Abstract
Problem
Analyses of the expression pattern of multiple cytokines are frequently required for characterization of the status of the immune system as it pertains to Th type bias and intrinsic levels of inflammation. Classically, analysis of cytokine expression patterns has been performed by enzyme-linked immunosorbent assays (ELISA) for each separate analyte. A new technology, Luminex MAP®, facilitates the simultaneous evaluation of multiple immune mediators with advantages of higher throughput, smaller sample volume, and lower cost. Validation of this technology has been limited to small sample sets, limited use of clinical study specimens, and use of non-commercial reagents.
Methods
Ninety-six specimens from women over the course of their respective pregnancies were evaluated for cytokine concentrations using commercially available ELISA kits and commercially available Luminex MAP® kits according to the manufacturers’ directions. Correlations between data sets were evaluated using Pearson’s correlation coefficient (r).
Results
Excellent correlations were demonstrated for IL-1β, IL-4, IL-5, IL-6, IL-10, IFN γ, and TNF α, in contrast to IL-12 p70 and IL-13.
Conclusions
Luminex multiplex technology has distinct advantages and is a valid alternative method to ELISA for the evaluation of the majority of cytokines tested and for the characterization of immune system status.
Keywords: Reproductive immunology, Clinical study, Cytokine, ELISA, Luminex, Multiplex analysis
1. Introduction
Characterization of the immune system has revealed a highly orchestrated, complex, integrative network of immune mediators that fine-tune both the magnitude and the class of immune response. Initial efforts to categorize immune responses and the functional status of the immune system focused on two opposing, polarized classes of immune responses, T helper type 1 (Th1) and type 2 (Th2), defined in part by their associated cytokine expression profiles. It is now clear that the balance of the immune system can be established anywhere along a continuum spanning the entire spectrum from Th1 to Th2 (Chaouat et al., 2003; Kidd, 2003; Raghupathy, 2001; Chtanova and Mackay, 2001). Additionally, it has recently become clear that a multitude of physiologic processes, some involving other non-immune systems such as the nervous and endocrine systems, can impact the balance or status of the immune system (Elenkov and Chrousos, 2002; von Hertzen, 2002; Schwarz et al., 2001; Besedovsky and del Rey, 2000; Ader et al., 2001; Piccinni et al., 2000; Kovalovsky et al., 2000; Marshall and Agarwal, 2000; Becher et al., 2000). Our appreciation of the complexity of the immune system, the interactions between the reproductive and immune systems, and the capacity of the immune system to be influenced by non-immune physiologic stimuli has driven the need to evaluate the expression profile of multiple cytokines to characterize the Th bias and the inflammatory state of the immune system, particularly in the context of reproductive biology.
Normal human pregnancies are distinguished by a Th2 bias (Poole and Claman, 2004; Kidd, 2003; Sacks et al., 2003; Suzuki and Okudaira, 2004; Shimaoka et al., 2000; Lim et al., 2000; Piccinni et al., 2000; Saito et al., 1999). The shift from Th1 cytokine production to Th2 cytokine production protects the semi-allogeneic fetus from rejection by the maternal immune system, but as gestation progresses a shift from a predominate Th2 response to a Th1 response occurs (Raghupathy et al., 2000). Poor reproductive outcomes such as preterm delivery and fetal loss have been associated with a Th1 bias (Balkanli-Kaplan et al., 2004; Kwak-Kim et al., 2003; Chaouat et al., 2003; Raghupathy et al., 2000; Kruse et al., 2000; Darmochwal-Kolarz et al., 1999), although this is not a uniform observation (Sakai et al., 2004; Zenclussen et al., 2002). This highly complex interaction between the reproductive system, pregnancy, other pathologic processes, and the immune system is gradually being elucidated. The careful characterization of the Th1/Th2 profile along with other cytokine measures of inflammation is likely to yield insights into potential interventions to prevent adverse reproductive outcomes (Kwak-Kim et al., 2003; Kidd, 2003).
Cytokines are small, soluble proteins that are immune mediators and together constitute the communication network for the immune system. Many of these immune mediators were first identified and characterized by biological assays. Although these assays represent the “gold standard”, they are cumbersome and labor intensive. Typically, cytokines have been evaluated by enzyme-linked immunosorbent assays (ELISA). This well-developed methodology requires significant sample volumes for each analyte, is labor intensive, and is limited to single analytes, and thus, is not amenable to multiplex analysis. The simultaneous evaluation of panels of cytokine mRNAs has been proposed as an alternative multiplex analysis methodology, but is complicated by the well-documented phenomenon of post-transcriptional regulation of cytokine expression or release (Fenton, 1992; Hoffmann et al., 2002; Kotlyarov and Gaestel, 2002; Clark, 2000; Schwenger and Sanderson, 1998; Bamford et al., 1996; Nimer and Uchida, 1995; Kruys and Huez, 1994). Therefore, the accurate evaluation of multiple immune mediators has been problematic due to the lack of a validated multiplex analysis methodology for protein expression.
Recently, several multiplex protein analysis technologies have been developed, including slide/microarray-based assays (Knight et al., 2004; Tam et al., 2002; Wiese et al., 2001) and flow cytometry-based assays (Fulton et al., 1997; Oliver et al., 1998; Swartzman et al., 1999; Cook et al., 2001). Flow cytometry-based systems are currently the most widely used multiplex biomarker analysis technology. The Luminex-100 system uses uniformly sized microspheres internally labeled with graded proportions of a red and a near infrared fluorophore, 658 and 712 nm, providing the capacity to interrogate and classify 100 discrete beads (Fulton et al., 1997; Oliver et al., 1998; Swartzman et al., 1999; Martins, 2002). This is in contrast to the BD CBA system that discriminates beads based upon fluorescence intensity from a single fluorophore, which limits the multiplexing capacity (Cook et al., 2001). In both technologies, the analysis is essentially an ELISA on a bead. Beads of a single identity are covalently coupled to a specific capture antibody for the analyte of interest. A second detection antibody is used to quantitate the amount of analyte captured on the bead. This secondary antibody is either directly conjugated to the phycoerythrin (PE) fluorophore or biotin, which is then reacted with streptavidin-phycoerythrin. Although these technologies are gaining prominence and now have multiple commercial vendors for the analysis kits, there are relatively few reports of direct comparison between this technology and commercial ELISA kits (Camilla et al., 2001; Carson and Vignali, 1999; de Jager et al., 2003; Hildesheim et al., 2002; Hutchinson et al., 2001; Kellar et al., 2001; Oliver et al., 1998; Prabhakar et al., 2002; Vignali, 2000; Biagini et al., 2003). Only one of these reports utilizes commercially produced multiplex kits (Hildesheim et al., 2002). Additionally, these reports are generally limited by small sample sizes or the use of spiked samples versus clinical specimens. In a group of clinical samples from pregnant patients over the course of their pregnancy and the postpartum period, we proposed that the Luminex-100 MAP technology would be equivalent to standard ELISAs for the analysis of a panel of cytokines contained in culture supernatants derived from the stimulation of whole blood clinical specimens.
2. Materials and methods
Phlebotomy samples were obtained from participants in an IRB-approved epidemiologic study tracking multiple parameters over the course of pregnancy. Because pregnancy is associated with large alterations in immune function and because the maternal/fetal immune response to microbial colonization has emerged as one of the major pathophysiological mechanisms leading to adverse reproductive health outcomes, the characterization of the cytokine expression profile is particularly relevant during the state of pregnancy. Serially enrolled female subjects had 10 ml of whole blood collected at each of four time points approximating the first trimester, second trimester, third trimester, and 6 weeks postpartum, unless fetal loss occurred or the patient was lost to follow-up. Using 12 well tissue culture plates, 0.5 ml of whole blood was placed into each well with RPMI 1640 (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen) penicillin/streptomycin (Invitrogen) at 1:100 dilution. Separate sets of wells for blood collected at each time point were supplemented with phytohemagglutinin (PHA) (Sigma, St. Louis, MO) at a final concentration of 5 µg/ml or lipopolysaccharide (LPS)-2 (Sigma) at a final concentration of 1 µg/ml to provide two different non-specific stimuli and cultured without stimulation resulting in three samples from each time point. Plates were maintained at 37 °C in 5% CO2 for 24 h. The plates were placed on ice, the contents of the wells for each time point and condition (unstimulated, PHA- and LPS-stimulated) were transferred to conical tubes. Samples were centrifuged at 4 °C for 10 min at 300 × g followed by distribution of the supernatant into 500 µl aliquots that were immediately frozen at −80 °C and maintained frozen until analysis. A total of 96 samples were analyzed by both ELISA and Luminex-100 MAP® methods.
2.1. ELISA analysis
Nine cytokines, including pro-inflammatory, Th1 and Th2 associated cytokines, were analyzed for each time point and for each of the three conditions. The cytokines analyzed were interleukin (IL)-1β, IL-4, IL-5, IL-6, IL-10, IL-12 p70, IL-13, interferon (IFN) γ, and tumor necrosis factor (TNF) α. Quantikine human ELISA kits were purchased from R&D systems (Minneapolis, MN, USA). Analyses were performed according to the manufacture’s protocol for each ELISA kit, assayed in triplicate, and read on a Molecular Devices microplate reader at 450 nm (Menlo Park, CA). Standard curves and individual well concentrations were determined using the Softmax 2.34 software (Molecular Devices, Menlo Park, CA). To keep experimental values within the linear region of the standard curves, samples were diluted from 1:5 to 1:100 as necessary for stimulated culture samples for IL-1β, IL-6, IL-10, TNF α, and IFN γ. The level of detection and %CV for each individual cytokine ELISA is summarized in Table 1.
Table 1.
Intra-assay and inter-assay precision for multiplex assays and ELISA for all nine cytokines
| Cytokine | %CV | %CV | %CV | Minimum detectable dose (pg/ml) | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||
| LINCOplex | Beadlyte | ELISA | LINCOplex | Beadlyte | ELISA | ||||
|
|
|
|
|||||||
| Intra-assay | Inter-assay | Intra-assay | Inter-assay | Intra-assay | Inter-assay | ||||
| IL-1β | 7.8 | 8.7 | 10.17 | 10.7 | 2.7 | 4.9 | 3.2 | 6 | 1 |
| IL-4 | 6.2 | 14.1 | 59.49 | 9.17 | 6.2 | 9.2 | 3.2 | 2 | 10 |
| IL-5 | 6.3 | 14.5 | 23.47 | 25.9 | 1.8 | 7 | 3.2 | 2 | 3 |
| IL-6 | 6.3 | 10.4 | 11.66 | 5.07 | 3.1 | 2.7 | 3.2 | 7 | 0.7 |
| IL-10 | 5.4 | 11.2 | 13.27 | 6.5 | 4.7 | 6.6 | 3.2 | 1 | 7.8 |
| IFN γ | 5.3 | 10.3 | 10.23 | 6.05 | 3.4 | 6 | 3.2 | 6 | 8 |
| TNF α | 5.8 | 9.4 | 10.17 | 6.23 | 4.9 | 7.6 | 3.2 | 8 | 1.6 |
| IL-12 p70 | 7.6 | 18.4 | 26.15 | 42.35 | 1.3 | 4.9 | 3.2 | 9 | 5 |
| IL-13 | 6 | 4.9 | 12.73 | 8.85 | 5 | 10.2 | 3.2 | 2 | 32 |
The values for the LINCOplex and ELISA are provided by the manufacturer and included here without further manipulation. The values for the Beadlyte kit represent the mean for values provided by the manufacturer from three samples. In all cases, the coefficient of variance (%CV) was calculated as the standard deviation/mean × 100. The minimum detectable dose for the ELISA and Beadlyte assays was provided by the respective manufacture by adding two standard deviations to the mean optical density value of twenty zero standard replicates and calculating the corresponding concentration. The minimum detectable dose for the Linoplex assay is set at the lowest standard concentration, per the manufacturer’s instruction.
2.2. Multiplex cytokine analysis
Multiple cytokine analysis kits were obtained from two sources, Linco Research Inc. (St. Charles, MO) and Upstate Cell Signaling Solutions (Lake Placid, NY). Millipore multiscreen 96 well filter plates (Bedford, MA) were used for all multiplex cytokine kits. Assays were run in triplicate according to the manufacturers’ protocol. Data was collected using the Luminex-100 system Version 1.7 (Luminex, Austin, TX). Data analysis was performed using the MasterPlex QT 1.0 system (MiraiBio, Alameda, CA). A five-parameter regression formula was used to calculate the sample concentrations from the standard curves. All 96 samples were analyzed with the LINCOplex kit (Linco Research Inc.) and due to volume constraints a smaller set of 48 samples was also analyzed with Beadlyte kits (Upstate, Lake Placid, NY). The level of detection and %CV for each individual cytokine for each of the multiplex kits is summarized in Table 1.
2.3. Statistical analysis
The correlation between data points for each sample generated by ELISA relative to the data points generated by each of the two multiplex kits was evaluated by Pearson’s correlation coefficient (r) (Sheskin, 2004) with two tailed p-values <0.05 considered significant. Each condition for each time point was considered an independent sample for these analyses. We set a threshold value for the correlation coefficient of 0.75 as being clinically meaningful. The sample set was chosen to provide a clinically relevant sized sample set covering a range of expected cytokine levels. The minimum sample size required to detect a population correlation of 0.75 for a two-sided test, α = 0.05 and a null hypothesis of ρ =0, is 12 with a sample size of 15 yielding 90% power to detect this correlation. The large sample size, with unstimulated and stimulated samples, covering a broad range of cytokine concentrations, supports the parametric nature of the data sets and the use of Pearson’s rather than the Spearman correlation coefficient (Sheskin, 2004). The figures were generated using Graph Pad Prism (Graph Pad Software, San Diego, CA) software programs with statistical analyses performed using SAS software (SAS Institute Inc., Cary, NC). Evaluation of potential outliers within the triplicate determinations for each sample was performed using the Grubbs formula at the 5% critical Z-value (Sheskin, 2004; Grubbs, 1969).
3. Results
Ninety-eight clinical samples were available for evaluation, however there was insufficient material in two samples to perform both multiplex and ELISA assays; therefore, 96 samples are included in this analysis. These 96 samples were obtained from 10 subjects with samples obtained from two to four of the time points noted above under all three culture conditions. Inclusion of all samples, stimulated and unstimulated, generally provided a broad range of concentrations for any particular cytokine, thus facilitating the stringent comparison of these analytical methodologies. Each sample was considered independent for the purposes of these analyses. Mean values of the replicate determinations are depicted on linear:linear scale plots with trend lines in place. Trend lines and Pearson’s coefficients are calculated both with and without the constraint of intersection with the origin. Optimally, a zero determination by one method should be recapitulated by another method’s determination, provided that the sensitivity and specificity of the two methods are comparable. For Luminex methods and ELISA, this has been addressed previously (Biagini et al., 2004; Earley et al., 2002; Kellar et al., 2001; Pickering et al., 2002b; Prabhakar et al., 2002; Vignali, 2000). Very slight decreases in Pearson’s correlation coefficients were seen with the above constraint; however, in no case did this result in a decrease below the threshold for a significant correlation. Although Pearson’s correlation coefficients >0.6 are widely considered indicative of a valid correlation between two independent variables, we employed a more rigorous r > 0.75 as our threshold for a clinically significant correlation for the complete sample set. Only one data set, IL-13, fell within the range of 0.75 > r > 0.6.
We evaluated the determination of concentration for nine cytokines, IL-1β, IL-4, IL-5, IL-6, IL-10, IL-12 p40, IL-13, IFN γ, and TNF α. Data sets from ELISA and LINCOplex cytokine kits demonstrated a high degree of correlation for all but IL-12 p70. Pearson’s correlation coefficient and the equations for the best-fit trend line are summarized for all cytokines in Table 2. Data generated with the Beadlyte multiplex cytokine kits yielded strong correlations for six of the nine cytokines, IL-1β, IL-4, IL-10, IL-13, IFN γ, and TNF α. Pearson’s correlation coefficient and the equations for the best-fit trend line are summarized for all cytokines in Table 3. Data for all cytokines were determined simultaneously using the multiplex kits and individually using the ELISA kits.
Table 2.
LINCOplex and ELISA correlations
| Cytokines | Pearson’s correlation (r) | Trendline |
|---|---|---|
| IL-1β | 0.8380 | y = 0.1033x − 0.2626 |
| IL-4 | 0.8686 | y = 12.151x + 33.603 |
| IL-5 | 0.8117 | y = 1462x − 0.206 |
| IL-6 | 0.9029 | y = 0.4209x + 1880 |
| IL-10 | 0.8204 | y = 2.0325x + 277.72 |
| IL-12 p70 | 0.002 (ns) | |
| IL-13 | 0.6193 | y = 0.7262x + 13.631 |
| INF γ | 0.9388 | y = 0.3278x + 16.974 |
| TNF α | 0.9377 | y = 0.1804x − 35.086 |
Pearson’s correlation coefficient (r) and best-fit trend line for individual cytokine results obtained by ELISA and LINCOplex kits (n = 96 each). All correlations had p < 0.0001 except for IL-12 for which there was not a significant correlation (ns).
Table 3.
Beadlyte and ELISA correlations
| Cytokine | Pearson’s correlation (r) | Trendline |
|---|---|---|
| IL-1β | 0.9347 | y = 1.0546x − 179.79 |
| IL-4 | 0.9409 | y = 4.7004x + 2.7327 |
| IL-10 | 0.8815 | y = 2.8154x + 147.74 |
| IL-13 | 0.8599 | y = 0.9672x − 6.8127 |
| IFN γ | 0.9256 | y = 1.3068x + 26.467 |
| TNF α | 0.9510 | y = 0.2631x + 17.966 |
| IL-12 p70 | 0.857a | |
| IL-5 | 0.453 ns | |
| IL-6 | 0.288 ns |
Pearson’s correlation coefficient (r) and best-fit trend line for individual cytokine results obtained by ELISA and Beadlyte kits (n = 48 each). All correlations had p < 0.0001 except for IL-6 and IL-5 for which there was not a significant correlation (ns).
Although IL-12 reached statistical significance, the correlation was not considered clinically significant.
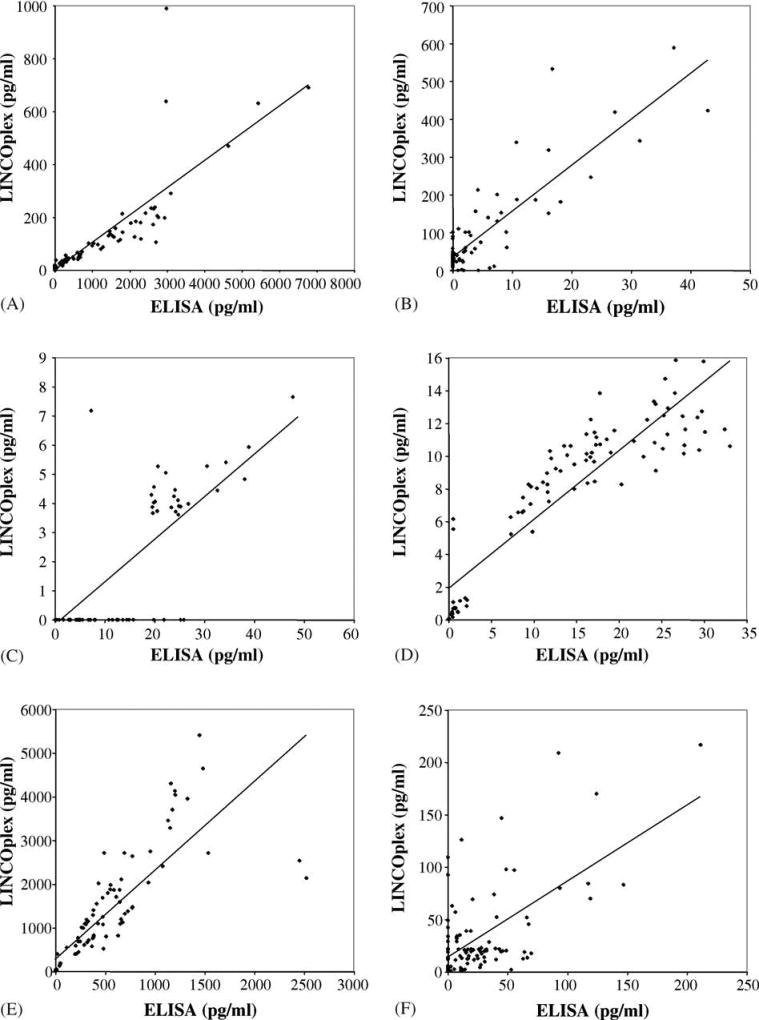
The correlative scatter plots of IL-1β, IL-6, and IL-10 determinations for the entire sample set (n = 96) by ELISA and Luminex using the LINCOplex cytokine kit are depicted in Fig. 1, panels A, D, and E, respectively. Analysis of unstimulated, PHA and LPS-stimulated sample subsets revealed statistically significant correlation between ELISA and Luminex determinations for all three cytokines in all subsets.
Fig. 1.
Scatter plots of ELISA and LINCOplex cytokine kit IL-1β, IL-4, IL-5, IL-6, IL-10 and IL-13 determinations. Panel A represents data obtained for IL-1β, r = 0.8380. Panel B represents data obtained for IL-4, r = 0.8686. Panel C, represents data obtained for IL-5, r = 0.8117. Panel D, represents data obtained for IL-6, r = 0.9029. Panel E represents data obtained for IL-10, r = 0.8204. Panel F represents data obtained for IL-13, r = 0.6193. Best-fit trend lines are depicted for each graph. All samples (n = 96) are included in these analyses.
IL-4, IL-5, and IL-13 are classical Th2-associated cytokines. Data comparing the R&D ELISA with the LINCOplex cytokine kit determinations for these three cytokines is depicted in Fig. 1 panels B, C, and F, respectively. IL-4 is arguably the most widely accepted Th2-associated cytokine. The Th2-associated cytokine IL-5, depicted in panel C, is widely acknowledged to be most representative of a polarized Th2 immune response but is generally expressed at low levels. IL-13, a third Th2 associated cytokine, is depicted in panel F. Analysis of unstimulated, PHA- and LPS-stimulated sample subsets revealed statistically significant correlation between ELISA and Luminex determinations in the subsets stimulated by PHA for all three cytokines. However, correlations between the two assays for the determination of IL-4 and IL-13 in the unstimulated and LPS-stimulated subsets did not reach statistical significance. This may be due to the disproportionate number of samples in which the cytokines were not detected. As these cytokines are primarily secreted by T lymphocytes, the low level of detectable cytokines in unstimulated and LPS-stimulated samples is not unanticipated.
Interferon γ, TNF α, and IL-12 are Th1-associated cytokines, although TNF α is less specific for a Th1 immune response than the other two cytokines. Data comparing the R&D ELISA with the LINCOplex cytokine kit determinations for these three cytokines are depicted in Fig. 2, panels A–C. IFN γ is the most widely accepted Th1-associated cytokine and demonstrates an excellent correlation, panel A. Similarly, an excellent correlation is seen for TNF α, panel B. Analysis of unstimulated, PHA- and LPS-stimulated sample subsets revealed statistically significant correlation between ELISA and Luminex determinations in the subsets stimulated by PHA for TNF α. However, correlations between the two assays for IFN γ determinations in the unstimulated and LPS-stimulated subsets did not reach statistical significance. IL-12 p70 failed to demonstrate a significant correlation between the determinations derived from R&D ELISA and the LINCOplex cytokine kit, depicted in Fig. 3, panel C.
Fig. 2.
Scatter plots of ELISA and multiplex cytokine kit Th1-associated cytokine determinations. Panel A represents data obtained for IFN γ, r = 0.9388. Panel B, represents data obtained for TNF α, r = 0.9377. Panel C represents data obtained for the LINCOplex kit determination of IL-12 p70, r = 0.002. Panel D represents data obtained for the Beadlyte kit determination of IL-12 p70, r = 0.8570. Best-fit trend lines are depicted for graphs of INF γ, TNF α, and Beadlyte IL-12 p70. All samples (n = 96 for Linco, n = 48 for Beadlyte) are included in these analyses.
Fig. 3.
Scatter plots of ELISA and Beadlyte multiplex cytokine kit determinations. Panel A represents data obtained for IL-1β, r = 0.9347. Panel B represents data obtained for IL-4, r = 0.9409. Panel C represents data obtained for IL-10, r = 0.8815. Panel D represents data obtained for IL-13, r = 0.8599. Panel E represents data obtained for IFN γ, r = 0.9256. Panel F represents data obtained for TNF α, r = 0.9510. Best-fit trend lines are depicted for each graph. All samples (n = 48) are included in these analyses.
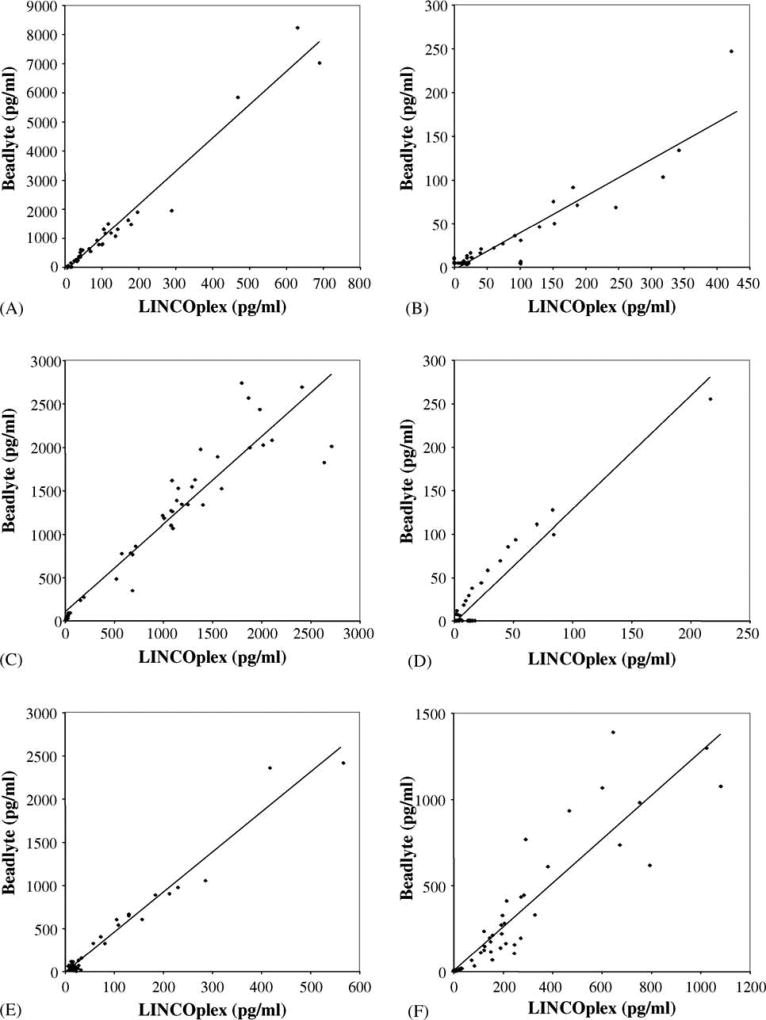
Due to limited available sample volume for some subjects, a smaller sample set (n = 48) was also analyzed using the Beadlyte kits for Luminex and provided a significant level of correlation with the ELISA determinations as above (Figs. 3 and 4). Although the determinations for IL-12 p70 (Fig. 3, panel D) reached statistical significance (r = 0.857) the plot demonstrates two distinct populations with essentially no bridging data points. Therefore, we conclude that the Pearson’s correlation coefficient is not representative of the level of correlation between these two data sets. A significant correlation between the Beadlyte and ELISA determination was observed for both IL-1β (Fig. 3, panel A) and for IL-10 (Fig. 3, panel C). For the Th2 cytokines, IL-4 (Fig. 3, panel B) has a high degree of correlation with a less robust, but significant correlation for IL-13, panel D. Fig. 3 panel F, represents the determinations for TNF α and Fig. 3, panel E, represents determinations for the prototypical Th1 cytokine, IFN γ, both of which have a similar high degree of correlation. Correlations with determinations for IL-5 and IL-6 were not significant. Due to the limited number of determinations for each subset, we did not perform subset analyses on the Beadlyte data set.
Fig. 4.
Scatter plots of LINCOplex cytokine and Beadlyte multiplex cytokine kit determinations. Panel A represents data obtained for IL-1β, r = 0.9824. Panel B represents data obtained for IL-4, r = 0.9147. Panel C represents data obtained for IL-10, r = 0.9410. Panel D represents data obtained for IL-13, r = 0.9565. Panel E represents data obtained for IFN γ, r = 0.9825. Panel F represents data obtained for TNF α, r = 0.9057. Best-fit trend lines are depicted for each graph. All samples analyzed with both kits (n = 48) are included in these analyses.
Cytokine determinations from the LINCOplex and Beadlyte kits that demonstrated a correlation with the data derived from ELISA were evaluated for their concordance. Fig. 4 depicts dot plots of the 48 data points from shared samples analyzed by both Luminex-100 multiplex cytokine kits. A high degree of correlation and concordance, summarized in Table 4, is seen between these two multiplex kits for IL-1β, IL-4, IL-10, IL-13, IFN γ, and TNF α.
Table 4.
LINCOplex and Beadlyte correlations
| Cytokine | Pearson’s correlation (r) | Trendline |
|---|---|---|
| IL-1β | 0.9824 | y = 11.449x − 163.49 |
| IL-4 | 0.9147 | y = 0.4203x − 4.1905 |
| IL-10 | 0.9410 | y = 1.0111x + 92.794 |
| IL-13 | 0.9565 | y = 1.3056x − 3.2762 |
| IFN γ | 0.9825 | y = 4.6358x − 19.813 |
| TNF α | 0.9057 | y = 1.2698x + 1.4662 |
Pearson’s correlation coefficient (r) and best-fit trend line for individual cytokine results obtained by LINCOplex and Beadlyte kits (n = 48 each). All correlations had p < 0.0001.
4. Discussion
This report evaluates nearly 100 clinically derived samples, covering a broad range of cytokine concentrations secondary to unstimulated, PHA-stimulated, and LPS-stimulated cultures, with commercially available ELISA and multiplex cytokine kits, the latter analyzed by Luminex-100 technology. Each of these commercially available kits have been extensively characterized and qualified by their respective manufacturers. This is the first report comparing these two cytokine analysis technologies on a large clinically derived sample set, in lieu of spiked samples. The panel of nine selected cytokines, IL-1β, IL-4, IL-5, IL-6, IL-10, IL-12 p70, IL-13, IFN γ, and TNF α, covers a range of non-specific, Th1-, and Th2-associated immune mediators. Two different vendors for multiplex cytokine kits were evaluated and yielded essentially identical results compared to ELISA determinations. We demonstrate excellent correlations between ELISA and Luminex determinations of cytokine levels in culture supernatants for seven (IL-1β, IL-4, IL-5, IL-6, IL-10, IFN γ, and TNF α) of the nine selected cytokines. In the case of IL-13, the degree of correlation falls within the generally acceptable range, but below our predetermined threshold for significance. Only for IL-12 there was a clear failure to establish a correlation between determinations obtained by the two analytical methods, regardless of the Luminex kit vendor. Indeed, ELISA and multiplex kit analyses for IL-12 revealed quite disparate results, suggesting that the optimal IL-12 p70 analysis methodology remains to be determined. Our data reported here, demonstrate that currently available cytokine multiplex kits and the Luminex-100 technology are suitable alternatives to the ELISA assay for most cytokines.
There is significant variation between the cytokine concentrations determined by ELISA and either multiplex kit, which is more prominent with the LINCOplex kit, even though the standards for each of the three manufacturers’ kits are standardized to cytokines from the National Institute of Biological Standards and Controls. This is not entirely surprising given that discrepancies are seen with ELISA kits from different vendors, likely representing differences in antibody pairs and sample diluent composition. Each antibody demonstrates optimal binding affinity at specific pH and salt concentration. The differences in antibody pairs and the necessity of a sample diluent compatible with all antibody pairs involved in the multiplex analysis may account for a significant variation in the determination of the cytokine concentrations. For IL-1β, IL-5, IL-6, IFN γ, and TNF α, the LINCOplex multiplex kit yielded cytokine concentrations that were from 2- to 10-fold lower than the ELISA determinations. For IL-10 and IL-4, the opposite was true, with the LINCOplex kit determinations being 2- and 12-fold higher, respectively. Although the Beadlyte cytokine concentration determinations more closely paralleled those obtained by ELISA, the determinations for IL-4 and IL-10 were higher than ELISA and the determinations for TNF α were lower, in a pattern that generally matched the LINCOplex kits. The relative sensitivity, as determined by a zero determination in one assay and a detectable concentration in the other, also varied. The evaluation of the cytokines IL-1β, IL-6, IL-10, and TNF α, demonstrated approximately equivalent relative sensitivity in the LINCOplex multiplex kit. However, the LINCOplex kit had higher relative sensitivity for IL-4, IL-13, and IFN γ, in contrast to IL-5, where the opposite was observed. Substantial differences in sensitivity were not readily apparent in the limited data set analyzed with the Beadlyte kits. The high degree of correlation seen for the majority of cytokines tested supports the use of Luminex technology for analysis of immune parameters but suggests that comparison of cytokine concentrations determined by multiplex technology using commercially available kits with determinations by ELISA might be problematic. Thus, we suggest that for any given study, consistent methodologies should be employed to facilitate data evaluation.
Given that the sample set included unstimulated culture samples and samples stimulated with either PHA or LPS, we evaluated individual subsets based on culture conditions. Stimulation with PHA targets primarily the lymphocyte compartment. LPS stimulation affects innate immune effector cell populations, primarily monocytes. The number of samples in these subsets, at most one third of the total, i.e. (n = 32), was small and the two stimuli have different cellular targets, resulting in a large proportion of subset samples with low or absent cytokine concentration determinations. Thus, in many of the subsets the correlations between ELISA and multiplex analyses did not reach statistical significance. The Th-associated cytokines, IL-4, IL-13, and IFN γ, generally had low or absent cytokine concentrations in unstimulated and LPS-stimulated cultures, leading to the observed poor correlation between the two analysis methodologies in these subsets. There was no significant difference in IL-5 concentrations with different culture stimuli in contrast to the other Th-associated cytokines, although concentrations were substantially lower across all subsets. For the other cytokines (IL-1β, IL-6, IL-10, and TNF α), the correlation between the determinations derived from multiplex and ELISA analyses remained statistically significant in all subsets, likely due to the broader cell populations responsible for secretion of these cytokines.
Our data are consistent with that previously reported using spiked samples or smaller clinically derived samples sets (Biagini et al., 2004; Camilla et al., 2001; Carson and Vignali, 1999; de Jager et al., 2003; Hildesheim et al., 2002; Hutchinson et al., 2001; Jiang et al., 2003; Kellar et al., 2001; Oliver et al., 1998; Prabhakar et al., 2002; Vignali, 2000). The Luminex technology has been used in several human clinical studies without direct comparison with ELISA or bioassays (Nelson et al., 2003; Jiang et al., 2003; Weber et al., 2003). Additionally, similar studies have been carried out using samples derived from various animal models (Keyes et al., 2003, 2002; Hutchinson et al., 2001). The multiplex analysis capability of the Luminex technology is being adapted to other assays including antibody screening (Biagini et al., 2004, 2003; Jones et al., 2002; Opalka et al., 2003; Pickering et al., 2002a,b; Martins, 2002), HLA typing (Pretl et al., 2003; Osowski et al., 2003; Chesterton et al., 2003; Brailey and Susskind, 2003), single nucleotide polymorphism or mutation analysis (Hutchings et al., 2003; Kube et al., 2003; Smith et al., 1998; Yang et al., 2001; Ye et al., 2001), and pathogen detection (Jenison et al., 2001; Earley et al., 2002; Dunbar et al., 2003; Cowan et al., 2004). The flexibility of this technology along with its inherent advantages suggests that additional applications will be forthcoming. It is not difficult to envision the ready application of this technology to evaluate parameters characterizing more than one physiologic system, e.g. reproductive endocrine, neuroendocrine, and/or immune systems.
Although this technology is relatively new, there are now commercial vendors for a number of multiplex analyses. The advantages of the multiplex cytokine assays over the standard ELISA assay include smaller sample volumes, higher throughput, and lower cost relative to equivalent ELISAs. In the case of our studies, triplicate multiplex analyses of all nine cytokines required 150 µl of sample while a comparable set of ELISAs required nearly 5.5 ml of sample. The performance of individual ELISA, requiring about 6 h each, versus approximately 5 h for the multiplex analysis results in a significantly higher throughput. The list prices of the multiplex kits are approximately 66% of the list price for a comparable set of ELISA plates. Any future widespread clinical application of this cytokine evaluation technology will require stringent establishment of normal values for any given kit, just as with ELISA kits from different vendors. Our results suggest that the Luminex-100 technology is a suitable alternative to the ELISA for most cytokines, provides a valid characterization of cytokine expression profile and T helper type bias, and may be superior to ELISA techniques commonly used in immunology.
Acknowledgments
This work was supported by ATPM/CDC TS-312. Christine McLaren, Ph.D., Professor and Director of the UCI Biostatistics Core Program, provided expertise and contributed statistical analyses.
References
- Ader R, Felten DL, Cohen N, editors. Psychoneuroimmunology. Academic Press; San Diego, CA: 2001. [Google Scholar]
- Balkanli-Kaplan P, Gucer F, Ali Yuce M. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004;114:239–240. doi: 10.1016/j.ejogrb.2003.11.021. (author reply 241) [DOI] [PubMed] [Google Scholar]
- Bamford RN, Battiata AP, Waldmann TA. J. Leukoc. Biol. 1996;59:476–480. doi: 10.1002/jlb.59.4.476. [DOI] [PubMed] [Google Scholar]
- Becher B, Prat A, Antel JP. Glia. 2000;29:293–304. [PubMed] [Google Scholar]
- Besedovsky HO, del Rey A. Z. Rheumatol. 2000;59(Suppl. 2):26–30. [Google Scholar]
- Biagini RE, Sammons DL, Smith JP, MacKenzie BA, Striley CA, Semenova V, Steward-Clark E, Stamey K, Freeman AE, Quinn CP, Snawder JE. Clin. Diagn. Lab. Immunol. 2004;11:50–55. doi: 10.1128/CDLI.11.1.50-55.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini RE, Schlottmann SA, Sammons DL, Smith JP, Snawder JC, Striley CA, MacKenzie BA, Weissman DN. Clin. Diagn. Lab. Immunol. 2003;10:744–750. doi: 10.1128/CDLI.10.5.744-750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailey P, Susskind BM. Hum. Immunol. 2003;64:S117. [Google Scholar]
- Camilla C, Mely L, Magnan A, Casano B, Prato S, Debono S, Montero F, Defoort JP, Martin M, Fert V. Clin. Diagn. Lab. Immunol. 2001;8:776–784. doi: 10.1128/CDLI.8.4.776-784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson RT, Vignali DA. J. Immunol. Methods. 1999;227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- Chaouat G, Ledee-Bataille N, Zourbas S, Ostojic S, Dubanchet S, Martal J, Frydman R. Am. J. Reprod. Immunol. 2003;50:177–186. doi: 10.1034/j.1600-0897.2003.00080.x. [DOI] [PubMed] [Google Scholar]
- Chesterton KA, Pretl K, Sholander JT, Leffell MS, Zachary AA. Hum. Immunol. 2003;64:S108. [Google Scholar]
- Chtanova T, Mackay CR. Adv. Immunol. 2001;78:233–266. doi: 10.1016/s0065-2776(01)78005-4. [DOI] [PubMed] [Google Scholar]
- Clark A. Arthritis Res. 2000;2:172–174. doi: 10.1186/ar83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EB, Stahl JL, Lowe L, Chen R, Morgan E, Wilson J, Varro R, Chan A, Graziano FM, Barney NP. J. Immunol. Methods. 2001;254:109–118. doi: 10.1016/s0022-1759(01)00407-0. [DOI] [PubMed] [Google Scholar]
- Cowan LS, Diem L, Brake MC, Crawford JT. J. Clin. Microbiol. 2004;42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmochwal-Kolarz D, Leszczynska-Gorzelak B, Rolinski J, Oleszczuk J. Eur. J. Obstet. Gynecol. Reprod. Biol. 1999;86:165–170. doi: 10.1016/s0301-2115(99)00065-2. [DOI] [PubMed] [Google Scholar]
- de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT. Clin. Diagn. Lab. Immunol. 2003;10:133–139. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. J. Microbiol. Methods. 2003;53:245–252. doi: 10.1016/s0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- Earley MC, Vogt RF, Jr, Shapiro HM, Mandy FF, Kellar KL, Bellisario R, Pass KA, Marti GE, Stewart CC, Hannon WH. Cytometry. 2002;50:239–242. doi: 10.1002/cyto.10140. [DOI] [PubMed] [Google Scholar]
- Elenkov IJ, Chrousos GP. Ann N.Y. Acad. Sci. 2002;966:290–303. doi: 10.1111/j.1749-6632.2002.tb04229.x. [DOI] [PubMed] [Google Scholar]
- Fenton MJ. Int. J. Immunopharmacol. 1992;14:401–411. doi: 10.1016/0192-0561(92)90170-p. [DOI] [PubMed] [Google Scholar]
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr Clin. Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- Grubbs FE. Technometrics: J. Stat. Phys. Chem. Eng. Sci. 1969;11:1–21. [Google Scholar]
- Hildesheim A, Ryan RL, Rinehart E, Nayak S, Wallace D, Castle PE, Niwa S, Kopp W. Cancer Epidemiol. Biomarkers Prev. 2002;11:1477–1484. [PubMed] [Google Scholar]
- Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. J. Leukoc. Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- Hutchings A, Fortinberry H, Jenkins S, Thomas JM. Hum. Immunol. 2003;64:S10. [Google Scholar]
- Hutchinson KL, Villinger F, Miranda ME, Ksiazek TG, Peters CJ, Rollin PE. J. Med. Virol. 2001;65:561–566. [PubMed] [Google Scholar]
- Jenison R, La H, Haeberli A, Ostroff R, Polisky B. Clin. Chem. 2001;47:1894–1900. [PubMed] [Google Scholar]
- Jiang B, Snipes-Magaldi L, Dennehy P, Keyserling H, Holman RC, Bresee J, Gentsch J, Glass RI. Clin. Diagn. Lab. Immunol. 2003;10:995–1001. doi: 10.1128/CDLI.10.6.995-1001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LP, Zheng HQ, Karron RA, Peret TC, Tsou C, Anderson LJ. Clin. Diagn. Lab. Immunol. 2002;9:633–638. doi: 10.1128/CDLI.9.3.633-638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellar KL, Kalwar RR, Dubois KA, Crouse D, Chafin WD, Kane BE. Cytometry. 2001;45:27–36. doi: 10.1002/1097-0320(20010901)45:1<27::aid-cyto1141>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Keyes K, Cox K, Treadway P, Mann L, Shih C, Faul MM, Teicher BA. Cancer Res. 2002;62:5597–5602. [PubMed] [Google Scholar]
- Keyes KA, Mann L, Cox K, Treadway P, Iversen P, Chen YF, Teicher BA. Cancer Chemother. Pharmacol. 2003;51:321–327. doi: 10.1007/s00280-003-0572-5. [DOI] [PubMed] [Google Scholar]
- Kidd P. Altern. Med. Rev. 2003;8:223–246. [PubMed] [Google Scholar]
- Knight PR, Sreekumar A, Siddiqui J, Laxman B, Copeland S, Chinnaiyan A, Remick DG. Shock. 2004;21:26–30. doi: 10.1097/01.shk.0000101668.49265.19. [DOI] [PubMed] [Google Scholar]
- Kotlyarov A, Gaestel M. Biochem. Soc. Trans. 2002;30:959–963. doi: 10.1042/bst0300959. [DOI] [PubMed] [Google Scholar]
- Kovalovsky D, Refojo D, Holsboer F, Arzt E. J. Neuroimmunol. 2000;109:23–29. doi: 10.1016/s0165-5728(00)00298-8. [DOI] [PubMed] [Google Scholar]
- Kruse N, Greif M, Moriabadi NF, Marx L, Toyka KV, Rieckmann P. Clin. Exp. Immunol. 2000;119:317–322. doi: 10.1046/j.1365-2249.2000.01123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V, Huez G. Biochimie. 1994;76:862–866. doi: 10.1016/0300-9084(94)90188-0. [DOI] [PubMed] [Google Scholar]
- Kube D, Mormann M, Tomiuk J, Rieth H, Hua TD, Kremsner PG, Vockerodt M. Genes Immun. 2003;4:459–468. doi: 10.1038/sj.gene.6364003. [DOI] [PubMed] [Google Scholar]
- Kwak-Kim JY, Chung-Bang HS, Ng SC, Ntrivalas EI, Mangubat CP, Beaman KD, Beer AE, Gilman-Sachs A. Hum. Reprod. 2003;18:767–773. doi: 10.1093/humrep/deg156. [DOI] [PubMed] [Google Scholar]
- Lim KJ, Odukoya OA, Ajjan RA, Li TC, Weetman AP, Cooke ID. Fertil. Steril. 2000;73:136–142. doi: 10.1016/s0015-0282(99)00457-4. [DOI] [PubMed] [Google Scholar]
- Marshall GD, Jr, Agarwal SK. Allergy Asthma Proc. 2000;21:241–246. doi: 10.2500/108854100778248917. [DOI] [PubMed] [Google Scholar]
- Martins TB. Clin. Diagn. Lab. Immunol. 2002;9:41–45. doi: 10.1128/CDLI.9.1.41-45.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KB, Grether JK, Dambrosia JM, Walsh E, Kohler S, Satyanarayana G, Nelson PG, Dickens BF, Phillips TM. Pediatr. Res. 2003;53:600–607. doi: 10.1203/01.PDR.0000056802.22454.AB. [DOI] [PubMed] [Google Scholar]
- Nimer SD, Uchida H. Stem Cells. 1995;13:324–335. doi: 10.1002/stem.5530130402. [DOI] [PubMed] [Google Scholar]
- Oliver KG, Kettman JR, Fulton RJ. Clin. Chem. 1998;44:2057–2060. [PubMed] [Google Scholar]
- Opalka D, Lachman CE, MacMullen SA, Jansen KU, Smith JF, Chirmule N, Esser MT. Clin. Diagn. Lab. Immunol. 2003;10:108–115. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osowski LD, Jakubek J, Woronkowicz M, Littleton N, KuKuruga D. Hum. Immunol. 2003;64:S97. [Google Scholar]
- Piccinni MP, Scaletti C, Maggi E, Romagnani S. J. Neuroimmunol. 2000;109:30–33. doi: 10.1016/s0165-5728(00)00299-x. [DOI] [PubMed] [Google Scholar]
- Pickering JW, Martins TB, Greer RW, Schroder MC, Astill ME, Litwin CM, Hildreth SW, Hill HR. Am. J. Clin. Pathol. 2002a;117:589–596. doi: 10.1309/lmch-c4q2-vfl9-3t1a. [DOI] [PubMed] [Google Scholar]
- Pickering JW, Martins TB, Schroder MC, Hill HR. Clin. Diagn. Lab. Immunol. 2002b;9:872–876. doi: 10.1128/CDLI.9.4.872-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole JA, Claman HN. Clin. Rev. Allergy Immunol. 2004;26:161–170. doi: 10.1385/CRIAI:26:3:161. [DOI] [PubMed] [Google Scholar]
- Prabhakar U, Eirikis E, Davis HM. J. Immunol. Methods. 2002;260:207–218. doi: 10.1016/s0022-1759(01)00543-9. [DOI] [PubMed] [Google Scholar]
- Pretl K, Chesterton KA, Sholander JT, Leffell MS, Zachary AA. Hum. Immunol. 2003;64:S108. [Google Scholar]
- Raghupathy R. Semin. Immunol. 2001;13:219–227. doi: 10.1006/smim.2001.0316. [DOI] [PubMed] [Google Scholar]
- Raghupathy R, Makhseed M, Azizieh F, Omu A, Gupta M, Farhat R. Hum. Reprod. 2000;15:713–718. doi: 10.1093/humrep/15.3.713. [DOI] [PubMed] [Google Scholar]
- Sacks GP, Redman CW, Sargent IL. Clin. Exp. Immunol. 2003;131:490–497. doi: 10.1046/j.1365-2249.2003.02082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Tsukaguchi N, Hasegawa T, Michimata T, Tsuda H, Narita N. Am. J. Reprod. Immunol. 1999;42:240–245. doi: 10.1111/j.1600-0897.1999.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Sakai M, Shiozaki A, Sasaki Y, Yoneda S, Saito S. J. Reprod. Immunol. 2004;61:133–143. doi: 10.1016/j.jri.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Schwarz MJ, Chiang S, Muller N, Ackenheil M. Brain Behav. Immun. 2001;15:340–370. doi: 10.1006/brbi.2001.0647. [DOI] [PubMed] [Google Scholar]
- Schwenger GT, Sanderson CJ. Leuk. Lymphoma. 1998;28:443–450. doi: 10.3109/10428199809058351. [DOI] [PubMed] [Google Scholar]
- Sheskin DJ. Handbook of Parametric and Nonparametric Statistical Procedures. Chapman & Hall/CRC Press; Boca Raton, FL: 2004. [Google Scholar]
- Shimaoka Y, Hidaka Y, Tada H, Nakamura T, Mitsuda N, Morimoto Y, Murata Y, Amino N. Am. J. Reprod. Immunol. 2000;44:143–147. doi: 10.1111/j.8755-8920.2000.440303.x. [DOI] [PubMed] [Google Scholar]
- Smith PL, WalkerPeach CR, Fulton RJ, DuBois DB. Clin. Chem. 1998;44:2054–2056. [PubMed] [Google Scholar]
- Suzuki S, Okudaira S. Arch. Gynecol. Obstet. 2004;270:260–262. doi: 10.1007/s00404-003-0549-y. [DOI] [PubMed] [Google Scholar]
- Swartzman EE, Miraglia SJ, Mellentin-Michelotti J, Evangelista L, Yuan PM. Anal. Biochem. 1999;271:143–151. doi: 10.1006/abio.1999.4128. [DOI] [PubMed] [Google Scholar]
- Tam SW, Wiese R, Lee S, Gilmore J, Kumble KD. J. Immunol. Methods. 2002;261:157–165. doi: 10.1016/s0022-1759(01)00572-5. [DOI] [PubMed] [Google Scholar]
- Vignali DA. J. Immunol. Methods. 2000;243:243–255. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- von Hertzen LC. J. Allergy Clin. Immunol. 2002;109:923–928. doi: 10.1067/mai.2002.124776. [DOI] [PubMed] [Google Scholar]
- Weber J, Sondak VK, Scotland R, Phillip R, Wang F, Rubio V, Stuge TB, Groshen SG, Gee C, Jeffery GG, Sian S, Lee PP. Cancer. 2003;97:186–200. doi: 10.1002/cncr.11045. [DOI] [PubMed] [Google Scholar]
- Wiese R, Belosludtsev Y, Powdrill T, Thompson P, Hogan M. Clin. Chem. 2001;47:1451–1457. [PubMed] [Google Scholar]
- Yang L, Tran DK, Wang X. Genome Res. 2001;11:1888–1898. doi: 10.1101/gr.190901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Li MS, Taylor JD, Nguyen Q, Colton HM, Casey WM, Wagner M, Weiner MP, Chen J. Hum. Mutat. 2001;17:305–316. doi: 10.1002/humu.28. [DOI] [PubMed] [Google Scholar]
- Zenclussen AC, Fest S, Busse P, Joachim R, Klapp BF, Arck PC. Am. J. Reprod. Immunol. 2002;48:245–251. doi: 10.1034/j.1600-0897.2002.01136.x. [DOI] [PubMed] [Google Scholar]