Abstract
Inflammation is associated with glenohumeral arthritis and rotator cuff tendon tears. Epigenetically, miRNAs tightly regulate various genes involved in the inflammatory response. Alterations in the expression profile of miRNAs and the elucidation of their target genes with respect to the pathophysiology could improve the understanding of their regulatory role and therapeutic potential. Here, we screened key miRNAs that mediate inflammation and linked with JAK2/STAT3 pathway with respect to the co-incidence of glenohumeral arthritis in patients suffering from rotator cuff injury (RCI). Human resected long head of the biceps tendons were examined for miRNA profile from two groups of patients: Group-1 included the patients with glenohumeral arthritis and massive rotator cuff tears and the Group-2 patients did not have arthritis or rotator cuff tears. The miRNA profiling revealed that 235 miRNAs were highly altered (fold change less than −3 and greater than +2 were considered). Data from the NetworkAnalyst program revealed the involvement and interaction between 3,430 different genes associated with inflammation out of which 284 genes were associated with JAK2/STAT3 pathway and interconnects 120 different pathways of inflammation. Around 1,500 miRNAs were found to play regulatory role associated with these genes of inflammatory responses and 77 miRNAs were found to regulate more than 10 genes. Among them 25 genes with <−10-fold change were taken to consideration which altogether constitute for the regulation of 102 genes. Targeting these miRNAs and the underlying regulatory mechanisms may advance our knowledge to develop promising therapies in the management of shoulder tendon pathology.
Keywords: Glenohumeral arthritis, Inflammation, Rotator cuff injury, Tendinopathy, miRNAs, miRNA regulators
Introduction
Hyper-activation and persistence of inflammation has been reported in several musculoskeletal diseases, especially rotator cuff injury (RCI). Inflammation and pain are the two major symptoms of the patients with RCI and the conventional treatment approaches are aiming in the management of these symptoms [1]. RCI can be both degenerative and traumatic in nature depending on the multifactorial etiology and these causative factors may be either extrinsic or intrinsic [2]. Interestingly, irrespective of the causative factors and severity, the inflammation is a prominent hallmark in most clinical cases and the sustainability of inflammation delays the response to medications and healing [3]. This was evident by the upregulation of proinflammatory cytokines, biomarkers and receptors in the tendon tissues even after the setting of clinical symptoms. The mechanism and driving force behind the prolonged inflammatory reactions are yet to be unveiled for a mechanically robust minimally vascular tissue like tendon [4].
The co-existence of clinical conditions like osteoarthritis is reported to exasperate the inflammatory reactions of joints [5]. The rheumatoid arthritis and glenohumeral arthritis has been reported to be the aggravating factors of inflammation in patients with degenerative RCI [6]. The cumulative effects of arthritis along with preexisting RCI demonstrate osteopenia, glenohumeral/acromioclavicular erosions and proximal humeral migration which offer challenges in repair pathways and therapeutic strategies of the rotator cuff [7]. The inflammation will be prolonged at cellular and molecular level for several months even after the setting of arthritis which creates a higher chance for reoccurrence of RCI. The molecular events leading to the persistence of inflammation in RCI regarding the presence/absence of arthritis is still unknown.
Cytokine release is considered to be the initial event in inflammation and most of the cytokine expression and action was reported to be regulated by STAT (signal transducers and activators of transcription) proteins [8]. Apart from cytokines, the association of JAK (Janus kinase) and the downstream molecules with STAT pathway activates inflammation by facilitating chemokine expression, differentiation and maturation of hematopoietic cells, stem cell activation, and production of reactive oxygen/nitrogen species [9]. The interplay between cytokine signals and STAT proteins are necessary for the execution of inflammatory responses associated with infection or injury. The extracellular interaction of ligand activates the JAK/STAT pathway by inducing conformational changes to the receptor which in turn triggers a phosphorylation cascade to downstream substrates including STATs. The phosphorylated STATs (activated) translocate to nucleus and assemble as dimeric or oligomeric complex at the specific enhancer sequences of the target inflammatory genes, thereby regulating their transcription. Seven STATs and four JAKs exist in mammalian system and each of which are recruited depending on tissue specificity and receptors and/or ligands involved [10].
The inflammatory signaling pathways and molecules are also regulated epigenetically by miRNAs. miRNAs like miR-29, miR-133a, miR-155, miR-221, miR-223, miR-652, etc. are well known for their active role in inflammatory diseases including arthritis [11]. Similarly, the specific roles of miR-9, miR-127, miR-125 were reported to trigger M1 macrophage polarization while miR-223, let-7c, miR-124, miR-132, miR-34a, miR-146a and miR-125a-5p mediates M2 polarization [12]. Immune cell differentiation and proliferation are also under the regulation of miRNAs. To cite, miR-106a, miR-20a and miR-17-5p mediates monocyte differentiation and miR-106a, miR-106b and miR-19 are actively involved in T cell differentiation and signaling [13]. miR-223, let-72, miR-147 and miR-9 regulate inflammation by targeting the downstream signaling molecules of TLR signaling pathway [14]. The miRNA mediated regulation of cytokines, which are associates with JAK/STAT pathway, was also well established. For example, miR-27a, miR-23a and miR-24a targets IFN-γ, miR-10a regulates TGF-β, miR-9 and miR-31 are the regulators of IL-2 [15]. The transcription factor NF-κβ, which regulate a battery of pro/ant-inflammatory genes were also reported to be regulated by miR-146, miR-125, and miR-21 [16] and miR-181a, miR-19a, and miR-124 regulates TNF-α [17], [18].
Even though limited number reports are available [19], [20], the miRNA mediated regulation of inflammation in human rotator cuff tendon has not been well established. The implications of miRNA mediated inflammatory responses (especially targeting JAK/STAT signaling) coinciding with arthritis and/or non-arthritis environments of RCI are still unknown. The goal of the study is to characterize miRNAs associated with JAK2/STAT3 pathway of inflammation and to identify their target genes involved in the pathophysiology of glenohumeral arthritis and its co-incidence with rotator cuff tears. The findings from this study could unravel their regulatory role and therapeutic potential. The present study compares the alterations in miRNA profile among the patients with and without glenohumeral arthritis and rotator cuff tears in combination with the integrative expression analysis of inflammatory genes associated with JAK/STAT pathway and their cross talk with other inflammatory pathways using the meta-analysis program NetworkAnalyst.
Materials and methods
Tissue collection and processing
The Institutional Review Board (IRB) of Creighton University approved the research protocol. All patient volunteers signed the consent form and the HIPPA form. The RCI patients were explained in layman terms about the details of the procedures and written informed consent was signed before the surgery. Eight RCI patients were recruited for the study over a 6-month period. Four patients were undergoing reverse shoulder arthroplasty to treat glenohumeral arthritis with massive rotator cuff tears (Group-1). The second group was undergoing arthroscopic biceps tenodesis surgery without rotator cuff tear of glenohumeral arthritis (Group-2). The grouping was based on severity of inflammation and presence of arthritis based on preoperative imaging. The biceps were tenotomized in all patients, resected, and sent for analysis. The tissue specimens were collected in UW (University of Wisconsin) solution at 4°C for transportation and temporary storage. One part of the tissues was fixed in 10% formalin for histology and another part was stored in RNA-later for RNA isolation.
Histology
The formalin fixed tissues were embedded in paraffin wax, sectioned along the horizontal axis (5μm thickness were cut using microtome, Leica, Germany) and heat-fixed on microscopic slides [21]. After deparaffinizing the sections in xylene and dehydrated with graded concentrations of ethanol Hematoxylin and Eosin (H&E) staining was carried out [22]. The H and E slides were mounted using xylene-based mounting media and imaged using an inverted microscope attached with CCD camera (Olympus BX51; Olympus America, Center Valley, PA) under bright field mode [23]. The histological evaluation of slides was performed qualitatively and independently by two blinded investigators which were confirmed by the third one.
Immunofluorescence
The antigen retrieval of deparaffinized sections was done in HIER (Heat Induced Epitope Retrieval) buffer at 95°C for 20 min. After washing in PBS, the sections were subjected to blocking using 0.25% Triton X-100 and 5% horse serum in PBS at room temperature for 2h. Primary antibody solution (1:50 diluted in blocking solution) was added and kept overnight at 4°C. After washing in PBS fluorochrome-conjugated corresponding secondary antibody (1:100 dilution) was added and incubated for 2h at room temperature followed by washing and mounting with 4′,6- diamidino-2-phenylindole (DAPI)-containing mounting media. The sections were viewed using the fluorescent microscope (Olympus BX51; Olympus America, Center Valley, PA) and the images were taken and merged using Olympus DP71 camera and associated software. All the antibodies were procured from SantaCruz Biotech and a negative control with secondary antibody alone was also maintained to fix the exposure [24]. The primary antibodies used were: mouse anti-human CD-68 and mouse anti-human CD-16. The donkey anti-mouse-FITC was used as the corresponding secondary antibodies [25].
RNA isolation from tendon
Around 200mg tissue from the proximal portion of the biceps tendons were minced to small pieces and Trizol reagent (1ml) was added followed by homogenization. After 10 min at room temperature Chloroform-isoamyl alcohol reagent (200μl) was added, centrifuged at 12,000 rpm for 15 min. 500μl isopropanol was added to the aqueous layer to precipitate RNA and again centrifuged at 12,000 rpm for 10 min to pellet down the RNA. The pellet was washed with 75% ethanol, dissolved in sterile RNase free water, quantified and stored at −80°C [26].
miRNA microarray
RNA integrity number (RIN) score was obtained by bioanalyzer before hybridizing the samples onto the miRNA microarray (miRNA 4.0 array). The RIN score of the isolated RNA was between (2.1 to 4.8) due to the tissue characteristic. MiRNA microarray analysis was conducted at Kansas University Medical Center and the raw data was analyzed using Expression Console software (Alegent). The analysis was performed in two separate batches (two specimens from each group) and compared for the consistency of results [27].
Gene identification using NetworkAnalyst
The interrelationship among the genes associated with inflammatory pathways was identified by using NetworkAnalyst program from published data base [27] – [28]. The major genes and associated regulators of JAK/STAT pathway of inflammation [29] were used as input in NetworkAnalyst to assess the cross talk of these genes with other signaling pathways of inflammation (Table 1; Fig. 1). The list of genes generated was assessed individually for their target miRNA from the microarray data.
Table 1.
The genes associated with JAK/STAT pathway of inflammation used as input to NetworkAnalyst.
| Activity | Genes |
|---|---|
| Janus Kinase Activity | JAK1, JAK2, JAK3, TYK2 |
| STAT Family | STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, STAT6 |
| Receptors | CSF1R, CSF2RB, EGFR, EPOR, F2R, GHR, IFNAR1, IFNGR1, IL10RA, IL2RA, IL2RG, IL4R, IL6ST, INSR, MPL, PDGFRA, SH2B1 |
| Phosphorylation | F2, F2R, IL20, PPP2R1A, PRLR, STAT1 |
| Regulators | HMGA1, SMAD3, SLA2, SPI1, STAT3, JUNB, SP1, USF1, PIAS1, YY1, SMAD1, SMAD5, CEBPB, CRK, GATA3, IRF1, IRF9, JUN, MYC, NFKB1, NR3C1, PPP2R1A, SMAD2, SMAD4 |
| STAT induced genes | CXCL9, IRF1, NOS2, A2M, BCL2L1, CDKN1A, CRP, FAS, MMP3, MYC, SOCS1, FCGR1A, IFNG, MYC, CCND1, CDKN1A, IL2RA, OSM, GATA3, GBP1, OAS1 |
| Negative Regulators | PIAS1, PIAS2, PTPN1, PTPRC, SOCS1, SOCS2, SOCS3, SOCS4, SOCS5 |
Fig. 1.
Determination of genes associated with JAK2 and STAT3 by NetworkAnalyst program.
Results
Histology
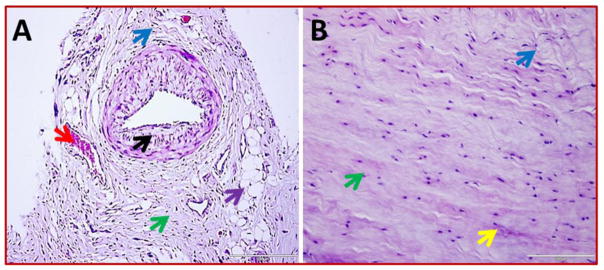
Group-1 consists of 4 patients with glenohumeral arthritis and a massive rotator cuff tear and Group-2 was included with 4 patients without arthritis. Altered physiological architecture of tendon tissue was observed on the patients of both the groups and disorganization of extracellular matrix was prominent as evident from histomorphological changes which was prominent in Group-1. The inflammation was prominent in Group-1 as confirmed by the presence of immune cells while the mild/negligible in Group-2. The MRI analysis of the patients was also provided a similar trend (data not shown in the article). Apart from ECM disorganization, the Group-2 patients displayed normal tendon cells as well as normal tissues towards the median side and the normal tenocytes were characterized by their less dense distribution of nuclei surrounding an intact ECM. But, the tenocytes were found to be clustered at the vicinity of ECM disorganization. Two patients of Group-1 showed fatty infiltration. Neoangiogenesis was also predominant in Group-1 while it was completely absent in Group-2 tendons which is a hallmark of inflammation and associated repair. The results are shown in Fig. 1.
Immunofluorescence
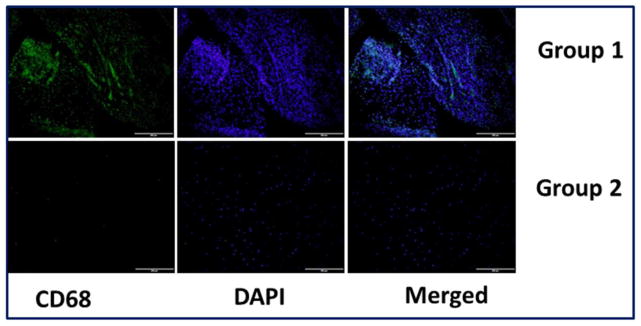
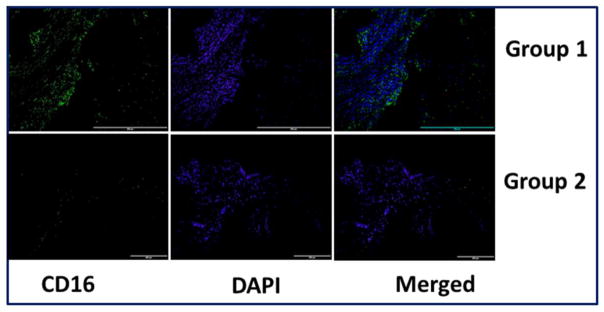
Immunofluorescence assessment of the tendon tissue specimens revealed the presence of CD68+ macrophages (Fig. 2) and CD16+ neutrophils (Fig. 3) predominantly in the Group-1 patients. This indicates inflammation associated with arthritis and the existence of macrophages and neutrophils was greater with respect to the severity of arthritis as evident from MRI analysis (data not shown in this article). All the Group-2 tendons showed absence of macrophages and neutrophils.
Fig. 2.
Representative images for H&E staining of biceps tendons – (A) Group-1 (represents four patient) and (B) Group-2 (represents four) showing difference in tissue organization. ECM disorganization and inflammation are more prominent in Group-1. The key features are indicated by colored arrows: green arrows - tendon cells, black arrows – inflammation, red arrows – angiogenesis, blue arrows - ECM disorganization, violet - fatty infiltration, and yellow arrows - normal ECM with dense collagen deposition. The Figs are shown in 400× magnification.
Fig. 3.
Representative images for CD68+expression in the tendon tissues of Group-1 and Group-2 patients by immunofluorescence. (A) Group-1 (represents four patient) and (B) Group-2 (represents four patient) patients. Group-1 tendons displayed higher density of macrophages than Group-2.
miRNA alterations Group-1 vs Group-2
The micro array profiling of whole miRNA showed several fold alterations in the expression status of miRNAs which has regulatory roles in inflammatory pathways and associated genes. 196 miRNAs were found to be down regulated with a fold change less than -3. Several miRNAs like hsa-miR-191-5p, hsa-miR-361-5p, hsa-miR-1273g-3p, hsa-miR-99b-5p, hsa-miR-145-5p, hsa-miR-99a-5p, and hsa-miR-100-5p were exhibited to have a fold change less than −50. Thirteen miRNAs were downregulated with a fold change between −50 and −20, thirty-one miRNAs possessed fold change between −20 and −10 while the rest of 144 miRNAs showed fold change in between −10 and −3 (Table 2). Thirty-nine miRNAs were upregulated and out of which twenty-six were in the fold change between 2- and 3-fold and nine were in between 3-fold and 4-fold change (Table 3). The hsa-miR-4467 (4.09), hsa-miR-6723-5p (4.57), hsa-miR-8071 (5.14), and hsa-miR-5001-5p (5.57) were the highly-upregulated ones.
Table 2.
Downregulated miRNAs – Group-1 vs Group-2 comparison
| miRNAs | FC |
|---|---|
| hsa-miR-191-5p | −71.26 |
| hsa-miR-361-5p | −69.26 |
| hsa-miR-1273g-3p | −64.17 |
| hsa-miR-99b-5p | −59.57 |
| hsa-miR-145-5p | −55.42 |
| hsa-miR-99a-5p | −51.68 |
| hsa-miR-100-5p | −51.6 |
| hsa-miR-23b-3p | −35.32 |
| hsa-miR-425-5p | −33.79 |
| hsa-miR-151a-3p | −32.1 |
| hsa-let-7a-5p | −31.63 |
| hsa-miR-22-3p | −30.84 |
| hsa-let-7d-5p | −29.9 |
| hsa-miR-146a-5p | −29.52 |
| hsa-miR-409-3p | −28.06 |
| hsa-miR-127-3p | −26.62 |
| hsa-miR-150-5p | −23.41 |
| hsa-miR-181a-5p | −21.34 |
| hsa-miR-4269 | −21.09 |
| hsa-miR-193b-3p | −20.35 |
| hsa-miR-30d-5p | −19.32 |
| hsa-miR-378c | −19.16 |
| hsa-miR-342-3p | −18.02 |
| hsa-miR-199a-3p | −17.88 |
| hsa-miR-199b-3p | −17.88 |
| hsa-miR-378i | −15.69 |
| hsa-miR-486-5p | −15.04 |
| hsa-miR-103a-3p | −14.87 |
| hsa-miR-134-5p | −14.82 |
| hsa-miR-31-5p | −14.44 |
| hsa-miR-23a-3p | −14.18 |
| hsa-miR-422a | −14.1 |
| hsa-miR-195-5p | −14.04 |
| hsa-miR-1246 | −14.01 |
| hsa-miR-26a-5p | −13.97 |
| hsa-miR-382-5p | −13.14 |
| hsa-let-7c-5p | −12.58 |
| hsa-miR-378f | −12.46 |
| hsa-miR-497-5p | −12.3 |
| hsa-miR-10b-5p | −12.27 |
| hsa-mir-361 | −12.13 |
| hsa-miR-199a-5p | −11.9 |
| miRNAs | FC |
| hsa-let-7e-5p | −11.7 |
| hsa-miR-30a-5p | −11.48 |
| hsa-miR-193a-5p | −10.93 |
| hsa-miR-3178 | −10.86 |
| hsa-miR-15a-5p | −10.66 |
| hsa-miR-574-3p | −10.54 |
| hsa-miR-451a | −10.45 |
| hsa-miR-500a-3p | −10.25 |
| hsa-miR-151a-5p | −10.22 |
| hsa-miR-21-5p | −10.11 |
| hsa-miR-125a-5p | −9.69 |
| hsa-miR-4532 | −8.88 |
| hsa-miR-29a-3p | −8.8 |
| hsa-miR-125b-2-3p | −8.66 |
| hsa-let-7i-5p | −8.38 |
| hsa-miR-370-3p | −8.38 |
| hsa-miR-28-5p | −8.31 |
| hsa-miR-130a-3p | −8.11 |
| hsa-miR-214-5p | −8.1 |
| hsa-miR-106b-5p | −8.08 |
| hsa-miR-3651 | −8.03 |
| hsa-miR-16-5p | −7.98 |
| hsa-let-7b-5p | −7.73 |
| hsa-miR-30c-5p | −7.67 |
| hsa-miR-132-3p | −7.55 |
| hsa-miR-663a | −7.43 |
| hsa-miR-25-3p | −7.23 |
| hsa-miR-339-5p | −7.22 |
| hsa-miR-146b-5p | −7.14 |
| hsa-miR-193a-3p | −7.13 |
| hsa-miR-532-3p | −7.12 |
| hsa-miR-378d | −7.01 |
| hsa-miR-744-5p | −6.94 |
| hsa-miR-193b-5p | −6.85 |
| hsa-mir-711 | −6.77 |
| hsa-miR-27b-5p | −6.64 |
| hsa-miR-3195 | −6.59 |
| hsa-miR-708-5p | −6.41 |
| hsa-miR-4674 | −6.29 |
| hsa-miR-491-5p | −6.22 |
| hsa-miR-130b-3p | −6.11 |
| hsa-miR-24-2-5p | −5.92 |
| miRNAs | FC |
| hsa-miR-652-3p | −5.88 |
| hsa-miR-339-3p | −5.8 |
| hsa-miR-221-3p | −5.72 |
| hsa-miR-381-3p | −5.67 |
| hsa-miR-139-5p | −5.65 |
| hsa-miR-4741 | −5.58 |
| hsa-miR-4443 | −5.45 |
| hsa-miR-874-3p | −5.31 |
| hsa-miR-21-3p | −5.23 |
| hsa-miR-324-3p | −5.2 |
| hsa-miR-4417 | −5.2 |
| hsa-miR-6722-3p | −5.15 |
| hsa-miR-154-5p | −5.1 |
| hsa-miR-362-5p | −5.1 |
| hsa-miR-665 | −5.1 |
| hsa-miR-1233-5p | −5.09 |
| hsa-miR-487b-3p | −5.06 |
| hsa-miR-99b-3p | −5.02 |
| hsa-miR-30a-3p | −5.01 |
| hsa-miR-28-3p | −4.94 |
| hsa-miR-27a-3p | −4.89 |
| hsa-miR-337-5p | −4.85 |
| hsa-miR-664b-5p | −4.82 |
| hsa-miR-6779-5p | −4.78 |
| hsa-miR-122-5p | −4.69 |
| hsa-miR-20a-5p | −4.67 |
| hsa-miR-7977 | −4.67 |
| hsa-miR-1271-5p | −4.61 |
| hsa-miR-17-3p | −4.6 |
| hsa-miR-6771-5p | −4.59 |
| hsa-miR-3687 | −4.52 |
| hsa-miR-378g | −4.51 |
| hsa-miR-342-5p | −4.46 |
| hsa-miR-671-5p | −4.45 |
| hsa-miR-4507 | −4.42 |
| hsa-miR-4651 | −4.41 |
| hsa-miR-324-5p | −4.31 |
| hsa-miR-4707-5p | −4.31 |
| hsa-miR-2110 | −4.28 |
| hsa-miR-151b | −4.2 |
| hsa-miR-664b-3p | −4.19 |
| hsa-miR-143-3p | −4.17 |
| hsa-miR-93-5p | −4.15 |
| hsa-miR-378a-3p | −4.14 |
| hsa-miR-6816-5p | −4.13 |
| hsa-miR-494-3p | −4.06 |
| hsa-miR-197-3p | −4.03 |
| hsa-miR-10a-5p | −4.02 |
| hsa-miR-432-5p | −4.02 |
| hsa-miR-532-5p | −4.02 |
| hsa-miR-7975 | −3.99 |
| hsa-miR-15b-5p | −3.98 |
| hsa-miR-345-5p | −3.98 |
| hsa-miR-409-5p | −3.93 |
| hsa-miR-1343-5p | −3.92 |
| hsa-miR-143-5p | −3.9 |
| hsa-miR-1587 | −3.9 |
| hsa-miR-433-3p | −3.89 |
| hsa-miR-664a-5p | −3.89 |
| hsa-miR-3197 | −3.89 |
| hsa-miR-379-5p | −3.88 |
| hsa-miR-3621 | −3.86 |
| hsa-miR-4433-3p | −3.83 |
| hsa-miR-140-3p | −3.81 |
| hsa-miR-20b-5p | −3.72 |
| hsa-mir-28 | −3.71 |
| hsa-miR-10b-3p | −3.7 |
| hsa-miR-6789-5p | −3.66 |
| hsa-miR-6798-5p | −3.63 |
| hsa-miR-1909-3p | −3.61 |
| hsa-miR-377-5p | −3.56 |
| hsa-mir-4281 | −3.54 |
| hsa-miR-17-5p | −3.53 |
| hsa-miR-24-3p | −3.53 |
| hsa-miR-155-5p | −3.49 |
| hsa-miR-4299 | −3.49 |
| hsa-miR-4505 | −3.46 |
| hsa-miR-455-3p | −3.45 |
| hsa-miR-92b-3p | −3.43 |
| hsa-miR-1268b | −3.41 |
| hsa-miR-185-5p | −3.39 |
| hsa-miR-224-3p | −3.38 |
| hsa-miR-4758-5p | −3.35 |
| hsa-miR-106b-3p | −3.33 |
| hsa-miR-127-5p | −3.33 |
| hsa-miR-4632-5p | −3.33 |
| hsa-miR-4649-5p | −3.32 |
| hsa-miR-4688 | −3.31 |
| hsa-miR-1225-5p | −3.3 |
| hsa-miR-6765-5p | −3.29 |
| hsa-miR-493-3p | −3.27 |
| hsa-miR-92b-5p | −3.26 |
| hsa-miR-3175 | −3.25 |
| hsa-miR-4646-5p | −3.25 |
| hsa-miR-654-3p | −3.23 |
| hsa-miR-1307-3p | −3.2 |
| hsa-miR-106a-5p | −3.18 |
| hsa-miR-503-5p | −3.17 |
| hsa-miR-6132 | −3.17 |
| hsa-miR-619-5p | −3.16 |
| hsa-miR-181a-3p | −3.14 |
| hsa-miR-629-5p | −3.12 |
| hsa-miR-378e | −3.11 |
| hsa-miR-139-3p | −3.09 |
| hsa-miR-181b-5p | −3.09 |
| hsa-miR-452-5p | −3.06 |
| hsa-miR-23b-5p | −3.04 |
| hsa-miR-937-5p | −3.03 |
| hsa-miR-378a-5p | −3 |
| hsa-miR-6126 | −3 |
Table 3.
Upregulated miRNAs – Group-1 vs Group-2 comparison
| miRNAs | FC |
|---|---|
| hsa-miR-4327 | 2.02 |
| hsa-miR-8072 | 2.04 |
| hsa-miR-498 | 2.05 |
| hsa-miR-1281 | 2.05 |
| hsa-miR-2861 | 2.05 |
| hsa-miR-7110-5p | 2.13 |
| hsa-miR-6775-5p | 2.16 |
| hsa-mir-6722 | 2.18 |
| hsa-miR-6831-5p | 2.27 |
| hsa-mir-320e | 2.29 |
| hsa-miR-6769b-5p | 2.29 |
| hsa-miR-4487 | 2.3 |
| miRNAs | FC |
| hsa-miR-6749-5p | 2.52 |
| hsa-miR-483-5p | 2.56 |
| hsa-miR-1229-5p | 2.56 |
| hsa-mir-6836 | 2.59 |
| hsa-miR-6127 | 2.73 |
| hsa-mir-550a-1 | 2.84 |
| hsa-mir-550a-2 | 2.84 |
| hsa-mir-550a-3 | 2.84 |
| hsa-miR-4745-5p | 2.86 |
| hsa-miR-7107-5p | 2.87 |
| hsa-miR-4481 | 3.01 |
| hsa-miR-6732-5p | 3.06 |
| miRNAs | FC |
| hsa-miR-8075 | 3.16 |
| hsa-miR-4668-5p | 3.73 |
| hsa-miR-297 | 3.81 |
| hsa-miR-6124 | 3.85 |
| hsa-miR-7150 | 3.86 |
| hsa-miR-6870-5p | 3.95 |
| hsa-miR-4467 | 4.09 |
| hsa-miR-6723-5p | 4.57 |
| hsa-miR-8071 | 5.14 |
| hsa-miR-5001-5p | 5.57 |
The key genes associated with JAK/STAT pathway of inflammation as reported elsewhere were evaluated [29] and were used as input in NetworkAnalyst to assess the cross talk of these genes with other signaling pathways of inflammation. The data revealed the involvement and interaction between 3430 different genes (Supplementary Table 1). From this gene repository, the interactions among JAK2/STAT3 signaling pathway and corresponding cross talk were chosen and the gene numbers narrow down to 284 (Table 4). These 284 genes are active members of 120 different pathways connected with inflammation. Each of the pathways and number of genes (hits) are displayed in (Supplementary Table 2). The 113 genes associated with the immune system were displayed to be closely linked to inflammation via JAK2/STAT3 signaling and around 1500 miRNAs were found to play regulatory roles associated with these genes (Supplementary Table 3).
Table 4.
Specific cross talk and interactions between genes of JAK2/STAT3 pathway of inflammation as revealed by NetworkAnalyst.
| Id | Label |
|---|---|
| P40763 | STAT3 |
| O60674 | JAK2 |
| P42224 | STAT1 |
| P18031 | PTPN1 |
| P12931 | SRC |
| P30154 | PPP2R1B |
| Q06124 | PTPN11 |
| P42229 | STAT5A |
| P51692 | STAT5B |
| P10912 | GHR |
| P0CG48 | UBC |
| P06241 | FYN |
| Q6IA86 | ELP2 |
| P07948 | LYN |
| Q9Y4K3 | TRAF6 |
| P00533 | EGFR |
| P04626 | ERBB2 |
| P29350 | PTPN6 |
| P52333 | JAK3 |
| Q14289 | PTK2B |
| P06213 | INSR |
| P78347 | GTF2I |
| P40189 | IL6ST |
| P51681 | CCR5 |
| Q9UM73 | ALK |
| Q05397 | PTK2 |
| P16473 | TSHR |
| P14784 | IL2RB |
| P07900 | HSP90AA1 |
| P11362 | FGFR1 |
| Q14469 | HES1 |
| Q5TA89 | HES5 |
| Q9UER7 | DAXX |
| O43318 | MAP3K7 |
| Q9UBE8 | NLK |
| Q13287 | NMI |
| Q13283 | G3BP1 |
| P51617 | IRAK1 |
| Q06187 | BTK |
| P05129 | PRKCG |
| P45983 | MAPK8 |
| P23458 | JAK1 |
| P31785 | IL2RG |
| P06454 | PTMA |
| P08670 | VIM |
| P63244 | GNB2L1 |
| P01019 | AGT |
| Q9Y6X2 | PIAS3 |
| Id | Label |
| P26358 | DNMT1 |
| Q13547 | HDAC1 |
| Q00653 | NFKB2 |
| P42345 | MTOR |
| Q14765 | STAT4 |
| Q9Y5S9 | RBM8A |
| Q14192 | FHL2 |
| P16333 | NCK1 |
| P62993 | GRB2 |
| P19174 | PLCG1 |
| Q99683 | MAP3K5 |
| P01584 | IL1B |
| Q13263 | TRIM28 |
| P38936 | CDKN1A |
| P22301 | IL10 |
| P20396 | TRH |
| P35372 | OPRM1 |
| P01282 | VIP |
| P69891 | HBG1 |
| P02741 | CRP |
| P15941 | MUC1 |
| O15392 | BIRC5 |
| P12314 | FCGR1A |
| P32455 | GBP1 |
| P05451 | REG1A |
| P15529 | CD46 |
| Q16666 | IFI16 |
| Q00978 | IRF9 |
| P15692 | VEGFA |
| P24385 | CCND1 |
| P50750 | CDK9 |
| Q8IZL8 | PELP1 |
| P17676 | CEBPB |
| P20749 | BCL3 |
| O15379 | HDAC3 |
| Q92769 | HDAC2 |
| Q9Y6K9 | IKBKG |
| O75177 | SS18L1 |
| O14543 | SOCS3 |
| Q99062 | CSF3R |
| P07949 | RET |
| P52294 | KPNA1 |
| Q04206 | RELA |
| O00570 | SOX1 |
| P61073 | CXCR4 |
| Q29980 | MICB |
| P05412 | JUN |
| P15260 | IFNGR1 |
| Id | Label |
| P35568 | IRS1 |
| P15927 | RPA2 |
| P29353 | SHC1 |
| P16471 | PRLR |
| P04150 | NR3C1 |
| P10275 | AR |
| P20823 | HNF1A |
| Q92783 | STAM |
| O75886 | STAM2 |
| P08887 | IL6R |
| P08238 | HSP90AB1 |
| P21802 | FGFR2 |
| P16410 | CTLA4 |
| P11142 | HSPA8 |
| Q99665 | IL12RB2 |
| Q9NPH3 | IL1RAP |
| Q92993 | KAT5 |
| P38398 | BRCA1 |
| Q96Q27 | ASB2 |
| Q15257 | PPP2R4 |
| Q07666 | KHDRBS1 |
| P03372 | ESR1 |
| Q05513 | PRKCZ |
| P04632 | CAPNS1 |
| Q8TE76 | MORC4 |
| Q9ULD0 | OGDHL |
| Q06520 | SULT2A1 |
| P46781 | RPS9 |
| Q9GZT8 | NIF3L1 |
| P62913 | RPL11 |
| P09769 | FGR |
| O43255 | SIAH2 |
| P07384 | CAPN1 |
| Q8N6P7 | IL22RA1 |
| P15822 | HIVEP1 |
| P04439 | HLA-A |
| P02760 | AMBP |
| O60232 | SSSCA1 |
| Q8IXJ9 | ASXL1 |
| P45984 | MAPK9 |
| P06748 | NPM1 |
| P38159 | RBMX |
| O43283 | MAP3K13 |
| P00966 | ASS1 |
| Q07820 | MCL1 |
| Q13950 | RUNX2 |
| Q16695 | HIST3H3 |
| P46527 | CDKN1B |
| Id | Label |
| P06401 | PGR |
| P08631 | HCK |
| Q9Y383 | LUC7L2 |
| P62158 | CALM1 |
| Q9BT73 | PSMG3 |
| O95661 | DIRAS3 |
| P52630 | STAT2 |
| P11309 | PIM1 |
| P21860 | ERBB3 |
| P10914 | IRF1 |
| P16885 | PLCG2 |
| P28482 | MAPK1 |
| O43293 | DAPK3 |
| P17948 | FLT1 |
| P35968 | KDR |
| P05362 | ICAM1 |
| P17181 | IFNAR1 |
| P24394 | IL4R |
| P08069 | IGF1R |
| Q09472 | EP300 |
| P02679 | FGG |
| Q8WXH5 | SOCS4 |
| O14512 | SOCS7 |
| Q13526 | PIN1 |
| P19438 | TNFRSF1A |
| P01579 | IFNG |
| Q8TAE8 | GADD45GIP1 |
| P32927 | CSF2RB |
| P27986 | PIK3R1 |
| O15524 | SOCS1 |
| P01100 | FOS |
| P08047 | SP1 |
| P01589 | IL2RA |
| Q9NSE2 | CISH |
| P01106 | MYC |
| P14210 | HGF |
| Q15672 | TWIST1 |
| Q9HBE4 | IL21 |
| Q99836 | MYD88 |
| P01011 | SERPINA3 |
| Q9NZQ7 | CD274 |
| Q12913 | PTPRJ |
| P21246 | PTN |
| Q01628 | IFITM3 |
| P41597 | CCR2 |
| Q96EY1 | DNAJA3 |
| Q16665 | HIF1A |
| P27695 | APEX1 |
| P35228 | NOS2 |
| P16871 | IL7R |
| Q9BYH8 | NFKBIZ |
| P06400 | RB1 |
| Q92793 | CREBBP |
| P51532 | SMARCA4 |
| Q9H0N5 | PCBD2 |
| Q6PD62 | CTR9 |
| O43248 | HOXC11 |
| P42226 | STAT6 |
| Q9NRF2 | SH2B1 |
| O14492 | SH2B2 |
| O14920 | IKBKB |
| P18146 | EGR1 |
| P58753 | TIRAP |
| Q13322 | GRB10 |
| P38484 | IFNGR2 |
| P50607 | TUB |
| Q01344 | IL5RA |
| P29590 | PML |
| P15172 | MYOD1 |
| Q15642 | TRIP10 |
| O14744 | PRMT5 |
| Q9P0J0 | NDUFA13 |
| P41159 | LEP |
| P54756 | EPHA5 |
| P51813 | BMX |
| P30101 | PDIA3 |
| P19838 | NFKB1 |
| P63000 | RAC1 |
| P19235 | EPOR |
| P19525 | EIF2AK2 |
| P42680 | TEC |
| Q15788 | NCOA1 |
| P08581 | MET |
| P62937 | PPIA |
| P15498 | VAV1 |
| P78324 | SIRPA |
| Q15797 | SMAD1 |
| Q05209 | PTPN12 |
| P07332 | FES |
| Q13309 | SKP2 |
| Q05655 | PRKCD |
| P04049 | RAF1 |
| P07947 | YES1 |
| P17706 | PTPN2 |
| P10721 | KIT |
| P28223 | HTR2A |
| O14503 | BHLHE40 |
| P30626 | SRI |
| Q92665 | MRPS31 |
| Q99590 | SCAF11 |
| O14874 | BCKDK |
| P55268 | LAMB2 |
| Q8IUQ4 | SIAH1 |
| Q96RT1 | ERBB2IP |
| P31146 | CORO1A |
| Q13011 | ECH1 |
| P53365 | ARFIP2 |
| P22736 | NR4A1 |
| P52272 | HNRNPM |
| Q8WW38 | ZFPM2 |
| Q9BVP2 | GNL3 |
| Q96G01 | BICD1 |
| Q8N960 | CEP120 |
| O43609 | SPRY1 |
| Q9BXP5 | SRRT |
| Q8NEM7 | FAM48A |
| P05112 | IL4 |
| P05113 | IL5 |
| P35225 | IL13 |
| P10415 | BCL2 |
| P40337 | VHL |
| Q96EB6 | SIRT1 |
| O60341 | KDM1A |
| Q71DI3 | HIST2H3C |
| P16070 | CD44 |
| Q8WTS6 | SETD7 |
| Q15717 | ELAVL1 |
| P61956 | SUMO2 |
| P36873 | PPP1CC |
| Q8TC07 | TBC1D15 |
| Q9BTW9 | TBCD |
| Q96PZ0 | PUS7 |
| P53367 | ARFIP1 |
| P30556 | AGTR1 |
| P43246 | MSH2 |
| Q9Y6Y0 | IVNS1ABP |
| P25105 | PTAFR |
| Q15911 | ZFHX3 |
| P43405 | SYK |
| P48357 | LEPR |
Hundreds of miRNAs obtained were exhibited multiple targets, where 77 miRNAs were found to regulate more than 10 genes (which altogether regulates 966 genes) where hsa-miR-21-5p was prominent with 22 target genes (Table 5). Among these hsa-miR-498 and hsa-miR-297 were upregulated ones. Neglecting the downregulated genes above −10FC the miRNA list was again narrowed down to 25 which altogether regulate 330 genes associated with inflammation (Table 6). On considering the fold change hsa-miR-145-5p (−55.42), hsa-miR-100-5p (−51.6), hsa-miR-23b-3p (-35.32), hsa-let-7d-5p (−31.63), hsa-miR-146a-5p (−29.52), hsa-miR-150-5p (−23.41), hsa-miR-181a-5p (−21.34) and hsa-miR-193b-3p (−20.35) were downregulated more than 20-fold suggesting their potential role in regulating inflammation. These miRNAs participate in the regulation of 102 genes.
Table 5.
The 77 miRNAs of Group 2 vs Group 1 tendons which were found to be regulating more than 10 target genes.
| miRNAs | FC | No: of Hits |
|---|---|---|
| hsa-miR-21-5p | −10.11 | 22 |
| hsa-miR-30a-5p | −11.48 | 21 |
| hsa-miR-23a-3p | −14.18 | 19 |
| hsa-miR-125a-5p | −9.69 | 18 |
| hsa-miR-155-5p | −3.49 | 18 |
| hsa-miR-16-5p | −7.98 | 17 |
| hsa-miR-744-5p | −6.94 | 17 |
| hsa-miR-181a-5p | −21.34 | 16 |
| hsa-miR-15a-5p | −10.66 | 16 |
| hsa-miR-93-5p | −4.15 | 16 |
| hsa-miR-15b-5p | −3.98 | 16 |
| hsa-miR-181b-5p | −3.09 | 16 |
| hsa-miR-145-5p | −55.42 | 15 |
| hsa-miR-132-3p | −7.55 | 15 |
| hsa-miR-339-3p | −5.8 | 15 |
| hsa-miR-665 | −5.1 | 15 |
| hsa-miR-23b-3p | −35.32 | 14 |
| hsa-miR-30d-5p | −19.32 | 14 |
| hsa-miR-195-5p | −14.04 | 14 |
| hsa-miR-130a-3p | −8.11 | 14 |
| hsa-miR-652-3p | −5.88 | 14 |
| hsa-miR-221-3p | −5.72 | 14 |
| hsa-miR-17-5p | −3.53 | 14 |
| hsa-miR-185-5p | −3.39 | 14 |
| hsa-miR-150-5p | −23.41 | 13 |
| hsa-miR-103a-3p | −14.87 | 13 |
| hsa-miR-382-5p | −13.14 | 13 |
| hsa-miR-497-5p | −12.3 | 13 |
| hsa-miR-25-3p | −7.23 | 13 |
| hsa-miR-140-3p | −3.81 | 13 |
| hsa-miR-92b-3p | −3.43 | 13 |
| hsa-let-7a-5p | −31.63 | 12 |
| hsa-let-7b-5p | −7.73 | 12 |
| hsa-miR-30c-5p | −7.67 | 12 |
| hsa-miR-130b-3p | −6.11 | 12 |
| hsa-miR-20a-5p | −4.67 | 12 |
| hsa-miR-342-5p | −4.46 | 12 |
| hsa-miR-143-3p | −4.17 | 12 |
| hsa-miR-10a-5p | −4.02 | 12 |
| miRNAs | FC | No: of Hits |
| hsa-miR-20b-5p | −3.72 | 12 |
| hsa-miR-377-5p | −3.56 | 12 |
| hsa-miR-654-3p | −3.23 | 12 |
| hsa-miR-503-5p | −3.17 | 12 |
| hsa-miR-100-5p | −51.6 | 11 |
| hsa-miR-146a-5p | −29.52 | 11 |
| hsa-miR-134-5p | −14.82 | 11 |
| hsa-miR-574-3p | −10.54 | 11 |
| hsa-miR-151a-5p | −10.22 | 11 |
| hsa-let-7i-5p | −8.38 | 11 |
| hsa-miR-106b-5p | −8.08 | 11 |
| hsa-miR-532-3p | −7.12 | 11 |
| hsa-miR-381-3p | −5.67 | 11 |
| hsa-miR-139-5p | −5.65 | 11 |
| hsa-miR-324-3p | −5.2 | 11 |
| hsa-miR-122-5p | −4.69 | 11 |
| hsa-miR-433-3p | −3.89 | 11 |
| hsa-miR-24-3p | −3.53 | 11 |
| hsa-miR-17-5p | −3.53 | 11 |
| hsa-miR-455-3p | −3.45 | 11 |
| hsa-miR-23b-5p | −3.04 | 11 |
| hsa-miR-193b-3p | −20.35 | 10 |
| hsa-miR-199a-3p | −17.88 | 10 |
| hsa-let-7c-5p | −12.58 | 10 |
| hsa-miR-199a-5p | −11.9 | 10 |
| hsa-miR-370-3p | −8.38 | 10 |
| hsa-miR-28-5p | −8.31 | 10 |
| hsa-miR-339-5p | −7.22 | 10 |
| hsa-miR-193b-5p | −6.85 | 10 |
| hsa-miR-708-5p | −6.41 | 10 |
| hsa-miR-491-5p | −6.22 | 10 |
| hsa-miR-874-3p | −5.31 | 10 |
| hsa-miR-337-5p | −4.85 | 10 |
| hsa-miR-24-3p | −3.53 | 10 |
| hsa-miR-92b-5p | −3.26 | 10 |
| hsa-miR-498 | 2.05 | 10 |
| hsa-miR-297 | 3.81 | 10 |
| Total | 966 |
Table 6.
Multiple target miRNAs of genes associated with JAK2/STAT3 pathway genes where ten or more target genes were considered.
| miRNAs | FC | No: of Hits |
|---|---|---|
| hsa-miR-145-5p | −55.42 | 15 |
| hsa-miR-100-5p | −51.6 | 11 |
| hsa-miR-23b-3p | −35.32 | 14 |
| hsa-let-7a-5p | −31.63 | 12 |
| hsa-miR-146a-5p | −29.52 | 11 |
| hsa-miR-150-5p | −23.41 | 13 |
| hsa-miR-181a-5p | −21.34 | 16 |
| hsa-miR-193b-3p | −20.35 | 10 |
| hsa-miR-30d-5p | −19.32 | 14 |
| hsa-miR-199a-3p | −17.88 | 10 |
| hsa-miR-103a-3p | −14.87 | 13 |
| hsa-miR-134-5p | −14.82 | 11 |
| hsa-miR-23a-3p | −14.18 | 19 |
| hsa-miR-195-5p | −14.04 | 14 |
| hsa-miR-382-5p | −13.14 | 13 |
| hsa-let-7c-5p | −12.58 | 10 |
| hsa-miR-497-5p | −12.3 | 13 |
| hsa-miR-199a-5p | −11.9 | 10 |
| hsa-miR-30a-5p | −11.48 | 21 |
| hsa-miR-15a-5p | −10.66 | 16 |
| hsa-miR-574-3p | −10.54 | 11 |
| hsa-miR-151a-5p | −10.22 | 11 |
| hsa-miR-21-5p | −10.11 | 22 |
| hsa-miR-498 | 2.05 | 10 |
| hsa-miR-297 | 3.81 | 10 |
| Total | 330 |
Discussion
Tendon disorders are mostly associated with pain and inflammation and a high proportion of patients are susceptible to other inflammatory diseases like glenohumeral arthritis and/or osteoarthritis. Coincidence of such clinical conditions aggravates clinical symptoms and hurdles the repair and regeneration of injured tendon. The histology of patients with tendinopathy (without arthritis) presents degenerative lesions even without classical symptoms of inflammation [30]. However, in molecular level the upregulation of inflammatory receptors and immunoglobulins and an increased infiltration of immune cells were found to be coupled with tendinopathies [31]. The molecular mechanism leading to inflammation and enhanced cell infiltration in an otherwise avascular tissue like rotator cuff tendon is poorly revealed. Also, how the tendinopathies deal with the coincidence of arthritis is yet to be explored. The role of cytokines like IL-6, TNF-α, IL-1β and IFN-γ and inflammatory mediators like prostaglandins were established in tendon tissue as well but their regulatory mechanisms are largely unknown [4], [32]. Moreover, the epigenetic regulation of inflammatory responses by miRNAs in shoulder tendinopathies is rare in the literature. Our previous article evaluates and describes the regulatory roles of several potential miRNAs associated with the ECM integrity of long head of the biceps with respect to glenohumeral arthritis. The miRNAs common to ECM regulation and inflammation signifies the interrelation of these two events in the pathogenesis of RCI [27]. The focus of this study was to elucidate and screen the miRNAs associated with inflammatory events regarding JAK2/STAT3 pathway using the data from same set of patients.
JAK2/STAT3 pathway is one of the predominantly operated signaling which ends up in the activation of a battery of pro-inflammatory genes. Members of JAK2/STAT3 form potential targets for anti-inflammatory therapies [33] and the pathway has been aggravated in inflammatory diseases like arthritis [34]. Limited reports are available on the execution of JAK2/STAT3 pathway in tendon tissue and the actual trigger for this pathway is largely unknown. JAK2/STAT3 pathway has also been reported to be regulated epigenetically by miRNAs as well as DNA and histone modifications. To cite, let-7 miRNA suppress SOCS3 expression and blocks STAT3 phosphorylation by JAK2 and subsequent downstream signaling in PDAC (pancreatic ductal adeno carcinoma) cell lines while the inhibition of let-7 resulted in IL-6 coupled activation of STAT3 [35]. But, to the best of our knowledge the reports regarding miRNA mediated regulation of inflammation in shoulder tendon and the correlation with arthritis are unavailable in the literature. Our attempt was to screen and elucidate the roles of miRNAs associated with inflammation regarding JAK2/STAT3 pathway based on the coincidence of glenohumeral arthritis. In the current study, 10 miRNAs were found to be crucial for the JAK2/STAT3 pathway in shoulder tendon which has implications on inflammation associated with arthritis.
hsa-miR-145-5p was one of the top ten downregulated miRNAs from our array data which has been reported to be key to innate immune response [36]. miR-145-5p was reported to be involved in phagocyte differentiation, migration and action, migration and proliferation of endothelial cells, smooth muscle cells and lymphocytes, TNF family of ligand mediated apoptosis by activating NF-κβ and MAPKs [37]. Our results also revealed the association of hsa-miR-145-5p with STATs and IFN-γ and corresponding receptors which are related to JAK2/STAT3 pathway of inflammation. Also, the presence of CD68+ macrophages, CD16+ neutrophils, and angiogenesis, which were completely absent in Group 2, in our Group 1 patients substantiates the role of hsa-miR-145-5p in aggravating inflammation of shoulder tendon tissue which can be correlated with arthritis.
hsa-miR-100-5p is another highly regulated miRNA whose targets were found to be NF-κβ, MAPK (MAPK8 and MAPK1), IL-6 receptor, PTK2 and so on. Another potent target of hsa-miR-100-5p is mTOR which signifies its role in mTOR pathway resulting in cell growth and proliferation. The downregulation of hsa-miR-100-5p in Group 1 patients can be a reason for their delayed repair responses and persistence of tissue damage. The direct involvement of hsa-miR-100-5p in inflammatory pathways is unknown and our results suggest that it can be linked to inflammation via NF-κβ and JAK2/STAT3 pathway through IL-6R.
hsa-miR-23b-3p has been established to be a regulator of inflammatory cytokines including NF-κβ and TNF-α and inhibits inflammation. hsa-miR-23b-3p is multifunctional and regulates pathways mediating cell proliferation, adhesion, differentiation and apoptosis [38], [39]. Contrastingly, hsa-miR-23b-3p exhibited a 35.2-fold decrease in Group 1 tendons, where the inflammation and ECM damage was severe, suggesting the existence of alternate routes of regulation. From our data, it was evident that IL-6R, STAT5 and EP300 are regulated by hsa-miR-23b-3p and its downregulation results in the upregulation of these genes. Being a histone modifier (due to histone acetyl transferase activity), EP300 can facilitate the transcription of a battery of pro-inflammatory genes.
let-7 (human lethal-7) miRNAs are highly conserved and common of all miRNAs which is a family of 13 miRNAs. hsa-let-7d-5p is involved in the regulation of cell cycle genes and its downregulation is linked with carcinogenesis [40]. The active role of hsa-let-7d-5p in inflammation is yet to be unveiled. IL5 and STAT5 form the target of hsa-let-7d-5p, as evident from our data, suggesting its role in inflammatory pathways. As with other highly regulated miRNAs the downregulation of hsa-let-7d-5p can be correlated with the severity of inflammation in Group 1 tendons.
hsa-miR-146a-5p is also a potent regulator of innate inflammatory responses and are found to be upregulated in cells challenged with TNF-α and lipopolysaccharide. hsa-miR-146a-5p inhibits inflammation by targeting interleukin-1-receptor-associated kinase-1 (IRAK1) and TNF receptor associated factor 6 (TRAF6), the modulators of NF-κβ [41], [42]. hsa-miR-146a-5p was found to mediate IRAK, TRAF6, NF-κβ, EGFR and ICAM1 in our data which substantiate these findings. Also, the hsa-miR-146a-5p mediated regulation of IL-1R and STATs in our results signifies the involvement of JAK/STAT pathway in RC tendon inflammation. Moreover, the downregulation of hsa-miR-146a-5p can be a potent reason for the severity of inflammation of Group 1 patients.
The main function of hsa-miR-150-5p was reported to be the regulation of angiogenesis [43]. It is also expressed in NK cells, and B and T cells of immune system and the high levels of hsa-miR-150-5p is associated with defect in immune system and sustenance of inflammation [44]. Our data shows several other targets of hsa-miR-150-5p of which NF-κβ is typical for inflammation. The action of hsa-miR-150-5p causes the inhibition of NF-κβ signaling and there by inflammation [45]. This may be the reason why the downregulation of hsa-miR-150-5p correlates with the severity of inflammation and enhanced angiogenesis in Group 1 tendons.
hsa-miR-181a-5p is involved in inflammation associated with cancer and its main target was found out to be IL-1a. The anti-inflammatory effects of hsa-miR-181a-5p has been established in lipopolysaccharide challenged macrophage and monocyte cell lines [46], [47]. Apart from IL-1a, TNF-α and IL-6 were also established to be the targets for hsa-miR-181a-5p suggesting its anti-inflammatory roles. The feedback regulation elicited by hsa-miR-181a-5p against TNF-α induced inflammation on targeting p300/CBP on hepatic epithelial cells reveals its mechanism of regulation at the molecular level. TLR4 and NF-κβ form other potential targets for hsa-miR-181a-5p and it has significant role in relieving oxidative stress associated with inflammation as well [46]. Moreover, hsa-miR-181a-5p is one of the prominent miRNAs reported to be regulated during exercise [48] and so has significant implications on tendinopathies. But, their regulatory roles in tendons, especially in shoulder tendons, are largely unknown. Our data revealed the IFN-γ, MAPK1, IL-5, and IRAK1 to be the targets for hsa-miR-181a-5p which can be linked to JAK2/STAT3 pathway through IFN-γ and MAPK1. The inflammation in Group 1 tendons can be a result of downregulation of hsa-miR-181a-5p and subsequent upregulation of these pro-inflammatory mediators.
hsa-miR-193b-3p was demonstrated to be associated with tumor suppression by targeting D1 cyclins in prostate cancer cells [49]. hsa-miR-193b-3p also targets collagen type-2, aggrecan and SOX5 and regulates chondrocyte metabolism [50]. To the best of our knowledge, the reports of hsa-miR-193b-3p mediated regulation of inflammatory responses are rare. Our data show genes associated with inflammatory signaling especially, MAPK1, MAPK8, IRF1, IRAK1 and NF-κβ are being targeted by hsa-miR-193b-3p as the miRNA is downregulated in Group 1 patients.
hsa-miR-498 was reported to be associated with rheumatoid cancer and arthritis as well as allergy [51], [52]. hsa-miR-498 has tumor suppressor function as it targets FOXO3 gene to inhibit cell proliferation [53]. Also, this miRNA activates smooth muscle cell proliferation mediated through VEGFR [54]. Even though, limited reports are available regarding the regulation of hsa-miR-498 in inflammatory signaling, it can be linked to inflammation because EP300 and IL1R1 form potential targets [54, 55]. The results from our analysis showed hsa-miR-498 was upregulated in Group 1 tendons and acts by JAK3, NF-κβ and MAPK1 showing their role in inflammation. Still, the upregulation of NF-κβ and MAPK1 by hsa-miR-498 can be less effective due to lesser fold degree of fold change (more than 10 times) when compared with miRNAs targeting the same. hsa-miR-297, another potentially upregulated miRNA in Group 1 tendons, is involved in multidrug resistance (MDR) in cancer cells by modulating MRP-2 (MDR associated protein −2) [56]. IFN-γ, IFNGR1, IRAK1, SOSC1, and Bcl2 are the target genes of inflammation mediated by hsa-miR-297 as derived from our data.
We are the first to report whole miRNA profile from shoulder tendon based on the coincidence of glenohumeral arthritis and 10 miRNAs (8 downregulated and 2 upregulated) were chosen from the pool of highly regulated miRNAs based on their potent target inflammatory genes about JAK2/STAT3 pathway. Inflammatory pathways other than JAK2/STAT3 (TREM-1 signaling, TLR signaling etc.) are also prevalent in shoulder tendons [25]. The compilation of all the inflammatory pathways, interconnecting genes and associated miRNAs may pool up bulk of information which is beyond the scope of this article. Interestingly, we have reported similar approach to screen the miRNAs associated with ECM disorganization and several miRNAs were found to be common for ECM disorganization and JAK2/STAT3 pathway [27]. However, integration of miRNAs associated with the pathological events and pathways could be appreciated for screening and identification of miRNAs with therapeutic potential for the management of RCI.
These miRNAs could be valuable as signature miRNAs in the pathophysiology of RCI and might help in the treatment strategies for the repair of rotator cuff. Obviously, additional studies are warranted to elucidate their therapeutic potential. In vitro and in vivo evaluations of these miRNAs using appropriate mimics and inhibitors need to be validated before extending these to therapeutic arena. The supplementation of the downregulated miRNAs either individually or in combination can benefit millions of RCI sufferers throughout the globe. Moreover, the lack of normal control specimen, variations in clinical history patients, and lesser RNA yield (being collagenous and lesser cellularity of tendons particularly of Group 2 makes RNA isolation and purification challenging from the available biceps tendon) form major hurdles to the study. Still, the study has thrown new insights into the key miRNA players in shoulder tendon inflammation by effectively correlating with coincidence and severity of glenohumeral arthritis.
Conclusion
The miRNAs were screened with respect to their targets of inflammation mediated by JAK2/STAT3 pathway on patients with RCI and glenohumeral arthritis and patients without glenohumeral arthritis or rotator cuff tears. The levels of hsa-miR-145-5p, hsa-miR-100-5p, hsa-miR-23b-3p, hsa-let-7d-5p, hsa-miR-146a-5p, hsa-miR-150-5p, hsa-miR-181a-5p and hsa-miR-193b-3p were predominantly downregulated in glenohumeral arthritis tendon where the severity of inflammation was greater. This suggests their regulatory roles in eliciting inflammatory responses by targeting key inflammatory genes JAK2/STAT3 and interconnecting pathways. Targeting these miRNAs and the knowledge of their regulatory mechanisms would be critical to develop promising therapies in the management of shoulder pathology.
Supplementary Material
Genes associated with JAK/STAT pathway of inflammation determined by NetworkAnalyst using 88 input genes.
Different pathways and the number of associated genes in which the genes of JAK2/STAT3 pathway of inflammation cross talk with.
miRNAs regulating the genes associated with JAK2/STAT3 pathway of inflammation as determined by miRNA array of Group 1 vs Group 2 tendons. The upregulated miRNAs are displayed in red fond.
Fig. 4.
Representative images for CD16+expression in the tendon tissues of Group-1 and Group-2 patients by immunofluorescence. (A) Group-1 (four patient) and (B) Group-2 (represents four patient) patients. Group-1 tendons displayed higher density of neutrophils than Group-2.
Acknowledgments
This work was supported by research grants R01 HL112597, R01 HL116042, and R01 HL120659 to DK Agrawal from the National Heart, Lung and Blood Institute, National Institutes of Health, USA, and Creighton University LB692 grant to MFD from the State of Nebraska. The content of this original research article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the State of Nebraska.
List of abbreviations
- DAPI
4′,6- diamidino-2-phenylindole
- ECM
extracellular matrix
- JAK
janus activated kinase
- miRNA
microRNA
- PBS
phosphate buffered saline
- RCI
rotator cuff injury
- RIN
RNA integrity number
- STAT
signal transducers and activators of transcription
References
- 1.Thankam FG, Dilisio MF, Agrawal DK. Immunobiological factors aggravating the fatty infiltration on tendons and muscles in rotator cuff lesions. Mol Cell Biochem. 2016 doi: 10.1007/s11010-016-2710-5. [DOI] [PubMed] [Google Scholar]
- 2.Pandey V, Jaap Willems W. Rotator cuff tear: A detailed update. Asia-Pac J Sports Med Arthrosc Rehabil Technol. 2015;2:1–14. doi: 10.1016/j.asmart.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itoi E. Rotator cuff tear: physical examination and conservative treatment. J Orthop Sci. 2013;18:197–204. doi: 10.1007/s00776-012-0345-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thankam FG, Dilisio MF, Dougherty KA, et al. Triggering receptor expressed on myeloid cells and 5’adenosine monophosphate-activated protein kinase in the inflammatory response: a potential therapeutic target. Expert Rev Clin Immunol. 2016:1–11. doi: 10.1080/1744666X.2016.1196138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks P. Inflammation as an important feature of osteoarthritis. Bull World Health Organ. 2003;81:689–690. [PMC free article] [PubMed] [Google Scholar]
- 6.Macaulay AA, Greiwe RM, Bigliani LU. Rotator Cuff Deficient Arthritis of the Glenohumeral Joint. Clin Orthop Surg. 2010;2:196. doi: 10.4055/cios.2010.2.4.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith AM. Rotator Cuff Repair in Patients with Rheumatoid Arthritis. J Bone Jt Surg Am. 2005;87:1782. doi: 10.2106/JBJS.D.02452. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan MH. STAT signaling in inflammation. JAK-STAT. 2013;2:e24198. doi: 10.4161/jkst.24198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray PJ. The JAK-STAT Signaling Pathway: Input and Output Integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 10.Harrison DA. The JAK/STAT Pathway. Cold Spring Harb Perspect Biol. 2012;4:a011205–a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy S, Benz F, Luedde T, Roderburg C. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg Nutr. 2015;4:24–33. doi: 10.3978/j.issn.2304-3881.2015.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essandoh K, Li Y, Huo J, Fan G-C. MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock Augusta Ga. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Jing Z, Cheng G. MicroRNAs: New Regulators of Toll-Like Receptor Signalling Pathways. BioMed Res Int. 2014;2014:1–14. doi: 10.1155/2014/945169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amado T, Schmolka N, Metwally H, et al. Cross-regulation between cytokine and microRNA pathways in T cells: Highlights-Review. Eur J Immunol. 2015;45:1584–1595. doi: 10.1002/eji.201545487. [DOI] [PubMed] [Google Scholar]
- 16.Olarerin-George AO, Anton L, Hwang Y-C, et al. A functional genomics screen for microRNA regulators of NF-kappaB signaling. BMC Biol. 2013;11:19. doi: 10.1186/1741-7007-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Chen X, Guan L, et al. MiRNA-181a Regulates Adipogenesis by Targeting Tumor Necrosis Factor-α (TNF-α) in the Porcine Model. PLoS ONE. 2013;8:e71568. doi: 10.1371/journal.pone.0071568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Qin Z, Li Q, et al. MicroRNA-124 negatively regulates LPS-induced TNF-α production in mouse macrophages by decreasing protein stability. Acta Pharmacol Sin. 2016;37:889–897. doi: 10.1038/aps.2016.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millar NL, Wei AQ, Molloy TJ, et al. Cytokines and apoptosis in supraspinatus tendinopathy. J Bone Jt Surg - Br. 2009;91–B:417–424. doi: 10.1302/0301-620X.91B3.21652. [DOI] [PubMed] [Google Scholar]
- 20.Jin-fang Zhang JG, Kai-ming Chan GL. A Mini-Review: MicroRNA in Tendon Injuries. J Stem Cell Res Ther. 2015 doi: 10.4172/2157-7633.1000303. [DOI] [Google Scholar]
- 21.Gupta GK, Agrawal T, DelCore MG, et al. Vitamin D deficiency induces cardiac hypertrophy and inflammation in epicardial adipose tissue in hypercholesterolemic swine. Exp Mol Pathol. 2012;93:82–90. doi: 10.1016/j.yexmp.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and Eosin Staining of Tissue and Cell Sections. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot4986. pdb.prot4986-pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- 23.Fong G, Backman LJ, Hart DA, et al. Substance P enhances collagen remodeling and MMP-3 expression by human tenocytes. J Orthop Res. 2013;31:91–98. doi: 10.1002/jor.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal T, Gupta GK, Agrawal DK. Vitamin D Supplementation Reduces Airway Hyperresponsiveness and Allergic Airway Inflammation in a Murine Model. Clin Exp Allergy. 2013 doi: 10.1111/cea.12102. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thankam FG, Dilisio MF, Dietz NE, Agrawal DK. TREM-1, HMGB1 and RAGE in the Shoulder Tendon: Dual Mechanisms for Inflammation Based on the Coincidence of Glenohumeral Arthritis. PLOS ONE. 2016;11:e0165492. doi: 10.1371/journal.pone.0165492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar NL, Gilchrist DS, Akbar M, et al. MicroRNA29a regulates IL-33-mediated tissue remodelling in tendon disease. Nat Commun. 2015;6:6774. doi: 10.1038/ncomms7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thankam FG, Boosani CS, Dilisio MF, et al. MicroRNAs Associated with Shoulder Tendon Matrisome Disorganization in Glenohumeral Arthritis. PLOS ONE. 2016;11:e0168077. doi: 10.1371/journal.pone.0168077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Benner MJ, Hancock REW. NetworkAnalyst - integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–W174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tedgui A. Cytokines in Atherosclerosis: Pathogenic and Regulatory Pathways. Physiol Rev. 2006;86:515–581. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- 30.Abate M, Gravare-Silbernagel K, Siljeholm C, et al. Pathogenesis of tendinopathies: inflammation or degeneration? Arthritis Res Ther. 2009;11:235. doi: 10.1186/ar2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millar NL, Hueber AJ, Reilly JH, et al. Inflammation Is Present in Early Human Tendinopathy. Am J Sports Med. 2010;38:2085–2091. doi: 10.1177/0363546510372613. [DOI] [PubMed] [Google Scholar]
- 32.Dakin SG, Dudhia J, Smith RKW. Resolving an inflammatory concept: The importance of inflammation and resolution in tendinopathy. Vet Immunol Immunopathol. 2014;158:121–127. doi: 10.1016/j.vetimm.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, He G, Hao Y, et al. The role of the JAK2-STAT3 pathway in pro-inflammatory responses of EMF-stimulated N9 microglial cells. J Neuroinflammation. 2010;7:54. doi: 10.1186/1742-2094-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao W, McCormick J, Connolly M, et al. Hypoxia and STAT3 signalling interactions regulate pro-inflammatory pathways in rheumatoid arthritis. Ann Rheum Dis. 2015;74:1275–1283. doi: 10.1136/annrheumdis-2013-204105. [DOI] [PubMed] [Google Scholar]
- 35.Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. 2015;5:17663. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Zhang M, He H, et al. MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med Genomics. 2013 doi: 10.1186/1755-8794-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu R-Q, Zhang D-F, Tu E, et al. The mucosal immune system in the oral cavity—an orchestra of T cell diversity. Int J Oral Sci. 2014;6:125–132. doi: 10.1038/ijos.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo X, Ranade K, Talker R, et al. microRNA-mediated regulation of innate immune response in rheumatic diseases. Arthritis Res Ther. 2013;15:210. doi: 10.1186/ar4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolenda T, Przybyła W, Teresiak A, et al. The mystery of let-7d– a small RNA with great power. Współczesna Onkol. 2014;5:293–301. doi: 10.5114/wo.2014.44467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kutty RK, Nagineni CN, Samuel W, et al. Differential regulation of microRNA-146a and microRNA-146b-5p in human retinal pigment epithelial cells by interleukin-1β, tumor necrosis factor-α, and interferon-γ. Mol Vis. 2013;19:737–750. [PMC free article] [PubMed] [Google Scholar]
- 42.Ye E-A, Steinle JJ. miR-146a Attenuates Inflammatory Pathways Mediated by TLR4/NF- κ B and TNF α to Protect Primary Human Retinal Microvascular Endothelial Cells Grown in High Glucose. Mediators Inflamm. 2016;2016:1–9. doi: 10.1155/2016/3958453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing Y, Fu J, Yang H, et al. MicroRNA expression profiles and target prediction in neonatal Wistar rat lungs during the development of bronchopulmonary dysplasia. Int J Mol Med. 2015 doi: 10.3892/ijmm.2015.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punga T, Le Panse R, Andersson M, et al. Circulating miRNAs in myasthenia gravis: miR-150-5p as a new potential biomarker. Ann Clin Transl Neurol. 2014;1:49–58. doi: 10.1002/acn3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sang W, Wang Y, Zhang C, et al. MiR-150 impairs inflammatory cytokine production by targeting ARRB-2 after blocking CD28/B7 costimulatory pathway. Immunol Lett. 2016;172:1–10. doi: 10.1016/j.imlet.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W, Li M, Xu N, et al. miR-181a Regulates Inflammation Responses in Monocytes and Macrophages. PLoS ONE. 2013;8:e58639. doi: 10.1371/journal.pone.0058639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynoso R, Laufer N, Hackl M, et al. MicroRNAs differentially present in the plasma of HIV elite controllers reduce HIV infection in vitro. Sci Rep. 2014 doi: 10.1038/srep05915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonevitsky AG, Maltseva DV, Abbasi A, et al. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 2013;13:9. doi: 10.1186/1472-6793-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaukoniemi KM, Rauhala HE, Scaravilli M, et al. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015;4:1417–1425. doi: 10.1002/cam4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takulla NF, Wolff MS, Schenkel AB. Caries management by risk assessment. N Y State Dent J. 2012;78:41–45. [PubMed] [Google Scholar]
- 51.Yan J-K, Wang W-Q, Ma H-L, Wu J-Y. Sulfation and Enhanced Antioxidant Capacity of an Exopolysaccharide Produced by the Medicinal Fungus Cordyceps sinensis. Molecules. 2012;18:167–177. doi: 10.3390/molecules18010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Wan Y, Guo Q, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai Y, Wang M, Zhang Q, Wang J. MicroRNA-498 is downregulated in non-small cell lung cancer and correlates with tumor progression. J Cancer Res Ther. 2015;11:107. doi: 10.4103/0973-1482.163859. [DOI] [PubMed] [Google Scholar]
- 54.Jin H, Li C, Ge H, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of Intracranial Aneurysm Rupture a case control study. J Transl Med. 2013;11:296. doi: 10.1186/1479-5876-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sánchez-Jiménez C, Carrascoso I, Barrero J, Izquierdo JM. Identification of a set of miRNAs differentially expressed in transiently TIA-depleted HeLa cells by genome-wide profiling. BMC Mol Biol. 2013;14:4. doi: 10.1186/1471-2199-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu K, Liang X, Shen K, et al. miR-297 modulates multidrug resistance in human colorectal carcinoma by down-regulating MRP-2. Biochem J. 2012;446:291–300. doi: 10.1042/BJ20120386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes associated with JAK/STAT pathway of inflammation determined by NetworkAnalyst using 88 input genes.
Different pathways and the number of associated genes in which the genes of JAK2/STAT3 pathway of inflammation cross talk with.
miRNAs regulating the genes associated with JAK2/STAT3 pathway of inflammation as determined by miRNA array of Group 1 vs Group 2 tendons. The upregulated miRNAs are displayed in red fond.