Abstract
Reactivation of fetal hemoglobin (HbF) is a promising approach for the treatment of β-hemoglobinopathies and the targeting of genes involved in HbF regulation is under intensive investigation. Here, we established a nonhuman primate (NHP) transplantation model to evaluate hematopoietic stem cell (HSC)-based gene editing strategies aimed at reactivating HbF. We first characterized the transient HbF induction to autologous HSC transplantation in pigtailed macaques, which was comparable in duration and amplitude to that of human patients. After validating function of the HbF repressor BCL11A in NHPs, we transplanted a pigtailed macaque with CD34+ cells electroporated with TALE nuclease mRNA targeting the BCL11A coding sequence. In vivo gene editing levels were low, but some BCL11A deletions were detected as late as 200 days post-transplantation. HbF production, as determined by F-cell staining and γ-globin expression, was slightly increased in this animal as compared to transplant controls. We also provided proof-of-concept results for the selection of edited NHP CD34+ cells in culture following integration of the P140K/MGMT cassette at the BCL11A locus. In summary, the NHP model described here will allow the testing of novel therapeutic approaches for hemoglobinopathies and should facilitate clinical translation.
Keywords: BCL11A, hemoglobinopathy, gene editing, nonhuman primate, fetal hemoglobin, hematopoietic stem cells, transplantation, TALENs
Introduction
β-hemoglobinopathies constitute a significant health problem worldwide, with approximately 276,000 children per year born with sickle cell disease (SCD) (two-thirds in Africa) and 42,000 born annually with β-thalassemias.1 Two approaches are currently in clinical use for treatment: (1) allogeneic bone marrow transplantation (BMT), a cellular therapy that is aimed to replace a patient’s defective hematopoietic stem cells (HSCs) with those of a normal donor; and (2) gene therapy, which to date has primarily been practiced as gene replacement or gene addition using viral vectors. Allogeneic BMT is limited by availability of matched donors and is associated with immunological complications (graft versus host disease/graft rejection), whereas gene replacement therapy is associated with genotoxic risks due to semi-random integration of the viral vector.
Rapid advancements in gene editing tools now offer the possibility to precisely correct mutations in a patient’s own HSCs and cure the disease through autologous transplantation of corrected HSCs. In the context of hemoglobinopathies, transplanted HSCs subsequently differentiate into red blood cells, producing therapeutic levels of hemoglobin. Although a single nucleotide substitution is responsible for SCD, β-thalassemia is caused by hundreds of mutations or deletions scattered throughout the beta-globin locus.2 Despite this heterogeneity in disease-causing mutations, a universal gene correction strategy is needed that would be applicable to all patients. One important predictor of the clinical severity of β-hemoglobinopathies is the level of expression of fetal hemoglobin (HbF, α2γ2),3 whereby γ-globin serves as a substitute for defective β-globin in β-thalassemia or as an anti-sickling agent in SCD. Although HbF is normally silenced after birth and replaced with adult hemoglobin (HbA), some individuals born with a benign genetic condition known as hereditary persistence in fetal hemoglobin (HPFH) continue to express high levels of HbF throughout adulthood (reviewed by Forget et al.4). Reactivation of HbF thus constitutes a promising approach for the treatment of hemoglobin disorders, for example, by targeting genes involved in HbF regulation.
Genome-wide association studies investigating individuals with HPFH have identified multiple causative genetic loci, among which figured the transcription factor BCL11A (B cell lymphoma/leukemia 11A).5 Recent molecular studies validated the role of BCL11A in the silencing of γ-globin expression in mouse and human cells, and knockdown of BCL11A in human cells was sufficient to increase HbF expression without altering erythroid differentiation of these cells.6 A direct link between BCL11A inactivation and amelioration of SCD symptoms was subsequently demonstrated in a murine model,7 confirming the therapeutic potential of this target.
The nonhuman primate (NHP) model is ideally suited for studying safety and efficacy of novel therapeutic gene therapy/gene editing approaches for the treatment of hemoglobinopathies for multiple reasons: (1) NHP HSCs express CD34 as well as other common cell surface markers homologous to human cell surface markers; therefore, many reagents used in the human setting, such as recombinant growth factors and antibodies, are cross-reactive with the NHP system; (2) the scale of cell populations collected and transplanted as well as the hematopoietic demand required in vivo in NHPs closely resembles that of human patients; and (3) the unique ability to follow differentiation of transplanted HSCs into hemoglobin-producing red blood cells. Our group previously demonstrated the multi-lineage engraftment of autologous HSCs modified by means of viral vectors8, 9, 10, 11 or zinc finger nucleases (ZFNs)12 in the macaque transplantation model. Levels of gene-modified cells were successfully increased post transplantation through in vivo selection using the P140K O6-methylguanine-DNA-methyltransferase (MGMT/P140K) system in NHPs8, 13 and human patients.14 In this report, we establish the NHP autologous transplantation model to evaluate gene editing strategies aimed at reactivating HbF expression. As proof of concept, we describe the transplantation of BCL11A-edited HSCs in the pigtailed macaque and present data on the long-term tracking of gene editing frequency and hemoglobin expression in vivo. These findings provide a foundation for the evaluation of additional gene editing strategies aimed at curing hemoglobinopathies in the NHP model.
Results
Transient Increase in HbF following Autologous HSC Transplantation in NHPs
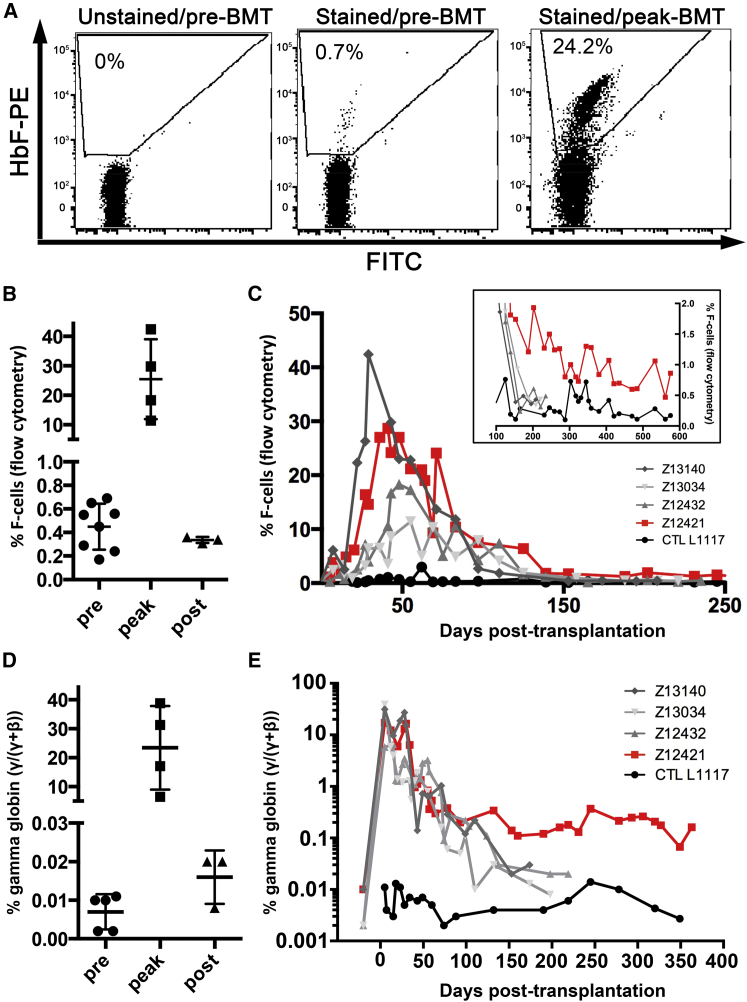
HbF (α2γ2) predominates during fetal life but is progressively replaced with the major adult hemoglobin A (HbA, α2β2) after birth to comprise about 97% of total hemoglobin. Hematologic disorders, such as leukemia, inherited bone marrow failure syndrome, and refractory anemia, have been shown to increase HbF expression in both adults and children,15 and BMT is also accompanied by a rapid and transient induction in HbF production in humans.16 In an effort to establish the NHP as transplantation model for the treatment of hemoglobinopathies, we first characterized the HbF induction in pigtailed macaques (Macaca nemestrina) in response to autologous BMT following myeloablative conditioning by total body irradiation (TBI). As shown in Figure S1, all animals showed a rapid drop in platelet, neutrophil, and lymphocyte counts after TBI and comparable hematopoietic recovery after BMT. We measured the HbF response in these animals by quantification of HbF-expressing cells (referred to as F-cells) in peripheral blood using intracellular antibody staining and flow cytometry analysis. F-cell quantification in peripheral blood was previously correlated with the HbF fraction detected in the hemolysate17 and thus provides a reliable measurement of the HbF response. Low F-cell frequencies were detected in all animals tested prior to transplantation, averaging 0.45% ± 0.07% (n = 7; Figures 1A and 1B) and were consistent with postnatal silencing of HbF in NHPs. After BMT, we observed a rapid increase in peripheral blood F-cells, which peaked at 29 to 45 days post-transplant (Figure 1C) and averaged frequencies of 25.5% ± 6.8% (n = 4; range, 11%–42%; Figure 1B). HbF production subsequently declined in all transplanted animals and returned to pre-transplantation levels at 150–200 days post transplantation (Figures 1B and 1C).
Figure 1.
Transient Increase in HbF following BMT in NHPs
(A) Representative flow plots of F-cell staining in Z12421 peripheral blood before or after BMT. (B) Fraction of F-cells in the peripheral blood of several pigtailed macaques before (pre), during (peak), and after BMT (∼200 days, post). Lines represent mean ± SD. (C) Time course of HbF response in transplanted pigtailed macaques, as determined by the frequency of F-cells in peripheral blood. Three control transplanted animals are shown in gray, one untransplanted control is shown in black, and the animal transplanted with BCL11A-edited HSCs is shown in red. Inset shows the long-term HbF response in the same animals. (D) γ-globin expression defined by γ/(γ + β) measured in several pigtailed macaques before (pre), during (peak), and after BMT (∼200 days, post). Lines represent mean ± SD. (E) Time course of γ-globin expression as defined in (D) in the same animals as (C).
As an independent measurement of HbF expression, we quantified hemoglobin transcripts (α-, β-, and γ-hemoglobin chains) by qPCR of peripheral blood RNA. We started by comparing blood obtained from an untransplanted control (A11225), from an animal transplanted with HSCs modified with a γ-globin SIN lentiviral vector (A09172),10 and from macaque cord blood (CB). As expected, α-globin expression was equivalent in all 3 blood specimens, but important differences were seen in γ-globin, which was highest in CB, intermediate in A09172, and lowest in A1125, as expected (Figure S2). β-globin expression was equivalent in A11225 and A09172 but was markedly reduced in CB, probably reflecting lower HbA (α2β2) relative to HbF (α2γ2) in this sample. Having validated this qPCR approach, we then analyzed blood from all transplanted animals described in Figure 1. Overall, α- and β-globin levels were comparable in all animals and substantially greater than γ-globin levels, consistent with profiles from adult blood (Figure S3). Transplantation and myeloablative conditioning by TBI initially resulted in a sharp decrease in α- and β-globin expression, which rapidly returned to pre-transplant levels after hematopoietic recovery. In parallel, γ-globin expression increased during the first month post-transplantation to reach levels comparable to α- and β-globins and gradually declined thereafter. The fraction of γ-globin among β-like globins (calculated as γ/(γ + β)) was comparable in all animals prior to transplantation (mean, 0.007% ± 0.002%; n = 5; Figure 1D) but increased about 3,000-fold after BMT to reach levels ranging from 6.5% to 39.9% (mean, 23.4% ± 7.2%; n = 4; Figure 1D). This surge in γ-globin was rapid and followed by a slow decline in expression that eventually return to near pre-transplant levels at about 200 days (mean, 0.016% ± 0.004%; n = 3; Figures 1D and 1E, gray lines). As expected, γ-globin expression in all transplanted animals showed a strong correlation with measurements of F-cell frequencies pre-transplantation and also at peaked levels post-transplantation (Figure S4), with Z13140 showing the strongest response and Z13034 showing the weakest. As expected, γ-globin expression remained low and mostly constant in the untransplanted control throughout the experiment (Figure 1D, black line). In summary, we characterized the HbF induction to autologous BMT and myeloablative conditioning in the NHP model; these findings will provide a valuable baseline for evaluating novel therapeutic approaches aimed at reactivating HbF production.
Conditions of stress erythropoiesis, such as hemolytic anemia and acute phlebotomy, also influence HbF production, as previously documented in baboons.18, 19 Measurement of peripheral blood F-cells in an untransplanted macaque undergoing repeated blood and bone marrow draws showed small and transient but reproducible increase in HbF for blood draws larger than 50 mL and for a 35-mL bone marrow aspirate (Figure S5); nevertheless, the increase in HbF seen under these conditions was modest (<1%) as compared to the effect reported for BMT (Figure 1).
TALEN-Mediated BCL11A Gene Editing in NHP CD34+ Cells
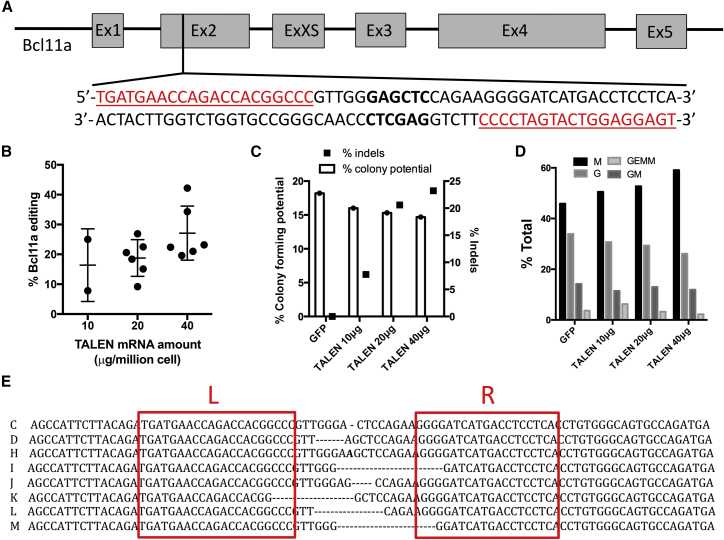
BCL11A was previously identified as a repressor of HbF6, 7 and thus constitutes a promising target for the treatment of β-hemoglobinopathies. Transplantation of autologous, BCL11A-edited HSCs in a patient suffering from SCD or β-thalassemia has the potential to augment HbF to therapeutic levels after engraftment and differentiation of HSCs into globin-producing cells. Nevertheless, successful treatment will require stable and long-term engraftment of an adequate number of BCL11A-edited HSCs. Toward this goal, we constructed a pair of transcription activator-like effector nucleases (TALENs) targeting exon 2 of BCL11A (Figure 2A), which would inactivate gene function upon repair of the resulting double-strand break by the non-homologous end joining pathway. The selected TALEN target site shows perfect sequence conservation among human, pigtailed macaque (M. nemestrina), and rhesus macaque (M. mulatta) so that this nuclease platform can be used across species. Using pre-established conditions for the electroporation of ZFN mRNA in NHP CD34+ cells,12 we defined the optimal TALEN mRNA concentration that would maximize BCL11A gene editing efficiency while minimizing cytotoxicity. We observed an mRNA dose-dependent increase in small nucleotide insertions or deletions (indels) introduced in BCL11A after electroporation of bone-marrow-derived NHP CD34+ cells (Figure 2B). 40 μg TALEN mRNA per million cells was optimal and resulted in 20%–35% indels (mean, 27.1% ± 3.7%; n = 6). The multilineage differentiation potential, determined by enumerating colony forming cells (CFCs) grown on methylcellulose media, was slightly reduced (<5%) in BCL11A-edited NHP CD34+ cells as compared to cells electroporated with GFP mRNA, as the dose of TALEN mRNA was increased from 10 to 40 μg per million cells (Figure 2C). This was accompanied by a small increase in the fraction of macrophage (M) colonies, whereas the fraction of granulocyte (G) colonies was reduced in the TALEN-treated cells (Figure 2D). BCL11A indels frequency was comparable between bulk NHP CD34+ cells and derived CFCs (25% in bulk versus 21% in CFCs; n = 43). Sequencing of the BCL11A target in edited CFCs revealed deletions ranging from 1 to 13 nt in length and one insertion of a single nucleotide (Figure 2E; n = 8). Approximately 2/3 of these mutations were null mutations, resulting in a translational frameshift, and are expected to inactivate BCL11A function. In summary, we demonstrated efficient BCL11A editing in NHP HSCs using TALEN mRNA electroporation, with minimal impact on the multilineage differentiation potential.
Figure 2.
TALEN-Mediated BCL11A Editing in NHP CD34+ Cells
(A) Schematic of BCL11A gene structure and TALEN target site. Grey boxes show exons (Ex) and lines show introns. Underlined red sequences show left and right TALEN DNA-binding sites. Sequence in bold shows the SacI restriction site used for assessment of BCL11A editing efficiency. (B) TALEN mRNA dose-dependent increase in BCL11A editing efficiency in NHP CD34+ cells (n = 6 donors). Dots represent results from independent experiments and lines show mean ± SD. (C) Effect of BCL11A TALEN mRNA electroporation on HSC colony-forming potential (left y axis) and BCL11A indels (right y axis). Results are from one representative experiment from (B). (D) Effect of BCL11A-TALEN mRNA electroporation on HSC differentiation potential. Fractions of each colony type obtained from (C) are shown. M, macrophages; G, granulocytes; GEMM, common myeloid progenitors; GM, granulocyte/macrophage progenitor. (E) Sequencing of TALEN target site in single colonies obtained from (C). Left (L) and right (R) boxes show TALEN-binding sites. Dashes show deletions and the bold letter shows insertion as compared to the reference sequence. In all experiments, BCL11A editing frequency was determined by SacI digestion assay as described in the Materials and Methods section.
BCL11A Editing Increases HbF Expression In Vitro in NHP CD34+-Derived Erythroblasts
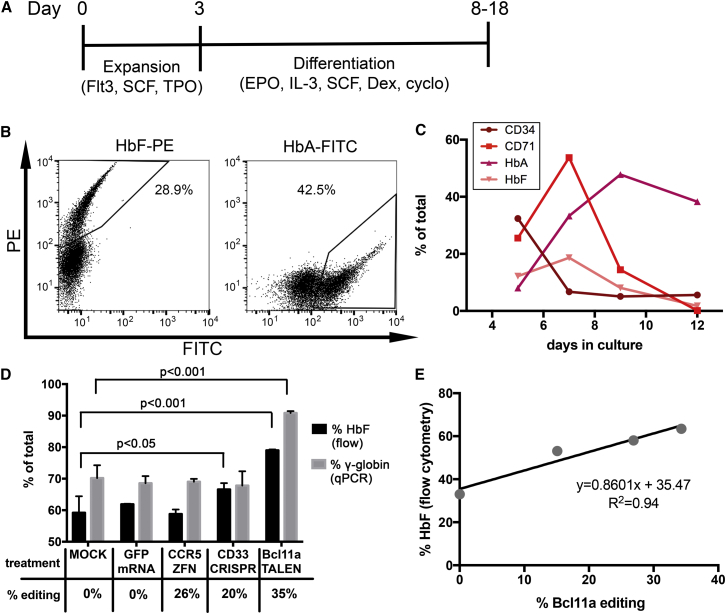
Recent molecular studies demonstrated that HbF expression is increased in vitro by inactivation of BCL11A in human cells.6, 20, 21 To determine if the HbF repressor function of BCL11A is conserved in NHPs, we compared hemoglobin expression in wild-type and BCL11A-edited CD34+ cell-derived erythroblasts. We first optimized conditions for efficient erythroid differentiation of NHP CD34+ cells following guidelines previously described for a two-phase liquid culture system involving cell expansion and differentiation6, 22 (Figure 3A). Erythroid differentiation was first validated in human cells using the marker glycophorin A (CD235a), which was expressed in over 80% of the cells after 10 days of culture (Figure S6). Several CD235a antibody clones were tested in NHP cells but did not cross-react (data not shown), so we instead relied on the transferrin receptor CD71, along with intracellular HbA and HbF expression as a differentiation marker for NHP cells (Figures 3B and 3C). Intracellular HbA and HbF expression generally peaked at 8–12 days post-differentiation and reached levels ranging from 20% to 50%. We started by comparing hemoglobin expression in differentiated, BCL11A-edited NHP cells (as described in Figure 2) with untreated cells (mock) or cells electroporated with GFP mRNA. HbF expression was evaluated both by flow cytometry and calculated as % HbF (HbF/(HbF + HbA)) and by qPCR as γ-globin expression (calculated as γ/(γ + β)). Bcl11 editing strongly increased HbF (p < 0.001) and γ-globin expression (p < 0.001) in TALEN-treated cells as compared to mock-treated cells, but no significant difference in expression was observed between mock and GFP mRNA electroporated cells (Figure 3D). To rule out an unspecific effect of DNA damage on HbF induction, we also treated cells with ZFN or CRISPR/Cas9-targeting non-hemoglobin genes (CCR5 and CD33, respectively), which did not robustly increase HbF expression (Figures 3D and S7 and Supplemental Materials and Methods). Although a small increase in % HbF was noted in cells treated with CD33 CRISPR/Cas9 as compared to mock-treated cells, no difference in γ-globin expression was observed between these two groups. A strong correlation between %HbF and Bcl11a indels was also noted in these cells (Figure 3E). In conclusion, these results validated BCL11A repressor function in NHPs and confirmed its therapeutic potential for HbF reactivation.
Figure 3.
Increased HbF Expression in BCL11A-Edited NHP Erythroblasts
(A) Schematic of the erythroid differentiation experiment design. (B) Representative flow cytometry plots for HbF or HbA staining in 10-day differentiated NHP cells. Fraction of stained population is given in each plot. (C) Time-dependent characterization of erythroid differentiation of NHP bone-marrow-derived CD34+ cells by flow cytometry analysis. (D) 0.5 million NHP CD34+ cells were untreated (mock) or electroporated with GFP mRNA (10 μg) or with the indicated mRNA-encoded nucleases (20 μg mRNA per reaction) and hemoglobin expression was measured by flow cytometry or qPCR in 10-day differentiated NHP cells. %HbF is defined as % F-cells normalized to the fraction of cells expressing HbF or HbA. % γ-globin expression is defined as γ/(γ + β) expression normalized to GAPDH. Editing efficiency was determined by the surveyor assay. Error bars show SDs for duplicate (%HbF) or triplicate (% γ-globin) measurements. Determination of statistical significance is described in the Supplemental Materials and Methods. (E) Correlation by linear regression analysis of %HbF (defined as above) relative to BCL11A editing efficiency (surveyor assay). Pearson’s correlation coefficient is given for a linear relationship between each dataset. In all panels, results are from one representative experiment.
Transplantation of BCL11A-Edited CD34+ Cells and Long-Term Follow-Up of Indels Frequency and Hemoglobin Expression in the NHP Model
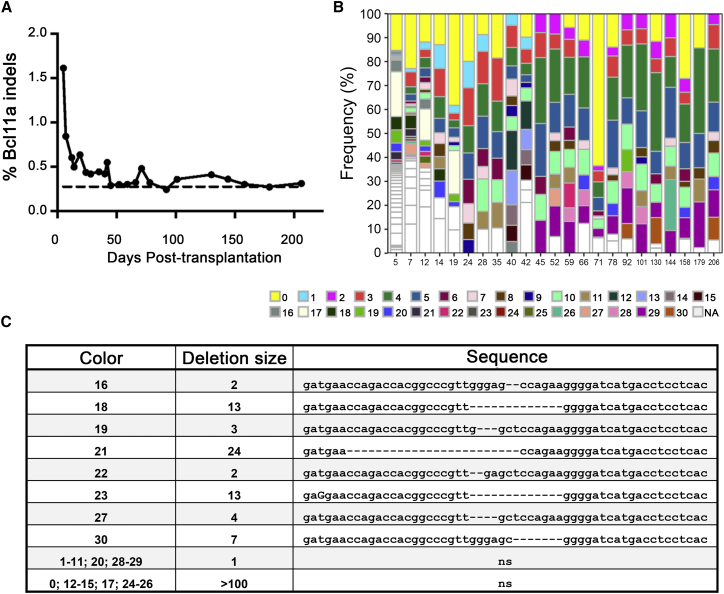
Using the HbF baseline established in Figure 1 from transplanted NHPs and following our validated gene editing approach to inactivate BCL11A (Figures 2 and 3), we infused a pigtailed macaque with autologous, BCL11A-edited CD34+ cells. We employed transplantation guidelines previously established by our laboratory,8, 9, 11, 12 and enriched 86 million CD34+ cells from G-CSF/stem cell factor (SCF)-primed bone marrow of animal Z12421 (purity >90%; Figure S8A). After overnight culture (day 1), 74 million cells were electroporated with BCL11A-TALEN mRNA at a dose of 16 μg per million cells. As transfection control, cells from the same animal were electroporated with GFP mRNA at a dose of 6 μg mRNA per million cells, resulting in 66% GFP+ cells at 24 hr post-electroporation (Figure S8B). Cell viability following treatment with BCL11A-TALEN mRNA was only slightly reduced as compared to GFP mRNA-treated cells at 24 hr post electroporation (76.6% versus 84.3%, as determined by forward/side scatter; Figure S8B). Fifty-one million cells treated with BCL11A-TALEN mRNA were infused in Z12421 (day 2) at a dose of 17 million cells per kg and hematopoietic recovery was closely monitored in this animal. Complete blood cell counts (CBCs) showed normal hematopoietic recovery in Z12421 as compared to other transplant controls (Figure S1, red lines). Deep sequencing analysis of the BCL11A target site in the infused product (TALEN-treated CD34+ cells) cultured in vitro for 4 and 10 days post-electroporation showed about 1.5% BCL11A indels frequency, which is considerably lower than results observed from small-scale electroporation experiments (Figure 2B; also see Discussion). From this analysis, we then focused on deletion events and detected about 50 unique signatures, ranging from 1 to 181 nt in length, with about 40% of these detected at both 4 and 10 days of culture (Figures S9A and S9B). Following infusion, approximately 1.5% BCL11A indels were detected in Z12421 peripheral blood at 5 days post-transplantation but indels frequency rapidly declined thereafter to reach a set point of 0.3%–0.4% by 50 days post-transplantation (Figure 4A). Despite being low, these levels of BCL11A editing are slightly above background, as determined from analysis of mock-treated cells from the same animal electroporated with GFP mRNA (Supplemental Materials and Methods). As shown in Figures 4B and 4C, several BCL11A deletions, ranging from 1 to over 100 nt in length, were detected in the peripheral blood of this animal at several time points and as late as 206 days post-transplantation, suggesting that some edited cells are persisting long-term in this animal. Unfortunately, the low levels of gene editing in vivo prevented further analysis of indels frequency in blood cell lineages or bone marrow cells.
Figure 4.
Tracking of BCL11A Mutations in Z12421 following Transplantation
(A) BCL11A indels frequency determined by Miseq sequencing of Z12421 peripheral blood after transplantation (day 0). Dashed line shows background level of BCL11A mutation established from mock-treated cells from the same animal (Supplemental Materials and Methods). (B) Tracking of deletion signatures in Z12421 from sequence analysis obtained from (A). Deletion signatures appearing at multiple time points are shown as colored boxes, whereas unique deletion signatures are shown in white. (C) Sequences of conserved deletion signatures in the BCL11A-TALEN target site following the legend from (B) with size of deletion (given in number of nucleotides). For simplicity, deletion of a single nucleotide or of more than 100 nt is not shown.
We next monitored HbF expression in the peripheral blood of Z12421 using flow cytometry and qPCR and compared results with baseline established from control transplants. As shown in Figure 1C, we initially observed a rapid increase in circulating F-cells in Z12421, similarly to control transplant animals, and F-cell frequency progressively returned to near basal levels at about 6 months. Long-term HbF levels were slightly higher in Z12421 than in controls, stabilizing at about 1% from 200 to 600 days, whereas all other animals showed less than 0.5% F-cells at 200 days (Figure 1C, inset). qPCR hemoglobin analysis in this animal confirmed these findings, with higher γ-globin expression (γ/(γ + β)) persisting for at least up to 360 days in Z12421 (Figure 1E). In summary, we documented a small increase in HbF expression in Z12421 peripheral blood as compared to control animals, which correlates with low levels of persisting BCL11A indels. Although no causal link between BCL11A editing and HbF induction was demonstrated in this animal, our study provides proof of feasibility for the use of the NHP model to evaluate new genome-editing treatments for hemoglobinopathies.
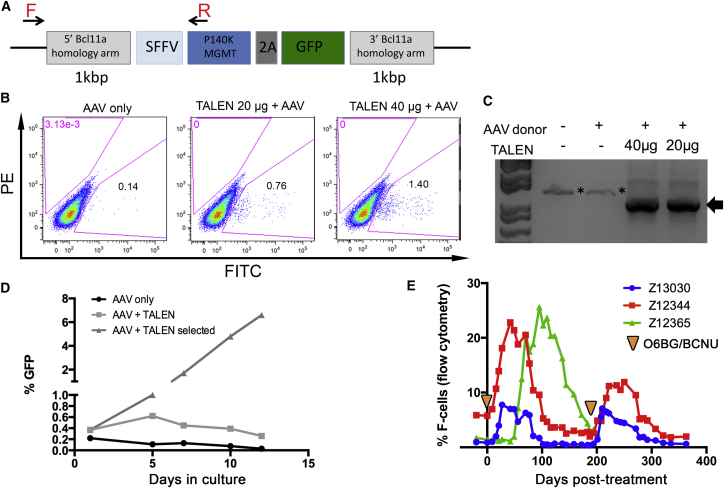
O6BG/BCNU-Mediated Selection of BCL11A-Modified NHP CD34+ Cells
Therapeutic levels of HbF expression can, in principle, be achieved by increasing the number of BCL11A-edited cells post-engraftment. Our group has previously used O6-benzylguanine (O6BG) and N,N’-bis(2-chloroethyl)-N-nitroso-urea (BCNU) in vivo to effectively increase levels of cells modified with the MGMT/P140K chemoselection cassette in large animals and human patients.8, 13, 14 The advances in genome editing now offer the possibility to integrate selection genes at precise sites by delivery of donor templates homologous to the target in combination with a nuclease. Efficient integration of homologous donor templates delivered by adeno-associated viral (AAV) vectors has been demonstrated in human CD34+ cells,23, 24, 25 so we treated NHP CD34+ with a combination of BCL11A-TALEN mRNA and a donor AAV vector containing both the MGMT/P140K selection cassette and a GFP reporter expressed from the SFFV promoter and flanked by BCL11A homologous sequences (Figure 5A). As shown in Figure 5B, we were able to detect targeted integration events based on GFP fluorescence (0.76% and 1.40% GFP+ cells in the AAV+TALEN group as compared to 0.14% GFP in the AAV-only group at 5 days post-treatment). These events were validated molecularly using an in-out PCR approach (Figures 5A and 5C), followed by sequencing of the resulting amplicon (data not shown). We demonstrated a rapid increase in GFP+ cells following one round of chemoselection with O6BG/BCNU in culture, which resulted in a 10-fold enrichment in GFP+ cells by 10 days and 17-fold by 12 days (Figure 5D). Although further optimization will be required to improve on the initial targeted integration efficiency prior to selection, our experiments provide proof of concept for the use of MGMT/P140K-based chemoselection to increase the frequency of BCL11A-edited CD34+ cells.
Figure 5.
Selection of BCL11A-Edited NHP CD34+ Cells In Vitro by O6BG/BCNU Treatment
(A) Schematic representation of the BCL11A AAV donor construct and position of the forward (F) and reverse (R) primers used in PCR analysis. 2A, Thosea asigna virus 2A self-cleaving peptide. (B) Representative flow cytometry results showing targeted integration events based on the frequency of GFP+ cells at 5 days post treatment using the conditions described on top. (C) PCR validation of targeted integration events described in (B) using primers designed inside and outside the donor cassette as described in (A). * denotes unspecific amplicons and the arrow shows the specific amplicon validated by sequencing. (D) Frequency of GFP+ cells following in vitro treatment with O6BG/BCNU (day 1) of NHP CD34+ cells treated with the indicated conditions. (E) HbF response to two rounds of O6BG/BCNU chemoselection in three transplanted pigtailed macaques. Animals Z13030, Z12344, and Z12365 underwent autologous BMT with CCR5 gene-edited CD34+ cells (subjects of separate study) and were allowed to recover for 221, 236, or 250 days, respectively. Two rounds of chemotherapy with O6BG/BCNU were subsequently administered in these animals at day 0 and 183 days thereafter (arrowheads).
The genotoxicity associated with O6BG/BCNU treatment is likely to cause hematopoietic stress and a corresponding induction in HbF, as was described above for the BMT procedure (Figure 1). We measured circulating F-cells in three pigtailed macaques that first underwent autologous HSC transplantation with CD34+ cells edited at the CCR5 locus (subjects of a separate study) and that were subsequently treated with two rounds of O6BG/BCNU. As shown in Figure 5E, we found a rapid increase in the frequency of F-cells in peripheral blood of Z13030 and Z12344, peaking at 30–40 days post-chemotherapy and with levels reaching 7.8% and 22.8%, respectively. In comparison, animal Z12365 showed a delayed HbF response, which peaked at about 100 days post-treatment and reached 25.6% F-cells. This delayed response correlated with a lag in hematopoietic recovery, as determined by CBCs (Figure S10). Circulating F-cells eventually returned to basal levels at about 100 days in Z13030 and Z12344 and at 200 days in Z12365. Interestingly, a second round of chemo-treatment promptly induced F-cell production again, with an amplitude comparable to the first round of treatment in Z13030, but with reduced amplitude in Z12344 (Figure 5E). Overall, this HbF response to O6BG/BCNU treatment is comparable, both in amplitude and time to recovery, to the response described for BMT (Figure 1) and will serve as baseline for future experiments aimed at increasing gene-edited cells post engraftment using in vivo chemoselection.
Discussion
This work establishes the NHP model to assess safety and efficacy of novel approaches for the treatment of hemoglobinopathies based on HSC editing and transplantation. Our group has previously used NHPs to validate the γ-globin lentiviral vector sGbG LV,10, 26 which is now being used in the clinical setting (NCT02186418). We have substantially expanded on these earlier findings by closely examining the hemoglobin response to BMT and providing proof of concept with the transplantation of a pigtailed macaque with autologous, BCL11A-edited HSCs. Although low levels of BCL11A indels were detected in this animal, several deletion signatures persisted for at least 200 days post-transplantation. These findings correlated with a small activation in long-term HbF expression in this animal as compared to control transplants, as evidenced by circulating F-cell frequency and γ-globin expression measured in peripheral blood. The low levels of BCL11A indels achieved in vivo did not allow us to establish a casual relation between BCL11A editing and HbF induction and prevented further analysis of editing in different blood lineages and bone marrow. Nevertheless, our study establishes the NHP as a model to evaluate new genome-editing strategies for hemoglobinopathies, which has notable advantages over the commonly used murine xenotransplantation model. NHPs closely reproduce parameters from a human patient with regards to scale and kinetics of hematopoietic repopulation and offer the unique ability to track differentiation into mature red blood cells, which is critical for studies of hemoglobin disorders.
We focused here on pigtailed macaques as the animal model, which are defective for the restriction factor TRIM5α27 and are thus permissible to genetic modification with lentiviral vectors in contrast to other NHPs that inhibit HIV-1. Pigtailed macaques are more adaptable to environmental changes as compared to the more commonly used rhesus macaques, and allow for greater volumes of blood sampling due to their larger body sizes. However, the advances of genomic information acquisition have particularly focused on rhesus, the genome of which was first sequenced in 2007,28 and a recent population study now allows for a better appreciation for genome-wide genetic variation in this species.29 This resource will prove advantageous for future genome engineering work. Our preliminary experiments have so far revealed no notable differences between pigtailed and rhesus macaques in hematopoietic composition, gene editing proficiency, or hemoglobin production profile (data not shown).
We chose to target the coding sequence of BCL11A because it was identified as a prime therapeutic candidate for reactivation of HbF (reviewed by Bauer et al.30). However, the non-selective inactivation of BCL11A may have undesirable effects in non-erythroid cell lineages because its function is required for lymphoid development,31, 32 neuronal morphogenesis,33 and dendritic cell fate.34 We observed no abnormal blood cell counts in Z12421 in the context of low frequency of BCL11A editing in peripheral blood. In the future, safer strategies will be needed to inactivate BCL11A exclusively in the erythroid lineage. As an example, a recent report described the use of an erythroid-specific promoter to express a short-hairpin RNA targeting BCL11A,35 which helped circumvent issues associated with impaired engraftment of human and mouse HSCs in a murine model when BCL11A expression was ubiquitously suppressed. Alternatively, BCL11A expression may be selectively knocked down in erythrocytes through inactivation of the BCL11A erythroid-specific enhancer motif identified in intron 2 of the gene.21, 36 Accordingly, BCL11A erythroid enhancer-edited human CD34+ cells using ZFNs achieved robust engraftment in immunodeficient mice.37 Our experiments documented a rapid drop in BCL11A editing frequency in animal Z12421 in the first 2 weeks post-transplantation but it is unclear whether this decrease is caused by inactivation of the BCL11A coding region or ex vivo manipulation of HSCs. This early decrease in BCL11A editing efficiency is comparable to effects reported by our group following transplantation of CD34+ cells treated with ZFNs targeting CCR5 using the same NHP model.12 Together, these results suggest that suboptimal conditions used for ex vivo HSC culture or editing may be responsible for low engraftment efficiency in our model, but an additional role for BCL11A cannot be ruled out. We are currently working on optimizing culture media and electroporation parameters to enhance HSCs engraftment post-editing.
Transplantation of large animals and, ultimately, human patients with gene-edited HSCs requires the efficient manipulation of hundreds of million cells with minimal toxicity to maintain their long-term repopulating potential. This requires a scalable, cGMP-compliant, electroporation device that maintains high performance when transitioning from a small cuvette system (<3 million cells), used for optimizing efficiency, to a large cuvette system (>50 million cells), used for transplantation experiments. In our study, we have documented a sharp decrease in BCL11A editing efficiency when scaling up conditions for TALEN mRNA electroporation, even though a control reaction using GFP mRNA carried out in parallel showed respectable transfection efficiency. It is possible that some adverse events, such as mRNA degradation, occurred during the experiment because efficient CCR5 editing was previously achieved by ZFN mRNA using the same electroporation platform.12 Generally, macro-scale devices suffer from the unsatisfactory transfection efficiency and/or cell viability due to use of the high voltage required by cuvettes using plate-like electrodes with relative large spacing. Electroporation devices that use micro-fluidic and flow-through systems may offer new possibilities, but are generally limited by low cell processing speed (reviewed by Lee et al.38).
One approach to increase the levels of gene editing after transplantation is to integrate a selection cassette at the site of the double-strand break by homology-directed repair, following co-delivery of the nuclease and a homologous donor template. Several reports have documented efficient target integration of donor cassettes in primary human cells by co-delivery of a nuclease and donor AAV.23, 25 As proof of principle, we demonstrated the targeted integration of the MGMT/P140K selection cassette in NHP HSCs, which could be selected for in vitro after O6BG/BCNU treatment. This approach will offer the benefit of selecting gene-edited/targeted cells post-engraftment, as was already shown by our laboratory for HSCs modified with MGMT/P140K lentiviral vectors in NHPs.8 As anticipated, we found that conditions of hematopoietic stress triggered by O6BG/BCNU-based chemoselection or by the transplantation procedure transiently increased HbF production in NHPs. The time required for F-cell levels to return to baseline was equivalent between chemoselection and BMT but the amplitude of HbF induction varied considerably between animal subjects. These results are consistent with findings from human patients undergoing allogenic BMT, in which both “low and high HbF responders” were identified.39 In a different study also involving allogenic BMT patients, maximum HbF expression occurred later (31–100 days)16 than the response documented here in NHPs (29–45 days). It is currently unclear whether this transient HbF induction is the result of differentiation of infused HSCs or of remaining HSCs that survived the conditioning regimen, but the amplitude of induction is likely dependent on genetic factors.39, 40
This work largely focuses on the targeting of the BCL11A coding region to reactivate HbF, but other promising approaches are being investigated that include the direct correction of β-globin SCD mutations25, 41, 42 or the engineering of a genetic alteration that recapitulates HPFH conditions.43, 44 The NHP model described here should serve as a valuable preclinical system to develop and assess novel genome editing strategies for the treatment of hemoglobinopathies.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH (“The Guide”). The protocol was approved by the Institutional Animal Care and Use Committees of the Fred Hutchinson Cancer Research Center and University of Washington, Protocol # 3235-01.
Animal Husbandry and Care
All animals were housed at and included in standard monitoring procedures prescribed by the Washington National Primate Research Center (WaNPRC). This included at least twice-daily observation by animal technicians for basic husbandry parameters (e.g., food intake, activity, stool consistency, and overall appearance) as well as daily observation by a veterinary technician and/or veterinarian. Animals were housed in cages approved by “The Guide” and in accordance with Animal Welfare Act regulations. Animals were fed twice daily and were fasted for up to 14 hr prior to sedation. Environmental enrichment included grouping in compound, large activity, or run-through connected cages, perches, toys, food treats, and foraging activities. If a clinical abnormality was noted by WaNPRC personnel, standard WaNPRC procedures were followed to notify the veterinary staff for evaluation and determination for admission as a clinical case. Animals were sedated by administration of ketamine HCl and/or Telazol and supportive agents prior to all procedures. Following sedation, animals were monitored according to WaNPRC standard protocols. WaNPRC surgical support staff are trained and experienced in the administration of anesthetics and have monitoring equipment available to assist in electronic monitoring of heart rate, respiration, and blood oxygenation; audible alarms and LCD readouts; monitoring of blood pressure, temperature, etc. For minor procedures, the presence or absence of deep pain was tested by the toe-pinch reflex. The absence of response (leg flexion) to this test indicates adequate anesthesia for this procedure. Similar parameters were used in cases of general anesthesia, including the loss of palpebral reflexes (eye blink). Analgesics were provided as prescribed by the Clinical Veterinary staff for at least 48 hr after the procedures and could be extended at the discretion of the clinical veterinarian based on clinical signs.
In Vitro Culture Conditions and Erythroid Differentiation
Human and NHP CD34+ cells were derived into erythroblasts using a previously described two-phase liquid culture system involving an expansion and differentiation phase.6 The expansion phase involved culturing cells in IMDM + 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 100 ng/mL each recombinant human SCF, thrombopoietin (TPO), and Flt-3 ligand and the differentiation phase involved culturing in the cytokines erythropoietin (EPO) (3 U/mL), dexamethasone (2 μM), interleukin-3 (IL-3) (5 ng/mL), cyclohexamide (1 μg/mL), and SCF (20 ng/mL). Two different FBS manufacturers were used to prepare the differentiation media, which were found to affect HbF basal levels, either resulting in higher (Clontech, cat#631106, lot#A15007) or lower HbF expression (Atlas Biologicals, Progeni serum, cat#PS-0500-A, lot#S13030611). Colony-forming assays were carried out by plating cells on methylcellulose media 1 day post-electroporation, as described previously.11 Differentiation was assessed by staining cells with fluorescein isothiocyanate (FITC)-conjugated CD71 (BD Biosciences, clone L01.1, NHP and human) and phycoerythrin (PE)-conjugated CD235a (BD Biosciences, clone GAR-2(HIR-2), human only). Hemoglobin expression was detected by intracellular staining with the PE-conjugated HbF antibody (Thermo Fisher Scientific, clone HBF-1) and FITC-conjugated hemoglobin β (Santa Cruz Biotechnology, clone 37-8) following fixation with the BD Perm/Wash kit (BD Biosciences). Hemoglobin mRNA transcript expression was measured as described in the Peripheral Blood Analyses section.
Ex Vivo CD34+ Cells Gene Editing and Transplantation of Pigtailed Macaque
Human CD34+ cells were isolated from mobilized peripheral blood using the donor program at the Fred Hutchinson Cancer Center. NHP CD34+ cells isolated from steady-state bone marrow using the Tissue Donor Program at the Washington National Primate Center. Cells were plated on methylcellulose media for evaluation of colony-forming units (CFUs), as described previously.12 Autologous hematopoietic stem cell transplants were conducted using our previously published protocols.12, 45 All transplanted control animals (Z13140, Z13034, and Z12432) were the focus of a separate study using lentiviral vector transduction for genetic modification. Animals Z13030, Z12344, and Z12365 underwent autologous BMT with CCR5 gene-edited CD34+ cells (subjects of a separate study) and were allowed to recover before treatment with two rounds of chemotherapy with O6BG/BCNU. Z12421 was transplanted with HSCs electroporated with BCL11A-TALEN mRNA (TriLink Biotechnologies) following guidelines established for electroporation of ZFNs.12 Briefly, CD34+ cells enriched from bone marrow were electroporated after overnight culture. TALEN mRNA was added to cells resuspended to 1 × 107 cells/mL in Cytoporation Media T (Harvard Apparatus, Holliston, MA) at a final concentration of 160 μg/mL for both TALEN mRNA. Electroporation was conducted using an AgilePulse Max machine and 5 mL, 6-mm gap width electroporation chambers (Harvard Apparatus), using a single 750-V pulse, 0.8 ms in duration. Following electroporation, cells were rested for 10 min, extracted from electroporation chambers, plated into fresh media, and recovered overnight in a 30°C, 3% CO2 incubator. The next day, cells were harvested, counted, resuspended to 5 × 106 cells/mL, and pulsed for 2 hr in 10 μM prostaglandin E2 (PGE2) on ice. Cells were then centrifuged, resuspended in Hank’s Balanced Salt Solution containing 2% autologous serum, and infused into the animal. During the 48-hr ex vivo culture period and prior to cell infusion, animals received a fractionated dose of 1,020 cGy total body irradiation. In small-scale electroporation reactions, 2-mm gap chambers were used (#BTX620, Harvard Apparatus) using a single 250 V/5 ms pulse delivered by the ECM 830 Square Wave Electroporation System (Harvard Apparatus). Cells were then recovered as described above. For in vivo chemotherapy, the first round of chemoselection drugs was administered at 7 to 8 months post transplantation using 2 × 120 mg/m2 O6BG (8 hr apart) and 15 mg/m2 BCNU with the first O6BG dose. The second round of treatment was administered 13 to 14 months post-transplantation using the same conditions as in the first round except that BCNU was increased to 20 mg/m2. For in vitro chemoselection, CD34+ cells were treated with 50 μM O6BG and 50 μM BCNU for about 3 hr.
Peripheral Blood Analyses
Blood draws were collected by venipuncture into heparin or EDTA collection tubes 1 to 2 times per week. White blood cell, platelet, neutrophil, and lymphocyte levels were measured by automated differential counting. To evaluate BCL11A editing frequency, total leukocytes were collected by hemolysis of whole blood and total genomic DNA was isolated by Blood Mini Kit (QIAGEN, Hilden, Germany) and processed as described below. For hemoglobin analysis by flow cytometry, unlysed blood was fixed for 10–12 min in a 0.05% glutaraldehyde solution, washed twice in PBS, permeabilized for 3–5 min in 0.01% Triton X-100, washed 2X in PBS, and stained with PE-conjugated HbF antibody (Thermo Fisher Scientific, clone HBF-1). For quantification of globin transcripts, whole RNA was isolated from unlysed blood using a combination of Trizol extraction and purification by RNeasy Mini Kit (QIAGEN, Hilden, Germany) and converted to cDNA using the Superscript III first strand synthesis kit (Thermo Fisher Scientific, Waltham, MA). Taqman reaction was run following the manufacturer’s protocol using the Applied Biosystems (Foster City, CA) 7500 Real-Time PCR system. Relative mRNA expression of alpha-, beta-, and gamma-globins was measured with established assays (HbA Rh02828921_qH, HbB Rh02809243_m1, and HbG Rh02576538_gH) and normalized to GAPDH (GAPDH Hs02758991_g1) by the comparative Ct method.
Nucleases and AAV Vector Generation
TALENs were assembled by GoldenGate cloning following guidelines from published work.46 The plasmid kit used for generation of TALENs was a gift from Daniel Voytas and Adam Bogdanove (Addgene kit # 1000000024). For generation of mRNA, TALEN-containing fragments were subcloned into the cloning vector pWNY.47 TALEN mRNA was generated using the mMESSAGE mMACHINE T7 transcription kit (#AM1344, Thermo Fisher Scientific) for small-scale preparations or was purchased as a large amount (Trilink Biotechnologies, San Diego, CA). ZFNs targeting CCR5 in NHP cells have previously been described.12 For CRISPR/Cas9, 20 μg purified Cas9 mRNA (Trilink Biotechnologies, San Diego, CA) was combined with 600 pmol chemically modified guide RNA (5′-GCATGTGACAGGTGAGGCAC-3′; Synthego, Redwood City, CA) targeting CD33 exon 2. The BCL11A donor insert was assembled by fusion PCR and cloned into the ClaI/SpeI restriction sites of plasmid pAAV-MCS (#VPK-411, Cell Biolabs, San Diego, CA). BCL11A donor serotype 5 AAV was prepared by the Kiem laboratory Vector Core services following a standard procedure involving transfection of packaging cells with a three-plasmid system.
Evaluation of Editing Efficiency
For semiquantitative measurement of BCL11A editing, the locus of interest was first recovered by PCR using primers described in Table S1 and digested with the SacI restriction enzyme (New England Biolabs) for at least 2 hr at 37°C. TALEN activity was then assessed by densitometry analysis of the ratio of PCR product resistant to SacI digestion over digested PCR product. Alternatively, the Surveyor Nuclease Detection Kit (Integrated DNA Technologies, Coralville, IA) was used to determine indels frequency by all nuclease platforms following the manufacturer’s instructions using primers in Table S1. Deep sequencing was also used to quantify BCL11A mutations using a first round of PCR amplification using primers Miseq BCL11A-F and –R (Table S1), followed by a second round of amplification using Illumina barcoded, pair-end Miseq primers for complete sequencing (Illumina, San Diego, CA). For validation of integration of Mgmt/P140K at the BCL11A target site, an in-out PCR approach was used with primers from Table S1.
Author Contributions
H.-P.K., O.H., and C.W.P. conceived the project and designed the experiments. O.H., Z.K.N., and S.R. performed the experiments. O.H. wrote the paper and assembled the figures. H.-P.K., C.W.P., Z.K.N., and S.R. reviewed and edited the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Veronica Nelson, Erica Curry, and Kelvin Sze for excellent support in our macaque studies; Helen Crawford and Bonnie Larson for their help with manuscript and figure preparations; Sowmya Reddy for help with the transplant; Willi Obenza and Morgan Giese for processing of macaque samples; and Martin Wohlfahrt and Don Gisch for preparation of the AAV vector. This work has been supported by grants R01 HL136135 and R01 HL115128 from the NIH, Bethesda, MD. H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the Jose Carreras/E. Donnall Thomas Endowed Chair for Cancer Research and recipient of the Endowed Chair for Cell and Gene Therapy.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, ten figures, and one table and can be found with this article online at https://doi.org/10.1016/j.omtm.2017.11.005.
Supplemental Information
References
- 1.Modell B., Darlison M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull. World Health Organ. 2008;86:480–487. doi: 10.2471/BLT.06.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orkin S.H., Kazazian H.H., Jr. The mutation and polymorphism of the human beta-globin gene and its surrounding DNA. Annu. Rev. Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- 3.Platt O.S., Brambilla D.J., Rosse W.F., Milner P.F., Castro O., Steinberg M.H., Klug P.P. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N. Engl. J. Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 4.Forget B.G. Molecular basis of hereditary persistence of fetal hemoglobin. Ann. N Y Acad. Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 5.Uda M., Galanello R., Sanna S., Lettre G., Sankaran V.G., Chen W., Usala G., Busonero F., Maschio A., Albai G. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. USA. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 7.Xu J., Peng C., Sankaran V.G., Shao Z., Esrick E.B., Chong B.G., Ippolito G.C., Fujiwara Y., Ebert B.L., Tucker P.W. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beard B.C., Trobridge G.D., Ironside C., McCune J.S., Adair J.E., Kiem H.P. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J. Clin. Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Younan P.M., Polacino P., Kowalski J.P., Peterson C.W., Maurice N.J., Williams N.P., Ho O., Trobridge G.D., Von Laer D., Prlic M. Positive selection of mC46-expressing CD4+ T cells and maintenance of virus specific immunity in a primate AIDS model. Blood. 2013;122:179–187. doi: 10.1182/blood-2013-01-482224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiem H.P., Arumugam P.I., Burtner C.R., Fox C.F., Beard B.C., Dexheimer P., Adair J.E., Malik P. Pigtailed macaques as a model to study long-term safety of lentivirus vector-mediated gene therapy for hemoglobinopathies. Mol. Ther. Methods Clin. Dev. 2014;1:14055. doi: 10.1038/mtm.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson C.W., Haworth K.G., Burke B.P., Polacino P., Norman K.K., Adair J.E., Hu S.L., Bartlett J.S., Symonds G.P., Kiem H.P. Multilineage polyclonal engraftment of Cal-1 gene-modified cells and in vivo selection after SHIV infection in a nonhuman primate model of AIDS. Mol. Ther. Methods Clin. Dev. 2016;3:16007. doi: 10.1038/mtm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson C.W., Wang J., Norman K.K., Norgaard Z.K., Humbert O., Tse C.K., Yan J.J., Trimble R.G., Shivak D.A., Rebar E.J. Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman primates. Blood. 2016;127:2416–2426. doi: 10.1182/blood-2015-09-672337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beard B.C., Sud R., Keyser K.A., Ironside C., Neff T., Gerull S., Trobridge G.D., Kiem H.P. Long-term polyclonal and multilineage engraftment of methylguanine methyltransferase P140K gene-modified dog hematopoietic cells in primary and secondary recipients. Blood. 2009;113:5094–5103. doi: 10.1182/blood-2008-09-176412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adair J.E., Beard B.C., Trobridge G.D., Neff T., Rockhill J.K., Silbergeld D.L., Mrugala M.M., Kiem H.P. Extended survival of glioblastoma patients after chemoprotective HSC gene therapy. Sci. Transl. Med. 2012;4:133ra57. doi: 10.1126/scitranslmed.3003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alter B.P. Fetal erythropoiesis in stress hematopoiesis. Exp. Hematol. 1979;7(Suppl 5):200–209. [PubMed] [Google Scholar]
- 16.Alter B.P., Rappeport J.M., Huisman T.H., Schroeder W.A., Nathan D.G. Fetal erythropoiesis following bone marrow transplantation. Blood. 1976;48:843–853. [PubMed] [Google Scholar]
- 17.Mundee Y., Bigelow N.C., Davis B.H., Porter J.B. Flow cytometric method for simultaneous assay of foetal haemoglobin containing red cells, reticulocytes and foetal haemoglobin containing reticulocytes. Clin. Lab. Haematol. 2001;23:149–154. doi: 10.1046/j.1365-2257.2001.00344.x. [DOI] [PubMed] [Google Scholar]
- 18.DeSimone J., Biel S.I., Heller P. Stimulation of fetal hemoglobin synthesis in baboons by hemolysis and hypoxia. Proc. Natl. Acad. Sci. USA. 1978;75:2937–2940. doi: 10.1073/pnas.75.6.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSimone J., Biel M., Heller P. Maintenance of fetal hemoglobin (HbF) elevations in the baboon by prolonged erythropoietic stress. Blood. 1982;60:519–523. [PubMed] [Google Scholar]
- 20.Wilber A., Hargrove P.W., Kim Y.S., Riberdy J.M., Sankaran V.G., Papanikolaou E., Georgomanoli M., Anagnou N.P., Orkin S.H., Nienhuis A.W. Therapeutic levels of fetal hemoglobin in erythroid progeny of β-thalassemic CD34+ cells after lentiviral vector-mediated gene transfer. Blood. 2011;117:2817–2826. doi: 10.1182/blood-2010-08-300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vierstra J., Reik A., Chang K.H., Stehling-Sun S., Zhou Y., Hinkley S.J., Paschon D.E., Zhang L., Psatha N., Bendana Y.R. Functional footprinting of regulatory DNA. Nat. Methods. 2015;12:927–930. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilber A., Tschulena U., Hargrove P.W., Kim Y.S., Persons D.A., Barbas C.F., 3rd, Nienhuis A.W. A zinc-finger transcriptional activator designed to interact with the gamma-globin gene promoters enhances fetal hemoglobin production in primary human adult erythroblasts. Blood. 2010;115:3033–3041. doi: 10.1182/blood-2009-08-240556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perumbeti A., Higashimoto T., Urbinati F., Franco R., Meiselman H.J., Witte D., Malik P. A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction. Blood. 2009;114:1174–1185. doi: 10.1182/blood-2009-01-201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirmaier A., Wu F., Newman R.M., Hall L.R., Morgan J.S., O’Connor S., Marx P.A., Meythaler M., Goldstein S., Buckler-White A. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 29.Xue C., Raveendran M., Harris R.A., Fawcett G.L., Liu X., White S., Dahdouli M., Rio Deiros D., Below J.E., Salerno W. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res. 2016;26:1651–1662. doi: 10.1101/gr.204255.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer D.E., Orkin S.H. Hemoglobin switching’s surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P., Keller J.R., Ortiz M., Tessarollo L., Rachel R.A., Nakamura T., Jenkins N.A., Copeland N.G. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y., Wang J., Khaled W., Burke S., Li P., Chen X., Yang W., Jenkins N.A., Copeland N.G., Zhang S. Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med. 2012;209:2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.John A., Brylka H., Wiegreffe C., Simon R., Liu P., Jüttner R., Crenshaw E.B., 3rd, Luyten F.P., Jenkins N.A., Copeland N.G. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 2012;139:1831–1841. doi: 10.1242/dev.072850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ippolito G.C., Dekker J.D., Wang Y.H., Lee B.K., Shaffer A.L., 3rd, Lin J., Wall J.K., Lee B.S., Staudt L.M., Liu Y.J. Dendritic cell fate is determined by BCL11A. Proc. Natl. Acad. Sci. USA. 2014;111:E998–E1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brendel C., Guda S., Renella R., Bauer D.E., Canver M.C., Kim Y.J., Heeney M.M., Klatt D., Fogel J., Milsom M.D. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J. Clin. Invest. 2016;126:3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang K.H., Smith S.E., Sullivan T., Chen K., Zhou Q., West J.A., Liu M., Liu Y., Vieira B.F., Sun C. Long-term engraftment and fetal globin induction upon BCL11A gene editing in bone-marrow-derived CD34+ hematopoietic stem and progenitor cells. Mol. Ther. Methods Clin. Dev. 2017;4:137–148. doi: 10.1016/j.omtm.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee W.G., Demirci U., Khademhosseini A. Microscale electroporation: challenges and perspectives for clinical applications. Integr. Biol. 2009;1:242–251. doi: 10.1039/b819201d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler K.J., Rea C.D., Rahbar S., Hill L.R., Blume K.G. Heterogeneous ontogeny of erythropoiesis after bone marrow ablation and allogeneic bone marrow grafting. Blood. 1983;61:167–170. [PubMed] [Google Scholar]
- 40.DeSimone J., Heller P., Amsel J., Usman M. Magnitude of the fetal hemoglobin response to acute hemolytic anemia in baboons is controlled by genetic factors. J. Clin. Invest. 1980;65:224–226. doi: 10.1172/JCI109654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoban M.D., Lumaquin D., Kuo C.Y., Romero Z., Long J., Ho M., Young C.S., Mojadidi M., Fitz-Gibbon S., Cooper A.R. CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Mol. Ther. 2016;24:1561–1569. doi: 10.1038/mt.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye L., Wang J., Tan Y., Beyer A.I., Xie F., Muench M.O., Kan Y.W. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: an approach for treating sickle cell disease and β-thalassemia. Proc. Natl. Acad. Sci. USA. 2016;113:10661–10665. doi: 10.1073/pnas.1612075113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trobridge G.D., Beard B.C., Gooch C., Wohlfahrt M., Olsen P., Fletcher J., Malik P., Kiem H.P. Efficient transduction of pigtailed macaque hematopoietic repopulating cells with HIV-based lentiviral vectors. Blood. 2008;111:5537–5543. doi: 10.1182/blood-2007-09-115022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(a) sequences. Mol. Ther. Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.