1097/PAI.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Cheng Y, Tai D, et al. Prognostic significance of BRMS1 expression in human melanoma and its role in tumor angiogenesis. Oncogene. 2011;30:896–906. doi: 10.1038/onc.2010.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Mayo MW, Nagji AS, et al. Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene. 2012;31:1143–1154. doi: 10.1038/onc.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Shen Y, Liu B, et al. Promoter methylation of BRMS1 correlates with smoking history and poor survival in non-small cell lung cancer patients. Lung Cancer. 2011;74:305–309. doi: 10.1016/j.lungcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Amin EB, Mayo MW, et al. CK2alpha’ Drives Lung Cancer Metastasis by Targeting BRMS1 Nuclear Export and Degradation. Cancer Res. 2016;76:2675–2686. doi: 10.1158/0008-5472.CAN-15-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balgkouranidou I, Chimonidou M, Milaki G, et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer. 2014;110:2054–2062. doi: 10.1038/bjc.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CONSORT diagram.
Expressional profiles of BRMS1 in histologic subtypes of lung adenocarcinoma. The expression of BRMS1 protein in different histologic subtypes of lung adenocarcinoma was assessed using immunohistochemistry.
Kaplan-Meier curves of overall survival based on the expression of BRMS1 mRNA in A) patients with stage I LUAD (Nagoya cohort, N=79) and B) LUAD patients without nodal metastasis (University of Michigan cohort, N=128).
The overall survival (OS) nomogram provides a graphical approach to calculate 5-year OS after resection based on a patient’s combination of clinicopathologic covariates. First, locate the patient’s age draw a line straight up to the points axis to derive the score associated with an age. Repeat for the other covariates on the nomogram. Add the scores for each covariate to determine the total score. Draw a vertical line from the total points axis to the 5-year OS axis to obtain the predicted probability.
Calibration plots for (A) overall survival nomogram and (B) disease-free survival nomogram. Values are optimism-corrected based on 500 bootstrap resamples. The predicted probabilities are calculated based on the respective nomogram, stratified in equally sized subgroups. In each subgroup, the average predicted probability (Predicted, x-axis) was plotted against the Kaplan-Meier estimate (Observed, y-axis). Calibration from an ideal nomogram lies along the 45-degree reference line. Vertical lines represent 95% confidence intervals of the Kaplan-Meier estimates. OS, overall survival; DFS, disease-free survival.