Abstract
The development of a methodology for the structural characterization at atomic detail of proteins conjugated to nanoparticles would be a breakthrough in nanotechnology. Solution and solid-state NMR spectroscopies are currently used to investigate molecules and peptides grafted onto nanoparticles, but the strategies used so far fall short in the application to proteins, which represent a thrilling development in theranostics. We here demonstrate the feasibility of highly-resolved multidimensional heteronuclear spectra of a large protein assembly conjugated to PEGylated gold nanoparticles. The spectra have been obtained by direct proton detection under fast MAS and allow for both a fast fingerprinting for the assessment of the preservation of the native fold and for resonance assignment. We thus demonstrate that the structural characterization and the application of the structure-based methodologies to proteins bound to gold nanoparticles is feasible and potentially extensible to other hybrid protein-nanomaterials.
Introduction
Protein-inorganic conjugate nanomaterials find increasing applications in nanotechnology, cell biology and medicine1–4. The intrinsic hybrid nature of these materials is the major strength of their use, but also hampers the structural characterization at atomic detail of the different components when assembled together. Among these hybrid nanomaterials, specific proteins conjugated to gold nanoparticles are well-established tools for immunohistochemistry and promising agents for photodynamic therapy, near-infrared optical imaging and drug delivery5–8. Highly stable and biocompatible gold nanoparticles (GNPs) are obtained by grafting long polyethylene glycol (PEG) chains onto the surface of the inorganic core in order to decrease the uptake by reticuloendothelial system cells and increase the circulation half-life9,10. The use of bifunctional PEG chains bearing a gold-reactive group at one end and a reactive moiety at the other allows for the synthesis of PEGylated gold nanoparticles functionalized with biologically relevant proteins11,12. The covalent linkage of the protein is often achieved through the formation of an amide bond with the ε-amino groups of at least one surface exposed lysine residue13. This simple and versatile synthetic strategy leads to the conjugation of the protein at different sites and to the possibility of binding several protein molecules per particle. Solution NMR investigations on functionalized nanoparticles have been already reported for small molecules and peptides covalently linked through flexible spacers14–16. More recently, also the potentiality of solid-state NMR has been used to characterize nanoparticles bearing relatively small functionalities on the surface17–19. Conversely, the use of SSNMR to obtain atomic structural details on proteins grafted onto GNPs has never been described before. Therefore, the functional integrity of the proteins conjugated to GNPs is usually assessed by biochemical assays, assuming that the structure is preserved if the functionality is preserved. However, structural biology tools can provide a wealth of information that can be applied to the proteins grafted onto nanoparticles. Heteronuclear single quantum coherence (HSQC)20 correlating nitrogen and amide proton on 15N isotopically enriched proteins or acquired at natural abundance is a relatively simple and rapid experiment that is an extremely faithful reporter of minor changes occurring in the chemical environment of the observed nuclei. It is thus widely used (i) to assess the protein folding, (ii) to observe structural changes, and (iii) to monitor chemical modifications or protein-protein and protein-ligand interactions21–24, and it may be ported to larger systems through tailored sequences, even if the achievable size is still low25,26. More recently, amide proton-nitrogen correlation spectra have been reported on crystalline protein samples at the solid state27–32. However, high quality solid state 2D 1H-15N CP-HSQC can be recorded only at high magnetic fields, under fast magic angle spinning (MAS) conditions (60 kHz), possibly on perdeuterated samples to abolish the detrimental effects of proton-proton dipolar coupling on spectral resolution.
The molecular weight is not a limiting factor for solid-state NMR, and also non-crystalline samples can provide high quality spectra33–37.
We here show that the solid-state version of the HSQC experiment can be used to structurally characterize proteins covalently linked to GNPs. We have synthesized protein-GNPs starting from commercially available PEGylated GNPs with a core size of 5 nm, and the therapeutically relevant protein E. coli asparaginase II (ANSII). This protein has been already investigated in the solid state both in its native form, after PEGylation (PEG-ANSII hereafter)38 and after conjugation with polysaccharides39. The conjugation with the protein affects neither the stability of the GNPs in solution, nor their maximum absorption wavelength. The protein is only a fraction of the whole sample filling the 1.3 mm rotor, and therefore proton detection under fast MAS is necessary to achieve enough sensitivity. Highly resolved SSNMR spectra from ANSII conjugated to GNPs (ANSII-GNPs, hereafter) have been obtained after rehydration of freeze-dried material.
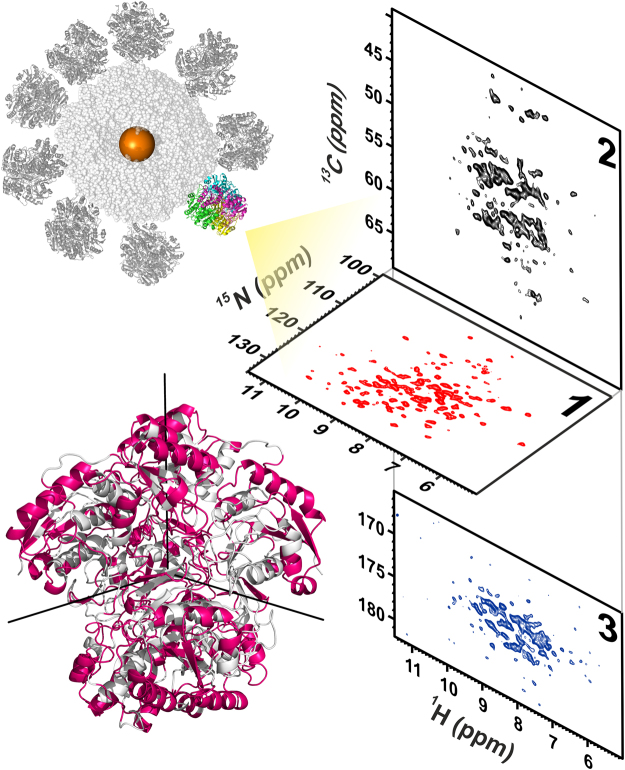
The 1D 1H NOESY of ANSII-GNPs shows a large group of signals in the region between 6.8 and 10.5 ppm, and one very intense peak around 3.6 ppm (Fig. 1). The chemical shift values in the 6.8–10.5 ppm range are typical for protein amides protons, while the intense peak observed at 3.6 ppm nicely matches with the methylene signals of the PEG forming the coating of the investigated nanoparticles. The large chemical shift dispersions of the amide protons and the presence of signals downfield of 8.5 ppm are positive markers of the protein folding and prompt the application of heteronuclear correlation experiments. The 2D 1H-15N CP-HSQC SSNMR of ANSII-GNPs (Fig. 2, panel 1) is of very good quality, and comparable to the 2D 1H-15N CP-HSQC spectra collected on PEG-ANSII and on the crystalline preparation of the free ANSII (Fig. 3). In the three spectra the resonances are largely superimposable, immediately demonstrating that the native three-dimensional structure of the protein is preserved after the conjugation with the nanoparticles and PEG38. Furthermore the spectra are sufficiently resolved so that 184 amide signals over a total of 224 visible cross-peaks can easily be assigned by comparison with the spectra of both the crystalline and PEGylated preparations of ANSII for which assignment is available (unpublished data), and by the analysis of the 1H-13C planes of 3D (H)CANH and 3D(H)CONH spectra acquired on the ANSII-GNPs (Fig. 2, panels 2 and 3).
Figure 1.
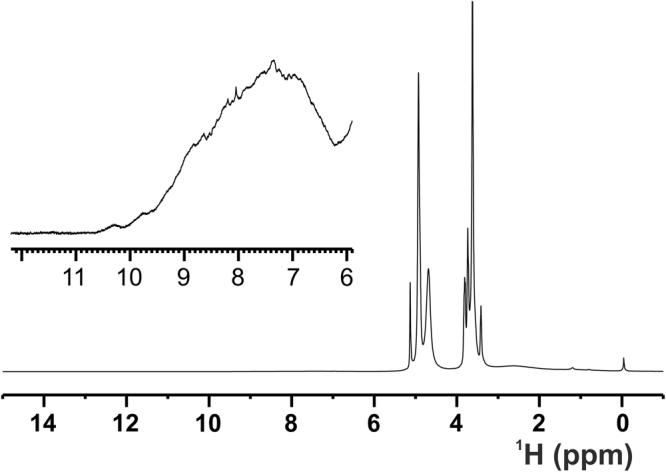
1D 1H NOESY NMR spectrum (mixing 20 ms) acquired at the solid-state on the ANSII-GNPs sample using a spectrometer operating at 800 MHz, 1H Larmor frequency, at ~282 K and MAS of 60 kHz. The sharp signals of the methylene group of the PEG chains and of the water are visible around 3.6 and 4.9 ppm, respectively. The top panel displays an enlargement, with a different intensity scale, of the region between 6 and 12 ppm, where the protein amide proton resonates. The high spreading of these signals is a positive marker of the protein folding.
Figure 2.
2D 1H-15N CP-HSQC SSNMR spectrum (1) and 2D 1H-13C planes of the 3D (H)CANH (2) and 3D(H)CONH (3) spectra obtained from ANSII-GNPs. The experiments were acquired on a spectrometer operating at 800 MHz, 1H Larmor frequency, at ~282 K and MAS of 60 kHz. Cartoon representation of tetrameric ANSII with highlighted in magenta the assigned residues in the 2D 1H-15N CP-HSQC SSNMR spectrum of ANSII-GNPs. The three C2 symmetry axes defining the D2 symmetry are displayed as black lines on the protein structure.
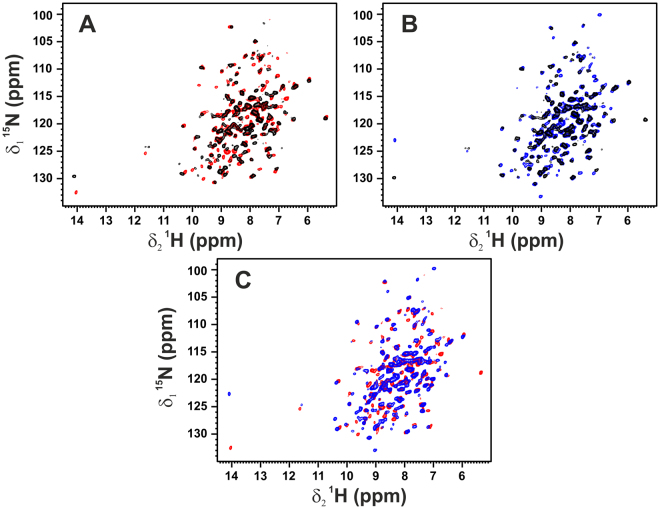
Figure 3.
2D 1H-15N CP-HSQC SSNMR spectrum of ANSII-GNPs (black) superimposed with the spectrum of crystalline ANSII (red) (panel A), and PEG-ANSII (blue) (panel B). The superimposition of the spectra of crystalline and PEG-ANSII is also reported (panel C). The spectrum of ANSII-GNPs was acquired on a 800 MHz spectrometer, while the spectra of crystalline ANSII and PEG-ANSII were acquired on a 850 MHz spectrometer. All the spectra were recorded at ~282 K and MAS of 60 kHz.
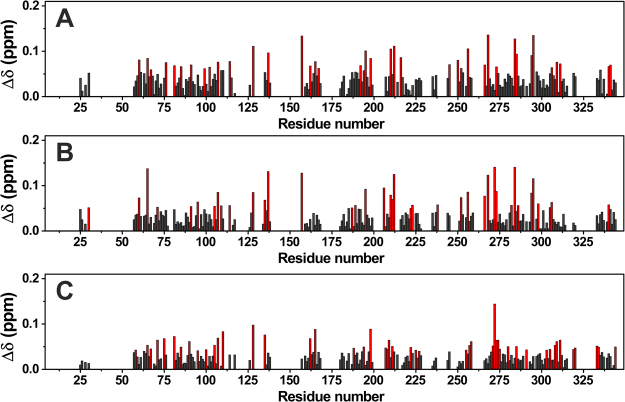
The effects of conjugation with the PEGylated nanoparticles were investigated by observing the differences in chemical shift of the resonances in the spectra of ANSII collected at the solid state for the different protein preparations (crystalline free ANSII, PEG-ANSII and ANSII-GNPs). The chemical shift perturbation analysis of the 2D 1H-15N CP-HSQC spectrum of ANSII-GNPs (Fig. 4) reveals that the largest variations involve the residues located on the protein surface or on loops. This finding is not surprising because the effect of packing forces in the crystalline free ANSII, and the chemical modifications in PEG-ANSII involve residues at the protein surface. For most of the residues the chemical shift values of ANSII conjugated to GNPs are in better agreement with those of the PEG-ANSII than with the crystalline free ANSII, and this remarks the similarity between PEG-conjugated samples. Interestingly, the chemical shift variations are larger for the PEG-ANSII, consistently with a lower level of conjugation of the lysine residues on the protein grafted onto the PEGylated GNPs. In the PEG-ANSII sample 7 lysine residues are conjugated on average, whereas virtually only a single one of the 19 exposed lysine residues at a time is reacted to the PEG-coated nanoparticle.
Figure 4.
Chemical-shift perturbations of PEG-ANSII (A) and ANSII-GNPs (B) with respect to the crystalline preparation, and between ANSII-GNPs and PEG-ANSII (C), according to the formula 40. The residues experiencing the highest perturbation are highlighted in red.
In summary, we demonstrate that highly resolved heteronuclear spectra can be recorded on protein-GNPs and assigned to obtain structural information at atomic detail. This new evolution of the NMR methodology opens new frontiers for the development of the structure-based strategies applied to nanoparticles and potentially to all nanomaterials functionalized with proteins. The accessibility to all the experiments previously used only for pure protein samples represents a breaking point for the engineering and GNPs applications. The simplicity of the procedures for sample preparation, the exquisite sensitivity of magnetic resonance to structural perturbations, and the possibility to obtain a full structural characterization of the proteins conjugated on the GNPs, are important features of this methodology.
An additional benefit is the possibility of monitoring protein-protein interactions involving the biomolecule immobilized onto the GNPs surface. Moreover, this strategy can be extended to GNPs of different sizes and shapes since they retain the same chemical and structural features. However, we should point out that the signal to noise ratio is obviously a function of the protein-GNPs mass ratio, which is strongly correlated to the surface to volume ratio of the nanoparticles. Finally, this methodology is also directly applicable to nanoparticles with a different core composition if they are stable under MAS conditions, and if their use is not prevented by a fast MAS inside high magnetic fields.
Methods
Expression and purification of ANSII [U- 2H-13C-15N]
Escherichia coli C41(DE3) cells were transformed with pET-21a(+) plasmid encoding ANSII gene. The cells were cultured in 2H13,C15,N-enriched Silantes OD2 medium supplied with 0.1 mg mL−1 of ampicillin, grown at 310 K, until A600 nm reached 0.6–0.8, then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside. They were further grown at 310 K overnight and then harvested by centrifugation at 7500 rpm (JA-10 Beckman Coulter) for 15 min at 277 K. The pellet was suspended in 10 mM Tris-HCl, pH 8.0, 15 mM EDTA, 20% sucrose buffer (60 mL per liter of culture) and incubated at 277 K for 20 min upon magnetic stirring. The suspension was centrifuged at 10000 rpm (F15–6 × 100 y Thermo Scientific) for 30 min and the supernatant discarded. The recovered pellet was re-suspended in H2O milli-Q (60 mL per liter of culture) and newly incubated at 277 K for 20 min under magnetic stirring. Again the suspension was centrifuged at 10000 rpm (F15–6 × 100 y Thermo Scientific) for 30 min. The pellet was discarded, whereas the supernatant treated with ammonium sulfate. Still under magnetic stirring solid ammonium sulfate was added in aliquots up to 50% saturation. The precipitate was removed by centrifugation, then further ammonium sulfate was added up to 90% saturation to trigger the precipitation of ANSII, which was recovered again by centrifugation. The precipitated ANSII was re-dissolved in a minimal amount of 20 mM Tris-HCl, pH 8.6 buffer and dialyzed extensively against the same buffer. ANSII was purified by anionic-exchange chromatography using a HiPrep Q FF 16/10 column (GE Healthcare Life Science). The protein was eluted in 20 mM Tris-HCl, pH 8.6 buffer with a linear 0–1 M NaCl gradient. Fractions containing pure ANSII were identified by Coomassie staining SDS-PAGE gels, then joined and dialyzed extensively against 50 mM phosphate, pH 7.5 buffer.
Conjugation of ANSII [U- 2H-13C-15N] to 5 nm gold nanoparticles and sample preparation for SSNMR
A sample of ANSII [U- 2H-13C-15N] has been conjugated to gold nanoparticles with core size of 5 nm functionalized with 5 kDa PEG chains and bearing NHS-ester (~1 NHS group/nm2) to react with the protein lysine residues (Cytodiagnostic). ANSII is a tetrameric assembly of 138 kDa formed of four identical subunits organized as a dimer of dimers. The molecular weight of the ANSII is of the same order of the monoclonal antibody used for the optimization of the conjugation strategy with these nanoparticles. The experimental conditions used in the protein-GNPs synthesis allow us to optimize the protein loading, avoid protein-mediated GNPs aggregation, and minimize the amount of protein needed for the reaction. 200 μL of 20 mg mL−1 ANSII [U- 2H-13C-15N] solution in 50 mM sodium phosphate, pH 7.5 buffer were diluted up to 400 μL with the Protein Re-suspension Buffer, then further diluted up to 900 μL with the Reaction Buffer. The solution was splitted in ten 90 μL aliquots which were directly transferred in ten vials containing lyophilized NHS-activated gold nanoparticles. The reaction mixtures were immediately mixed by pipetting, incubated at room temperature for 2 hours, then quenched with 10 μL (per vial) of Quencher Solution. Protein-GNPs nanoparticles were purified from the unreacted protein by multistep washing on a centrifugal device with membrane nominal pore size of 10 nm (MWCO Microsep Advanced Centrifugal Device, Pall Corporation). Three washings were performed using 20 mM Tris-HCl, pH 8.0 buffer until all the free protein was reduced (see Figure S1), and at the end the volume was reduced down to 500 μL, splitted in 10 new 1.5 mL eppendorfs, frozen with liquid nitrogen and lyophilized. The freeze-dried sample, corresponding approximately to about 0.2 mg of protein, was packed on 1.3 mm rotor for a preliminary NMR analysis, but rehydration was needed to collect well resolved spectra.
NMR measurements
All SSNMR spectra were recorded at ~282 K on Bruker AvanceIII HD spectrometer operating at 800 MHz 1H Larmor frequency, equipped with a 1.3 mm HCN probe-head, at MAS frequency of 60 kHz, using the standard parameters reported in literature33,34.
The nonselective 90° pulses were set to 2.05 μs (1H), 4 μs (15N), and 2.8 μs (13C).
1H-15N forward cross-polarization (CP) was achieved using a contact time of 1 ms (ωH = 50 kHz, ωN = 10 kHz) with a 70–100 linearly ramped contact pulse on 1H. The 15N-1H back CP was achieved using a contact time of 0.4 ms, (ωH = 50 kHz, ωN = 10 kHz) with a 100–70 linearly ramped contact pulse on 1H. The CP-HSQC was acquired with 2048 scans and with 156 increments in the indirect 15N dimension.
The 1H-13C forward CP had contact time of 3.5 ms for H-Cα and 2 ms for the H-CO transfer, respectively, with 70–100 linearly ramped contact pulses on 1H (ωH = 40 kHz, ωC = 20 kHz). CA-N CP and CO-N steps had contact time of 6 ms and 5 ms, respectively, with a 90–100 linearly ramped contact pulse of mean rf-field amplitude of about 40 kHz on 13C and a constant-amplitude spin lock of about 20 kHz on 15N.
All the spectra were with the Bruker TopSpin 3.2 software package and analysed with the program CARA (ETH Zürich).
Electronic supplementary material
Acknowledgements
This work has been supported by Fondazione Cassa di Risparmio di Firenze, MIUR PRIN 2012SK7ASN, by the Horizon 2020 programme of the European Union, iNEXT (# 653706) and West-Life (# 675858). The authors acknowledge the support and the use of resources of Instruct-ERIC, a landmark ESFRI project, and specifically the CERM/CIRMMP Italy center. ER was supported by FIRC (grant number 17941) and by PhosAgro/UNESCO/IUPAC Green Chemistry for Life grant. The authors acknowledge the support of the University of Florence CERM-TT, Recombinant Proteins JOYNLAB.
Author Contributions
E.R., M.F. and C.L. designed the research and wrote the manuscript; S.G. expressed and purified the protein and performed the conjugation of the protein with the gold nanoparticles; L.C., E.R. and M.F. performed the N.M.R. experiments; L.C. performed the spectral analysis and resonance assignment.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18109-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Enrico Ravera, Email: ravera@cerm.unifi.it.
Marco Fragai, Email: fragai@cerm.unifi.it.
Claudio Luchinat, Email: claudioluchinat@cerm.unifi.it.
References
- 1.Brown SD, et al. Gold Nanoparticles for the Improved Anticancer Drug Delivery of the Active Component of Oxaliplatin. J. Am. Chem. Soc. 2010;132:4678–4684. doi: 10.1021/ja908117a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Gold nanoparticles with a monolayer of doxorubicin-conjugated amphiphilic block copolymer for tumor-targeted drug delivery. Biomaterials. 2009;30:6065–6075. doi: 10.1016/j.biomaterials.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 3.Willner I, Willner B. Biomolecule-based nanomaterials and nanostructures. Nano Lett. 2010;10:3805–3815. doi: 10.1021/nl102083j. [DOI] [PubMed] [Google Scholar]
- 4.Khlebtsov B, Zharov V, Melnikov A, Tuchin V, Khlebtsov N. Optical amplification of photothermal therapy with gold nanoparticles and nanoclusters. Nanotechnology. 2006;17:5167–5179. doi: 10.1088/0957-4484/17/20/022. [DOI] [Google Scholar]
- 5.Cheng Y, et al. Highly efficient drug delivery with gold nanoparticle vectors for in vivo photodynamic therapy of cancer. J. Am. Chem. Soc. 2008;130:10643–10647. doi: 10.1021/ja801631c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Jong WH, Borm PJ. Drug delivery and nanoparticles: Applications and hazards. Int. J. Nanomedicine. 2008;3:133–149. doi: 10.2147/IJN.S596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Lasers Med. Sci. 2008;23:217–228. doi: 10.1007/s10103-007-0470-x. [DOI] [PubMed] [Google Scholar]
- 8.Mout R, et al. General Strategy for Direct Cytosolic Protein Delivery via Protein–Nanoparticle Co-engineering. ACS Nano. 2017;11:6416–6421. doi: 10.1021/acsnano.7b02884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abuchowski A, McCoy JR, Palczuk NC, van Es T, Davis FF. Effect of covalent attachment of polyethylene glycol on immunogenicity and circulating life of bovine liver catalase. J. Biol. Chem. 1977;252:3582–3586. [PubMed] [Google Scholar]
- 10.Lipka J, et al. Biodistribution of PEG-modified gold nanoparticles following intratracheal instillation and intravenous injection. Biomaterials. 2010;31:6574–6581. doi: 10.1016/j.biomaterials.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Olivier J-C, Huertas R, Lee HJ, Calon F, Pardridge WM. Synthesis of pegylated immunonanoparticles. Pharm. Res. 2002;19:1137–1143. doi: 10.1023/A:1019842024814. [DOI] [PubMed] [Google Scholar]
- 12.Choi CHJ, Alabi CA, Webster P, Davis ME. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc. Natl. Acad. Sci. USA. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abuchowski A, et al. Cancer therapy with chemically modified enzymes. I. Antitumor properties of polyethylene glycol-asparaginase conjugates. Cancer Biochem. Biophys. 1984;7:175–186. [PubMed] [Google Scholar]
- 14.Han HD, et al. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salorinne K, et al. Conformation and dynamics of the ligand shell of a water-soluble Au102 nanoparticle. Nat. Commun. 2016;7:10401. doi: 10.1038/ncomms10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W, et al. Control of Protein Orientation on Gold Nanoparticles. J. Phys. Chem. C. 2015;119:21035–21043. doi: 10.1021/acs.jpcc.5b07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bower PV, Louie EA, Long JR, Stayton PS, Drobny GP. Solid-State NMR Structural Studies of Peptides Immobilized on Gold Nanoparticles. Langmuir. 2005;21:3002–3007. doi: 10.1021/la040092w. [DOI] [PubMed] [Google Scholar]
- 18.Abraham A, Mihaliuk E, Kumar B, Legleiter J, Gullion T. Solid-State NMR Study of Cysteine on Gold Nanoparticles. J. Phys. Chem. C. 2010;114:18109–18114. doi: 10.1021/jp107112b. [DOI] [Google Scholar]
- 19.Berruyer P, et al. Three-Dimensional Structure Determination of Surface Sites. J. Am. Chem. Soc. 2017;139:849–855. doi: 10.1021/jacs.6b10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. doi: 10.1016/0009-2614(80)80041-8. [DOI] [Google Scholar]
- 21.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 22.Yao J, Dyson HJ, Wright PE. Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett. 1997;419:285–289. doi: 10.1016/S0014-5793(97)01474-9. [DOI] [PubMed] [Google Scholar]
- 23.Orts J, et al. NMR-Based Determination of the 3D Structure of the Ligand–Protein Interaction Site without Protein Resonance Assignment. J. Am. Chem. Soc. 2016;138:4393–4400. doi: 10.1021/jacs.5b12391. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa A, Konuma T, Yanaka S, Sugase K. Quantitative analysis of protein-ligand interactions by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016;96:47–57. doi: 10.1016/j.pnmrs.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 26.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevelkov V, Rehbein K, Diehl A, Reif B. Ultrahigh Resolution in Proton Solid-State NMR Spectroscopy at High Levels of Deuteration. Angew. Chem. Int. Ed. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- 28.Öster C, et al. Characterization of Protein–Protein Interfaces in Large Complexes by Solid-State NMR Solvent Paramagnetic Relaxation Enhancements. J. Am. Chem. Soc. 2017;139:12165–12174. doi: 10.1021/jacs.7b03875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiyama Y. Fast magic-angle sample spinning solid-state NMR at 60–100kHz for natural abundance samples. Solid State Nucl. Magn. Reson. 2016;78:24–36. doi: 10.1016/j.ssnmr.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Zhang R, Mroue KH, Ramamoorthy A. Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy. Acc. Chem. Res. 2017;50:1105–1113. doi: 10.1021/acs.accounts.7b00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fricke P, et al. Backbone assignment of perdeuterated proteins by solid-state NMR using proton detection and ultrafast magic-angle spinning. Nat. Protoc. 2017;12:764–782. doi: 10.1038/nprot.2016.190. [DOI] [PubMed] [Google Scholar]
- 32.Ishii Y, Tycko R. Sensitivity Enhancement in Solid State 15N NMR by Indirect Detection with High-Speed Magic Angle Spinning. J. Magn. Reson. 2000;142:199–204. doi: 10.1006/jmre.1999.1976. [DOI] [PubMed] [Google Scholar]
- 33.Zhou DH, Shah G, Mullen C, Sandoz D, Rienstra CM. Proton-detected solid-state NMR spectroscopy of natural-abundance peptide and protein pharmaceuticals. Angew. Chem. Int. Ed Engl. 2009;48:1253–1256. doi: 10.1002/anie.200801029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbet-Massin E, et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J. Am. Chem. Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurauskas V, et al. Sensitive proton-detected solid-state NMR spectroscopy of large proteins with selective CH3 labelling: application to the 50S ribosome subunit. Chem. Commun. 2016;52:9558–9561. doi: 10.1039/C6CC04484K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struppe, J. et al. Expanding the horizons for structural analysis of fully protonated protein assemblies by NMR spectroscopy at MAS frequencies above 100 kHz. Solid State Nucl. Magn. Reson., 10.1016/j.ssnmr.2017.07.001 (2017). [DOI] [PMC free article] [PubMed]
- 37.Ravera, E., Cerofolini, L., Martelli, T., Louka, A., Fragai, M. & Luchinat, C. (1) H-detected solid-state NMR of proteins entrapped in bioinspired silica: a new tool for biomaterials characterization. Scientific reports6, 27851 (2017). [DOI] [PMC free article] [PubMed]
- 38.Ravera E, et al. Solid-State NMR of PEGylated Proteins. Angew. Chem. Int. Ed Engl. 2016;55:2446–2449. doi: 10.1002/anie.201510148. [DOI] [PubMed] [Google Scholar]
- 39.Giuntini S, et al. Characterization of the Conjugation Pattern in Large Polysaccharide-Protein Conjugates by NMR Spectroscopy. Angew. Chem. Int. Ed Engl. 2017;56:14997–15001. doi: 10.1002/anie.201709274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grzesiek S, et al. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.