Abstract
The pretranslocation complex of the ribosome can undergo spontaneous fluctuations of messenger RNA and transfer RNAs (tRNAs) between classical and hybrid states, and occupation of the hybrid tRNA positions has been proposed to precede translocation. The classical and hybrid state tRNA positions have been extensively characterized when the ribosome is stalled along the messenger RNA by either the absence or delayed addition of elongation factor G (EF-G), or by the presence of antibiotics or GTP analogs that block translocation. However, during multiple ongoing elongation cycles when both EF-G and ternary complexes are present, EF-G can bind to the pretranslocation complex much faster than the timescale of the classic-hybrid transitions. Using single-molecule fluorescence resonance energy transfer between adjacent tRNAs and between A-site tRNA and ribosomal protein L11, we found that the tRNAs do not fluctuate between the hybrid and classical states, but instead adopt a position with fluorescence resonance energy transfer efficiencies between those of the stalled classical and hybrid states.
Introduction
During protein synthesis, transfer RNAs (tRNAs) successively occupy three positions on the ribosome: A (aminoacyl), P (peptidyl), and E (exit) sites. After an aminoacylated tRNA enters the A site, a new peptide bond is formed as the nascent peptide chain is transferred from the P-site tRNA to the A-site tRNA. The resulting pretranslocation (PRE) complex of the ribosome undergoes elongation factor G (EF-G)-catalyzed translocation (1, 2, 3, 4, 5) to a posttranslocation (POST) complex, which shifts the messenger RNA (mRNA) by one three-base codon relative to the ribosome, and moves the tRNAs from the A and P sites to the P and E sites, respectively. Binding of the next aminoacylated tRNA into the A site initiates a subsequent cycle of elongation. The PRE complex formed by ribosomes stalled in the absence of EF-G fluctuates between so-called “classical” and “hybrid” states (6, 7, 8).
In these earlier works, the structural distinction between classical and hybrid states was focused on the tRNA positions within the ribosome’s active site: “classical” tRNA positions referred to the A/A and P/P locations of bound tRNAs, where the first and second letters represent binding sites on the 30S and 50S subunits, respectively, and a canonical hybrid state, in which tilted tRNAs bound in mixed A/P and P/E positions. The hybrid state was proposed to be an intermediate between the classical PRE state and the POST complex in which the tRNAs are bound at P/P and E/E sites. Concurrent and subsequent structural studies showed that EF-G catalysis of tRNA-mRNA translocation was also accompanied by large-scale motions within the ribosome as a whole, including 6° (9) to 9° (10, 11) rotations of the 30S subunit with respect to the 50S subunit, and of the head region of the 30S subunit relative to its body (12, 13). These studies were supported by fluorescence resonance energy transfer (FRET) measurements in both single-molecule (14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24) and ensemble (25, 26) experiments. Further structural studies identified additional distinct states of the PRE complex containing tRNAs in somewhat different and/or intermediate locations from the canonical hybrid A/P and P/E sites, which have been identified as ap/ap:pe/E (27), ap/P:pe/E (28), and A/P∗:P/E (12).
It was proposed that tRNA tilting, rotations of the large and small subunits, and motions of protein L1 could be considered as coupled, defining two main global states: classical states with unrotated large and small subunits and nontilted tRNAs, and hybrid states with rotated subunits and tilted tRNAs (29). However, more recent evidence demonstrates that tRNA tilting, subunit rotations, and L1 stalk motions are temporally distinct processes (18, 25, 26, 30) that are not tightly coupled.
Before the work presented here, single molecule fluorescence resonance energy transfer (smFRET) studies that monitored changes in tRNA positions during EF-G-catalyzed translocation have been conducted mainly by adding EF-G to stalled PRE complexes, formed in the absence of EF-G, that were equilibrated between the classical and canonical hybrid states, with a significant fraction displaying reversible fluctuations between these two states (13, 21, 31, 32, 33, 34). These studies supported the notion that the classical-hybrid transition in PRE complexes plays an important role during translocation. One study that did observe multicodon elongation with EF-G present in the solution (22) observed extremely slow fluctuations in the PRE state, but monitored subunit rotations rather than tRNA positions.
Once EF-G binds to the ribosome, tRNA and subunit fluctuations diminish or stop (21, 22). Our recent study employing smFRET to monitor ongoing translation of a full-length protein gave results indicating that A-site-bound peptidyl-tRNA in PRE complexes did not fluctuate and had, at most, a minor fraction (<6%) in the hybrid position (35). This result raised the interesting question of whether the translocation pathway during ongoing polypeptide synthesis might differ in important ways from the translocation pathway resulting from EF-G addition to stalled PRE complexes. However, it did not resolve that question because it employed a time resolution similar to the measured rate of classical-hybrid tRNA fluctuations (which could have prevented their detection) and at one EF-G·GTP concentration.
In this work, we overcome these limitations in studying elongation in ribosomes that are not stalled. We use smFRET between two ribosome-bound tRNAs, and between a tRNA and L11, a ribosomal protein located near the A site, to monitor translocation during ongoing translation of a model polypeptide at much higher time resolution, at varying levels of EF-G·GTP, and during the synthesis of peptides of various lengths (dipeptide, tripeptide, hexapeptide, heptapeptide). These results show that an EF-G·GTP concentration of 2 μM is sufficient to rapidly place an incoming tRNA in a position between the canonical classical and hybrid states before its translocation into the P site. We discuss differences in our results from those of previous studies, and the implications of our work for translocation within cells.
Materials and Methods
In brief, 70S initiation complexes were formed using a 5′-biotinylated mRNA (Dharmacon RNAi Tech; sequences in Supporting Materials and Methods), and fluorescence-labeled ternary complexes (TCs) were preformed from elongation factor thermo unstable (EF-Tu), GTP, and charged tRNAs labeled with either Cy3 or Cy5 at dihydrouridine positions in the D loop (36). For stalled experiments, the unlabeled TCs, (as well as the labeled Phe TC in the particular case of tRNA-tRNA FRET experiments), 2 μM EF-G, and 3 mM GTP were added to the initiation complexes and incubated for 5 min, followed by immobilization of the resulting POST complexes on a streptavidin/biotin-PEG-coated glass surface (21, 37). Unbound reaction components including EF-G were then washed out of the channel. Labeled Val TC was then added in the absence of EF-G to form a new PRE complex, with image stacks recorded either during the addition or after washing away unbound labeled TCs. For ongoing experiments, the initiation complexes alone were immobilized on the slide, and all cognate TCs (labeled and unlabeled) were added in the presence of 2 μM EF-G and 3 mM GTP during recording.
Cy3 and Cy5 fluorescence intensities (allowing calculation of FRET between Cy3 and Cy5) were collected using the alternating laser excitation (ALEX) mode to switch between 532 nm and 640 nm lasers on an objective-type total internal reflection fluorescence microscope (21). Except as otherwise indicated, stalled experiments were recorded with ALEX at 35 ms integration time per frame, resulting in a net time resolution of 70 ms, whereas ongoing translation experiments were recorded solely with 532 nm excitation, giving 35 ms time resolution. Most translation experiments were carried out with buffer TAM15 (15 mM MgAc2, 50 mM Tris-HCl (pH 7.5), 30 mM NH4Cl, 70 mM KCl, and 1 mM DTT) at 23°C. More extensive details of materials preparation, the experimental setup, FRET measurements, alternative buffers, swapped labels, and data analysis are described in the Supporting Materials and Methods.
Results
Labeled complexes
We examined the dynamics of FRET between A-site tRNA and L11 or between adjacent tRNAs in the A- and P-sites, or P- and E-sites, during ongoing polypeptide elongation experiments when EF-G is continuously present from the start of elongation. For this purpose, we used ribosomes programmed with mRNA-6,7 (Table 1) and four fluorescence-labeling schemes: A) Val-tRNAVal(Cy3) and Cy5-L11 [denoted V7(Cy3)-L11(Cy5)], B) Phe-tRNAPhe(Cy5) and Val-tRNAVal(Cy3) [F6(Cy5)-V7(Cy3)], C) Phe-tRNAPhe(Cy3) and Val-tRNAVal(Cy5) [F6(Cy3)-V7(Cy5)], and D) Phe-tRNAPhe(Cy3) and Cy5-L11 [F6(Cy3)-L11(Cy5)]. Experiments on ribosomes programmed with mRNA-2,3 tested dynamics of FRET between A-site bound tRNA and L11 earlier in elongation with three labeling schemes: E) Val-tRNAVal(Cy3) and Cy5-L11 [V3(Cy3)-L11(Cy5)], F) Val-tRNAVal(Cy5) and Cy3-L11 [V3(Cy5)-L11(Cy3)], and G) and Phe-tRNAPhe(Cy3) and Cy5-L11 [F2(Cy3)-L11(Cy5)].
Table 1.
Summary of smFRET results
| mRNA | Labeling scheme | Fluorescent Labeling Scheme | FRET Pair | FRET Efficiencies |
(Eongoing – Ehybrid)/(Eclassical – Ehybrid) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stalled-PRE Classic | Stalled-PRE Hybrid | INTongoing | |||||||||||||||||
| mRNA-6,7 | A | AUG | UAU | UAU | UAU | UAU | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | V7(Cy3)-L11(Cy5) | 0.73 ± 0.01 (Fig 1C) | 0.49 ± 0.02 (Fig 1C) | 0.65 ± 0.01 (Fig 1F) | 0.67 |
| fMet | Tyr | Tyr | Tyr | Tyr | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| B | AUG | UAU | UAU | UAU | UAU | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | F6(Cy5)-V7(Cy3) | 0.68 ± 0.01 (Fig 2C) | 0.39 ± 0.01 (Fig 2C) | 0.58 ± 0.01 (Fig 2F) | 0.66 | |
| fMet | Tyr | Tyr | Tyr | Tyr | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| C | AUG | UAU | UAU | UAU | UAU | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | F6(Cy3)-V7(Cy5) | 0.73 ± 0.01 (Fig S1C) | 0.40 ± 0.01 (Fig S1C) | 0.57 ± 0.02 (Fig S1F) | 0.52 | |
| fMet | Tyr | Tyr | Tyr | Tyr | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| D | AUG | UAU | UAU | UAU | UAU | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | F6(Cy3)-L11(Cy5) | 0.73 ± 0.01 (Fig S2C) | 0.43 ± 0.01 (Fig S2C) | 0.69 ± 0.01 (Fig S2F) | 0.87 | |
| fMet | Tyr | Tyr | Tyr | Tyr | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| mRNA-2,3 | E | AUG | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | V3(Cy3)-L11(Cy5) | 0.73 ± 0.01 (Fig S1J) | 0.46 ± 0.02 (Fig S1J) | 0.66 ± 0.01 (Fig S1M) | 0.74 |
| fMet | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| F | AUG | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | V3(Cy5)-L11(Cy3) | 0.70 ± 0.01 (Fig S1Q) | 0.49 ± 0.03 (Fig S1Q) | 0.63 ± 0.02 (Fig S1T) | 0.67 | |
| fMet | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
| G | AUG | UUC | GUG | CGU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | UAU | F2(Cy3)-L11(Cy5) | 0.74 ± 0.01 (Fig S2L) | 0.43 ± 0.01 (Fig S2L) | 0.68 ± 0.01 (Fig S2O) | 0.81 | |
| fMet | Phe | Val | Arg | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | Tyr | |||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||||||
mRNA-6,7 and mRNA-2,3 displayed in the table were used to program unlabeled ribosomes (B and C), L11(Cy5) ribosomes (A, D, E, and G), and L11(Cy3) ribosomes (F) for smFRET experiments using the FRET pairs listed in the table, corresponding to the italicized (Cy5) or underlined (Cy3) codons in the sequences. PRE complexes were stalled by the absence of EF-G and used to form two-peaked FRET distributions fit with a double Gaussian function, giving the high and low FRET peaks corresponding to the classical and hybrid tRNA positions. The peak centers from aggregated data, as well as SEs of the peaks from multiple replicate experiments, are displayed in the table. Experiments were also conducted during ongoing translation in the presence of 2 μM EF-G, which yielded single-peaked FRET distributions of aggregate data, whose values are displayed in the table along with SEs from multiple replicate experiments. The single peaks (corresponding to INTongoing) had FRET efficiencies in between the stalled classical and hybrid values, as expressed in the ratio (Eongoing – Ehybrid)/(Eclassical – Ehybrid).
smFRET in stalled PRE complexes
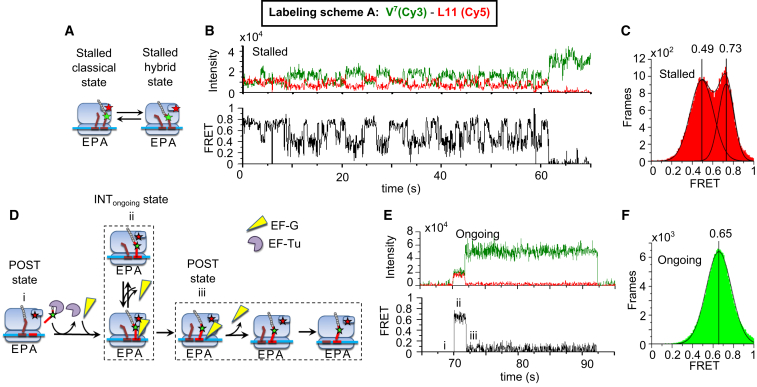
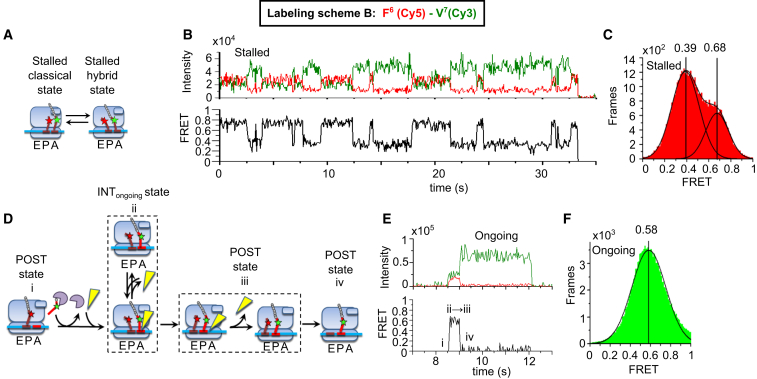
Stalled PRE complexes formed in the absence of EF-G·GTP displayed high and low values for both tRNA-L11 FRET and tRNA-tRNA FRET, associated with the classical and hybrid tRNA positions, respectively, consistent with previous work by ourselves and others (8, 21, 23, 31) and observed with labeling schemes A–G. To examine the stalled V7(Cy3)-L11(Cy5) (labeling scheme A, Fig. 1, A–C) and F6(Cy5)-V7(Cy3) (labeling scheme B, Fig. 2, A–C) Val-tRNAVal(Cy3) states, TC was added to a sample containing stalled POST-6 complexes (with P-site bound fMYYYYF-tRNAPhe) in the absence of EF-G, forming the stalled PRE-7 complex. FRET efficiency distribution for each FRET pair had two clear peaks that were fit with double Gaussian functions to provide the classical and hybrid FRET efficiencies for tRNA-L11 (0.73 ± 0.01 and 0.49 ± 0.02) and for tRNA-tRNA (0.68 ± 0.01 and 0.39 ± 0.01, Table 1, schemes A and B). Hidden Markov analysis of the fluctuating complexes, using HaMMy software (38), identified two states with essentially the same FRET values identified by fitting Gaussian components. Similar results were obtained for the stalled PRE-6, PRE-3, and PRE-2 complexes formed using labeling schemes D–G (Figs. S2–S6; Table 1). These classical and hybrid states had average dwell times in the range of 0.4–1.7 s (Table S1), measured by fitting exponential decays to their dwell time distributions (Fig. S7).
Figure 1.
Stalled and ongoing tRNA-L11 FRET measurements. L11(Cy5) ribosomes were programmed with mRNA-6,7 (Table 1) and underwent FRET with Val-tRNAVal(Cy3). (A) Schematic of classical/hybrid equilibrium of a stalled ribosome. The green and red stars represent Cy3 fluorophore and Cy5 fluorophore, respectively. (B) Representative FRET recording. (C) Frames of fluctuating stalled traces formed a two-peaked distribution fit with a double Gaussian function (n = 592 molecules). (D–F) 2 μM EF-G was present during ongoing translation from codons 1–7. (D) Reaction scheme for ongoing translation. (E) A representative trace with the following FRET states: i) the POST state after the ribosome has translated through the fifth (Tyr) and sixth (Phe) codons, with peptidyl-tRNAPhe residing in the P site; ii) INTongoing; and iii) the POST complex after translocation. (F) Frames of traces during ongoing translation formed a peaked PRE-state distribution fit with a single Gaussian component (n = 3664 molecules).
Figure 2.
Stalled and ongoing tRNA-tRNA FRET measurements. Unlabeled ribosomes were programmed with mRNA-6,7 (Table 1) and Phe-tRNAPhe(Cy5) at codon 6 underwent FRET with Val-tRNAVal(Cy3) at codon 7. (A) Schematic of classical/hybrid equilibrium of a stalled ribosome. The green and red stars represent Cy3 fluorophore and Cy5 fluorophore, respectively. (B) Representative FRET recording. (C) Frames of fluctuating stalled traces formed a two-peaked distribution fit with a double Gaussian function (n = 531 molecules). (D–F) 2 μM EF-G was present during ongoing translation from codons 1–7. (D) Reaction scheme for ongoing translation. The yellow triangle represents EF-G; the purple disc represents EF-Tu. (E) A representative trace with the following FRET states: i) the POST state after the ribosome has translated through the fifth (Tyr) and sixth (Phe) codons, with peptidyl-tRNAPhe residing in the P site; (ii, iii) INTongoing and the initial POST state, which have similar FRET efficiencies (21); and iv) the POST state after dissociation of tRNAPhe(Cy5) from the E-site. (F) Frames of traces during ongoing translation formed a peaked PRE state distribution fit with a single Gaussian component (n = 2359 molecules).
smFRET in PRE complexes formed during ongoing translation at 2 μM EF-G
tRNA-L11
In marked contrast to the results obtained with the stalled PRE-7 complex formed using V7(Cy3)-L11(Cy5) in the absence of EF-G, the PRE-7 complex formed in the presence of EF-G during ongoing translation after six elongation cycles displayed a single, dominant, high-FRET efficiency centered at 0.65 ± 0.01 (Fig. 1 F; Table 1, scheme A) on binding of Val-tRNAVal(Cy3) to the transient POST-6 complex (Fig. 1 E). This state was followed by a period with much lower FRET efficiency (<0.15), as expected for translocation of peptidyl-tRNAVal(Cy3) to the P site. No FRET signal was observed when Phe-tRNAPhe, cognate to codon 6, was omitted from the reaction mixture, demonstrating that the observed FRET events depended on translation from codon 1 to codon 7, which is cognate to Val-tRNAVal(Cy3).
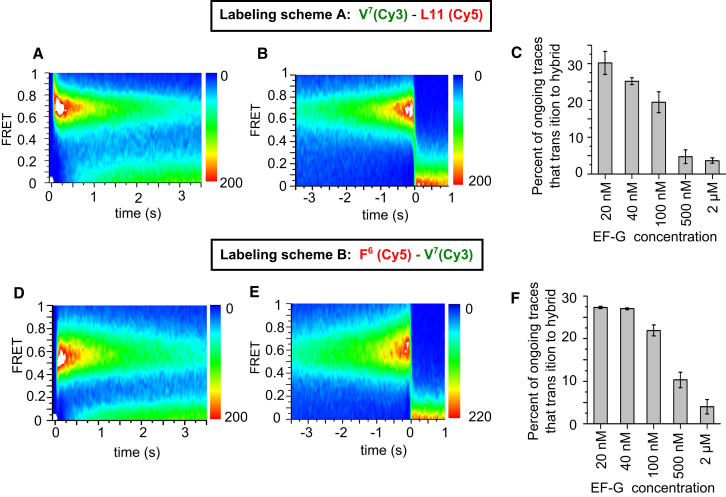
Importantly, contour plots derived from ongoing traces synchronized to the start (Fig. 3 A) or end (Fig. 3 B) of the high FRET events show a single, dominant state throughout their duration, with less than 6% of traces showing transitions to either the lower (0.49) or higher (0.73) efficiency characteristic of the stalled PRE-7 complex (Table S1). This low number of transitions implies a fluctuation rate slower than 0.03 s−1, rather than the 0.6–2.3 s−1 that we observed in stalled ribosomes (Fig. 1 B), consistent with earlier results of ours showing that binding of EF-G suppresses classical-hybrid fluctuations established in stalled PRE complexes by at least 5–60 fold (21, 39). In addition to this PRE-7 complex, measuring tRNA-L11 FRET during ongoing translation gave very similar results: almost no fluctuations for the PRE-6, PRE-3, and PRE-2 complexes earlier in elongation, with complexes formed using labeling schemes D–G (Figs. S2–S6; Table 1).
Figure 3.
Dynamics of tRNA-L11 and tRNA-tRNA FRET. (A–C) L11(Cy5) ribosomes were programmed with mRNA-6,7 (Table 1) and underwent FRET with Val-tRNAVal(Cy3). (D–F) Unlabeled ribosomes were programmed with mRNA-6,7 (Table 1), and Phe-tRNAPhe(Cy5) at codon 6 underwent FRET with Val-tRNAVal(Cy3) at codon 7, all in the presence of 2 μM EF-G during ongoing translation. FRET recordings were shifted in time to synchronize their beginnings ((A and D) presynchronized) or terminations ((B and E) postsynchronized) and overlaid to form contour plots. The color maps indicate the number of frames recorded at each FRET value and time. (C and F) Lowering EF-G concentration increased the percentage of fluctuating PRE complexes during ongoing translation; mean ± SE.
The nonfluctuating FRET state (which we term INTongoing) was also clearly shown in histograms formed from the average FRET efficiencies of FRET events in each trace during ongoing synthesis (Fig. S8, A and B, rather than from each movie frame as in Figs. 1, C and F and 2, C and F), which provides a complementary method of analyzing the data. Although histograms from frames (rather than event averages) better reflect the measurements' signal/noise ratios, the histograms of event averages reduce the frame-to-frame noise that might have caused an artefactual merger of FRET distributions arising from a wide bimodal population of PRE complexes. However, the peak centers remain the same, indicating that is not the case. In addition, negative cross-correlation of donor and acceptor intensities is a mark of fluctuating distance between tRNA and L11 in stalled PRE complexes. Lack of this negative cross-correlation in the PRE-7 complex formed during ongoing polypeptide synthesis provided additional evidence for the lack of distance fluctuations at the 35 ms camera timescale, and is further supported by time lag analysis of the decay of the stalled and ongoing cross-correlation signals (Fig. S8 E; Table S4).
The brief FRET events during ongoing translation (Fig. 1 E; Fig. S3 E, S4 E, S5 E, and S6 E) terminated upon translocation and dissociation of E-site tRNA. Duration histograms of the PRE states are shown in Fig. S7 and Table S2. In principle, the disappearance of FRET might also occur because of photobleaching of the Cy5 acceptor fluorophore. To check whether Cy5 photobleaching was a substantial contributor to the termination of the FRET events, a control experiment was performed alternating the excitation wavelength between 532 nm (to excite the Cy3 probes and measure FRET) and 640 nm to directly excite Cy5 (ALEX excitation, Fig. S1, A–D) (40). This procedure showed that the majority (>83%) of the FRET events at 2 μM EF-G terminated because of translocation rather than photobleaching (see Supporting Materials and Methods, Photobleaching). Because ALEX results in a two-fold decrease in time resolution of the experiment, subsequent data were collected without ALEX to obtain higher time resolution adequate for detecting fast fluctuations or transitions.
To distinguish whether the dominant single tRNA-L11 FRET state in ongoing translation (INTongoing) results from immediate binding of the TC to the previous POST state or from the presence of EF-G when the TC binds, we paused elongation at the POST-6 complex in the V7(Cy3)-L11(Cy5) experiment by waiting 5 min before injecting Val-tRNAVal(Cy3)·EF-Tu·GTP and EF-G·GTP together. We found the pause to have essentially no effect on the FRET results obtained, indicating that suppression of fluctuations and the intermediate FRET efficiency during ongoing translation do not arise as a consequence of the short lifetime of the POST-6 complex, but rather depend on EF-G being present when Val-TC is added to form PRE-7 (Fig. S1, E–G). Thus, when EF-G is present before PRE complex formation, the canonical classical-hybrid transitions found in stalled PRE complexes (Fig. 1, A–C) are almost completely absent (Table S3).
When FRET recordings were initiated before the addition of TC in the absence of EF-G, the initial PRE state was mainly classical and the partition between classical and hybrid emerged gradually during the next 5–15 s (Fig. S8, I–K). This contrasts strongly with the time course and distribution of FRET values in the presence of EF-G, which entered the state with FRET efficiency in between classical and hybrid values within 100 ms (Fig. 1; Fig. S3– S6).
tRNA-tRNA
The PRE-7 complex formed in the presence of EF-G during ongoing translation using F6(Cy5)-V7(Cy3), labeling scheme B, displays a single, dominant FRET efficiency (Fig. 2, D–F; Table 1, scheme B) that is in between the values found for the classical and hybrid tRNA positions, with minimal detectable fluctuations (Table S3), paralleling the tRNA-L11 results for INTongoing reported above using V7(Cy3)-L11(Cy5). As expected, FRET efficiency declines and becomes zero after translocation and dissociation of tRNAPhe from the ribosome. That the tRNA-tRNA FRET efficiency in the transient POST-7 complex before tRNAPhe dissociation is indistinguishable from the FRET efficiency of the corresponding PRE-7 complex is not surprising. We have previously shown that the tRNA-tRNA FRET efficiency in a POST complex formed on addition of EF-G to a stalled PRE complex has an intermediate value between those found for the classical and hybrid PRE complexes (21).
As mentioned, ALEX experiments ruled out a major component of Cy5 photobleaching in tRNA-L11 experiments. For tRNA-tRNA experiments, we could provide an additional safeguard against premature photobleaching by swapping the labeling positions. In labeling scheme C, F6(Cy3)-V7(Cy5) (Fig. S3, D and E), binding of tRNAVal(Cy5) caused reduction of Cy3 fluorescence due to energy transfer, and both Cy3 and Cy5 fluorescence decreased after translocation and dissociation of the tRNAPhe(Cy3). Photobleaching of the Cy5 after formation of the PRE state would have caused return of the high Cy3 fluorescence intensity, in contrast to its observed disappearance. Thus, the essentially identical results of FRET efficiency and virtually no fluctuations obtained using F6(Cy5)-V7(Cy3) (scheme B) and F6(Cy3)-V7(Cy5) (scheme C, Fig. S3; Table 1, scheme C) show that the stable PRE FRET state terminates upon translocation and dissociation of the E-site tRNA.
As discussed for tRNA-L11 FRET, event histograms for tRNA-tRNA FRET also exclude a merged bimodal population of classical and hybrid states during ongoing translation (Fig. S8, C and D), and cross-correlation analysis excludes fluctuations at or slower than the 35 ms frame rate (Fig. S8 G). The nonfluctuating INTongoing conformation was also observed in experiments with a commonly used alternative buffer containing a lower (4.5 mM) Mg2+ concentration and polyamines (Supporting Materials and Methods (41)), thus it is not unique to the TAM15 buffer (Fig. S9).
PRE complex dwell times during ongoing translation
In the presence of 2 μM EF-G, all of the FRET pairs examined had dwell time distributions with both fast and slow phases, similar to what has been observed by others (24, 42). Both phases had the same intermediate FRET value for all complexes studied. Dwell times for the fast phase (110–510 ms) and slow phase (1.2–2.5 s) (Fig. S7; Table S2), correspond to translocation rates of 2–9 s−1 and 0.4–0.8 s−1, respectively, measured at 23°C. The fast-phase rates are reasonably consistent with in vivo rates of protein synthesis (12–21 s−1) measured at 37°C (43), because the 14°C increase in temperature should result in ≥5-fold increase in translation rate, based on ensemble reconstituted, cell-free protein synthesis rates measured at 25°C (0.3 s−1 (44)) and 37°C (1.5 s−1 (45)). These rates are comparable to those previously reported for single-molecule reconstituted experiments: 0.57 s−1 at 30°C (35), 0.45 s−1 at room temperature (46), and 0.1−1 s−1 at room temperature (47).
smFRET in PRE-7 complexes formed during ongoing translation at decreasing EF-G
The experiments displayed in Figs. 1, 2, and S2–S6 demonstrate nearly complete absence of fluctuations or classical-hybrid transitions (Table S1) in the presence of 2 μM EF-G. The proportion of ribosomes exhibiting fluctuations increased as EF-G concentration was decreased to 20 nM (Fig. 3, C and F), which is attributable to delayed binding of EF-G to the PRE complex at low EF-G, thereby permitting temporary partitioning between the classical and hybrid positions. This interpretation is supported by the dynamics of those complexes that did fluctuate, which showed similar gradual establishment of classical-hybrid FRET values and fluctuations (Fig. S10). Although fluctuating PRE complexes at low EF-G concentrations translocated from either classical or hybrid positions as previously observed (21, 22, 23), the proportion of translocation from the hybrid tRNA positions increased as EF-G concentration was lowered (Fig. S10).
Discussion
Here we demonstrate that the presence of EF-G during ongoing polypeptide synthesis, even at a concentration (2 μM) well below that present in the bacterial cell [10–20 μM (48, 49)], is sufficient to suppress fluctuations of tRNAs within PRE complexes during translocation. A single, dominant PRE-state intermediate during ongoing translation, termed INTongoing, is detected by either tRNA-L11 or tRNA-tRNA FRET (Table 1). We further show that such suppression is common within PRE-2 and PRE-3 complexes shortly after polypeptide synthesis is initiated, and within PRE-6 and PRE-7 complexes when the ribosome has moved out of the initial phase (50, 51). tRNA-L11 and tRNA-tRNA FRET fluctuations appear as EF-G concentration is reduced below 500 nM, until between 20 and 40 nM EF-G, these fluctuations mimic those seen previously in stalled complexes (14, 19, 21, 30, 31, 33) and here. This negative dependence of active site fluctuations on EF-G concentration provides strong evidence that such fluctuation will only occur if there is sufficient time between PRE complex formation on binding of the cognate TC and binding of EF-G to initiate translocation. At 2 μM, EF-G binds to the ribosome at a rate of ∼60–300 s−1 (52, 53), suggesting that the onset of active site fluctuations in stalled PRE complexes requires times greater than 15 ms before EF-G binding.
A possible caveat to the conclusion that EF-G suppresses fluctuations and induces formation of a stable intermediate FRET state during translocation would be that bound EF-G strongly accelerates active site fluctuations so that, rather than occurring at rates of 0.5–2 s−1 (14, 19, 21, 30, 31, 33), they occur at rates markedly faster than our experimental frame rate of 35 ms. If this were true, our FRET results (Table 1) during ongoing synthesis would correspond to two or more rapidly equilibrating states, rather than to a single state. Although we cannot formally rule out this possibility, the cross-correlation analysis of Fig. S8 indicates that such an acceleration would require vastly different dynamics than have ever been observed in a stalled ribosome.
In earlier studies, Sharma et al. (25, 26) have shown that saturating EF-G accelerates the counterclockwise rotation of the 30S subunit with respect to the 50S subunit within the PRE complex by 5-fold, affording a rate constant of 12–14 s−1 under conditions (22°C, 15 mM Mg2+) closely approximating our own. As mentioned, the putative tRNA-tRNA and tRNA-L11 fluctuations would have to proceed considerably more rapidly for us not to detect them. However, recent evidence strongly suggests that rotation of the 30S subunit with respect to the 50S subunit, or of the head region of the 30S subunit relative to the body, are both uncoupled from (and proceed more rapidly than) tRNA motions in either the stalled or translocating PRE complex (25, 26). Because subunit rotations do not necessarily require fluctuations of the tRNA positions, they would be consistent with the stable intermediate tRNA conformation we observed.
This hypothesis raises the question of whether INTongoing corresponds to a known or novel structure. The tRNA-tRNA FRET efficiency values reported in Table 1, 0.57 and 0.58, lead to apparent distances of ∼53 Å between the Cy3 and Cy5 dyes bound to the two tRNA D loops. These values are compatible with D-loop:D-loop tRNA-tRNA distances of 46–50 Å observed in three high-resolution structures of ribosomal complexes containing two tRNAs and bound EF-G before formation of the POST state (12, 27, 28). Those complexes were stabilized by the antibiotics neomycin and/or fusidic acid in conformations corresponding to neither the canonical classical or hybrid states formed in the absence of EF-G. An intermediate tRNA-tRNA FRET state was also formed on addition of EF-G to a stalled PRE complex in (24).
In contrast, the tRNA-L11 FRET efficiencies (0.63–0.69, Table 1) of INTongoing correspond to distances of 49–51 Å between the Cy3 and Cy5 dyes bound to the peptidyl-tRNA D loop and position 87 of L11, which are inconsistent with distances of 77–81 Å estimated from the chimeric or hybrid state structures cited above (12, 27, 28). Instead, INTongoing tRNA-L11 FRET efficiencies fall in between those found for the stalled classical and hybrid PRE complexes (but closer to the classical value). They are inconsistent with a putative translocation state identified in an L11-peptidyl-tRNA FRET study of Adio et al. (23), which has a FRET efficiency of 0.4, considerably lower than the FRET efficiencies determined for the stalled classical (0.8) and hybrid (0.6) complexes. In that study, for which translocation was initiated from a stalled PRE complex, no transient state was demonstrated on addition of native EF-G and GTP, but the 0.4 FRET state was demonstrated using a variety of protocols that stabilized translocation states, including the use of lower activity mutant variants of EF-G, the replacement of GTP by the nonhydrolyzable GTP analog GTPγS, and the addition of fusidic acid. This approach contrasts quite sharply with our use of native EF-G and GTP during ongoing translation in the absence of antibiotics. In addition, we labeled L11 at position 87, within the C-terminal domain, which interacts strongly with 23S ribosomal RNA (54, 55), whereas Adio et al. labeled position 38 in the more flexible N-terminal domain that is held less tightly within the 50S structure, and may be freer to move during translocation.
Other than the continuous presence of EF-G during ongoing translation in the experiments presented here, it is unclear which of these further differences contribute to the discrepancy between our tRNA-L11 results and these other reports. The disparity between tRNA-L11 smFRET results summarized in Table 1 and the prior structural and smFRET results discussed above suggests that INTongoing has tRNA positions similar to the chimeric states, but a tRNA-L11 distance that has not been previously observed in structural or single molecule studies. We speculate that it most likely corresponds to an early stage of the translocation process that precedes the larger separation between L11 and peptidyl tRNA that has been seen in earlier work. On the other hand, given the known mobility of L11 within the ribosome structure (56), we cannot exclude the possibility that INTongoing corresponds to a later stage of translocation involving a larger displacement of the C-terminal domain of L11 from the positions identified in previous reports.
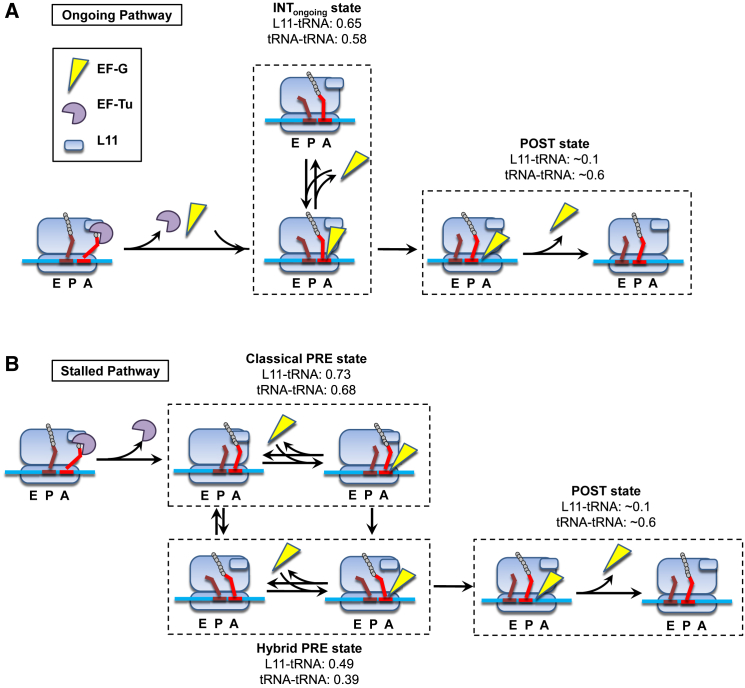
Fig. 4 A is a scheme for ongoing protein synthesis that illustrates the main conclusions we draw from our results. During ongoing protein synthesis in the presence of ≥2 μM EF-G, cognate aminoacyl-tRNA binds transiently to an A/T site (Figs. S3, D, E, and H, and S6, D, E, and H) before moving directly to the INTongoing state without detectable sampling of the canonical classical and hybrid states characteristic of stalled PRE complexes. It then translocates into the P site. EF-G is known to undergo rapid, reversible, nonproductive binding events before the final association that results in translocation (52, 53, 57), leading to the inclusion of the reversible dissociation and rebinding of EF-G to the PRE state in Fig. 4 A. The rapid phase of PRE-state dwell times, 110–510 ms (Table S2), is comparable to the average 200 ms residence time of EF-G on the ribosome during the final binding event that catalyzes translocation (57) at this temperature, indicating that the rapidly translocating ribosomes experience limited numbers of nonproductive interactions. Such interactions should be much more prevalent for the slowly translocating subpopulation having dwell times of 1.2–2.5 s. Despite this difference, rapidly and slowly translocating ribosomes had the same intermediate FRET value for each FRET pair, suggesting that the FRET value of INTongoing results from EF-G interacting with the PRE complex during both rapid on-off events and the productive final interaction. This is in accord with earlier work demonstrating the effects of transient EF-G binding on ribosome dynamics (21).
Figure 4.
Schematics of divergent translocation pathways. In the ongoing translation pathway (A), newly formed PRE complexes occupy INTongoing in the presence of EF-G, and quickly translocate. In the stalled pathway (B), relaxation of newly formed PRE complexes to a reversible distribution between stalled classical and hybrid tRNA positions in the absence of EF-G or at low EF-G concentrations. EF-G can interact with either state to promote translocation.
We note that our study of the tRNA positions does not address large and small subunit rotations or fluctuations of their relative positions, which have been observed in many structural studies. As mentioned, the structural studies have required inhibitors or absence of EF-G to pause or stall translation. Chen et al. (22) have reported smFRET signals that monitored subunit rotations and fluctuations during ongoing translation at 200–500 nM EF-G, but these fluctuations were extremely slow, with dwell times greater than 10 s. Presumably, the very slow rotational fluctuations result from delayed EF-G binding or dramatic slowing by EF-G. Thus EF-G may also prevent the initiation of subunit rotational fluctuations. Given the high concentrations of EF-G in the cell, the translocation pathway of the tRNAs including INTongoing (Fig. 4 A) is likely to be the dominant one, physiologically. However, an alternative pathway in which peptidyl-tRNA fluctuates between canonical classical and hybrid states in PRE complexes before translocation (Fig. 4 B) could be physiologically important when local EF-G concentration within the cell is atypically low.
Author Contributions
R.M.J. performed experiments and analyzed data. R.M.J., C.C., B.S.C., and Y.E.G. designed research and wrote the article.
Acknowledgments
This work was supported by National Institutes of Health grants GM080376 and GM118139 to B.S.C. and Y.E.G. C.C. was supported by an American Heart Association Postdoctoral Fellowship (12POST8910014). R.M.J. was supported by a National Institutes of Health Predoctoral Fellowship (F30AI114187).
Editor: Karin Musier-Forsyth.
Footnotes
Supporting Materials and Methods, ten figures, and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30981-5.
Contributor Information
Chunlai Chen, Email: chunlai@biomed.tsinghua.edu.cn.
Barry S. Cooperman, Email: cooprman@pobox.upenn.edu.
Yale E. Goldman, Email: goldmany@upenn.edu.
Supporting Material
References
- 1.Cukras A.R., Southworth D.R., Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol. Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 2.Fredrick K., Noller H.F. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 3.Gavrilova L.P., Kostiashkina O.E., Spirin A.S. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 4.Gavrilova L.P., Spirin A.S. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett. 1971;17:324–326. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- 5.Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem. 1969;244:1533–1539. [PubMed] [Google Scholar]
- 6.Bretscher M.S. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. doi: 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 7.Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard S.C., Kim H.D., Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 10.Dunkle J.A., Wang L., Cate J.H.D. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan S., Donohue J.P., Noller H.F. Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. USA. 2014;111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brilot A.F., Korostelev A.A., Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. USA. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer N., Konevega A.L., Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 14.Fei J., Kosuri P., Gonzalez R.L., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Munro J.B., Altman R.B., Blanchard S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fei J., Bronson J.E., Gonzalez R.L., Jr. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc. Natl. Acad. Sci. USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fei J., Richard A.C., Gonzalez R.L., Jr. Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat. Struct. Mol. Biol. 2011;18:1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munro J.B., Altman R.B., Blanchard S.C. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish P.V., Ermolenko D.N., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornish P.V., Ermolenko D.N., Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C., Stevens B., Cooperman B.S. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Petrov A., Puglisi J.D. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adio S., Senyushkina T., Rodnina M.V. Fluctuations between multiple EF-G-induced chimeric tRNA states during translocation on the ribosome. Nat. Commun. 2015;6:7442. doi: 10.1038/ncomms8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasserman M.R., Alejo J.L., Blanchard S.C. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belardinelli R., Sharma H., Rodnina M.V. Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 2016;23:342–348. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- 26.Sharma H., Adio S., Rodnina M.V. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Reports. 2016;16:2187–2196. doi: 10.1016/j.celrep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J., Lancaster L., Noller H.F. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramrath D.J.F., Lancaster L., Spahn C.M.T. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank J., Gonzalez R.L., Jr. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu. Rev. Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munro J.B., Altman R.B., Blanchard S.C. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc. Natl. Acad. Sci. USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H.D., Puglisi J.D., Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro J.B., Altman R.B., Blanchard S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B., Ho J., Lin Q. A microfluidic approach for investigating the temperature dependence of biomolecular activity with single-molecule resolution. Lab Chip. 2011;11:274–281. doi: 10.1039/c0lc00157k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ning W., Fei J., Gonzalez R.L., Jr. The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc. Natl. Acad. Sci. USA. 2014;111:12073–12078. doi: 10.1073/pnas.1401864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenblum G., Chen C., Cooperman B.S. Quantifying elongation rhythm during full-length protein synthesis. J. Am. Chem. Soc. 2013;135:11322–11329. doi: 10.1021/ja405205c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur J., Raj M., Cooperman B.S. Fluorescent labeling of tRNA dihydrouridine residues: mechanism and distribution. RNA. 2011;17:1393–1400. doi: 10.1261/rna.2670811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKinney S.A., Joo C., Ha T. Analysis of single-molecule FRET trajectories using hidden markov modeling. Biophys. J. 2006;19:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C., Greenberg M.J., Shuman H. Kinetic schemes for post-synchronized single molecule dynamics. Biophys. J. 2012;102:L23–L25. doi: 10.1016/j.bpj.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapanidis A.N., Laurence T.A., Weiss S. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 41.Dinos G., Kalpaxis D.L., Nierhaus K.H. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Pulk A., Blanchard S.C. Allosteric control of the ribosome by small-molecule antibiotics. Nat. Struct. Mol. Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young R., Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem. J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi S., Tsuji K., Okahata Y. Traveling Time of a translating ribosome along messenger RNA monitored directly on a quartz crystal microbalance. J. Am. Chem. Soc. 2012;134:6793–6800. doi: 10.1021/ja300993d. [DOI] [PubMed] [Google Scholar]
- 45.Underwood K.A., Swartz J.R., Puglisi J.D. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol. Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 46.Wen J.-D., Lancaster L., Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uemura S., Aitken C.E., Puglisi J.D. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurland C.G., Hughes D., Ehrenberg H. American Society for Microbiology; Washington DC: 1995. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. [Google Scholar]
- 49.Munishkin A., Wool I.G. The ribosome-in-pieces: binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc. Natl. Acad. Sci. USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenson T., Hauryliuk V. Does the ribosome have initiation and elongation modes of translation? Mol. Microbiol. 2009;72:1310–1315. doi: 10.1111/j.1365-2958.2009.06741.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen C., Stevens B., Goldman Y.E. Allosteric vs spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc. Natl. Acad. Sci. USA. 2011;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katunin V.I., Savelsbergh A., Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 53.Seo H.-S., Kiel M., Cooperman B.S. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004;43:12728–12740. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- 54.Wimberly B.T., Guymon R., Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 55.Conn G.L., Draper D.E., Gittis A.G. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 56.Kavran J.M., Steitz T.A. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 2007;371:1047–1059. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 57.Chen C., Cui X., Goldman Y.E. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. USA. 2016;113:7515–7520. doi: 10.1073/pnas.1602668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.