Abstract
Cellular pathways controlling chemotaxis, growth, survival, and oncogenesis are activated by receptor tyrosine kinases and small G-proteins of the Ras superfamily that stimulate specific isoforms of phosphatidylinositol-3-kinase (PI3K). These PI3K lipid kinases phosphorylate the constitutive lipid phosphatidylinositol-4,5-bisphosphate (PIP2) to produce the signaling lipid phosphatidylinositol-3,4,5-trisphosphate (PIP3). Progress has been made in understanding direct, moderate PI3K activation by receptors. In contrast, the mechanism by which receptors and Ras synergistically activate PI3K to much higher levels remains unclear, and two competing models have been proposed: membrane recruitment versus activation of the membrane-bound enzyme. To resolve this central mechanistic question, this study employs single-molecule imaging to investigate PI3K activation in a six-component pathway reconstituted on a supported lipid bilayer. The findings reveal that simultaneous activation by a receptor activation loop (from platelet-derived growth factor receptor, a receptor tyrosine kinase) and H-Ras generates strong, synergistic activation of PI3Kα, yielding a large increase in net kinase activity via the membrane recruitment mechanism. Synergy requires receptor phospho-Tyr and two anionic lipids (phosphatidylserine and PIP2) to make PI3Kα competent for bilayer docking, as well as for subsequent binding and phosphorylation of substrate PIP2 to generate product PIP3. Synergy also requires recruitment to membrane-bound H-Ras, which greatly speeds the formation of a stable, membrane-bound PI3Kα complex, modestly slows its off rate, and dramatically increases its equilibrium surface density. Surprisingly, H-Ras binding significantly inhibits the specific kinase activity of the membrane-bound PI3Kα molecule, but this minor enzyme inhibition is overwhelmed by the marked enhancement of membrane recruitment. The findings have direct impacts for the fields of chemotaxis, innate immunity, inflammation, carcinogenesis, and drug design.
Introduction
Receptor-Ras-PI3K-PIP3 signaling is central to an array of essential pathways. Localized PIP3 signals are generated at the leading-edge membrane of chemotaxing cells, including leukocytes migrating toward a site of infection or inflammation (1). PIP3 signals also play central roles in cell growth and survival pathways (2). Many, perhaps most, human cancers are linked to excessive PIP3 production arising from oncogenic mutations in receptor, Ras, or PI3K components (2, 3, 4, 5, 6).
Previous mechanistic studies have shown that receptor tyrosine kinase (RTK) activation of the dominant class IA PI3K family modulates the interactions between the two subunits of the PI3K heterodimer (5, 7, 8, 9, 10). This modulation occurs when receptor phospho-Tyr residues, located in a flexible cytoplasmic loop, bind to one or both inhibitory SH2 domains of the p85 regulatory subunit (Fig. 1). The resulting phospho-Tyr binding displaces the SH2s from the p110 catalytic subunit, triggering a conformational change that exposes lipid binding surfaces and activates the kinase domain, thereby generating modest levels of membrane binding and kinase activity.
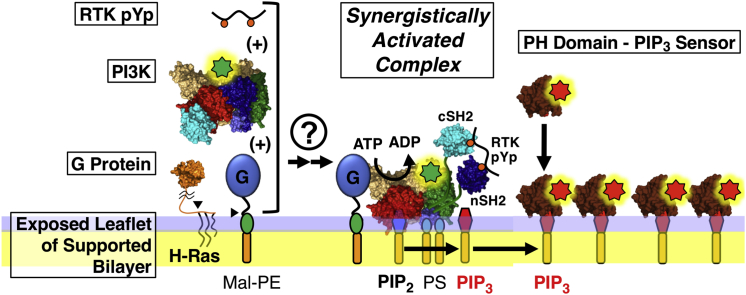
Figure 1.
Molecular schematic of the PI3K complex synergistically activated by receptor and G-protein in the single-molecule kinase activity assay. Shown is a schematic depiction of the fully active PI3Kα complex assembled on the supported lipid bilayer. In this complex, PI3Kα is synergistically activated by both 1) an RTK-derived phospho-Tyr peptide (RTK-pYp) based on the activation loop of the platelet-derived growth factor receptor and phosphorylated at specific Tyr residues, and 2) a small G-protein (H-Ras) anchored to maleimide lipid. The arrowhead indicates native Cys 181, which is a palmitoylation site in wild-type H-Ras and is sufficient for functional, covalent coupling to maleimide lipid in the in vitro single-molecule kinase assay. Single-molecule assays monitor stable docking of fluorescent PI3Kα molecules (green star) to the membrane, and PI3Kα-catalyzed production of the product PIP3 signaling lipid. Each product PIP3 molecule is detected by the binding of fluorescent GRP PH domain (red star) added in excess as a PIP3 sensor. The assay monitors the initial rate of PIP3 production when <2% of the substrate PIP2 population is converted to PIP3. At these low levels of PIP3, product rebinding to PI3Kα is negligible; thus, sequestration of the product PIP3 by the sensor protein is non-perturbing. To see this figure in color, go online.
In cells and in vitro, receptors and G-proteins have been observed to act in concert to synergistically stimulate PI3Ks and PIP3 production (11, 12, 13). In cells, some stimuli may simultaneously activate multiple, parallel G-protein responses that stimulate multiple PI3K populations to produce additive PIP3 signals, making it difficult to ascertain whether synergy requires direct, simultaneous binding of receptor and G-protein to PI3K (12, 13). In vitro, it has recently been demonstrated that the simultaneous presence of receptor phospho-Tyr and G-protein does indeed generate synergistic activation of class IA PI3Ks (11), consistent with the simultaneous binding of both ligands to PI3K at their distinct binding sites previously identified by crystallographic studies (6, 10, 14).
Two competing models have been proposed for the molecular mechanism by which G-protein amplifies the modest PI3K activation triggered by receptor alone to generate dramatic, synergistic activation. In the membrane recruitment model, G-protein enhances PI3K binding to the membrane, yielding increased membrane density of the active lipid kinase (15, 16). In the enzyme activation model, G-protein increases the specific activity (or turnover number) of each membrane-bound, G-protein-associated, kinase molecule (14, 17). The two mechanisms are not mutually exclusive, so one could dominate or both could contribute. Current evidence is inconclusive. For example, Ras isoforms do recruit PI3K to the membrane in cells (15). On the other hand, Ras binding does trigger PI3K conformational changes detected in crystallographic and hydrogen-deuterium exchange studies that could, in principle, modulate its enzyme activity (11, 14). Previous studies take opposite sides or conclude that the issue is an open question (11, 13, 14, 16). To date, the two models have not been resolved for any G-protein-PI3K regulatory pair.
To address these mechanistic questions, this study employs single-molecule total internal reflection fluorescence microscopy (TIRFM) to investigate a representative receptor-Ras-PI3K-PIP3 signaling module reconstituted from four protein/peptide components and two lipid components on a supported lipid bilayer: 1) A widely employed soluble phospho-Tyr peptide (pYp) is used to mimic the flexible, regulatory phospho-Tyr loop of an RTK receptor (7, 8, 9, 10, 18, 19). This peptide possesses phospho-Tyr at one or both conserved phosphorylation sites and is derived from the platelet-derived growth factor receptor, an RTK central to chemotaxis and inflammation. 2) The membrane-anchored, small G-protein H-Ras is employed as a representative Ras isoform. H-Ras is central to leukocyte transmigration and is linked to at least 65 oncogenic mutations (http://cancer.sanger.ac.uk/cosmic; https://www.cancer.gov/research/key-initiatives/ras/about) (20, 21). More broadly, mutations in this and other Ras isoforms are linked to >25% of human cancers (http://cancer.sanger.ac.uk/cosmic; https://www.cancer.gov/research/key-initiatives/ras/about) (17). 3) PI3Kα is the most prevalent and oncogenic PI3K isoform (6, 22). This heterodimeric lipid kinase is composed of catalytic (p110α) and regulatory (p85α) subunits, and is activated by both RTKs and Ras isoforms (6, 7). PI3Kα is involved in leukocyte chemotaxis and inflammation (1, 23) and is linked to 255 oncogenic mutations (http://cancer.sanger.ac.uk/cosmic; https://www.cancer.gov/research/key-initiatives/ras/about) (3, 15, 22, 24). 4–6) The anionic lipids phosphatidylserine (PS) and PIP2 serve as PI3Kα lipid binding targets; the latter is also the substrate for PIP3 production (7, 25). 7) General receptor for phosphoinositide (GRP1) pleckstrin homology (PH) domain is a high-affinity PIP3 sensor employed to specifically detect product PIP3 even in the presence of high PIP2 densities (26).
The findings reveal strong, direct synergy between simultaneous receptor phospho-Tyr and H-Ras activators. As previously observed, receptor phospho-Tyr residues are required for modest activation of PI3Kα lipid binding and PIP3 production (9, 10). When membrane-bound H-Ras is also present, the resulting receptor-G-protein synergy drives much stronger PI3Kα activation, and the findings show that this synergy is generated by the membrane recruitment mechanism. The enzyme activation mechanism does not contribute to this activation; instead, the findings reveal that H-Ras binding significantly inhibits the specific enzyme activity of membrane-bound PI3Kα.
Methods
Reagents
Except where noted otherwise, fluors, synthetic lipids, synthetic phospho-Tyr peptide containing the activation loop sequence of platelet-derived growth factor receptor (an RTK), and other reagents and materials were obtained as previously described from the same suppliers (18, 19). In addition, the maleimide headgroup phospholipid dioleolyl-phosphoenthanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide] (DOPE-MCC or Mal-PE) was obtained from Avanti Polar Lipids (Alabaster, AL). Functional PI3Kα and GRP1 PH domain were obtained and labeled with fluorophore by a gentle, enzymatic procedure at their Sfp labeling tag, as previously described (18, 19).
Preparation of supported lipid bilayers possessing membrane-anchored H-Ras
A published method was employed to express and purify unlipidated H-Ras from Escherichia coli (27) (Fig. S1). After confirmation of identity by mass spectroscopy analysis, the desired nucleotide (generally non-hydrolyzable GTP analog GMPPNP, or, where indicated, GTP or GDP) was loaded into H-Ras by nucleotide exchange using a standard, high-EDTA exchange method (27). The bilayer lipid mixture employed was DOPE/DOPS/DOPIP2/DOPE-MCC in a mole ratio of 73:25:1:1. Our previously described procedures (18, 19, 26, 28, 29) were used to deposit a homogeneous bilayer composed of these lipids on ultra-clean glass, yielding a supported lipid bilayer (28). The purified, nucleotide-loaded H-Ras was covalently coupled to the headgroups of DOPE-MCC via its native membrane-anchoring Cys residues 181 and 184 or, where noted, via only Cys 181 (see Table S1). After washing the bilayer to remove all remaining free H-Ras, the surface density and diffusion speed of anchored H-Ras were measured by single-molecule TIRFM using a fluor-tagged, antibody to detect and count each membrane-bound H-Ras molecule (see Fig. S2; Table S1). Functional analysis revealed that the membrane-anchored H-Ras is active in PI3Kα recruitment and kinase assays and exhibits the expected nucleotide specificity for activation by GTP or GTP analog).
Single-molecule TIRFM measurements
TIRFM experiments were carried out at 21.5 ± 0.5°C on an objective-based TIRFM instrument, as described previously (26, 28). The instrument utilized a Nikon (Melville, NY) TE2000U inverted TIRF microscope; a Nikon Apochromat 60×, NA 1.49 TIRF oil immersion objective; and a CNI-Laser 300 mW, 532 nm, diode-pumped solid-state laser model MGLIII-532-300 mW. The laser power exiting the objective was reduced by pre-microscope neutral density filters to 2.3 mW for observation (power measured in epi mode before switching to TIRFM). Sample fluorescence emerging from the 600 nm shortpass emission filter was captured by an Evolve electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ).
TIRFM supported bilayers were first washed with TIRF assay buffer (100 mM KCl, 20 mM HEPES (pH 6.9), 15 mM NaCl, 5 mM glutathione, 2.0 mM EGTA, 1.9 mM Ca2+, and 0.5 mM Mg2+, where this Ca2+/Mg2+ buffering system yields 10 μM free Ca2+ and 0.5 mM free Mg2+), then imaged before and after adding a concentrated mixture of bovine serum albumin (BSA) and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) to final concentrations of 100 μg/mL and 0.05%, respectively. After this addition, only a few dim, rapidly dissociating fluorescent contaminants were typically observed on the bilayer before protein addition, and they were easily eliminated from the data, as described below. Occasionally, the contaminant level was excessive and the reagents (starting with the lipids) were remade.
After confirmation of minimal contamination, stabilizers, proteins, and nucleotides were added to the bilayer as needed and were equilibrated for 3 min. These added components included a low level of BSA (100 μg mL−1 BSA final concentration) to block sticky surfaces that could absorb the dilute proteins (30). Other additions were included as appropriate, including CHAPS (0.05%) to prevent PI3K aggregation, phospho-Tyr peptide (saturating) for PI3K activation, guanosine nucleotide (1 mM) for maintaining H-Ras in its nucleotide-loaded state (in experiments employing this protein), ATP (1 mM) as a PI3K substrate for phosphorylation of PIP2, and GRP PH domain (800 pM) as a sensor for PIP3 detection. Where needed, aliquots of PI3K were thawed on ice and diluted into stabilizing buffer (125 mM NaCl, 20 mM HEPES (pH 7.2), 25% glycerol, 4 mM TCEP, 0.05% CHAPS, and 100 μg mL−1 BSA) that maximizes their stability on ice until they are diluted, just before use, to the final concentration (2 nM) in the buffer above the membrane.
To minimize contributions from small numbers of immobile unfolded proteins bound to a low density of membrane defects, a bleach pulse of ∼5-fold higher power than that used for imaging was applied for ∼15 s; then fluorescence was allowed to return to a steady state for at least 60 s before data acquisition. For each sample, a set of two to four movie streams were acquired at a frame rate of 20 frames/s and a spatial resolution of 4.2 pixels/μm on the home-built instrument, using NIS Elements Basic Research (Nikon).
Single-molecule diffusion tracking
As in our previous studies (26, 28, 31), diffusion trajectories of single-protein molecules were tracked and quantitated using the Particle Tracker plugin for ImageJ (32), yielding a per-frame quantitation of particle position and brightness. The resulting data were then imported into Mathematica for further analysis. Only particles possessing fluorescence intensities within a defined range were included in the analysis, thereby eliminating bright fluorescent contaminants/protein aggregates and dim, non-protein contaminants. Additional displacement-based exclusions removed immobile particles, rapidly dissociating particles, and overlapping tracks for which particle identity is lost. All exclusions were described and validated previously (26, 28, 31).
PI3K membrane binding densities
To investigate the mechanisms of PI3Kα activation by receptor-derived pYp and/or membrane-anchored H-Ras, we employed our recently published single-molecule methods to quantify the effects of activators on PI3Kα surface density and lipid kinase activity (18, 19). Binding measurements focused on stable PI3Kα complexes bound to the membrane for at least five consecutive 20 ms movie frames, yielding tracks lasting ≥100 ms.
To quantify the average density of PI3Kα on the membrane surface in a given TIRF movie, the number of single-particle tracks (defined as described above) in a given field of view was determined for each movie frame, then averaged over all frames. Bleaching of individual tracks or the bulk population was not a major issue, since the average residence time on the membrane before dissociation was short compared to the average bleach time; as a result, fluorescent proteins dissociate from the membrane before bleaching and are replaced from the bulk population, which lies predominantly outside the TIRF excitation field. Under our experimental conditions, the average time to bleach of the Alexa 555 fluor was ≥25 s (26, 28), which is at least 10-fold longer than the average bound-state lifetime of any labeled protein in this study.
PI3K kinase activities
A single-molecule kinase assay was employed to quantify PI3Kα lipid kinase activity and PIP3 production, as previously described (19). The method counts all single molecules of product PIP3 generated by the PI3K lipid kinase reaction, using a saturating concentration of fluor-tagged, GRP-PH domain (800 pM) to bind and detect each PIP3 molecule generated on the membrane surface. After PI3K was added to the chamber (see Single-Molecule TIRFM Measurements above), ATP (1 mM) was added from a buffered stock (assay buffer containing 100 mM ATP and 82.5 mM Mg2+) to start the kinase reaction. Subsequently, fluor-tagged PH domain tracks were quantified as previously described at five time points (18, 19). To determine the PI3Kα specific activity, the resulting net rate of PIP3 production is divided by the average density of PI3Kα determined by the above binding assay (with appropriate correction for the PI3K fluorescence labeling efficiency).
Statistics
PI3Kα is a large, 208 kDa heterodimeric enzyme with nine structural domains and retains function well when stored in frozen, single thaw aliquots. However, thawed aliquots exhibit variability in activity that represent the major limitation on precision in studies of membrane binding, kinase activity, and specific activity. Thus, large numbers of replicates were carried out on multiple days to allow rigorous statistical analysis. Specifically, for each measured parameter, n means were determined, where each mean averaged 3+ replicates carried out on the same day. Error bars represent the standard error of these n means determined on 5–15 different days (n = 5–15).
Results
Mimicking the cellular signaling pathway, and production of membrane-anchored H-Ras
The single-molecule approach enables use of near-physiological protein concentrations and target lipid densities designed to approximate those found in the cellular pathway. Thus, this study employed a PI3Kα concentration (2 nM) of the same order as its physiological concentration (estimated 3 nM (33)), together with a pYp level (saturating) to simulate PI3Kα bound to the regulatory loop of its phospho-activated RTK receptor. The H-Ras coupling protocol yields a surface density of monomeric H-Ras (1 per μM2; see Fig. S2; Table S1) ∼30-fold lower than the surface density reported in cells (34, 35). However, the in vitro system presented here fully loads membrane-anchored H-Ras with GTP or non-hydrolyzable GTP analog, yielding a surface density of active, monomeric H-Ras similar to that expected for moderate H-Ras signals in cells where only a fraction of the membrane-anchored population is activated by GTP loading (36). Moreover, the specific kinase activity (turnover number) of PI3Kα bound to pYp and H-Ras on the membrane surface is the same, within error, at different H-Ras surface densities (Fig. S4). These findings are consistent with a simple picture in which, as the H-Ras surface density increases, the density of membrane-bound PI3Kα and its net kinase activity increase linearly, whereas the specific activity of membrane-bound PI3Kα remains unchanged because its activation mechanism is density independent. The fluorescent GRP1 PH domain employed to detect PIP3 molecules generated by PI3Kα is present in the in vitro single-molecule kinase assays at levels (800 pM) sufficient to fully bind all product PIP3 molecules, just as sufficient PH domain proteins are present in cells to bind all product PIP3 (37). Finally, the supported bilayer densities of the anionic background lipid PS (mole fraction 25%) and the target-substrate lipid PIP2 (mole fraction 1%) are similar to those estimated for the plasma membrane cytoplasmic leaflet (38, 39).
Our previous single-molecule TIRFM studies have characterized the membrane binding and two-dimensional diffusion of the PI3Kα and GRP PH domain proteins on supported bilayers, revealing that PI3Kα possesses extensive bilayer contacts involving multiple lipids, whereas GRP PH domain binds a single PIP3 and diffuses like a single lipid, since the lipid drag against the bilayer largely controls the diffusion speed. This study carries out the first single-molecule analysis, to our knowledge, of PI3K regulation by H-Ras, employing a recently developed approach (27) to covalently anchor functional H-Ras to the supported lipid bilayer. The membrane coupling procedure targeted two native Cys residues that are located near the C-terminus of the hyper-variable region (HVR) (Fig. S1) and are palmitoylated in native H-Ras, where they serve as plasma membrane anchors. Here, both of these Cys residues were employed as supported bilayer anchors by covalently coupling them to a maleimidyl-modified phospholipid headgroup.
Single-molecule TIRFM was used to characterize the density and diffusion speed of the maleimide-lipid-anchored H-Ras on the supported bilayer surface (Fig. S2; Table S1). The measured density was ∼1 H-Ras/μm2 (Table S1). Analysis of the two-dimensional diffusion tracks revealed two subpopulations of membrane-anchored monomeric H-Ras with fast and slow diffusion speeds, corresponding to the frictional drag of one and two coupled lipids (26, 40), respectively (Table S1). The latter H-Ras subpopulation coupled to two lipids via C181 and C184 was found to be the larger, predominant subpopulation (Table S1). The resulting membrane-anchored H-Ras retains the native 14 residue, unstructured tether between membrane-anchored C181 and the folded GTPase domain (Figs. 1 and S1).
Synergistic activation of PI3Kα by receptor-derived phospho-Tyr peptide and H-Ras
To probe the activation of PI3Kα lipid kinase, we employed our previously described single-molecule TIRFM assay of lipid kinase activity (18, 19). This assay directly monitors the number of individual product PIP3 molecules produced as a function of time by catalytically active, membrane-bound PI3Kα (Figs. 1 and 2).
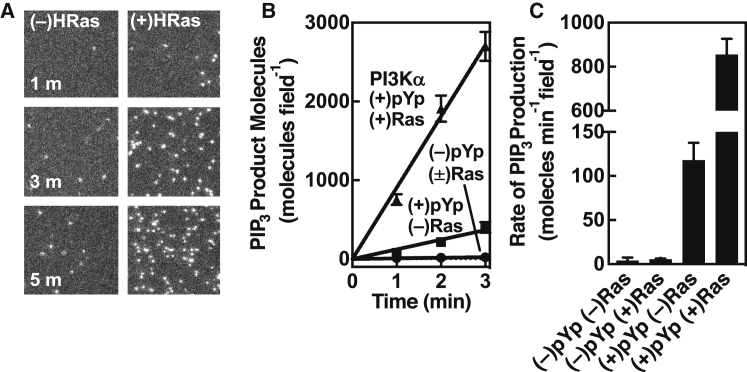
Figure 2.
Effect of receptor-derived pYp and H-Ras on net PI3Kα kinase activity and PIP3 production. (A) TIRFM images of pYp-activated PI3Kα lipid kinase activity on a target bilayer surface. Shown is the accumulation of product PIP3 lipid detected by the fluor-labeled PIP3 sensor (GRP PH domain), with and without membrane-anchored H-Ras. Each image is a square section (121 pixels or 29.0 μm per side) of the monitored TIRFM area (256 pixels or 61.4 μm per side). (B) Time course plotting the increasing number of PIP3 molecules per field under four activating conditions, illustrating strong synergy between the pYp peptide and H-Ras in activating PI3Kα kinase and PIP3 production. (C) The average slopes of these time courses, yielding average net rates of PIP3 production. Each average rate was generated from three replicates on each of nine or more different days (n = 27–36). In all cases in (A)–(C), H-Ras is loaded with non-hydrolyzable GTP analog GMPPNP.
Fig. 2 reveals strong, direct synergy in PI3Kα activation between the receptor-mimicking pYp and membrane-bound H-Ras loaded with non-hydrolyzable GTP analog. Notably, in the absence of pYp, H-Ras alone yields little or no activation of PI3Kα-catalyzed PIP3 production. In contrast, saturating pYp alone triggers an ∼30-fold increase (p < 0.002) in the net rate of PIP3 production, indicating moderate PI3Kα activation in the absence of H-Ras. Together, however, pYp and H-Ras synergistically speed net PIP3 production by nearly an order of magnitude (8 ± twofold) (p < 0.001) more than pYp alone, or ∼200-fold (p < 0.001) more than H-Ras alone. Controls show that the additional PI3Kα activation triggered by H-Ras in the presence of pYp requires membrane-anchored H-Ras, is GTP-regulated, and is specific (Fig. S3). Additional controls show that the pYp and H-Ras activators have no effect on the binding of the PIP3 sensor GRP1 PH domain to PIP3 on the supported bilayer surface (Fig. S5), confirming that the assay accurately measures the effects of these activators on PI3Kα lipid kinase activity.
Synergy dramatically increases the membrane density of stably bound PI3Kα molecules
To investigate the unknown mechanism by which H-Ras dramatically amplifies the moderate PI3Kα activation observed for pYp alone, we measured the effect of both activators on fluor-labeled PI3Kα membrane binding and specific kinase activity.
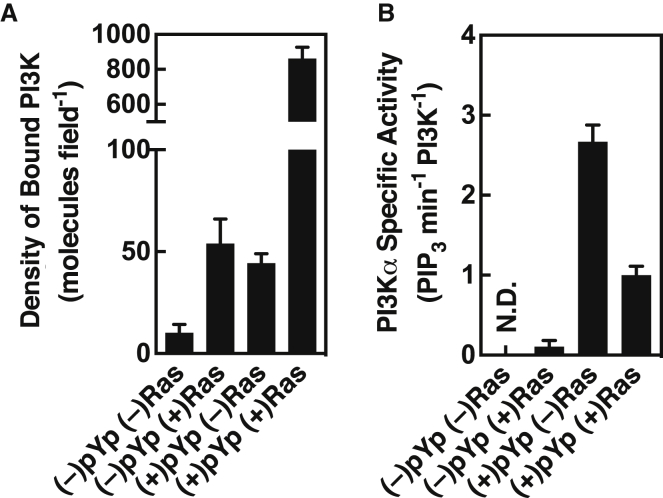
Fig. 3 A shows that either pYp or H-Ras alone triggers moderate densities of long-lived PI3Kα molecules on the membrane surface, whereas the two activators together exhibit strong synergy and dramatically increase the surface density of kinetically stable PI3Kα complexes. Measurements focused on stable PI3Kα diffusion tracks lasting ≥100 ms. Addition of saturating pYp increases the density of PI3Kα 4 ± twofold (p < 0.001) on supported bilayers lacking H-Ras, comparable to our recently published result on membranes of nearly identical lipid composition (19). Similarly, in the absence of pYp, supported bilayers possessing membrane-anchored H-Ras yielded 5 ± twofold (p < 0.001) higher PI3Kα surface density than bilayers lacking H-Ras. Strikingly, when both pYp and membrane-anchored H-Ras were present, the PI3Kα surface density was 80 ± 30-fold (p < 0.001) greater than in the absence of both activators, or 20 ± 2.5-fold (p < 0.001) greater than pYp alone.
Figure 3.
Effect of pYp and H-Ras on PI3Kα membrane recruitment and on the specific kinase activity of the membrane-bound enzyme. (A) Surface density of fluor-labeled PI3Kα measured by single-molecule TIRFM under the indicated activation conditions. Each density was an average over three replicates on each of five different days (n = 15). (B) Specific activity of membrane-bound PI3Kα under each activating condition, calculated as the ratio of net PIP3 production (Fig. 2C) to the kinase surface density (A). This specific activity ratio could not be determined for the (–)pYp (–)Ras condition due to the low density of kinase on the bilayer surface. In all cases, in both (A) and (B), H-Ras is loaded with non-hydrolyzable GTP analog GMPPNP.
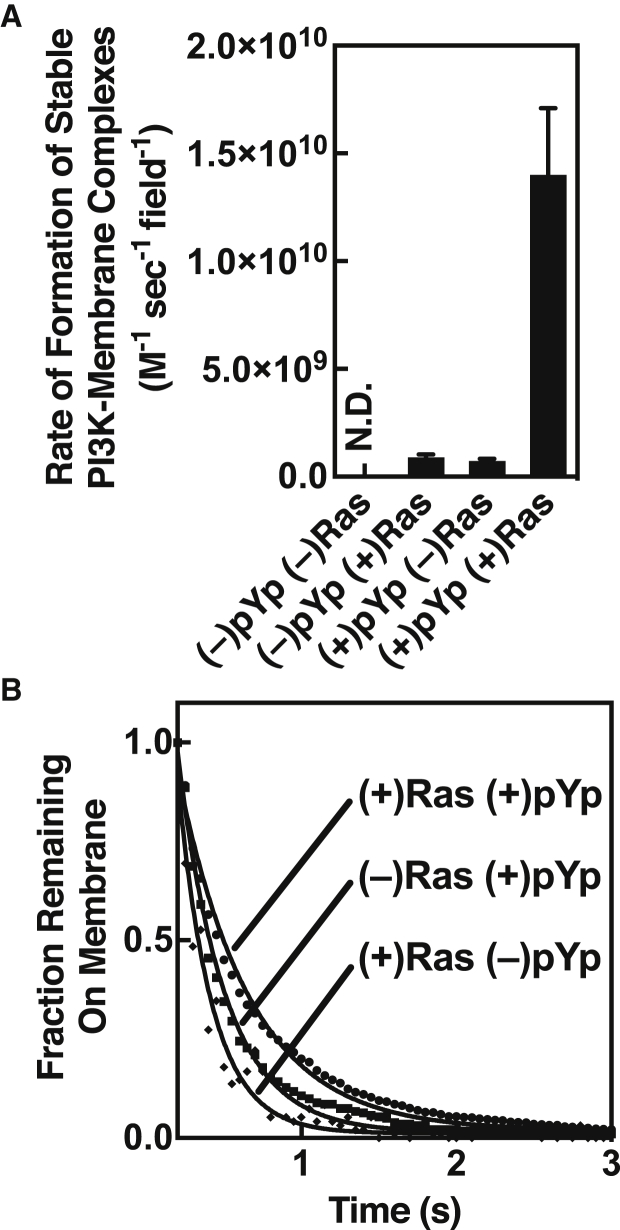
The strong synergy observed between pYp and H-Ras in driving increased equilibrium levels of PI3Kα on the membrane surface arises both from faster formation of stable PI3Kα membrane complexes and slower dissociation. Fig. 4 A shows that the apparent formation rates of stable PI3Kα single-particle tracks with bound-state lifetimes exceeding 100 ms are equally slow, within error, for either activator alone. By contrast, relative to pYp alone, combined peptide and H-Ras greatly increases the formation rate of stable tracks 19 ± fivefold (p < 0.008) and reproducibly decreases their dissociation rate by a small but significant factor (1.4 ± 0.3-fold; p = 0.003). Together, these kinetic effects fully account for the 20 ± 2.5-fold (p < 0.001) greater density of membrane-bound PI3Kα observed when pYp and H-Ras act synergistically.
Figure 4.
Effect of pYp and H-Ras on PI3Kα association-dissociation kinetics. (A) Relative appearance rates of stable, fluor-PI3Kα single-particle diffusion tracks were measured under different activating conditions. These appearance rates were used to calculate the formation rate of active, stably bound PI3Kα complexes on the membrane surface as follows. The number of PI3Kα tracks with bound-state lifetimes exceeding 100 ms in a TIRFM movie was determined, then divided by movie time and bulk PI3Kα concentration to yield an operationally defined appearance rate. Each rate was generated from groups of three replicate movies acquired on each of five different days (n = 15). (B) Off rates of fluor-PI3Kα from the membrane surface, calculated by binning the bound-state lifetimes of stable PI3Kα single-particle diffusion tracks at least 150 ms in length and fitting with a single-exponential decay. Each decay curve was generated from groups of three replicate movies acquired on each of five different days (n = 15). The resulting exponential decay constants were: 2.9 ± 0.6 s−1 for (–)pYp (+)Ras, 2.5 ± 0.6 s−1 for (+)pYp (–)Ras, and 1.9 ± 0.2 s−1 for (+)pYp (+)Ras. No kinetic data could be obtained for (–)pYp (–)Ras due to its very low density on the bilayer surface. In all cases, in both (A) and (B), H-Ras is loaded with non-hydrolyzable GTP analog GMPPNP.
Synergy significantly decreases the specific activity of membrane-bound PI3Kα molecules
Surprisingly, although pYp and membrane-anchored H-Ras synergize to increase the density of stably bound PI3Kα molecules on the supported bilayer surface, this synergy also decreases the specific lipid kinase activity of the membrane-bound enzyme. Fig. 3 B presents specific lipid kinase activities calculated by determining the ratio of the net rate of PI3Kα-catalyzed PIP3 production to the surface density of PI3Kα (Figs. 2 C and 3 A). The specific kinase activity of the average membrane-bound molecule activated by pYp and H-Ras together is, unexpectedly, 2.6 ± 0.6-fold (p < 0.001) lower than observed for activation by pYp alone, indicating that the formation of the H-Ras-PI3K complex significantly inhibits the catalytic activity of membrane-bound, pYp-activated PI3Kα.
Overall, the findings reveal that the 8 ± twofold (p < 0.001) faster net PIP3 production observed for synergistic pYp and H-Ras activation, relative to pYp alone, arises from both the 20 ± 2.5-fold (p < 0.001) greater PI3Kα membrane recruitment noted above, combined with the 2.6 ± 0.6-fold (p < 0.001) slower specific kinase activity. Notably, H-Ras alone yields moderate levels of PI3Kα surface density but fails to generate any detectable kinase activation. Thus, both in the absence and presence of H-Ras (10, 12, 18, 19), pYp plays an essential role in making PI3Kα competent for lipid association and stimulating lipid kinase activity. These results fully support the currently held view that association of receptor phospho-Tyr residues with the inhibitory SH2 domains of the PI3K p85 regulatory domain is required to make the p110 catalytic domain accessible for PIP2 binding and catalysis (5, 7, 8, 9, 10). The observation that H-Ras significantly inhibits the specific kinase activity of pYp-activated PI3Kα emphasizes that the contribution of H-Ras to synergistic G-protein-receptor activation arises purely from a membrane recruitment mechanism, with no contribution from an enzyme activation mechanism.
Discussion
The findings presented here show a strong, direct synergy between receptor-derived pYp and monomeric, membrane-anchored H-Ras in activating net PIP3 production by PI3Kα lipid kinase. Previously, synergy between receptor and Ras activation of specific PI3K isoforms, including PI3Kα, has been proposed by studies in cells, where it is difficult to rule out indirect synergies arising from multi-pathway activation (12, 13). A recent in vitro study observed direct, synergistic activation of class IA PI3K kinase activity by simultaneously added pYp and H-Ras (11). Notably, however, this study did not quantify the effect of activators on PI3K membrane density, nor on the specific kinase activity of the membrane-bound enzyme; thus, it was not able to resolve the long-standing controversy between the membrane recruitment and enzyme activation models for the mechanism of synergistic activation.
The findings presented here resolve this long-standing controversy and reveal that H-Ras contributes to synergistic PI3Kα activation via a membrane-recruitment mechanism, without any detectable contribution from enzyme activation. Relative to pYp alone, H-Ras interactions both speed the formation of kinetically stable, membrane-bound pYp-PI3Kα complex by a factor of 19 ± fivefold (p < 0.008), and slightly but significantly slow the dissociation of PI3Kα from membrane by 1.4 ± 0.3-fold (p = 0.003). At the same time, binding to H-Ras significantly decreases, rather than increases, the specific kinase activity of the membrane-bound pYp-PI3Kα complex by 2.6 ± 0.6-fold (p < 0.001). Thus, the H-Ras contribution to synergistic PI3Kα activation arises purely from its ability to dramatically increase the surface density of active lipid kinase. Notably, H-Ras alone drives only a modest increase in PI3Kα surface density and triggers no measurable kinase activation, reiterating the known requirement for receptor phospho-Tyr binding to the PI3K inhibitory SH2 domains to make the enzyme competent for bilayer and PIP2 binding, as well as catalytic activity (5, 7, 8, 9, 10, 12, 18, 19).
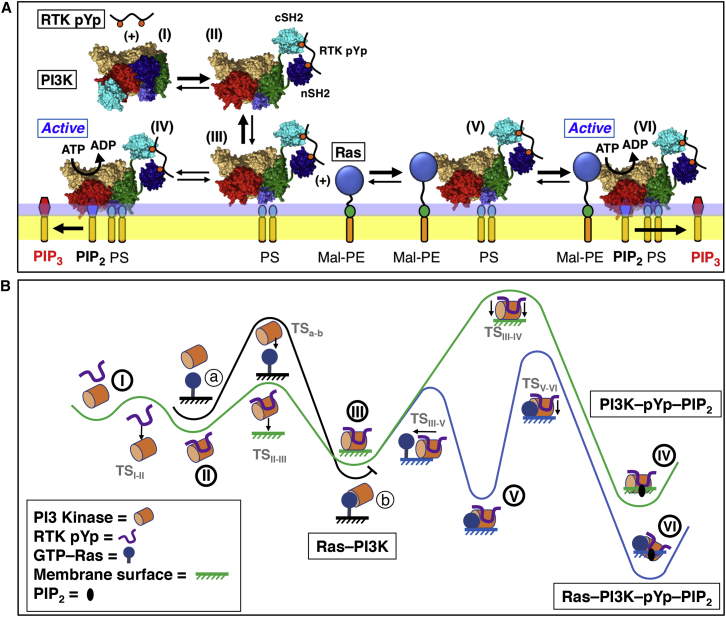
Fig. 5, A and B, presents the simplest kinetic scheme consistent with these data, and the corresponding reaction-coordinate free-energy profile, able to explain the observed kinetic and thermodynamic contrasts between PI3Kα activation by the two activators, alone and in synergy. In the presence of receptor-derived phospho-Tyr peptide, the pYp first binds to the SH2 domains of the free kinase (I), triggering a conformational change that displaces the inhibitory SH2 domains from the catalytic subunit, thereby activating the lipid binding surfaces of PI3Kα (5, 7, 8, 9, 10, 12, 18, 19) to yield the docking-competent free kinase (II) (Fig. 5 A). It has long been established that this specific pYp binding to the SH2 domains requires both sequence motifs and phospho-Tyr residues on the receptor-derived peptide (41, 42). The resulting pYp-PI3Kα complex is then hypothesized to bind via an electrostatic mechanism to the anionic membrane surface, where negative charge is provided mainly by PS, yielding a transient, weakly bound surface state (III). This transient state (III) is hypothesized to undergo two-dimensional diffusion (analogous to the electrostatic surface search of PH domains for PIP3 (27)) on the membrane surface. Usually it dissociates, but sometimes it binds PIP2 and penetrates more deeply into the bilayer to yield a stably bound, kinase-active state (IV). In the presence of H-Ras, the pYp-PI3Kα complex can encounter and bind H-Ras, either via its free state in solution (II) or via its transient surface state (III). The resulting binding of pYp-PI3Kα to both H-Ras and the membrane surface in the quasi-stable state (V) slows dissociation of the transient state (III) from the membrane. Moreover, the H-Ras-bound surface state (V) is proposed to catalyze the transition to the more deeply penetrating, stable, kinase-active state bound to PIP2 (VI), thereby speeding the formation rate of this stable, active state. Thus, the pYp and H-Ras activators act together in synergy to create a pathway to the stable, active state with lower activation barriers and enhanced thermodynamic stability, as illustrated by comparing the synergistic path, I-II-III-V-VI, to the pYp-only path, I-II-III-IV, in Fig. 5 B. Notably, the net stabilization of the final H-Ras-pYp-PI3Kα-PIP2 complex (path I–VI) is less than expected for simple additive stabilization by membrane and H-Ras binding (path I–IV, plus path III–V). This observation suggests that the interaction between H-Ras and PI3Kα in the H-Ras-pYp-PI3Kα-PIP2 complex perturbs the optimal PI3Kα bilayer docking geometry achieved in the pYp-PI3Kα-PIP2 complex lacking H-Ras (5). Additional evidence for this perturbation is the lower specific kinase activity of the H-Ras-pYp-PI3Kα-PIP2 complex relative to the pYp-PI3Kα-PIP2 complex.
Figure 5.
Working model for the mechanism of synergistic PI3Kα activation by receptor-derived pYp and H-Ras. Shown are two different depictions of the same mechanistic model in which pYp peptide and H-Ras synergistically activate PI3Kα by enhancing its membrane recruitment. (A) Kinetic scheme depicting structural cartoons for each state in the hypothesized multi-step activation reaction, with dominant rates highlighted in bold. Activation begins with the binding of free PI3Kα in solution (I) to the receptor-derived pYp (RTK pYp), present in excess. One or both inhibitory SH2 domains bind pYp and dissociate from the PI3Kα catalytic domain, exposing its PIP2 binding and catalytic sites (9, 10). The resulting free pYp-activated PI3Kα complex (II) docks to the membrane, yielding an inactive pYp-PI3Kα complex transiently bound to the membrane surface in a shallow surface-bound state (III). This transient surface complex (III) rapidly dissociates and returns to solution (II) unless converted to a stable, membrane-bound pYp-PI3Kα complex by additional membrane interactions, such as deeper penetration into the bilayer (IV). The resulting stable, membrane-bound pYp-PI3Kα complex (IV) is fully active and displays the maximum specific kinase activity. Alternatively, if present, membrane-anchored H-Ras can rapidly bind to the transient surface complex (III) and catalyze its conversion to the stably bound, deeply penetrating, active Ras-pYp-PI3Kα complex (V). Such catalytic assistance explains the faster rate of formation of the Ras-bound stable, active complex (V) compared to Rasless stable, active complex (IV). Notably, the Ras-bound complex (V) exhibits two unexpected properties, leading to the hypothesis that Ras binding perturbs the optimal membrane docking geometry displayed by pYp-PI3Kα in the Rasless complex (IV). First, the rate of Ras-bound complex (V) dissociation from membrane is only slightly slower than that of Rasless complex (IV), indicating that the former complex does not possess the full kinetic stability expected for simple additivity of the binding free energies for pYp-PI3Kα binding to H-Ras plus pYp-PI3Kα binding to membrane. Second, the specific kinase activity of the Ras-bound complex (VI) is, surprisingly, significantly lower than that of the Rasless complex (IV). These unexpected features are both consistent with a Ras-bound state (VI) in which structural constraints prevent the optimal interactions of pYp-PI3Kα with Ras and the membrane simultaneously (see text). Further single-molecule studies will test this working model and determine the rate constants of individual steps. (B) Reaction coordinate depiction of the kinetic scheme shown in (A), using the same numbering scheme for intermediates (I) through (VI). Also shown is the unproductive path a-to-b for the absence of pYp, wherein PI3Kα is able to bind weakly to Ras but the lack of phospho-Tyr binding prevents release of the inhibitory SH2 domains bound to the catalytic domain. As a result, the SH2 domains continue to block PI3K membrane binding and kinase activation. To see this figure in color, go online.
Conclusions
Overall, the findings presented here reveal that simultaneous activation of PI3Kα by a receptor activation loop and H-Ras generates strong, synergistic, activation of PI3Kα, yielding a large increase in net kinase activity via a membrane recruitment mechanism. The findings provide important mechanistic insights into the receptor-Ras synergy that strongly activates PI3K in leukocyte chemotaxis, innate immunity, and inflammation, as well as in carcinogenesis. These insights also have implications for drug design targeting PI3K-catalyzed PIP3 production in carcinogensis or inflammation. The findings suggest that drugs designed to block activation of the PI3K SH2 domains by receptor phospho-Tyr residues will provide the strongest PI3K inhibition, since phospho-Tyr occupancy of the SH2 domains is required for kinase activation by receptor alone, or by synergistic receptor-Ras activation. On the other hand, the findings indicate that drugs designed to block the interaction between H-Ras and PI3K should provide nearly the same degree of PI3K inhibition, since H-Ras dominates the synergistic activation. Moreover, the finding that H-Ras binding actually inhibits, rather than activates, the PI3Kα lipid kinase alleviates the potential concern that drug binding to the Ras binding domain might prevent Ras association but inadvertently activate PIP3 production via allosteric kinase regulation. Planned studies will further test the model of Fig. 5 and determine the rate constants for each step in Fig. 5 A. In addition, it is important to ascertain whether this simple synergy mechanism observed for the H-Ras/PI3Kα regulatory pair is generalizable to all G protein-PI3K pairings, or whether specialized activation mechanisms exist for specific pairings of PI3K isoforms with G proteins of the Ras, Rho, and Gβγ families.
Author Contributions
Conception, J.J.F.; Experimental design, T.C.B., B.P.Z., and J.J.F.; Data collection, T.C.B.; Data analysis, T.C.B., B.P.Z., and J.J.F.; Data interpretation, T.C.B., B.P.Z., and J.J.F.; Other essential materials, advice, and context, R.L.W. and G.R.M.
Acknowledgments
We gratefully acknowledge Olga Perisic for expert PI3K purification and Jay Groves for kindly providing the H-Ras expression plasmid.
Funding for this work was provided by the National Institutes of Health (R01 GM063235 to J.J.F.), the Medical Research Council (MC U105184308 to R.L.W.), the AstraZeneca/LMB Blue Skies Initiative (MC A024-5PF9G to R.L.W.), and St. Catharine’s College (G.R.M.).
Editor: Arne Gericke.
Footnotes
Five figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31030-5.
Supporting Material
References
- 1.Swaney K.F., Huang C.H., Devreotes P.N. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denley A., Gymnopoulos M., Vogt P.K. Requirement of phosphatidylinositol(3,4,5)trisphosphate in phosphatidylinositol 3-kinase-induced oncogenic transformation. Mol. Cancer Res. 2009;7:1132–1138. doi: 10.1158/1541-7786.MCR-09-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 5.Burke J.E., Perisic O., Williams R.L. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc. Natl. Acad. Sci. USA. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke J.E., Perisic O., Williams R.L. Allosteric activation of PI3Kα by oncogenic mutations. Oncotarget. 2013;4:180–181. doi: 10.18632/oncotarget.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vadas O., Burke J.E., Williams R.L. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 2011;4:re2. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 8.Burke J.E., Williams R.L. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS) Adv. Biol. Regul. 2013;53:97–110. doi: 10.1016/j.jbior.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hon W.C., Berndt A., Williams R.L. Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene. 2012;31:3655–3666. doi: 10.1038/onc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellor P., Furber L.A., Anderson D.H. Multiple roles for the p85α isoform in the regulation and function of PI3K signalling and receptor trafficking. Biochem. J. 2012;441:23–37. doi: 10.1042/BJ20111164. [DOI] [PubMed] [Google Scholar]
- 11.Siempelkamp B.D., Rathinaswamy M.K., Burke J.E. Molecular mechanism of activation of class IA phosphoinositide 3-kinases (PI3Ks) by membrane-localized HRas. J. Biol. Chem. 2017;292:12256–12266. doi: 10.1074/jbc.M117.789263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke J.E., Williams R.L. Synergy in activating class I PI3Ks. Trends Biochem. Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Yang H.W., Shin M.G., Heo W.D. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacold M.E., Suire S., Williams R.L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 15.Denley A., Kang S., Vogt P.K. Oncogenic signaling of class I PI3K isoforms. Oncogene. 2008;27:2561–2574. doi: 10.1038/sj.onc.1210918. [DOI] [PubMed] [Google Scholar]
- 16.Castellano E., Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellano E., Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziemba B.P., Swisher G.H., Falke J.J. Regulation of a coupled MARCKS-PI3K lipid kinase circuit by calmodulin: single-molecule analysis of a membrane-bound signaling module. Biochemistry. 2016;55:6395–6405. doi: 10.1021/acs.biochem.6b00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziemba B.P., Burke J.E., Falke J.J. Regulation of PI3K by PKC and MARCKS: single-molecule analysis of a reconstituted signaling pathway. Biophys. J. 2016;110:1811–1825. doi: 10.1016/j.bpj.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber K.S., Ostermann G., Weber C. Dual role of H-Ras in regulation of lymphocyte function antigen-1 activity by stromal cell-derived factor-1α: implications for leukocyte transmigration. Mol. Biol. Cell. 2001;12:3074–3086. doi: 10.1091/mbc.12.10.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolly L.A., Massoll N., Franco A.T. Immune suppression mediated by myeloid and lymphoid derived immune cells in the tumor microenvironment facilitates progression of thyroid cancers driven by Hras(G12V) and Pten loss. J. Clin. Cell. Immunol. 2016;7:451. doi: 10.4172/2155-9899.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorpe L.M., Yuzugullu H., Zhao J.J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kölsch V., Charest P.G., Firtel R.A. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan T.L., Cantley L.C. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gericke A., Leslie N.R., Ross A.H. PtdIns(4,5)P2-mediated cell signaling: emerging principles and PTEN as a paradigm for regulatory mechanism. Adv. Exp. Med. Biol. 2013;991:85–104. doi: 10.1007/978-94-007-6331-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight J.D., Lerner M.G., Falke J.J. Single molecule diffusion of membrane-bound proteins: window into lipid contacts and bilayer dynamics. Biophys. J. 2010;99:2879–2887. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin W.C., Iversen L., Groves J.T. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight J.D., Falke J.J. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalb E., Frey S., Tamm L.K. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim. Biophys. Acta. 1992;1103:307–316. doi: 10.1016/0005-2736(92)90101-q. [DOI] [PubMed] [Google Scholar]
- 30.Nair P.M., Salaita K., Groves J.T. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat. Protoc. 2011;6:523–539. doi: 10.1038/nprot.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziemba B.P., Knight J.D., Falke J.J. Assembly of membrane-bound protein complexes: detection and analysis by single molecule diffusion. Biochemistry. 2012;51:1638–1647. doi: 10.1021/bi201743a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sbalzarini I.F., Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Hein M.Y., Hubner N.C., Mann M. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 34.Gureasko J., Galush W.J., Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan X., Tamgüney T.M., Chu S. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc. Natl. Acad. Sci. USA. 2015;112:7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haugh J.M., Huang A.C., Lauffenburger D.A. Internalized epidermal growth factor receptors participate in the activation of p21ras in fibroblasts. J. Biol. Chem. 1999;274:34350–34360. doi: 10.1074/jbc.274.48.34350. [DOI] [PubMed] [Google Scholar]
- 37.Corbin J.A., Dirkx R.A., Falke J.J. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrell J.E., Jr., Huestis W.H. Phosphoinositide metabolism and the morphology of human erythrocytes. J. Cell Biol. 1984;98:1992–1998. doi: 10.1083/jcb.98.6.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagelberg C., Allan D. Restricted diffusion of integral membrane proteins and polyphosphoinositides leads to their depletion in microvesicles released from human erythrocytes. Biochem. J. 1990;271:831–834. doi: 10.1042/bj2710831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziemba B.P., Falke J.J. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013;172-173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitman M., Kaplan D.R., Roberts T.M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 42.Carpenter C.L., Auger K.R., Cantley L.C. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.