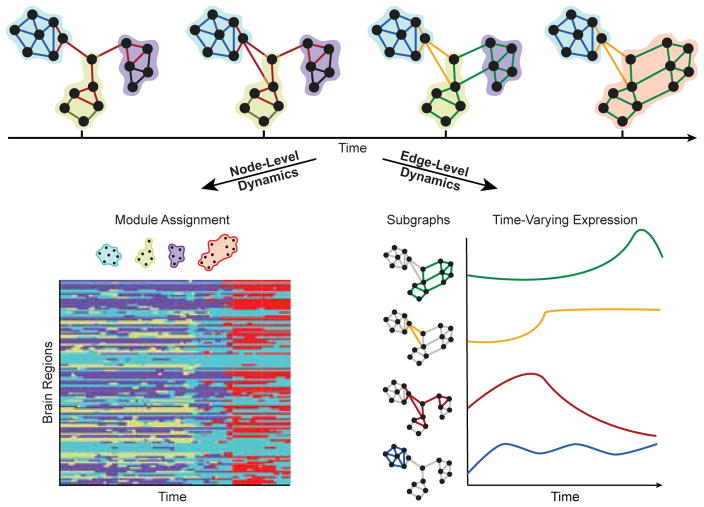
FIG. 2. Dynamic network modules and subgraphs.
(Top) Network science enables investigators to study dynamic architecture of complex brain networks in terms of the collective organization of nodes and of edges. Clusters of strongly interconnected nodes are known as modules, and clusters of edges whose strengths, or edge weights, vary together in time are known as subgraphs. Nodes and edges of the same module or subgraph are shaded by color. Each module represents a collection of nodes that are highly interconnected to one another and sparsely connected to nodes of other modules, and each node may only be a member of a single module. Each subgraph is a recurring pattern of edges that link information between nodes at the same points in time, and each edge can belong to multiple subgraphs. (Bottom Left) Dynamic community detection assigns nodes to time-varying modules. Nodes may shift their participation between modules over time based on the demands of the system. (Bottom Right) Non-negative matrix factorization pursues a parts-based decomposition of the dynamic network into subgraphs and time-varying coefficients, which quantify the level of expression of each subgraph over time.