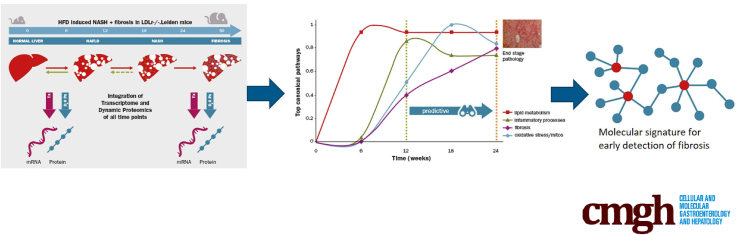
Abstract
Background & Aims
The incidence of nonalcoholic steatohepatitis (NASH) is increasing. The pathophysiological mechanisms of NASH and the sequence of events leading to hepatic fibrosis are incompletely understood. The aim of this study was to gain insight into the dynamics of key molecular processes involved in NASH and to rank early markers for hepatic fibrosis.
Methods
A time-course study in low-density lipoprotein–receptor knockout. Leiden mice on a high-fat diet was performed to identify the temporal dynamics of key processes contributing to NASH and fibrosis. An integrative systems biology approach was used to elucidate candidate markers linked to the active fibrosis process by combining transcriptomics, dynamic proteomics, and histopathology. The translational value of these findings were confirmed using human NASH data sets.
Results
High-fat-diet feeding resulted in obesity, hyperlipidemia, insulin resistance, and NASH with fibrosis in a time-dependent manner. Temporal dynamics of key molecular processes involved in the development of NASH were identified, including lipid metabolism, inflammation, oxidative stress, and fibrosis. A data-integrative approach enabled identification of the active fibrotic process preceding histopathologic detection using a novel molecular fibrosis signature. Human studies were used to identify overlap of genes and processes and to perform a network biology-based prioritization to rank top candidate markers representing the early manifestation of fibrosis.
Conclusions
An early predictive molecular signature was identified that marked the active profibrotic process before histopathologic fibrosis becomes manifest. Early detection of the onset of NASH and fibrosis enables identification of novel blood-based biomarkers to stratify patients at risk, development of new therapeutics, and help shorten (pre)clinical experimental time frames.
Keywords: Systems Biology, Metabolic Syndrome, Liver Disease, Diagnosis
Abbreviations used in this paper: ALT, alanine aminotransferase; AST, aspartate aminotransferase; DEG, differentially expressed genes; ECM, extracellular matrix; HFD, high-fat diet; IPA, Ingenuity Pathway Analysis; LDLr-/-, low-density lipoprotein receptor knock out; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; THBS1, thrombospontin-1
Graphical abstract
See editorial on page 65.
Summary.
This article presents a predictive molecular signature that marks the early onset of fibrosis in a translational nonalcoholic steatohepatitis mouse model. Overlap of genes and processes with human nonalcoholic steatohepatitis and a list of top candidate biomarkers for early fibrosis are described.
Nonalcoholic fatty liver disease (NAFLD) has become the most common chronic liver disease in developed countries.1 This increasing prevalence is associated closely with the incidence of obesity, insulin resistance, and dyslipidemia, all of which are risk factors for NAFLD.2, 3, 4, 5 NAFLD is associated with 26% higher overall health care costs, mainly from associated cardiometabolic diseases,6 and is projected to become the primary indication for liver transplantation within the next several years.7 NAFLD encompasses a spectrum of liver diseases ranging from the relatively benign hepatic steatosis to nonalcoholic steatohepatitis (NASH), the progressive form of NAFLD.
NASH is characterized by the presence of hepatocellular damage and inflammation,8 which in concert can drive the development of fibrosis.9 Recently, liver fibrosis was recognized to be strongly associated with long-term overall mortality, independently of other histologic features of NAFLD or NASH.10, 11 There is currently no method to identify which patient will progress from NAFLD and/or NASH to fibrosis. In addition, NASH and liver fibrosis are clinically silent, with hardly any symptoms, which means that detection often does not occur until the advanced stages of disease.
The molecular and cellular mechanisms involved in the pathogenesis of NAFLD and NASH have not been elucidated completely yet, but it is clear that disease progression is the result of complex and dynamic interactions between many processes, such as lipid metabolism, inflammation, oxidative stress, and fibrosis. However, the current body of knowledge relies mostly on results from studies that investigate these processes at a single time point (generally end point pathology) rather than investigating their interplay and dynamics over time. Information on the temporal dynamics and interaction between various molecular and pathologic processes has been shown to provide insight into early disease manifestations and allow detection of the onset of progressive disease.12
Animal models of NAFLD and NASH can be used for time-resolved studies and are suitable to provide crucial information on the processes that contribute to disease development. In the current study, we investigated the development of NASH in a time-resolved manner in high-fat-diet–fed low-density lipoprotein-receptor knockout (LDLr-/-.Leiden) mice, which develop NASH and hepatic fibrosis in the context of obesity, dyslipidemia, and insulin resistance, as is typical for NASH patients.13 Dynamic proteomic analyses that involve deuterated water labeling and tandem mass spectrometry were used to measure the formation of new collagens representing the active fibrosis process.14, 15, 16 RNA sequencing was used to generate a genetic time-resolved profile of processes involved in the development of NASH. This allowed identification of the dynamics of key molecular processes involved in the development of NASH and fibrosis. An integrative systems biology approach was used to investigate the molecular processes involved in the active fibrosis process by combining transcriptomics, dynamic proteomics, and histopathology. To gain insight into the translational value of these findings, the LDLr-/-.Leiden NASH mouse was compared with NASH patients on the molecular level. In addition, network biology-based ranking was performed using databases containing data from human cohort studies to identify candidate markers that represent the early manifestation of fibrosis.
Materials and Methods
Animals and Housing
Animal experiments were approved by an independent Animal Care and Use Committee and were in compliance with European Community specifications for the use of laboratory animals.
Time-Course Study
Twelve-week-old male LDL-receptor knockout mice were obtained from the breeding facility of TNO Metabolic Health Research (Leiden, The Netherlands). Animals received either standard rodent chow (Sniff-R/M-V1530 with 33 kcal% protein, 58 kcal% carbohydrate, and 9 kcal% fat; Uden, The Netherlands) (N = 45) or a high-fat diet (HFD) (D12451; Research Diets, Inc, New Brunswick, NJ; with 20 kcal% protein, 35 kcal% carbohydrate, and 45 kcal% lard fat) (N = 75) for a total of 30 weeks. Mice were group-housed in the specified pathogen free animal facility of TNO Metabolic Health Research, in a temperature-controlled room on a 12-hour light/dark cycle with ad libitum access to food and water. All interventions were performed during the light cycle. Groups were sacrificed after 6, 12, 18, 24, and 30 weeks on the diets. Blood samples were collected via the tail vein for EDTA plasma isolation after a 5-hour fast at 6-week intervals. A subset of mice (chow, n = 6; HFD, n = 15) was sacrificed every 6 weeks. This subset was matched to the remaining mice for body weight and the biochemical parameters of plasma cholesterol, triglycerides, blood glucose, and insulin. One group of mice (n = 15) was sacrificed before the start of the diets to define the starting condition (time [t] = 0). In the 18-week and 24-week groups, 1 animal died before sacrificing, which was not included in the analyses (resulting in HFD, n = 14 for these 2 time points). One week before sacrifice, all mice received an intraperitoneal injection with deuterated water (35 μL/g body weight) followed by 8% deuterated water in the drinking water until sacrifice to allow for dynamic proteomics analyses. Animals were terminated by CO2 asphyxiation, and a terminal blood sample (for EDTA plasma) was collected by cardiac puncture. Liver and adipose tissue depots were isolated. Tissues were partly fixed in formalin and paraffin-embedded for histologic analysis and partly snap frozen in liquid nitrogen and stored at -80°C for RNA isolation and dynamic protein profiling.
Biochemical Analysis of Circulating Factors
Total plasma cholesterol and triglycerides were measured with enzymatic assays (Roche Diagnostics, Almere, The Netherlands). Blood glucose level was measured immediately during blood sampling using a hand-held glucose analyzer (FreeStyle Lite, Abbot Laboratories, Hoofddorp, the Netherlands). Plasma insulin level was determined by enzyme-linked immunosorbent assay (ultrasensitive mouse insulin enzyme-linked immunosorbent assay; Mercodia, Uppsala, Sweden). Plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were measured using a spectrophotometric activity assay (Reflotron-Plus; Roche Diagnostics). HOmeostatic Model Assessment for Insulin Resistance was used to evaluate insulin resistance (fasting plasma insulin [μg/L] × fasting plasma glucose [mmol/L]/22.5).17
Intrahepatic Lipid Analysis
Liver lipids were analyzed by high-performance thin-layer chromatography as described previously.18 Briefly, lipids were extracted from liver homogenates using methanol and chloroform following the Bligh and Dyer19 method, after which they were separated by high-performance thin-layer chromatography on silica gel plates as described previously.20 Lipid spots were stained with color reagent, and triglycerides, cholesteryl esters, and free cholesterol were quantified using TINA software version 2.09 (Raytest, Straubenhardt, Germany).
Histologic Analysis
For histologic analysis of liver, 3-μm–thick cross-sections of the median lobe were stained with H&E. NAFLD was scored blindly by a board-certified liver pathologist using a general scoring system for rodent models, which is based on the human NASH Activity Score grading criteria.21 Briefly, 2 cross-sections per mouse were examined and the level of microvesicular and macrovesicular steatosis was expressed as a percentage of the cross-sectional area. Hepatocellular hypertrophy (hepatocyte size > 1.5× normal diameter) was determined and expressed as the percentage of the total liver slide area. Hepatic inflammation was assessed by counting the number of inflammatory foci per field at a magnification of 100× in 5 nonoverlapping fields per specimen, expressed as the average number of foci per mm2 field. Fibrosis was assessed histochemically by Picro-Sirius Red staining (Chroma; WALDECK-GmbH, Munster, Germany). Collagen content was quantified using ImageJ Software (National Institutes of Health, Bethesda, MD) by assessment of the area of liver tissue that was stained positively (expressed as the percentage of total tissue area). In addition, the development of fibrosis was assessed by a liver pathologist to quantify the percentage of perisinusoidal fibrosis (expressed as the percentage of perisinusoidal fibrosis relative to the total perisinusoidal area).
Mouse Hepatic Gene Expression Analysis
Total RNA was extracted from the liver at all time points (n = 6 for chow group/time point and n = 12 for HFD group/time point), with Ambion RNAqueous total RNA isolation kit (Thermo Fisher Scientific, Inc, Waltman, MA). The RNA concentration was determined spectrophotometrically using Nanodrop 1000 (Isogen Life Science, De Meern, The Netherlands), and RNA quality was assessed using the 2100 Bioanalyzer (Agilent Technologies, Amstelveen, The Netherlands). Strand-specific messenger RNA sequencing libraries for the Illumina (San Diego, CA) platform were generated and sequenced at BaseClear BV (Leiden, The Netherlands). The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2500 with a single-read 50-cycle sequencing protocol, 15 million reads per sample, and indexing. Differentially expressed genes (DEGs) were determined at weeks 6, 12, 18, and 24 using the DEseq-method with statistical cut-off false discovery rate of less than 0.001.22 DEGs were used as an input for pathway analysis through Ingenuity Pathway Analysis (IPA) suite (www.ingenuity.com, accessed 2016).
Dynamic Proteomics
A dynamic proteomics platform described previously23, 24 was applied to quantify the fractional synthesis rates of a large numbers of proteins via stable isotope labeling and liquid chromatography–mass spectrometry–based mass isotopomer analysis. Briefly, mice were labeled with deuterated water for 7 days, frozen liver tissue (chow, n = 3; HFD, n = 4) was subjected to sequential protein extraction to fractionate cellular, guanidine-soluble extracellular matrix (ECM) proteins and residual insoluble ECM proteins, and protein fractional synthesis rates (fraction of each protein that had been newly synthesized during the 7-day labeling period) were calculated using mass isotopomer analyses as described previously.25
Translational Aspects of LDLr-/-.Leiden NASH Mouse Model
To gain insight into the translational value of the LDLr-/-.Leiden NASH mouse model, a comparison was made at the molecular level between the LDLr-/-.Leiden mouse and data from NASH patients. The human gene expression data set (GSE48452) was downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) including 12 control liver samples (group C), 16 healthy obese samples (group H), 9 steatosis samples (group S), and 17 NASH samples (group N). This data set was derived from a study performed by the laboratory of Dr J. Hampe (Kiel, Germany). From this data set, samples were used that were obtained before the patients underwent a gastric bypass surgery.26 Gene expression levels were measured using the Affymetrix Human Gene 1.1 ST array (transcript version) (Affymetrix, Inc, Santa Clara, CA). The probe-level, background-subtracted, expression values were used as input for the lumi package27 of the R/Bioconductor (http://www.bioconductor.org; http://www.r-project.org) to perform quality control and quantile normalization. Differentially expressed probes were identified using the limma package of R/Bioconductor,28, 29 and calculated values of P < .01 were used as the threshold for significance. These differentially expressed probes were used as input for pathway analysis through IPA suite (www.ingenuity.com, accessed 2017).
Feature Selection for Molecular Fibrosis Signature
Identification and ranking of features (genes and proteins) for the molecular fibrosis signature was obtained by calculating a rank/composite score based on 3 approaches: correlation analyses (Pearson and Spearman) to link differentially expressed genes to newly formed proteins; weighted association of genes and proteins to key disease processes (direct and indirect biological link); and the presence of genes/proteins in a biomarker database (Integrity Biomarker Module; Thomson Reuters, London) for NASH and hepatic fibrosis, or other fibrotic diseases.
The approaches in more detail were as follows: the Pearson correlation coefficient was calculated for all normalized gene counts (expressed as Reads Per Kilobase per Million mapped reads30) per subject and fractional synthesis rates from the dynamic proteome analysis.25 Because the Pearson correlation method is prone to induce a bias in feature selection because of the presence of potential outliers, Spearman rank correlation also was performed on the same data set. Features were selected for the fibrosis signature when both the Pearson and Spearman correlation coefficient were greater than 0.9 (P < .01). The feature list of the fibrosis signature resulted in 232 genes and 8 proteins that were used as the seed list for the association to key disease processes (direct biological link). The Path Explorer tool (IPA; Qiagen, Redwood City, CA) was used to calculate the shortest path between 232 signature genes and defined 4 key processes. This algorithm connects predefined molecules such as the fibrosis signature to other molecules or processes using the curated knowledge from Ingenuity Knowledge Base (Qiagen). Because not all genes could be linked directly to these processes, the biological context of the remaining genes and proteins was determined by building an induced modules networks using databases within ConsensusPathDB (http://cpdb.molgen.mpg.de/), indicated as the indirect biological link. The interactions from ConsensusPathDB were visualized in Gephi using ForceAtlas2, a continuous graph layout algorithm31 for network visualization. Final prioritization was obtained by identification of which of the 232 genes and 8 proteins were documented in the Integrity biomarker module (accessed March 2017) and used as a biomarker in human studies (clinical trials and observational studies) related to NASH, hepatic fibrosis, or other fibrotic diseases.
Statistical Analysis
In vivo data are presented as means ± SD. The significance of differences between chow and HFD animals in continuous variables were tested using a 2-way analysis of variance with the Bonferroni post hoc test. Statistical differences between HFD and chow-fed animals were tested using the Student t test. Differences with a P value less than .05 were considered significantly different.
Results
HFD Feeding Induces Obesity, Hypercholesterolemia, Hypertriglyceridemia, and Insulin Resistance
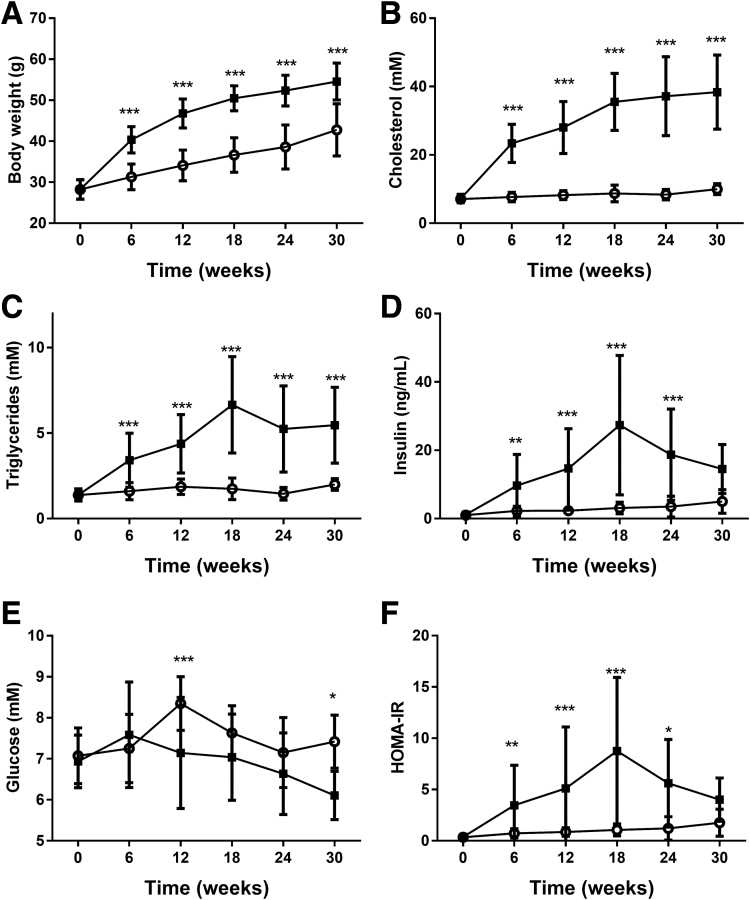
At the start of the experiment, mice from both groups had an equal average body weight of 28.2 ± 2.4 g (chow group) and 28.3 ± 2.4 g (HFD group). Body weight increased in HFD-fed mice relative to chow-fed mice, and was statistically significant after 6 weeks of HFD treatment (HFD, 40.4 ± 3.2 vs chow, 31.3 ± 3.1 g; P < .001). This difference in body weight was sustained until the end of the study at week 30 (Figure 1A). This body weight increase was reflected by increased weights of various white adipose tissue depots (data not shown). HFD feeding resulted in an obese phenotype with obesity-associated hypercholesterolemia, hypertriglyceridemia, and hyperinsulinemia (Figure 1B–D). All parameters showed a strong increase from week 6 until week 18 and remained at this increased level until the end of the study. Although blood glucose levels were not increased significantly at all time points (Figure 1E), the HOMA index indicated that HFD-fed mice became insulin resistant from week 6 until week 24 (Figure 1F), indicating appropriate β-cell compensation.
Figure 1.
Effect of HFD and chow on (A) body weight, and plasma levels of (B) cholesterol, (C) triglycerides, (D) insulin, and (E) glucose, and (F) HOMA index.Black solid squares indicate HFD; open circles indicate chow diet. *P < .05, **P < .01, ***P < .001 vs chow. HOMA-IR, HOmeostatic Model Assessment for Insulin Resistance.
HFD Feeding Induces NAFLD, Which Progresses to NASH Over Time
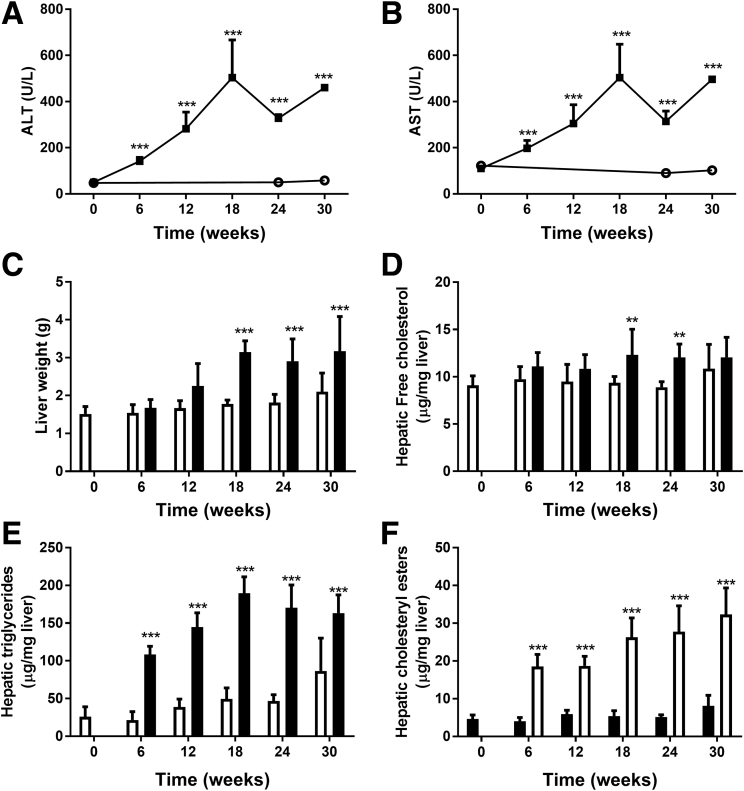
In parallel with the development of an obese phenotype, plasma levels of liver damage markers ALT and AST increased significantly upon HFD feeding. Plasma levels of ALT and AST were increased rapidly and significantly from 6 weeks onward in HFD vs chow mice (Figure 2A and B). Liver weight in HFD-fed animals was increased significantly relative to chow-fed animals after week 18 (Figure 2C). Biochemical analysis of intrahepatic lipids showed that free cholesterol in the liver was increased significantly at weeks 18 and 24 in HFD-fed animals (Figure 2D). Liver triglyceride levels reached a maximum at week 18 and remained at this level up until week 30 (Figure 2E). Cholesteryl esters already were increased significantly at week 6 and remained increased significantly at all later time points (Figure 2F).
Figure 2.
Effect of HFD and chow diet on liver characteristics such as liver damage enzymes (A) ALT and (B) AST, (C) liver weight, and (D) liver lipids free cholesterol, (E) triglycerides, and (F) cholesteryl esters.Black solid squares indicate HFD; open circles indicate chow diet. **P < .01, ***P < .001 vs chow.
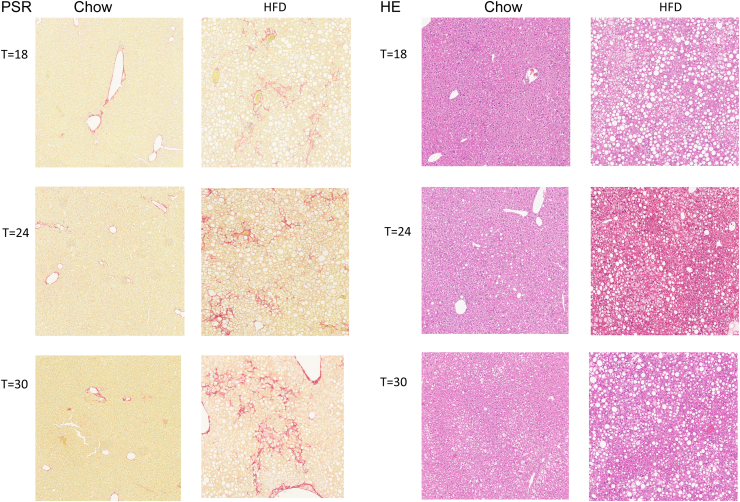
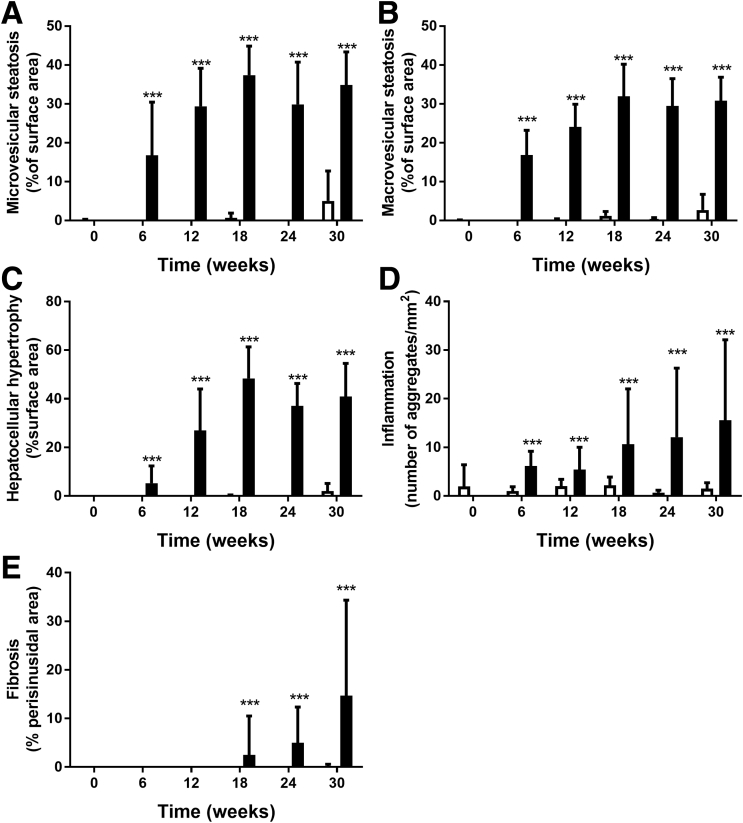
Histopathologic analysis of hepatic steatosis (both microvesicular and macrovesicular), hepatocellular hypertrophy, hepatic inflammation, and hepatic fibrosis showed the development of NASH with fibrosis on prolonged HFD feeding (Figure 3). In HFD-fed animals both macrovesicular and microvesicular steatosis were pronounced at week 6 and increased until week 18 (Figure 4A and B). Mild liver cell hypertrophy was detectable at week 6, and strongly increased until week 18, after which no further increase was observed (Figure 4C). In contrast, the number of inflammatory aggregates in the liver in HFD mice was comparable at weeks 6 and 12, and showed a strong but variable increase starting at week 18 (Figure 4D). Histopathologic liver fibrosis was not present at weeks 6 and 12, but became detectable at week 18 and showed a gradual increase up until week 30 (Figure 4E).
Figure 3.
Histologic figures of H&E staining (top panels) and Picro-Sirius Red (PSR) staining (bottom panels)of relevant time points (t) shows the development of NASH and fibrosis in the HFD-fed LDLr-/-.Leiden mice.
Figure 4.
Pathologic features of NASH after HFD and chow diet determined by the level of (A) microvesicular steatosis, (B) macrovesicular steatosis vacuolation, (C) hepatocellular hypertrophy, (D) hepatic inflammation, and the level of (E) perisinusoidal fibrosis.Black bars indicate HFD, white bars indicate chow diet. ***P < .001 vs chow.
Transcriptome Analysis Showed Dynamics of Key Processes Involved in NASH and Fibrosis
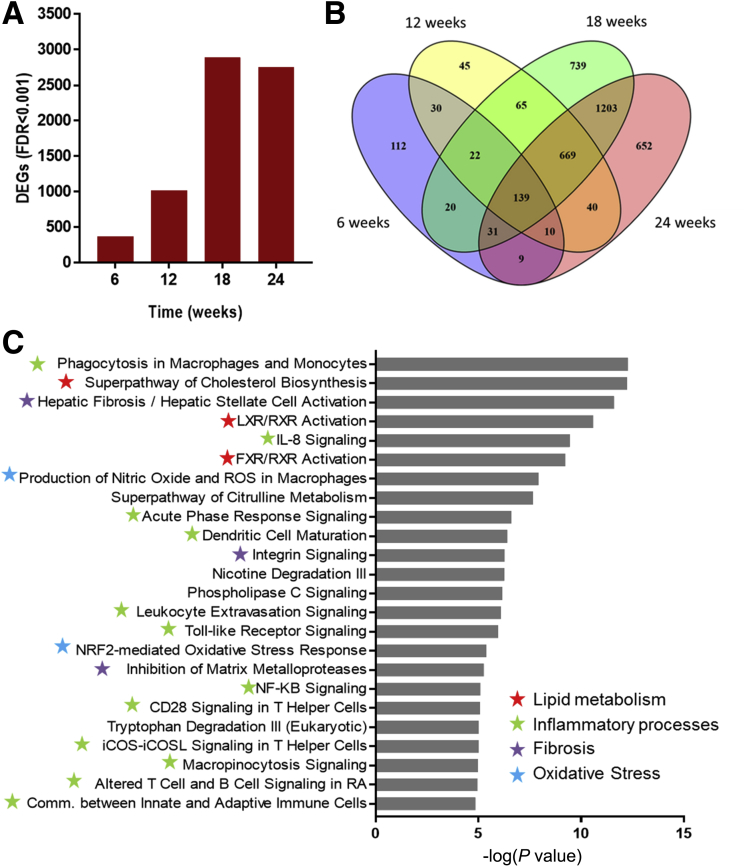
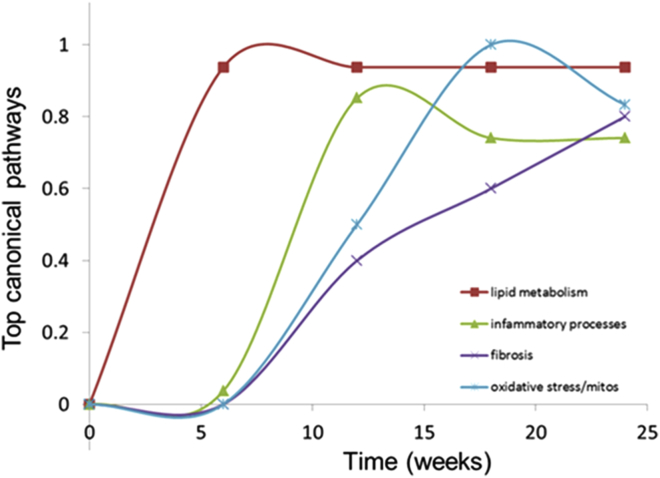
To unravel the molecular processes affected during the development of NASH and fibrosis and to provide insight into their time-resolved patterns of regulation during disease progression, next-generation sequencing of hepatic gene expression was performed. HFD feeding substantially increased the number of DEGs compared with chow feeding, ultimately leading to 2888 and 2753 DEGs (false discovery rate < 0.001) at weeks 18 and 24, respectively (Figure 5A). Analysis of the degree of overlap between the different time points shows that the majority of genes expressed at weeks 18 and 24 are shared. In addition, a large proportion of the genes that are differentially expressed at week 12 remain differentially regulated at weeks 18 and 24, as shown in the Venn diagram (Figure 5B). Gene set enrichment analysis indicated a clear modulation of pathways related to NASH and hepatic fibrosis at week 24 after HFD treatment, as exemplified by expression changes of genes in lipid metabolism pathways and a strong activation of genes in the hepatic fibrosis/hepatic stellate cell activation and integrin signaling pathways. In addition, among the top canonical pathways, 13 inflammation-related pathways and oxidative stress response pathways were activated by the HFD treatment compared with chow (Figure 5C). Integration of expression data from all time points clearly showed a time-resolved response of the main categories of processes that play a role in the development of NASH and hepatic fibrosis, namely lipid metabolism, inflammation, oxidative stress, and fibrosis (Figure 6). The process of lipid metabolism is the first to be activated (from week 6 onward), while the inflammatory, oxidative stress, and fibrotic response were activated from week 12 onward. These data show early response genes and processes from all main categories that already were expressed differentially in week 12.
Figure 5.
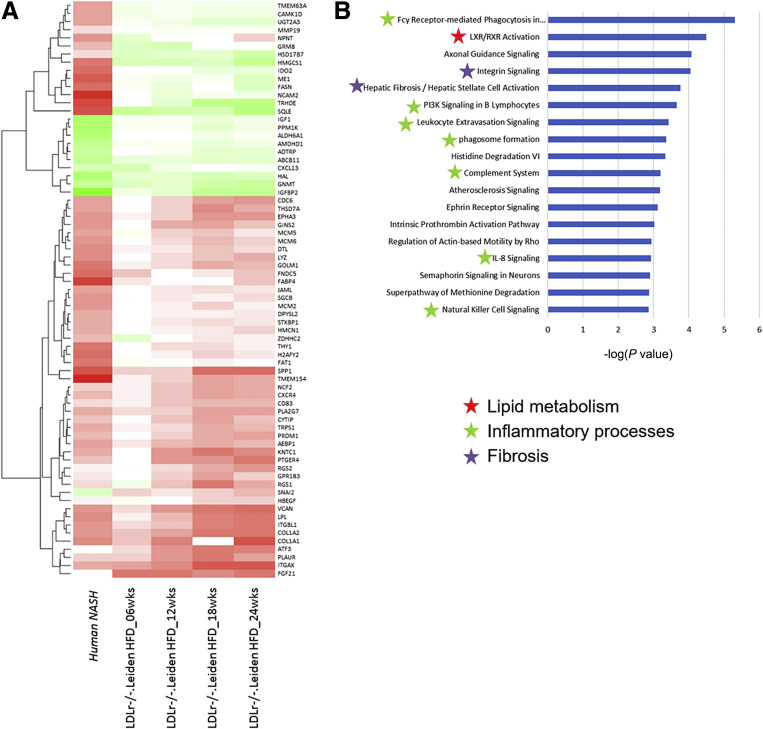
(A) Effect of HFD on the number of differentially expressed genes as measured by RNA sequencing technology. Visualization of overlapping genes per time point represented in a (B) Venn diagram and (C) enrichment analysis of the top 25 enriched canonical pathways, values are expressed as -log(P value). Red stars indicate pathways related to lipid metabolism, green stars are related to inflammatory processes, blue stars are related to oxidative stress, and purple stars are related to extracellular matrix processes.
Figure 6.
Graphic visualization of temporal dynamics of key processes involved in the development of NASH and fibrosis as determined by time-resolved enrichment analysis of the top canonical pathways.Red line, lipid metabolism; green line, inflammatory processes; blue line, oxidative stress; purple line, extracellular matrix processes.
Dynamic Proteomic Analyses Uncovers Early Synthesis of Extracellular Matrix Proteins
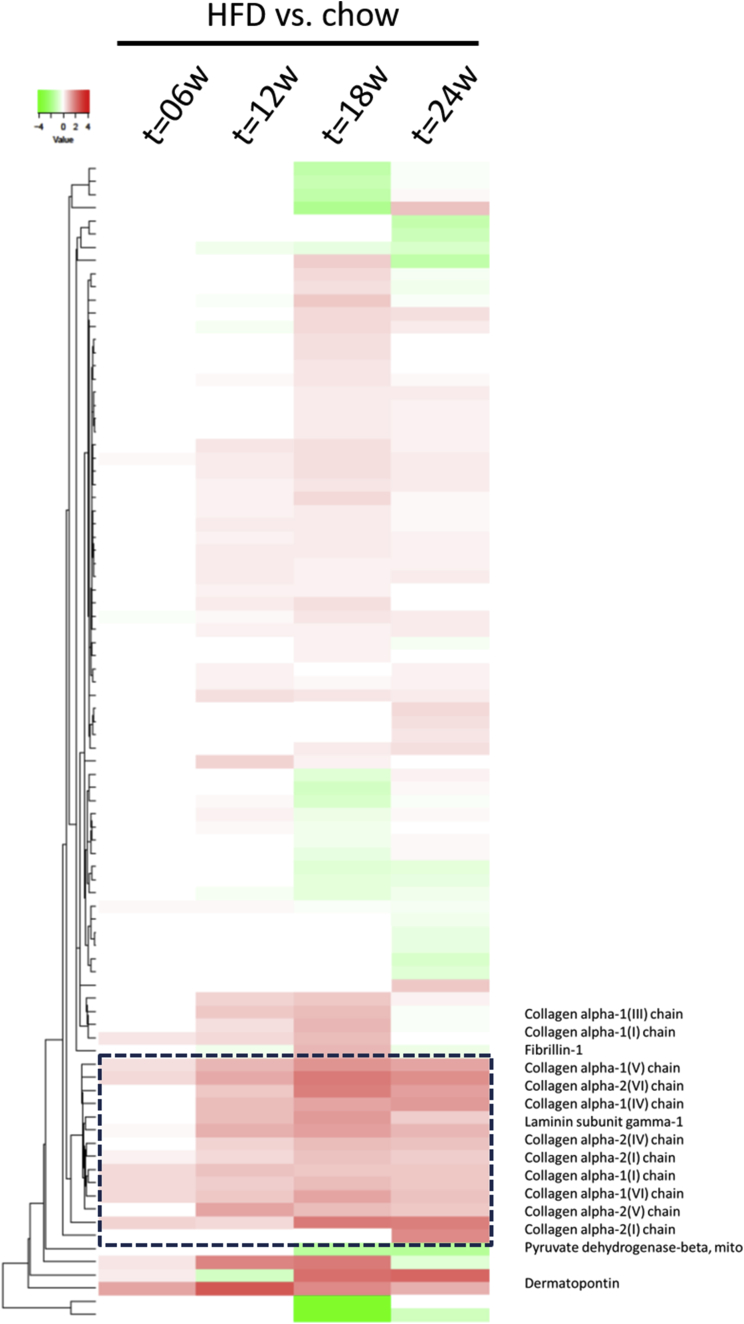
Next, we investigated whether these pronounced effects of HFD feeding on gene expression also were reflected on the protein level by measuring protein turnover rates, using deuterated water-labeled. This dynamic protein analysis was performed, using the guanidine-soluble and guanidine-insoluble proteins from liver, to provide insight into the proteins that were synthesized during the last week before sacrifice (expressed as a fractional synthesis value; ie, the fraction of each protein that was newly synthesized during the 7-day labeling period). The guanidine-soluble fraction contained many extracellular matrix proteins, of which the synthesis rate was increased significantly at an early time point (week 6 or 12) and that remained high during the progression of liver disease (week 24); these included biglycan, collagen1α1, collagen1α2, collagen6α1, fumarylacetoacetase, keratin type I cytoskeletal 18, keratin type II cytoskeletal 8, and nidogen-1. The guanidine-insoluble fraction also contained several extracellular matrix proteins, of which the synthesis rate was significantly different from chow-fed mice at an early stage of disease and remained different during the progression of liver disease, including collagen1α1, collagen3α1, collagen4α1, collagen4α2, collagen6α1, collagen6α2, laminin subunit γ1, and several tubulins (data not shown). To visualize the protein synthesis rate, a heatmap was generated based on fold-change differences (Figure 7). The most predominant difference was seen in a cluster of proteins involved in ECM deposition and fibrosis, which already was abundant after 12 weeks of HFD treatment (Figure 7, between the dashed lines). These data show increased extracellular matrix synthesis already after 12 weeks of HFD feeding by dynamic protein profiling analysis, a time point at which histopathologic fibrosis was not detectable yet.
Figure 7.
Heatmap visualization of the effect of HFD on significant liver proteins synthesized the week before sacrifice as measured by dynamic protein profiling using deuterated water labeling technique. The black box with dashed lines indicates the set of ECM proteins. Green indicates down-regulation, red indicates up-regulation.
LDLr-/-.Leiden NASH Mouse Model Shares Genes and Processes With NASH Patients
To determine the translational value of the molecular changes in the LDLr-/-.Leiden NASH mouse model a comparison analysis was performed using data from human NASH patients (GSE48452). A total of 123 genes (mapped cross-species) were selected that were differentially expressed between NASH patients and healthy controls as previously determined by Teufel et al.32 From these 123 genes, 71 genes were identified to be expressed in a time-dependent manner in HFD-fed LDLr-/-.Leiden mice, with the majority of genes being regulated in the same direction as in human beings (Figure 8A). Because analysis on individual gene level may overlook common disease mechanisms, we compared gene set enrichments between LDLr-/-.Leiden mice at week 24 with NASH patients. Interestingly, the previously identified key processes in mice (Figures 5C and 6) involved in the development of NASH also were enriched in the top 18 pathways in human NASH patients (Figure 8B). This indicates that the LDLr-/-.Leiden mouse model can be used to study key processes related to NASH and generate data that reflect the human situation.
Figure 8.
Heatmap visualization of individual genes compared with their controls. Human NASH indicates the gene response of human NASH patients compared with health control subjects. LDLr-/-.Leiden mice indicates the gene response of HFD-fed mice compared with chow at the corresponding time point. (A) Green indicates down-regulation, red indicates up-regulation. (B) Visualization of the enrichment analysis of the top 25 enriched canonical pathways, values are expressed as -log(P value). Red stars indicate pathways related to lipid metabolism, green stars are related to inflammatory processes, and purple stars are related to extracellular matrix processes.
Feature Selection to Generate an Early Fibrosis Signature and Rank Candidate Biomarkers
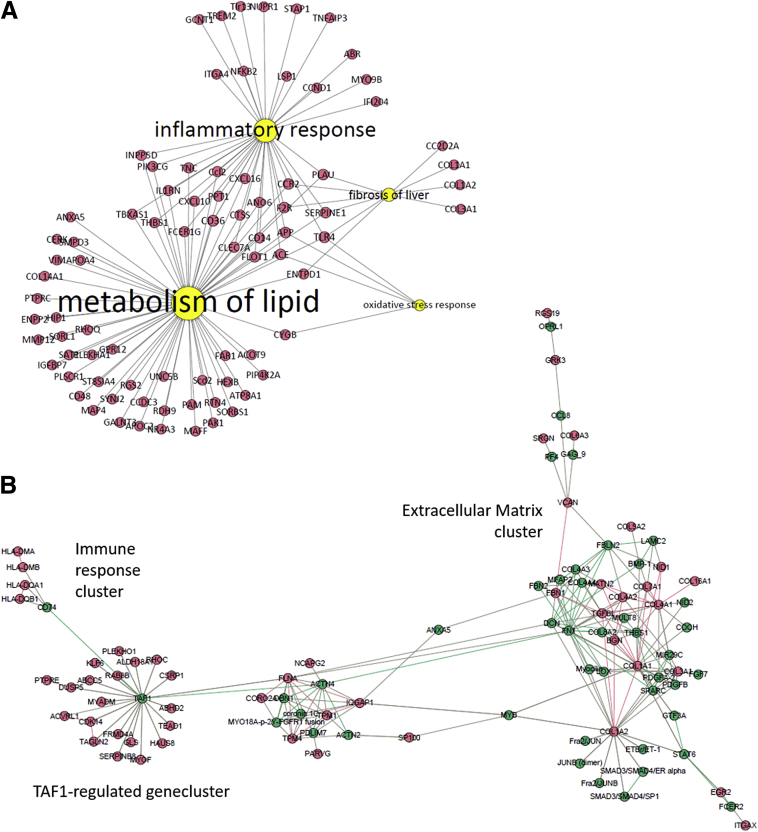
An increased expression of fibrosis-related genes and molecular processes as well as synthesis of new matrix proteins was detected already at week 12 and preceded histopathologic detection. Next, a data integrative genomics-proteomics approach was applied to select and prioritize the specific molecular features that enable early detection of hepatic fibrosis. First, the HFD-induced differentially expressed 2753 genes from week 24, a time point at which hepatic fibrosis was abundant, were compared with the differentially expressed genes at week 12. This resulted in a total of 568 differentially expressed genes that were up-regulated at weeks 24 and 12. Next, a selection of 33 newly formed proteins were identified that were statistically different compared with chow animals at both weeks 24 and 12 (P < .05). Correlation analysis of these 568 DEGs with 33 statistically different proteins at week 24 resulted in a list of 232 genes that were strongly correlated with 8 proteins (R2 > 0.9; P < .01) (Supplementary Table 1). This set of genes and proteins was designated as the molecular fibrosis signature, of which the biological relevance was investigated further. By using the Path Explorer tool including the Shortest Path algorithm, 88 DEGs were identified to be linked directly to the 4 major processes of lipid metabolism, inflammatory response, oxidative stress, and fibrosis (Figure 9A). To determine relations between the remaining genes and the dynamic proteins, an induced-modules networks (Figure 9B) was generated connecting 144 genes and 5 proteins. This indicated clusters of genes/proteins, of which one was highly related to the ECM and the other indicated genes under control of TAF1, a transcription factor that regulates cell proliferation by affecting the transforming growth factor-β signaling pathway. Next, the relevance of these genes and proteins were calculated (composite score) based on their connection directly or indirectly to one or more key biological processes, whether they were documented in the literature as a biomarker for NASH, hepatic fibrosis, or another fibrotic disease, and based on the fold-change in HFD condition compared with control chow at week 12 (Supplementary Table 2). An overview of the top 20 most relevant genes and proteins and the calculated composite scores are shown in Table 1. To illustrate the relevance of these genes and proteins the correlation between gene expression and histologic grade of fibrosis as measured by Picro-Sirius Red staining was determined. This further strengthened the relationship between signature gene expression and hepatic fibrosis.
Figure 9.
(A) Network visualization of the direct link between features of the molecular signature associated with key processes, lipid metabolism, inflammation, oxidative stress, and fibrosis, and (B) the indirect link via induced-modules network.Yellow nodes indicate key processes, red nodes indicate genes and proteins from the signature, and green nodes indicate nodes from the induced modules network.
Table 1.
Overview of the Top 20 Most Relevant Genes and Proteins Based on Their Calculated Composite Scores and Correlations With Histopathologic Fibrosis Score
| Gene name | Expression change, logFC | Documented biomarker for NASH | Documented biomarker for hepatic fibrosis | Composite score | Correlation Sirius Red | Ensemble gene ID |
|---|---|---|---|---|---|---|
| SERPINE1 | 3.0 | Y | Y | 16.1 | 0.858 | ENSMUSG00000037411 |
| CCL2 | 2.0 | Y | Y | 11.2 | 0.853 | ENSMUSG00000035385 |
| COL1A1 | 2.4 | Y | Y | 10.5 | 0.862 | ENSMUSG00000001506 |
| THBS1 | 2.1 | Y | Y | 9.3 | 0.961 | ENSMUSG00000040152 |
| CXCL10 | 2.1 | Y | Y | 9.3 | 0.788 | ENSMUSG00000034855 |
| CCR2 | 1.7 | Y | Y | 9.3 | 0.975 | ENSMUSG00000049103 |
| CD14 | 1.9 | Y | Y | 8.6 | 0.851 | ENSMUSG00000051439 |
| IL1RN | 2.5 | Y | N | 8.4 | 0.749 | ENSMUSG00000026981 |
| TNC | 1.4 | Y | N | 6.5 | 0.890 | ENSMUSG00000028364 |
| SMPD3 | 2.7 | N | N | 6.5 | 0.880 | ENSMUSG00000031906 |
| PLAU | 1.3 | N | Y | 6.3 | 0.882 | ENSMUSG00000021822 |
| COL3A1 | 1.8 | Y | Y | 6.3 | 0.927 | ENSMUSG00000026043 |
| APOA4 | 2.6 | Y | N | 6.3 | 0.808 | ENSMUSG00000032080 |
| MMP12 | 5.2 | N | N | 6.2 | 0.842 | ENSMUSG00000049723 |
| ACE | 1.0 | Y | Y | 6.1 | 0.870 | ENSMUSG00000020681 |
| COL1A2 | 1.7 | N | Y | 6.1 | 0.914 | ENSMUSG00000029661 |
| TLR4 | 0.7 | Y | Y | 5.5 | 0.786 | ENSMUSG00000039005 |
| ITGAX | 2.2 | Y | N | 5.4 | 0.850 | ENSMUSG00000030789 |
| VCAN | 1.9 | N | N | 4.9 | 0.910 | ENSMUSG00000021614 |
| CLEC7A | 1.8 | N | N | 4.7 | 0.885 | ENSMUSG00000079293 |
N, no; Y, yes.
Discussion
The development of NASH and hepatic fibrosis is a long-term progressive process. The sequence of molecular events that contribute to the development of NASH and fibrosis is largely unknown. This is partly due to the late diagnosis of NASH and fibrosis because their clinical symptoms do not become manifest until an advanced stage of disease. Therefore, it is difficult to study the early processes involved in disease development in human beings. Animal models of NASH allow time-resolved analysis of events that shows crucial information on early processes contributing to disease development. For such analysis the translational aspects of the mouse model used are a prerequisite.
To study mechanisms of disease development, a wide variety of animal models for NASH and fibrosis are available, which all have their specific advantages and disadvantages.33, 34 None of these resembles the complete spectrum of molecular processes involved in the development of NASH and fibrosis in human beings.32 However, to study NASH and fibrosis in a more physiological setting, HFD-induced models better represent human disease development, although the degree of liver injury and fibrosis is less severe than in chemically induced (eg, carbon tetrachloride) fibrosis.35
HFD-fed LDLr-/-.Leiden mice develop characteristics of the metabolic syndrome indicated by obesity, hypercholesterolemia, hypertriglyceridemia, and insulin resistance. As a consequence of HFD feeding, liver damage occurred, as indicated by increased levels of ALT and AST relative to chow-fed controls. Furthermore, liver damage was confirmed by histopathologic analysis, which showed a gradual increase of steatosis, cellular hypertrophy, and inflammation over time. More importantly, LDLr-/-.Leiden mice also developed hepatic perisinusoidal fibrosis. These results show that the LDLr-/-.Leiden mouse model is a suitable model for NASH with associated hepatic fibrosis in the context of obesity, dyslipidemia, and insulin resistance,20 as is typical for NASH patients.1 By using a systems biology approach we have provided a time-dependent sequence of key molecular processes involved in the development of NASH and fibrosis. This approach allowed us to unravel the mechanisms of disease development, enabled early identification of disease processes leading to hepatic fibrosis, and can guide the development of tools for the discovery of blood-based biomarkers for fibrosis.
To determine the translational value of our findings we compared mouse-derived data with human data using a publicly available NASH patient data set (Gene Expression Omnibus dataset: GSE48452). In a previous study, Teufel et al32 identified a panel of 123 genes differentially expressed in NASH patients compared with healthy controls. Although Teufel et al32 reported only few (range, 1–18) overlapping genes between several NASH mouse models and NASH patients, we showed in the current study that the majority (71) of these 123 genes also could be detected in our LDLr-/-.Leiden mouse model. Because an analysis of the individual gene level may depreciate common disease mechanisms, we compared gene set enrichments between HFD-fed LDLr-/-.Leiden mice at week 24 with NASH patients. These data illustrate the overlap between NASH patients and HFD-fed LDLr-/-.Leiden mice on NASH-related processes. Results obtained in this LDLr-/-.Leiden NASH mouse model on these key processes therefore might have clinical relevance.
We identified the temporal dynamics of key molecular processes involved in the development of NASH, namely lipid metabolism, inflammatory response, oxidative stress, and fibrosis. This was supported by time-resolved histopathologic observations showing similarities to human disease development. Furthermore, these data support the multiple-hit hypothesis, which considers multiple processes acting together to induce NASH and fibrosis.36 This includes triglyceride accumulation and associated lipotoxicity followed by, at least in part, a proinflammatory reaction and oxidative stress response, and a profibrotic process leading to the synthesis of new extracellular matrix and deposition of collagens. The clinical symptoms of this profibrotic process do not become manifest until an advanced stage of disease, at which time disease development is difficult to treat. Therefore, it is important to identify profibrotic processes at an early time point at which pathologic fibrosis is not present yet.
Data integrative approaches were used to correlate a subset of differentially expressed genes to the active formation of newly formed collagen, which was synthesized in the week before animals were sacrificed. This resulted in the identification of a molecular fibrosis signature associated with key disease processes, which can be detected at the molecular level before histopathologic fibrosis becomes manifest. This shows a molecular readout that can be used as a molecular diagnostic tool for the detection of early hepatic fibrosis. In a clinical setting, the use of molecular diagnostics already is used to perform prognostic risk assessments for several diseases including hepatocellular carcinoma37, 38 and breast cancer.39 To our knowledge, molecular diagnostics based on a combination of transcriptomics and dynamic proteomics constitute a novel approach that allows early diagnosis of hepatic fibrosis. This tissue-specific molecular signature may lead to the discovery of novel blood-based biomarkers for early detection of fibrosis. The application of advanced -omics technology in the search for novel biomarkers for hepatocellular carcinoma was described earlier.40 A network biology-based ranking including prior knowledge from databases and the selected genes and proteins from our tissue-specific molecular signature was used to generate a list of candidate blood-based biomarkers.
The set of genes included in the molecular fibrosis signature consists of markers already known to be related to existing hepatic fibrosis as well as novel markers. For example, thrombospontin-1 (THBS1) has been reported to be part of a gene signature implicated in human chronic liver disease.41 Our data show that THBS1 is also part of our molecular fibrosis signature and strongly correlates with collagen1a1 synthesis (R2 > 0.95; P < .01). Furthermore, we show that THBS-1 expression strongly correlates with the histologic grade of fibrosis at week 24 (R2 > 0.96; P < .01). On the other hand, sphingomyelin phosphodiesterase 3, which catalyzes the hydrolysis of sphingomyelin to form ceramide and phosphocholine, is as far as we know not been reported in human beings in relation to fibrosis before but also correlates strongly to the amount of histopathologic fibrosis (R2 > 0.88; P < .01). These data indicate the relevance of the signature for developing novel biomarker assays and future diagnostics for early detection of hepatic fibrosis.
Moylan et al42 published a set of 64 genes that differentiate between patients with mild NAFLD (fibrosis stages, 0–1) and severe NAFLD (fibrosis stages, 3–4). These 64 genes are categorized in several biological processes involved in NAFLD including inflammation, metabolism, and cellular stress responses including oxidative stress and also ECM formation. The presence of these biological processes in human NAFLD patients shows similarities with the key molecular processes as defined in our mouse data set. In addition, similarities were found on the single gene level as exemplified by the abundance of insulin-like growth factor binding protein 7, versican, and fibrilin 1. Differences may be explained by the fact that our molecular fibrosis signature was generated based on the correlation between genes and ECM proteins, thereby emphasizing the fibrotic process, whereas the data set of 64 genes from Moylan et al42 includes genes involved in multiple NAFLD- and fibrosis-related processes. The mouse molecular fibrosis signature reflects specific aspects of the human fibrosis processes and therefore can contribute to translational application of the signature.
In summary, our results show time-resolved regulation of key molecular processes involved in the development of NASH and hepatic fibrosis in HFD-fed LDLr-/-.Leiden mice. We have identified a molecular fibrosis signature that marks the active fibrosis process and can be detected before pathologic fibrosis is present. These data have translational value and can facilitate further development of candidate blood-based biomarkers for the early detection of hepatic fibrosis.
Footnotes
Author contributions Lars Verschuren, Arianne van Koppen, Anita M. van den Hoek, Reinout Stoop, and Roeland Hanemaaijer were responsible for the study concept and design; Lars Verschuren, Arianne van Koppen, Martine C. Morrison, Anita M. van den Hoek, Ivana Bobeldijk-Pastorova, and Roeland Hanemaaijer were responsible for writing the manuscript; Arianne van Koppen, Joanne Verheij, Adalberto Costessi, John Sagartz, Carine Beysen, Claire Emson, Kelvin Li, and Scott M. Turner were responsible for the acquisition of data; Lars Verschuren, Arianne van Koppen, Hiroshi Nagabukuro, Adalberto Costessi, Martien P. M. Caspers, Tim J. van den Broek, Cornelis Kluft, Carine Beysen, Claire Emson, Kelvin Li, Alain J. van Gool, Roel Goldschmeding, and Guido Hanauer were responsible for the analysis and interpretation of data; Hiroshi Nagabukuro, Joanne Verheij, Adalberto Costessi, John Sagartz, Cornelis Kluft, DS, Carine Beysen, Kelvin Li, Alain J. van Gool, Roel Goldschmeding, Ivana Bobeldijk-Pastorova, Martine C. Morrison, Anita M. van den Hoek, Reinout Stoop, Scott M. Turner, Guido Hanauer, and Roeland Hanemaaijer were responsible for critical revision of the manuscript; Lars Verschuren, Arianne van Koppen, Martien P. M. Caspers, Tim J. van den Broek, Cornelis Kluft, and Reinout Stoop were responsible for the statistical analysis; and Hiroshi Nagabukuro, Adalberto Costessi, Carine Beysen, Kelvin Li, Cornelis Kluft, Alain J. van Gool, and Scott M. Turner provided technical support.
Conflicts of interest The authors disclose no conflicts.
Funding This work was generated by a consortium that was supported by grant 114025001 from ZonMW, and the TNO research program “Predictive Health Technologies.”
Supplementary Material
Supplementary Table 1.
List of Newly Synthesized Proteins as Determined by Dynamic Protein Profiling Ranked by the Number of Correlating Genes (R2 > 0.95; P < .05)
| Newly synthesized ECM proteins | Correlating genes, n |
|---|---|
| Collagen α-2(I) chain | 150 |
| s_Collagen α-2(I) chain | 82 |
| s_Collagen α-1(I) chain | 70 |
| Collagen α-1(III) chain | 66 |
| s_Collagen α-1(VII) chain | 49 |
| Hydroxymethylglutaryl-CoA synthase, mitochondrial | 43 |
| Laminin subunit γ-1 | 42 |
| Collagen α-1(I) chain | 36 |
| s_Keratin, type II cytoskeletal 8 | 22 |
| s_Collagen α-1(VI) chain | 19 |
| 3-ketoacyl-CoA thiolase, mitochondrial | 12 |
| s_Dermatopontin | 11 |
| s_ATP synthase subunit α, mitochondrial | 10 |
| Tubulin β-4A chain | 9 |
| s_Keratin, type I cytoskeletal 18 | 7 |
| Tubulin β-4B chain | 7 |
| s_Betaine--homocysteine S-methyltransferase 1 | 5 |
| Collagen α-1(VI) chain | 5 |
| Collagen α-2(IV) chain | 5 |
| s_Biglycan | 4 |
| s_Peptidyl-prolyl cis-trans isomerase A | 4 |
| s_60S ribosomal protein L35 | 3 |
| s_Superoxide dismutase [Cu-Zn] | 3 |
| Collagen α-2(VI) chain | 3 |
| s_Nidogen-1 | 2 |
| s_Carbamoyl-phosphate synthase [ammonia], mitochondrial | 1 |
| s_3-ketoacyl-CoA thiolase, mitochondrial | 0 |
| s_60-kilodalton heat shock protein, mitochondrial | 0 |
| s_Fumarylacetoacetase | 0 |
| s_Histone H3.2 | 0 |
| Collagen α-1(IV) chain | 0 |
| Microsomal glutathione S-transferase 1 | 0 |
| Tubulin β-5 chain | 0 |
NOTE. Proteins marked with an “s” in front of the protein name were detected in the guanidine-soluble fraction. The top 8 proteins are included in the molecular signature and were used for further feature selection.
Supplementary Table 2.
All 232 Genes Included in the Molecular Fibrosis Signature Correlated With the Onset of Fibrosis and Their Ranking Based on the Calculated Composite Scores
| Gene name | Expression change (logFC) | Documented biomarker for NASH | Documented biomarker for hepatic fibrosis | Composite score | ENSMUS-id | Reference NASH | Reference hepatic fibrosis, or other fibrotic disease |
|---|---|---|---|---|---|---|---|
| SERPINE1 | 3.0 | Y | Y | 16.1 | ENSMUSG00000037411 | Sookoian, Atherosclerosis 2011;218:378 | Armendariz-Borunda, J Invest Med 2008;56:944 |
| CCL2 | 2.0 | Y | Y | 11.2 | ENSMUSG00000035385 | Leach, 84th European Atherosclerosis Society (EAS) Congress (May 29 to June 1, Innsbruck, Austria) 2016, abstr 083 | Page, Am J Gastroenterol 2011;106:abstr 331 |
| COL1A1 | 2.4 | Y | Y | 10.5 | ENSMUSG00000001506 | Dattaroy, Am J Physiol Gastrointest Liver Physiol 2015;308:G298 | Decaris, PLoS One 2015;10:e0123311 |
| THBS1 | 2.1 | Y | Y | 9.3 | ENSMUSG00000040152 | Smalling, Am J Physiol Gastrointest Liver Physiol 2013;305:G364 | Smalling, Am J Physiol Gastrointest Liver Physiol 2013;305:G364 |
| CXCL10 | 2.1 | Y | Y | 9.3 | ENSMUSG00000034855 | Wada, Digestive Disease Week (May 21–24, San Diego, CA) 2016, abstr Sa1670 | Andersen, Eur J Clin Microbiol Infect Dis 2011;30:761 |
| CCR2 | 1.7 | Y | Y | 9.3 | ENSMUSG00000049103 | Sookoian, Atherosclerosis 2011;218:378 | Estrabaud, 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 7–11, Boston, MA) |
| CD14 | 1.9 | Y | Y | 8.6 | ENSMUSG00000051439 | Krakora, Conference on Retroviruses and Opportunistic Infections (CROI) (February 22–25, Boston, MA) 2016, abstr 5 | Estrabaud, 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 7–11, Boston, MA) |
| IL1RN | 2.5 | Y | N | 8.4 | ENSMUSG00000026981 | Yang, PLoS One 2015;10:e0131664 | |
| TNC | 1.4 | Y | N | 6.5 | ENSMUSG00000028364 | Sookoian, Atherosclerosis 2011;218:378 | Hisatomi, Intern Med (Tokyo) 2009;48:1501 |
| SMPD3 | 2.7 | N | N | 6.5 | ENSMUSG00000031906 | DePianto, Thorax 2015;70:48 | |
| PLAU | 1.3 | N | Y | 6.3 | ENSMUSG00000021822 | Andersen, Eur J Clin Microbiol Infect Dis 2011;30:761 | |
| COL3A1 | 1.8 | Y | Y | 6.3 | ENSMUSG00000026043 | Sookoian, Atherosclerosis 2011;218:378 | Decaris, PLoS One 2015;10:e0123311 |
| APOA4 | 2.6 | Y | N | 6.3 | ENSMUSG00000032080 | Shores, Digestive Disease Week (May 16–19, Washington, DC) 2015, abstr Su1036 | |
| MMP12 | 5.2 | N | Y | 6.2 | ENSMUSG00000049723 | Andersen, Eur J Clin Microbiol Infect Dis 2011;30:761 | |
| ACE | 1.0 | Y | Y | 6.1 | ENSMUSG00000020681 | Sookoian, Atherosclerosis 2011;218:378 | Granzow, Hepatology 2014;60:334 |
| COL1A2 | 1.7 | N | Y | 6.1 | ENSMUSG00000029661 | Decaris, PLoS One 2015;10:e0123311 | |
| TLR4 | 0.7 | Y | Y | 5.5 | ENSMUSG00000039005 | Sharifnia, Am J Physiol Gastrointest Liver Physiol 2015;309:G270 | Li, J Hepatol 2009;51:750 |
| ITGAX | 2.2 | Y | N | 5.4 | ENSMUSG00000030789 | Sookoian, Atherosclerosis 2011;218:378 | |
| VCAN | 1.9 | N | N | 4.9 | ENSMUSG00000021614 | Estany, 107th International Conference of the American Thoracic Society (May 13–18, Denver, CO) 2011, abstr | |
| CLEC7A | 1.8 | N | N | 4.7 | ENSMUSG00000079293 | ||
| COL6A3 | 1.1 | Y | Y | 4.3 | ENSMUSG00000048126 | Baker, PLoS One 2010;5:e9570 | Decaris, PLoS One 2015;10:e0123311 |
| PIK3CG | 1.1 | N | N | 4.3 | ENSMUSG00000020573 | DePianto, Thorax 2015;70:48 | |
| VIM | 1.1 | N | Y | 4.2 | ENSMUSG00000026728 | Ando, 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 7–11, Boston, MA) | |
| EGR2 | 3.2 | N | N | 4.2 | ENSMUSG00000037868 | ||
| CD36 | 0.8 | Y | N | 4.1 | ENSMUSG00000002944 | Garcia-Monzon, Eur J Clin Invest 2014;44:65 | Kang, Nat Med 2015;21:37 |
| TNFAIP3 | 1.5 | Y | N | 4.1 | ENSMUSG00000019850 | Sookoian, Atherosclerosis 2011;218:378 | |
| TREM2 | 2.8 | N | N | 3.8 | ENSMUSG00000023992 | ||
| COL4A2 | 0.8 | Y | Y | 3.5 | ENSMUSG00000031503 | Abdelmalek, 64th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 1–5, Washington, DC) | Attallah, J Immunoassay Immunochem 2007;28:155 |
| INPP5D | 0.8 | N | Y | 3.5 | ENSMUSG00000026288 | Katsounas, Hepatology 2010;52:abstr 609 | |
| CCDC3 | 1.2 | N | N | 3.5 | ENSMUSG00000026676 | DePianto, Thorax 2015;70:48 | |
| KLF6 | 1.2 | Y | N | 3.4 | ENSMUSG00000000078 | Nobili, J Pediatr Gastroenterol Nutr 2014;58:632 | |
| DEFB1 | 2.4 | N | N | 3.4 | ENSMUSG00000044748 | Han, 107th International Conference of the American Thoracic Society (May 13–18, Denver, CO) 2011, abstr | |
| CD48 | 1.2 | N | Y | 3.3 | ENSMUSG00000015355 | Utsunomiya, World J Gastroenterol 2007;13:383 | |
| NR4A3 | 2.3 | N | N | 3.3 | ENSMUSG00000028341 | ||
| ENPP2 | 0.7 | Y | Y | 3.2 | ENSMUSG00000022425 | Arendt, Hepatology 2015;61:1565 | Nakagawa, Clin Chim Acta 2011;412:1201 |
| APP | 0.4 | Y | N | 3.1 | ENSMUSG00000022892 | Mendoza, Exp Mol Pathol 2015;98:65 | Yang, Am J Respir Crit Care Med 2014;190:1263 |
| TAGLN | 1.1 | N | Y | 3.1 | ENSMUSG00000032085 | Bracht, J Proteome Res 2015;14:2278 | |
| STAP1 | 2.1 | N | N | 3.1 | ENSMUSG00000029254 | ||
| LSP1 | 1.1 | N | N | 3.1 | ENSMUSG00000018819 | DePianto, Thorax 2015;70:48 | |
| FBN1 | 1.0 | N | Y | 3.0 | ENSMUSG00000027204 | Ippolito, Toxicol Sci 2016;149:67 | |
| APOC2 | 0.7 | Y | Y | 3.0 | ENSMUSG00000002992 | Baker, PLoS One 2010;5:e9570 | Cheung, J Viral Hepat 2009;16:418 |
| CCND1 | 1.0 | N | Y | 3.0 | ENSMUSG00000070348 | Sarfraz, BMC Infect Dis (online) 2009;9:125 | |
| SYNJ2 | 2.0 | N | N | 3.0 | ENSMUSG00000023805 | ||
| GPR12 | 1.9 | N | N | 2.9 | ENSMUSG00000041468 | ||
| CTSS | 0.6 | N | N | 2.9 | ENSMUSG00000038642 | Marmai, Am J Physiol Lung Cell Mol Physiol 2011;301:L71 | |
| MAFF | 1.9 | N | N | 2.9 | ENSMUSG00000042622 | ||
| CYGB | 0.6 | Y | N | 2.8 | ENSMUSG00000020810 | Thuy, Am J Pathol 2015;185:1045 | |
| COL4A1 | 0.9 | N | Y | 2.8 | ENSMUSG00000031502 | Estrabaud, 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 7–11, Boston, MA) | |
| F2R | 0.6 | N | N | 2.8 | ENSMUSG00000048376 | ||
| PTPRC | 0.9 | N | Y | 2.8 | ENSMUSG00000026395 | Utsunomaiya, World J Gastroenterol 2007;13:383 | |
| NUPR1 | 1.7 | N | N | 2.7 | ENSMUSG00000030717 | ||
| TBXAS1 | 0.8 | N | N | 2.7 | ENSMUSG00000029925 | ||
| UNC5B | 0.8 | N | Y | 2.7 | ENSMUSG00000020099 | Utsunomiya, World J Gastroenterol 2007;13:383 | |
| ENTPD1 | 0.8 | N | N | 2.7 | ENSMUSG00000048120 | ||
| DUSP5 | 1.6 | N | Y | 2.6 | ENSMUSG00000034765 | Ahmad, J Transl Med (online) 2012;10:41 | |
| FSTL1 | 0.8 | N | N | 2.6 | ENSMUSG00000022816 | Murphy, Am J Pathol 2016;186:600 | |
| ANO6 | 0.8 | N | N | 2.6 | ENSMUSG00000064210 | ||
| FCER1G | 0.8 | N | N | 2.6 | ENSMUSG00000058715 | ||
| TGFBI | 0.5 | Y | Y | 2.6 | ENSMUSG00000035493 | Decaris, PLoS One 2015;10:e0123311 | |
| COL14A1 | 0.8 | N | Y | 2.5 | ENSMUSG00000022371 | Bracht, J Proteome Res 2015;14:2278 | |
| MYOF | 1.5 | N | N | 2.5 | ENSMUSG00000048612 | ||
| HAUS8 | 1.4 | N | N | 2.4 | ENSMUSG00000035439 | ||
| TPM1 | 0.7 | N | N | 2.4 | ENSMUSG00000032366 | Deng, PLoS One 2013;8:e68352 | |
| ARHGAP25 | 1.4 | N | Y | 2.4 | ENSMUSG00000030047 | Utsunomiya, World J Gastroenterol 2007;13:383 | |
| MX1 | 1.4 | N | Y | 2.4 | ENSMUSG00000000386 | PLoS One 2015;10:e0130899 | |
| GCNT1 | 1.4 | N | N | 2.4 | ENSMUSG00000038843 | ||
| IFIT3 | 1.3 | N | Y | 2.3 | ENSMUSG00000074896 | Ibrahim, PLoS One 2016;11:e0154512 | |
| GALNT3 | 1.3 | N | N | 2.3 | ENSMUSG00000026994 | ||
| MATN2 | 1.3 | N | N | 2.3 | ENSMUSG00000022324 | ||
| CXCL16 | 0.7 | N | N | 2.3 | ENSMUSG00000018920 | ||
| SERPINB8 | 1.3 | N | N | 2.3 | ENSMUSG00000026315 | ||
| TNFRSF11A | 1.2 | N | N | 2.2 | ENSMUSG00000026321 | Boorsma, International Conference of the American Thoracic Society (May 16–21, San Diego, CA) 2014, abstr A1252 | |
| PLSCR1 | 1.2 | N | N | 2.2 | ENSMUSG00000032369 | ||
| TLR13 | 1.2 | N | N | 2.2 | ENSMUSG00000033777 | ||
| ABR | 1.1 | N | N | 2.1 | ENSMUSG00000017631 | ||
| CD52 | 1.1 | N | Y | 2.1 | ENSMUSG00000000682 | Utsunomiya, World J Gastroenterol 2007;13:383 | |
| FGL2 | 1.1 | N | Y | 2.1 | ENSMUSG00000039899 | Foerster, J Hepatol 2010;53:608 | |
| NFKB2 | 1.1 | N | N | 2.1 | ENSMUSG00000025225 | ||
| RTN4 | 0.5 | N | Y | 2.1 | ENSMUSG00000020458 | Wen, Dis Markers 2015;2015:419124 | |
| ITGA4 | 1.1 | N | N | 2.1 | ENSMUSG00000027009 | ||
| ACOT9 | 1.0 | N | N | 2.0 | ENSMUSG00000025287 | ||
| FLOT1 | 0.5 | N | N | 2.0 | ENSMUSG00000059714 | ||
| IFIT2 | 1.0 | N | Y | 2.0 | ENSMUSG00000045932 | Ibrahim, PLoS One 2016;11:e0154512 | |
| COL16A1 | 1.0 | N | N | 2.0 | ENSMUSG00000040690 | ||
| ST8SIA4 | 1.0 | N | N | 2.0 | ENSMUSG00000040710 | ||
| ALDH18A1 | 1.0 | N | N | 2.0 | ENSMUSG00000025007 | ||
| CSRP1 | 1.0 | N | N | 2.0 | ENSMUSG00000026421 | ||
| SORL1 | 1.0 | N | N | 2.0 | ENSMUSG00000049313 | ||
| PPT1 | 0.5 | N | N | 2.0 | ENSMUSG00000028657 | ||
| BGN | 0.5 | N | Y | 2.0 | ENSMUSG00000031375 | 47th Annual Meeting of the European Association of the Study of the Liver (EASL) (April 18–22, Barcelona, Spain) 2012, abstr 105 | |
| MLKL | 0.9 | Y | N | 1.9 | ENSMUSG00000012519 | Gautheron, 67th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 11–16, Boston) 20 | |
| NID1 | 0.9 | N | N | 1.9 | ENSMUSG00000005397 | ||
| CDK14 | 0.9 | N | N | 1.9 | ENSMUSG00000028926 | ||
| ADCY7 | 0.9 | N | N | 1.9 | ENSMUSG00000031659 | ||
| HEXB | 0.9 | N | N | 1.9 | ENSMUSG00000021665 | ||
| PAK1 | 0.8 | N | N | 1.8 | ENSMUSG00000030774 | ||
| IGFBP7 | 0.4 | Y | N | 1.8 | ENSMUSG00000036256 | ||
| PLEKHA1 | 0.8 | N | N | 1.8 | ENSMUSG00000040268 | ||
| ANXA5 | 0.8 | N | N | 1.8 | ENSMUSG00000027712 | ||
| TEAD1 | 0.8 | N | N | 1.8 | ENSMUSG00000055320 | ||
| RGS2 | 0.8 | N | N | 1.8 | ENSMUSG00000026360 | ||
| CERK | 0.8 | N | N | 1.8 | ENSMUSG00000035891 | ||
| SCD2 | 0.8 | N | N | 1.8 | ENSMUSG00000025203 | ||
| SORBS1 | 0.7 | N | N | 1.7 | ENSMUSG00000025006 | ||
| CARD10 | 0.7 | N | N | 1.7 | ENSMUSG00000033170 | Huang, PLoS One 2014;9:e107055 | |
| PLEKHO1 | 0.7 | N | N | 1.7 | ENSMUSG00000015745 | ||
| SRGN | 0.7 | N | N | 1.7 | ENSMUSG00000020077 | ||
| UBA7 | 0.7 | N | Y | 1.7 | ENSMUSG00000032596 | Ahmad, J Transl Med (online) 2012;10:41 | |
| ACVRL1 | 0.7 | N | N | 1.7 | ENSMUSG00000000530 | Chrobak, 19th Annual Congress of the European Respiratory Sociecy (ERS) (September 12–16, Vienna, Austria) 2009, abstr | |
| MYO9B | 0.7 | N | N | 1.7 | ENSMUSG00000004677 | ||
| PIP4K2A | 0.7 | N | N | 1.7 | ENSMUSG00000026737 | ||
| ABCC5 | 0.7 | N | N | 1.7 | ENSMUSG00000022822 | ||
| RHOC | 0.7 | N | N | 1.7 | ENSMUSG00000002233 | ||
| SAT1 | 0.7 | N | N | 1.7 | ENSMUSG00000025283 | ||
| RHOQ | 0.6 | N | N | 1.6 | ENSMUSG00000024143 | ||
| RAB8B | 0.6 | N | N | 1.6 | ENSMUSG00000036943 | ||
| GLS | 0.6 | N | N | 1.6 | ENSMUSG00000026103 | ||
| HIP1 | 0.6 | N | N | 1.6 | ENSMUSG00000039959 | ||
| FAR1 | 0.6 | N | N | 1.6 | ENSMUSG00000030759 | ||
| CC2D2A | 0.6 | N | N | 1.6 | ENSMUSG00000039765 | ||
| MYADM | 0.6 | N | N | 1.6 | ENSMUSG00000068566 | ||
| ATP8A1 | 0.6 | N | N | 1.6 | ENSMUSG00000037685 | ||
| SP100 | 0.6 | N | N | 1.6 | ENSMUSG00000026222 | ||
| ARMCX3 | 0.6 | Y | N | 1.6 | ENSMUSG00000049047 | Higuera, 67th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) (November 11–16, Boston) 2016 | |
| ABHD2 | 0.6 | N | N | 1.6 | ENSMUSG00000039202 | ||
| ITM2C | 0.5 | N | N | 1.5 | ENSMUSG00000026223 | DePianto, Thorax 2015;70:48 | |
| PAM | 0.4 | N | N | 1.4 | ENSMUSG00000026335 | ||
| MAP4 | 0.4 | N | N | 1.4 | ENSMUSG00000032479 | ||
| RGS19 | 1.0 | N | N | 1.0 | ENSMUSG00000002458 | ||
| DENND1C | 1.0 | N | N | 1.0 | ENSMUSG00000002668 | ||
| CLCN5 | 0.5 | N | N | 1.0 | ENSMUSG00000004317 | ||
| PRG4 | 0.7 | N | N | 1.0 | ENSMUSG00000006014 | ||
| CRIP1 | 1.1 | N | N | 1.0 | ENSMUSG00000006360 | ||
| APOBEC3 | 0.9 | N | N | 1.0 | ENSMUSG00000009585 | ||
| FXYD5 | 1.0 | N | N | 1.0 | ENSMUSG00000009687 | ||
| TMEM86A | 0.9 | N | N | 1.0 | ENSMUSG00000010307 | ||
| MCOLN2 | 1.4 | N | N | 1.0 | ENSMUSG00000011008 | ||
| FCGR1 | 0.9 | N | N | 1.0 | ENSMUSG00000015947 | ||
| PPFIBP1 | 0.6 | N | N | 1.0 | ENSMUSG00000016487 | ||
| IKZF1 | 0.9 | N | N | 1.0 | ENSMUSG00000018654 | ||
| RCN3 | 0.6 | N | N | 1.0 | ENSMUSG00000019539 | ||
| SLC6A8 | 1.3 | N | N | 1.0 | ENSMUSG00000019558 | ||
| NUDT4 | 0.4 | N | N | 1.0 | ENSMUSG00000020029 | ||
| MYO1G | 1.1 | N | N | 1.0 | ENSMUSG00000020437 | ||
| GPR137B | 1.1 | N | N | 1.0 | ENSMUSG00000021306 | ||
| SLC17A4 | 1.0 | N | N | 1.0 | ENSMUSG00000021336 | ||
| SEMA4D | 1.4 | N | N | 1.0 | ENSMUSG00000021451 | ||
| LRRC14B | 0.9 | N | N | 1.0 | ENSMUSG00000021579 | ||
| SAMD4 | 0.8 | N | N | 1.0 | ENSMUSG00000021838 | ||
| PARVG | 1.3 | N | N | 1.0 | ENSMUSG00000022439 | ||
| ST6GAL1 | 0.7 | N | N | 1.0 | ENSMUSG00000022885 | ||
| MS4A6B | 1.2 | N | N | 1.0 | ENSMUSG00000024677 | ||
| MS4A6D | 1.5 | N | N | 1.0 | ENSMUSG00000024679 | ||
| MAGED2 | 0.8 | N | N | 1.0 | ENSMUSG00000025268 | ||
| CPEB1 | 0.7 | N | N | 1.0 | ENSMUSG00000025586 | ||
| SGK3 | 0.5 | N | N | 1.0 | ENSMUSG00000025915 | ||
| COL5A2 | 1.4 | N | N | 1.0 | ENSMUSG00000026042 | ||
| GPR35 | 2.1 | N | N | 1.0 | ENSMUSG00000026271 | ||
| FAM129A | 0.6 | N | N | 1.0 | ENSMUSG00000026483 | ||
| TAGLN2 | 0.8 | N | N | 1.0 | ENSMUSG00000026547 | ||
| FRMD4A | 0.6 | N | N | 1.0 | ENSMUSG00000026657 | ||
| ZEB2 | 0.4 | N | N | 1.0 | ENSMUSG00000026872 | ||
| UAP1L1 | 1.6 | N | N | 1.0 | ENSMUSG00000026956 | ||
| DNAJC10 | 0.6 | N | N | 1.0 | ENSMUSG00000027006 | ||
| EHD4 | 0.7 | N | N | 1.0 | ENSMUSG00000027293 | ||
| ARHGEF2 | 0.9 | N | N | 1.0 | ENSMUSG00000028059 | ||
| CORO2A | 1.1 | N | N | 1.0 | ENSMUSG00000028337 | ||
| TTC39A | 2.5 | N | N | 1.0 | ENSMUSG00000028555 | ||
| ANXA3 | 0.7 | N | N | 1.0 | ENSMUSG00000029484 | ||
| OASL2 | 1.4 | N | N | 1.0 | ENSMUSG00000029561 | ||
| IQGAP1 | 1.0 | N | N | 1.0 | ENSMUSG00000030536 | ||
| MVP | 0.5 | N | N | 1.0 | ENSMUSG00000030681 | ||
| TRIM30A | 0.8 | N | N | 1.0 | ENSMUSG00000030921 | ||
| FLNA | 1.0 | N | N | 1.0 | ENSMUSG00000031328 | ||
| CTPS2 | 0.6 | N | N | 1.0 | ENSMUSG00000031360 | ||
| RASA3 | 0.6 | N | N | 1.0 | ENSMUSG00000031453 | ||
| SLC25A4 | 0.7 | N | N | 1.0 | ENSMUSG00000031633 | ||
| TPM4 | 0.5 | N | N | 1.0 | ENSMUSG00000031799 | ||
| TAGAP | 1.6 | N | N | 1.0 | ENSMUSG00000033450 | ||
| CHST11 | 1.7 | N | N | 1.0 | ENSMUSG00000034612 | ||
| FAM124A | 0.9 | N | N | 1.0 | ENSMUSG00000035184 | ||
| SSC5D | 2.4 | N | N | 1.0 | ENSMUSG00000035279 | ||
| ADAMTS2 | 1.3 | N | N | 1.0 | ENSMUSG00000036545 | ||
| H2-AA | 1.1 | N | N | 1.0 | ENSMUSG00000036594 | ||
| CPZ | 1.6 | N | N | 1.0 | ENSMUSG00000036596 | ||
| ABCC12 | 2.7 | N | N | 1.0 | ENSMUSG00000036872 | ||
| H2-DMA | 0.9 | N | N | 1.0 | ENSMUSG00000037649 | ||
| TMEM237 | 0.8 | N | N | 1.0 | ENSMUSG00000038079 | ||
| RFTN1 | 0.8 | N | N | 1.0 | ENSMUSG00000039316 | ||
| PRSS23 | 0.8 | N | N | 1.0 | ENSMUSG00000039405 | ||
| MYO9A | 0.6 | N | N | 1.0 | ENSMUSG00000039585 | ||
| ENC1 | 0.8 | N | N | 1.0 | ENSMUSG00000041773 | ||
| PTPRE | 1.2 | N | N | 1.0 | ENSMUSG00000041836 | ||
| NCAPG2 | 1.3 | N | N | 1.0 | ENSMUSG00000042029 | ||
| 2010003K11RIK | 1.7 | N | N | 1.0 | ENSMUSG00000042041 | ||
| SLC35F2 | 1.7 | N | N | 1.0 | ENSMUSG00000042195 | ||
| GRK3 | 1.0 | N | N | 1.0 | ENSMUSG00000042249 | ||
| FILIP1L | 0.6 | N | N | 1.0 | ENSMUSG00000043336 | ||
| BDH1 | 0.5 | N | N | 1.0 | ENSMUSG00000046598 | ||
| ARHGAP30 | 0.7 | N | N | 1.0 | ENSMUSG00000048865 | ||
| THEMIS | 1.8 | N | N | 1.0 | ENSMUSG00000049109 | ||
| AMZ1 | 1.4 | N | N | 1.0 | ENSMUSG00000050022 | ||
| SELENON | 0.7 | N | N | 1.0 | ENSMUSG00000050989 | ||
| PLEKHM3 | 0.8 | N | N | 1.0 | ENSMUSG00000051344 | ||
| WDFY4 | 0.9 | N | N | 1.0 | ENSMUSG00000051506 | ||
| TCEAL8 | 0.9 | N | N | 1.0 | ENSMUSG00000051579 | ||
| ZFP608 | 0.8 | N | N | 1.0 | ENSMUSG00000052713 | ||
| SLFN5 | 0.7 | N | N | 1.0 | ENSMUSG00000054404 | ||
| CLCA3A1 | 0.7 | N | N | 1.0 | ENSMUSG00000056025 | ||
| FAM105A | 0.8 | N | N | 1.0 | ENSMUSG00000056069 | ||
| PGM3 | 0.7 | N | N | 1.0 | ENSMUSG00000056131 | ||
| RDH9 | 1.2 | N | N | 1.0 | ENSMUSG00000056148 | ||
| GLIPR1 | 1.4 | N | N | 1.0 | ENSMUSG00000056888 | ||
| GM5431 | 0.8 | N | N | 1.0 | ENSMUSG00000058163 | ||
| TNFRSF19 | 0.8 | N | N | 1.0 | ENSMUSG00000060548 | ||
| H2-EB1 | 1.2 | N | N | 1.0 | ENSMUSG00000060586 | ||
| CD200R4 | 1.4 | N | N | 1.0 | ENSMUSG00000062082 | ||
| CLIP2 | 0.9 | N | N | 1.0 | ENSMUSG00000063146 | ||
| CD300LB | 1.6 | N | N | 1.0 | ENSMUSG00000063193 | ||
| SP140 | 1.0 | N | N | 1.0 | ENSMUSG00000070031 | ||
| CSF2RB2 | 1.1 | N | N | 1.0 | ENSMUSG00000071714 | ||
| 1810058I24RIK | 0.4 | N | N | 1.0 | ENSMUSG00000073155 | ||
| H2-AB1 | 1.0 | N | N | 1.0 | ENSMUSG00000073421 | ||
| IFI204 | 0.8 | N | N | 1.0 | ENSMUSG00000073489 | ||
| WIPF1 | 0.8 | N | N | 1.0 | ENSMUSG00000075284 | ||
| SLFN1 | 1.4 | N | N | 1.0 | ENSMUSG00000078763 | ||
| MS4A6C | 1.4 | N | N | 1.0 | ENSMUSG00000079419 | ||
| H2-DMB1 | 1.2 | N | N | 1.0 | ENSMUSG00000079547 | ||
| AI662270 | 0.8 | N | N | 1.0 | ENSMUSG00000087107 | ||
| ITPRIPL2 | 0.8 | N | N | 1.0 | ENSMUSG00000095115 | ||
| SOWAHC | 0.7 | N | N | 1.0 | ENSMUSG00000098188 | ||
| KCTD12 | 0.9 | N | N | 1.0 | ENSMUSG00000098557 |
logFC, logarithmic conversion of the fold change (FC).
References
- 1.Wong R.J., Aguilar M., Cheung R., Perumpail R.B., Harrison S.A., Younossi Z.M., Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y., Jung H.-S., Cho J., Zhang Y., Yun K.E., Lazo M., Pastor-Barriuso R., Ahn J., Kim C.-W., Rampal S., Cainzos-Achirica M., Zhao D., Chung E.C., Shin H., Guallar E., Ryu S. Metabolically healthy obesity and the development of nonalcoholic fatty liver disease. Am J Gastroenterol. 2016;111:1133–1140. doi: 10.1038/ajg.2016.178. [DOI] [PubMed] [Google Scholar]
- 3.Katsiki N., Mikhailidis D.P., Mantzoros C.S. Non-alcoholic fatty liver disease and dyslipidemia: an update. Metabolism. 2016;65:1109–1123. doi: 10.1016/j.metabol.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui M.S., Fuchs M., Idowu M.O., Luketic V.A., Boyett S., Sargeant C., Stravitz R.T., Puri P., Matherly S., Sterling R.K., Contos M., Sanyal A.J. Severity of nonalcoholic fatty liver disease and progression to cirrhosis are associated with atherogenic lipoprotein profile. Clin Gastroenterol Hepatol. 2015;13:1000–1008.e3. doi: 10.1016/j.cgh.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dongiovanni P., Rametta R., Meroni M., Valenti L. The role of insulin resistance in nonalcoholic steatohepatitis and liver disease development – a potential therapeutic target? Expert Rev Gastroenterol Hepatol. 2016;10:229–242. doi: 10.1586/17474124.2016.1110018. [DOI] [PubMed] [Google Scholar]
- 6.Baumeister S.E., Völzke H., Marschall P., John U., Schmidt C.O., Flessa S., Alte D. Impact of fatty liver disease on health care utilization and costs in a general population: a 5-year observation. Gastroenterology. 2008;134:85–94. doi: 10.1053/j.gastro.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Charlton M.R., Burns J.M., Pedersen R.A., Watt K.D., Heimbach J.K., Dierkhising R.A. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011;141:1249–1253. doi: 10.1053/j.gastro.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 8.Rinella M.E. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 9.Fujii H., Kawada N. Inflammation and fibrogenesis in steatohepatitis. J Gastroenterol. 2012;47:215–225. doi: 10.1007/s00535-012-0527-x. [DOI] [PubMed] [Google Scholar]
- 10.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., Mills P.R., Keach J.C., Lafferty H.D., Stahler A., Haflidadottir S., Bendtsen F. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulai P.S., Singh S., Patel J., Soni M., Prokop L.J., Younossi Z., Sebastiani G., Ekstedt M., Hagstrom H., Nasr P., Stal P., Wong V.W.-S., Kechagias S., Hultcrantz R., Loomba R. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. doi: 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleemann R., van Erk M., Verschuren L., van den Hoek A.M., Koek M., Wielinga P.Y., Jie A., Pellis L., Bobeldijk-Pastorova I., Kelder T., Toet K., Wopereis S., Cnubben N., Evelo C., van Ommen B., Kooistra T. Time-resolved and tissue-specific systems analysis of the pathogenesis of insulin resistance. PLoS One. 2010;5:e8817. doi: 10.1371/journal.pone.0008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang W., Lindeman J.H., Menke A.L., Koonen D.P., Morrison M., Havekes L.M., van den Hoek A.M., Kleemann R. Metabolically induced liver inflammation leads to NASH and differs from LPS- or IL-1β-induced chronic inflammation. Lab Invest. 2014;94:491–502. doi: 10.1038/labinvest.2014.11. [DOI] [PubMed] [Google Scholar]
- 14.Holmes WE, Angel TE, Li KW, Hellerstein MK. Chapter seven - dynamic proteomics: in vivo proteome-wide measurement of protein kinetics using metabolic labeling. In: Christian MM, ed. Methods in enzymology. Volume 561. Academic Press, Cambridge, MA, 2015:219–276. [DOI] [PubMed]
- 15.Decaris M.L., Emson C.L., Li K., Gatmaitan M., Luo F., Cattin J., Nakamura C., Holmes W.E., Angel T.E., Peters M.G., Turner S.M., Hellerstein M.K. Turnover rates of hepatic collagen and circulating collagen-associated proteins in humans with chronic liver disease. PLoS One. 2015;10:e0123311. doi: 10.1371/journal.pone.0123311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decaris M.L., Li K.W., Emson C.L., Gatmaitan M., Liu S., Wang Y., Nyangau E., Colangelo M., Angel T.E., Beysen C., Cui J., Hernandez C., Lazaro L., Brenner D.A., Turner S.M., Hellerstein M.K., Loomba R. Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology. 2017;65:78–88. doi: 10.1002/hep.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 18.Kupke I.R., Zeugner S. Quantitative high-performance thin-layer chromatography of lipids in plasma and liver homogenates after direct application of 0.5-microliter samples to the silica-gel layer. J Chromatogr. 1978;146:261–271. doi: 10.1016/s0378-4347(00)81892-7. [DOI] [PubMed] [Google Scholar]
- 19.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 20.Morrison M.C., Mulder P., Salic K., Verheij J., Liang W., van Duyvenvoorde W., Menke A., Kooistra T., Kleemann R., Wielinga P.Y. Intervention with a caspase-1 inhibitor reduces obesity-associated hyperinsulinemia, non-alcoholic steatohepatitis and hepatic fibrosis in LDLR-/-.Leiden mice. Int J Obes. 2016;40:1416–1423. doi: 10.1038/ijo.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang W., Menke A.L., Driessen A., Koek G.H., Lindeman J.H., Stoop R., Havekes L.M., Kleemann R., van den Hoek A.M. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9:e115922. doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:1–12. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decaris M.L., Gatmaitan M., FlorCruz S., Luo F., Li K., Holmes W.E., Hellerstein M.K., Turner S.M., Emson C.L. Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol Cell Proteomics. 2014;13:1741–1752. doi: 10.1074/mcp.M113.037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didangelos A., Yin X., Mandal K., Baumert M., Jahangiri M., Mayr M. Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price J.C., Holmes W.E., Li K.W., Floreani N.A., Neese R.A., Turner S.M., Hellerstein M.K. Measurement of human plasma proteome dynamics with 2H2O and liquid chromatography tandem mass spectrometry. Anal Biochem. 2012;420:73–83. doi: 10.1016/j.ab.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Ahrens M., Ammerpohl O., von Schönfels W., Kolarova J., Bens S., Itzel T., Teufel A., Herrmann A., Brosch M., Hinrichsen H., Erhart W., Egberts J., Sipos B., Schreiber S., Häsler R., Stickel F., Becker T., Krawczak M., Röcken C., Siebert R., Schafmayer C., Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 28.Verschuren L., Wielinga P.Y., Kelder T., Radonjic M., Salic K., Kleemann R., van Ommen B., Kooistra T. A systems biology approach to understand the pathophysiological mechanisms of cardiac pathological hypertrophy associated with rosiglitazone. BMC Med Genomics. 2014;7:1–11. doi: 10.1186/1755-8794-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 30.Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 31.Jacomy M., Venturini T., Heymann S., Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teufel A., Itzel T., Erhart W., Brosch M., Wang X.Y., Kim Y.O., von Schönfels W., Herrmann A., Brückner S., Stickel F., Dufour J.-F., Chavakis T., Hellerbrand C., Spang R., Maass T., Becker T., Schreiber S., Schafmayer C., Schuppan D., Hampe J. Comparison of gene expression patterns between mouse models of nonalcoholic fatty liver disease and liver tissues from patients. Gastroenterology. 2016;151:513–525.e0. doi: 10.1053/j.gastro.2016.05.051. [DOI] [PubMed] [Google Scholar]
- 33.Ibrahim S.H., Hirsova P., Malhi H., Gores G.J. Animal models of nonalcoholic steatohepatitis: eat, delete, and inflame. Dig Dis Sci. 2016;61:1325–1336. doi: 10.1007/s10620-015-3977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs A, Warda A-S, Verbeek J, Cassiman D, Spincemaille P. An overview of mouse models of nonalcoholic steatohepatitis: from past to present. Current protocols in mouse biology. John Wiley & Sons, Inc, Hoboken, NJ, 2016. [DOI] [PubMed]
- 35.Hebbard L., George J. Animal models of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2011;8:35–44. doi: 10.1038/nrgastro.2010.191. [DOI] [PubMed] [Google Scholar]
- 36.Buzzetti E., Pinzani M., Tsochatzis E.A. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Lemmer E.R., Friedman S.L., Llovet J.M. Molecular diagnosis of chronic liver disease and hepatocellular carcinoma: the potential of gene expression profiling. Semin Liver Dis. 2006;26:373–384. doi: 10.1055/s-2006-951604. [DOI] [PubMed] [Google Scholar]
- 38.Kim J.U., Cox I.J., Taylor-Robinson S.D. The quest for relevant hepatocellular carcinoma biomarkers. Cell Mol Gastroenterol Hepatol. 2017;4:283–284. doi: 10.1016/j.jcmgh.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van 't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A.M., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Schreiber G.J., Kerkhoven R.M., Roberts C., Linsley P.S., Bernards R., Friend S.H. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 40.Nwosu Z.C., Megger D.A., Hammad S., Sitek B., Roessler S., Ebert M.P., Meyer C., Dooley S. Identification of the consistently altered metabolic targets in human hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2017;4:303–323.e1. doi: 10.1016/j.jcmgh.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalling R.L., Delker D.A., Zhang Y., Nieto N., Mcguiness M.S., Liu S., Friedman S.L., Hagedorn C.H., Wang L. Genome-wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease. Am J Physiol Gastrointest Liver Physiol. 2013;305:G364–G374. doi: 10.1152/ajpgi.00077.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moylan C.A., Pang H., Dellinger A., Suzuki A., Garrett M.E., Guy C.D., Murphy S.K., Ashley-Koch A.E., Choi S.S., Michelotti G.A., Hampton D.D., Chen Y., Tillmann H.L., Hauser M.A., Abdelmalek M.F., Diehl A.M. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59:471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]