Abstract
By mole, cholesterol is the most abundant component of animal cell plasma membranes. Many membrane proteins have been shown to be functionally dependent on cholesterol, several of which have also been shown to bind cholesterol at well-defined locations on their membrane-facing surface. In this work, a combination of coarse-grained “Martini” and all-atom simulations are used to identify two, to our knowledge, new cholesterol-binding sites on the A2A adenosine receptor, a G-protein-coupled receptor that is a target for the treatment of Parkinson’s disease. One of the sites is also observed to bind cholesterol in several recent, high-resolution crystal structures of the protein, and in the simulations, interacts with cholesterol only when bound to the inverse agonist ZM241385. Cataloguing cholesterol-binding sites is a vital step in the effort to understand cholesterol-dependent function of membrane proteins. Given that cholesterol content in plasma membranes varies with cell type and on administration of widely prescribed pharmaceuticals, such as statins, understanding cholesterol-dependent function is an important step toward exploiting membrane compositional variation for therapeutic purposes.
Introduction
Of the hundreds of different lipids of distinct chemistry found in the plasma membrane of an animal cell, cholesterol comprises the largest fraction, ∼20% by mole, averaged across various cell types (1, 2). Locally, the composition may be significantly higher (3), with the envelope of an influenza virus containing 50 mol% cholesterol (1), presumably reflecting the location of viral budding. This is in stark contrast to the composition of other cellular membranes. The endoplasmic reticulum, for example, has a composition of perhaps 1 mol% cholesterol (4). Beyond animal cell membranes, sterols and sterol-like molecules are present in every cell type that is bounded by a lipid membrane, including ergosterol (yeast) and hopanoids (plant cells and bacteria) (4).
Cholesterol-binding motifs have been identified in a diverse array of membrane proteins (5). The amyloid precursor protein C99, which is cleaved into pathogenic Aβ-peptide in Alzheimer’s disease, has a “GXXXG” cholesterol-binding motif (6). The cholesterol recognition amino acid consensus (CRAC) motif was originally identified on a benzodiazepine receptor that transports cholesterol between mitochondrial membranes (7), and has been found in many transmembrane segments (8, 9), including G-protein-coupled receptors (GPCRs) (10, 11) and ion channels (12). A similar motif, but with the direction reversed (CARC motif) has also been reported in both GPCRs and ion channels (9, 12, 13). Yet another motif (cholesterol consensus motif, or CCM) was first identified in a cholesterol-bound crystal structure of the β2-adrenergic receptor, a GPCR (14).
In many proteins that contain these motifs, cholesterol-dependent function is observed (9, 12, 13). Because of their importance as drug targets (15), significant efforts in this area have focused on GPCRs, which are also the focus of this work. Rhodopsin has long been known to respond to changes in the membrane (16, 17), including cholesterol in bilayers and sterol derivatives in micelle environments. Chattopadhyay and coworkers (11) have published extensive biochemical data showing that cyclic adenosine monophosphate signaling induced by the seratonin1A receptor is reduced upon cholesterol depletion in cells, and hypothesized the mechanism to be an interaction mediated by the CRAC motif (10). More recently, the same group have reported similar results for a bitter taste receptor (18). Using a G-protein-coupling assay inside giant unilamellar vesicles reconstituted with seratonin1A, Malmstadt and coworkers have shown that increasing cholesterol enhances ligand binding and accelerates GTP turnover (19). In contrast to the preceding results, which generally observe enhancement of function in the presence of cholesterol, a recent publication reported inhibition of function with increasing cholesterol (20).
Despite the existence of cholesterol interaction motifs on these proteins, the mechanism by which cholesterol exerts its functional effects is largely unknown. It may be that cholesterol interaction motifs serve as sites of allosteric regulation (21), perhaps ensuring that the protein signals with maximal efficiency in specific, cholesterol-rich locations within the cell. A different, but not mutually exclusive hypothesis is that the receptor responds to cholesterol-induced changes in curvature stress (16, 17, 22). Disentangling these two mechanisms is challenging, in part because hypotheses for direct cholesterol interaction sites are limited to those identified by structural and/or sequence analysis. One can imagine developing mutant sequences designed to disrupt cholesterol interactions, but it is very possible that there are locations not encompassed by the known motifs; for example, sites not observed in any crystal structure or comprised of residues located on adjacent transmembrane segments, making them more challenging to identify via bioinformatic analysis, or sites which are specific to a particular ligation state. If a receptor has multiple binding sites, only some of which are known, directed mutagenesis to disrupt cholesterol interactions followed by cholesterol-dependent functional assays may produce complex, confounding results.
Computational methods to predict cholesterol interaction sites, in a blind and unbiased way, are therefore of great value. Both coarse-grained (CG) and all-atom simulations have had a significant impact on efforts to understand integral membrane protein-lipid interactions. (The discussion is limited here to GPCRs; see Hedger and Sansom (23) for a comprehensive recent review.) Using Martini simulations, Chattopadhyay and coworkers (24) have identified cholesterol interactions on serotonin receptors, in some cases mediated by the CRAC motif. Several groups have used all-atom simulations for the same problem, starting with Grossfield and coworkers (25, 26), focusing on rhodopsin. Lee and Lyman (27) used relatively short all-atom simulations to predict cholesterol-binding sites on the A2A adenosine receptor (A2AR); although the more recent, much longer trajectories reported by Cang et al., (28) on the β2-adenosine receptor suggest that binding at the CCM requires a slow, concerted rearrangement of side chains. Also studying β2-adenosine receptor, Neale et al., (29) postulated a role for an anionic headgroup as an allosteric modulator of protein function, in which interaction with the ionic lock stabilizes the active state.
Here, we present two, to our knowledge, new cholesterol interaction sites that are identified on the A2AR by extensive, unbiased molecular simulation, including both CG Martini simulations and 12 μs of all-atom simulations obtained on the “Anton2” special purpose supercomputer. One of these two sites is identical to the location of a cholesterol in several high-resolution crystal structures (30, 31, 32). A2AR is a ubiquitously expressed GPCR that stimulates cyclic adenosine monophosphate production (33) and is a target for several therapeutic applications, including Parkinson’s disease (34, 35). Robinson and coworkers (36) have shown A2AR to be sensitive to cholesterol in several systems, including micelles, where sterol derivatives enhance ligand binding, and in CHO and HEK cells, where results similar to those reported by Chattopadhyay and coworkers (11) for the seratonin1A receptor are observed (A. Robinson, personal communication). The Malmstadt laboratory has also obtained evidence for cholesterol enhancement of A2AR activity in giant unilamellar vesicles using a G-protein-coupling assay, and recently reported ligand-dependent partitioning between different lipid phases, with liganded receptor preferring the liquid-ordered phase (19). There is also a recent report demonstrating the opposite effect, namely that cholesterol inhibits ligand binding to A2AR (20), which is not supported by our data. Thus, a more complete understanding of cholesterol-A2AR interactions is important to both pharmacological development targeting A2AR and to resolve the general mechanism of membrane protein regulation by direct cholesterol interaction.
The use of Martini to identify lipid-binding sites is general, and readily applicable to any membrane protein (37). Cholesterol exhaustively samples the protein surface in unbiased Martini simulations of A2AR in a lipid/cholesterol mixture, producing statistically converged cholesterol density in localized, reproducible locations on the protein surface. Individual cholesterols interact frequently with the protein. The duration of interaction events is found to have a power-law distribution, indicating a broad distribution of cholesterol-protein interaction strengths. Thus, although no single event is sufficient to identify an interaction site, analysis of the ensemble of events admits “scoring” all the residues on the membrane-facing surface according to their propensity to interact with cholesterol. The CG model therefore provides rapid, blind predictions for cholesterol interaction sites, which are then available for follow-on all-atom simulations and experimental study. In our results, the two highest ranked locations from the Martini analysis, located in the inner and outer leaflets between helices 5 and 6, bind cholesterol for extended periods of time in 12 μs unbiased all-atom simulations, both when the protein is bound to an inverse-agonist, and when no ligand is bound. The all-atom results provide detailed structural information on the cholesterol interaction motifs, enabling comparison to high-resolution crystallographic data and providing targets for mutagenesis.
Materials and Methods
Martini simulation details
The initial coordinates for the protein were based on a high-resolution structure of A2AR bound to the inverse agonist ZM241385 (PDB: 4eiy) (31). A fusion protein (cytochrome b562) present between residues 208 and 219, as well as the cocrystallized ZM241385 ligand, were removed. The structure was then converted to a CG representation using the martinize.py script, and the system was built with the insane.py script (38) (both scripts freely available from http://cgmartini.nl/). The secondary structure of the protein was restrained by an elastic network among the backbone sites (39) (cutoff distance of 0.9 nm with a force constant of 500 kJ/mol−1 nm−2), ensuring that the inactive conformation was simulated. The simulated system consists of one protein embedded in a palmitoyloleoylphosphatidylcholine (POPC)/cholesterol (70:30) mixture: protein (305 residues), POPC bilayer (168 lipids) including cholesterols (72 cholesterol (40)), and aqueous phase (6119 water beads and 9 neutralizing sodium ions). The lipids and protein were modeled with the Martini CG force field for biomolecules (41, 42). Nonbonded interactions were cut off at a distance rcut = 1.2 nm. The Lennard-Jones potential is shifted from rshift = 0.9 nm to rcut. The electrostatic potential is shifted from rshift = 0.0 nm to rcut.
All CG simulations were performed using the GROMACS simulation package version 5.0.6 (43). The system was energy minimized via steepest descent followed by a 1-ns integration at constant temperature, pressure, and particle number using a 10 fs timestep (leap-frog algorithm). The integration timestep was then increased to 20 fs for a 200 μs production simulation, with configurations stored every 500 ps. The protein/bilayer (A2A/POPC and cholesterol) and the aqueous phase (water and sodium ions) were coupled independently to external temperature baths at 300 K, using a Bussi-Donadio-Parinello thermostat (44) with a relaxation time of 1.0 ps. The pressure was coupled (using the Parinello-Rahman barostat (45)) to an external bath at 1.0 bar using a relaxation time of 12.0 ps, a semi-isotropic pressure scheme, and compressibility set to 3 × 10−4 bar−1.
All-atom simulation details
Initial coordinates were based on the same high-resolution, ZM241385-bound structure as the Martini simulations (31). Initial coordinates for residues 209–219 were obtained by aligning to the structure of the thermostabilized mutant bound to ZM241385 (3 pwh), published by Tate and coworkers (46), and in which the missing residues are resolved. The apo structure was created by simply deleting the ligand. A symmetric lipid bilayer containing 504 dipalmitoyl phosphatidyl choline, 132 dioleoyl phosphatidyl choline, and 273 cholesterol randomly distributed and solvated with ∼55,000 tip3p (47) waters was prepared using the CHARMMGUI (48), heated to 295 K, and run for 50 ns with NAMD v2.9 under semi-isotropic constant temperature, pressure, and particle number conditions using the Langevin piston and temperature coupling method. The protein was embedded in the membrane by aligning to the OPM database (49) structure of A2AR (also based on PDB: 4eiy) and deleting any lipid with a significant steric clash. The only components from the crystal structure that were included were one sodium ion and the ZM241385 ligand (in the ligand bound case); no cholesterols from the crystal structure were included. Ten chloride ions were added to neutralize the system. The protein and lipids were modeled with the CHARMM36 force field (50, 51) and the ligand with the CHARMM general force field (52) with atoms typed by the ParamChem server (53).
Several equilibration steps were run with NAMD v2.9 (54). The initial configuration was relaxed by 4000 steps of steepest descent. The protein backbone was then restrained using a force constant of 2 kcal/mol/Å2 as the system was heated to 295 K, reassigning velocities from a Maxwell-Boltzmann distribution every four timesteps, with the simulation cell volume allowed to change semi-isotropically via a Langevin piston with a damping timescale of 0.1 ps and period 0.2 ps (55). An additional 20,000 equilibration steps were performed, rescaling velocities every 100 steps to enforce a temperature of 295 K. Finally, the Langevin equation was integrated for 1 ns with a 1.0 fs timestep, followed by 10 ns with a 2.0 fs timestep, and all covalently bonded hydrogens constrained by SHAKE (56). During all relaxation steps electrostatics were computed with the particle-mesh Ewald method (57) on a 1 Å grid, with a tolerance of 10−6 and fourth order interpolation. Lennard-Jones interactions were cut off at 10 Å and shifted to zero at 12.5 Å ensuring both continuous potential and force.
The equilibrated binary restart files were converted to dms format for production simulation on Anton2. Force field information was added using Viparr v4.5.34. Integration was performed under constant pressure (1 atm), temperature (295 K), and particle number with the multigrator (58) method, with temperature (295 K) controlled by a Nose-Hoover (59) chain coupled every 24 timesteps and pressure by the Martyna-Tobias-Klein barostat (pressure 1 atm, semi-isotropic) coupled every 480 timesteps (60). Electrostatics were computed using the k-space Gaussian split Ewald method (61), with long-range interactions computed every third timestep. Lennard-Jones interactions were cut off at 11 Å or greater. (Full details are available in the simulation configuration file in the Supporting Material.) Both ZM241385 bound and apo simulations were run for 6 μs with a 2.5 fs timestep (hydrogens constrained by M-SHAKE (62)), and simulation configurations were stored every 240 ps.
Results
Cholesterol exhaustively samples the protein surface during Martini simulation, obtaining converged densities at preferred loci of interaction
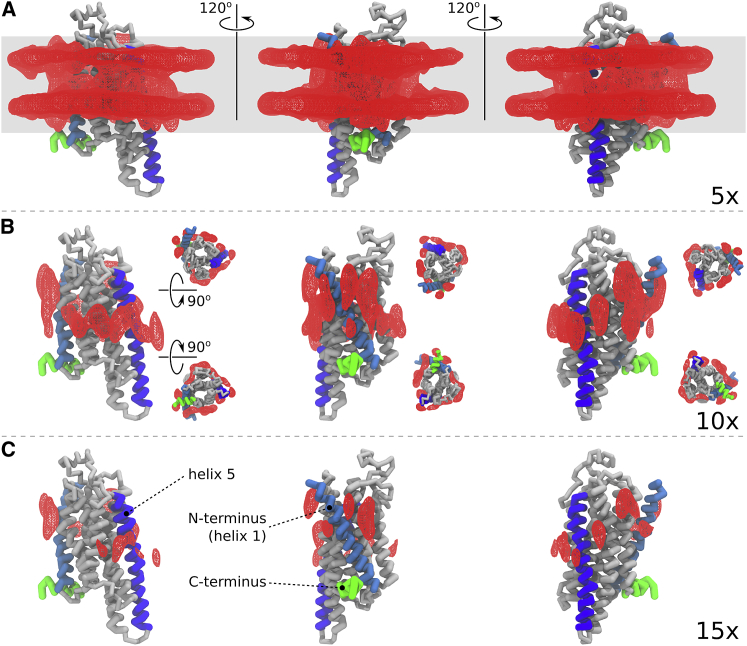
Over the course of the 200 μs Martini simulation, the 72 cholesterols completely sample the membrane-facing surface of the protein, and identify regions of preferred interaction. This is shown in Fig. 1, in which isosurfaces of cholesterol density are rendered. The densities were computed on a 0.2 nm resolution grid using the VolMap plugin to VMD (63), with each cholesterol site contributing equally to voxel densities. The isosurface corresponding to five times the bulk cholesterol density (top row) shows that the protein surface is completely sampled by cholesterol. Furthermore, because the density of cholesterol near the protein is greater than the bulk, the analysis indicates that the local protein environment is enriched in cholesterol, consistent with a model in which cholesterol forms a nearby annular pool (13). A Voronoi analysis of the first two solvation shells around the protein is reported in Table 1, based on centers of mass of lipids and α-carbon atoms of the protein, projected onto the bilayer midplane (64) (see Fig. S1). The analysis indicates that this enrichment is confined to the first solvation shell around the protein, which is enriched on average by slightly more than two cholesterols and depleted by two POPC, relative to the bulk concentration. The second shell shows the reverse trend, being depleted by ∼1.5 cholesterols and enriched by 1.5 POPC. Note that the enrichment in the first shell is also asymmetric, with the inner leaflet slightly more enriched in cholesterol than the outer leaflet.
Figure 1.
Cholesterol density as observed in Martini simulations. Red wire frame shows isosurface of cholesterol density at 5× (A), 10× (B), and 15× (C) the bulk density of cholesterol in a 200 μs Martini simulation of A2AR. Three views are rendered to show the entire surface, rotated by 120°; helix 1 (cyan), helix 5 (blue), and helix 8 (green) are shown to orient the viewer. Top and bottom views are shown in the middle panel. To see this figure in color, go online.
Table 1.
Number of Cholesterol in First and Second Solvation Shells in Inner and Outer Leaflets
| First Shell | Second Shell | |
|---|---|---|
| Cholesterol (outer) | 7.1 (0.2) | 7.7 (0.1) |
| Cholesterol (inner) | 8.3 (0.2) | 7.6 (0.1) |
| POPC (outer) | 14.4 (0.3) | 19.9 (0.1) |
| POPC (inner) | 13.2 (0.3) | 20.1 (0.1) |
The SEs computed from five nonoverlapping blocks are in parentheses.
Rendering isosurfaces at higher cholesterol density (middle and lower panels) yields a more stringent criterion, and reveals locations on the protein surface that may be specific interaction sites. At 10 times the bulk density, preferred loci of cholesterol interactions begin to emerge. At this level, comparison of the density between nonoverlapping trajectory segments demonstrates convergence of the sampling of the protein surface by cholesterol (Fig. S2). Although inaccuracies in the force field cannot be ruled out, predictions based on these data do obtain a high level of statistical confidence within the limitations of the model. At 15 times the bulk density, tightly localized interaction regions are clearly identified.
Although the cholesterol density indicates converged sampling, it does not provide a sufficient basis for prediction of cholesterol interaction sites. This is because any cutoff in cholesterol density for defining such sites is likely to be arbitrary. One could, for example, raise or lower the cutoff to eliminate or add cholesterol interaction sites to provide better agreement with experimental data. A “blind” algorithm to rank residues according to their propensity to interact with cholesterol is therefore preferred. A ranking based on the analysis of all observed individual cholesterol interaction is described below, after a discussion of the nature of cholesterol interaction events.
Individual cholesterol interaction events are frequent, with a broad distribution of interaction times
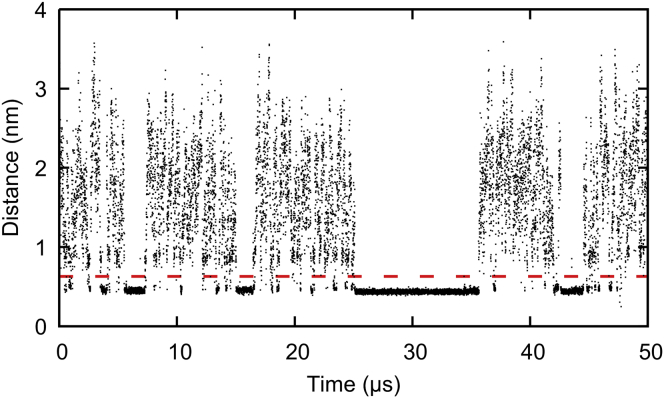
Several observables were considered as candidates to identify individual interaction events, including cholesterol diffusivity and statistical analysis of individual trajectories. As shown in Fig. 2, the distance between individual cholesterols and the protein clearly identifies binding events, visible as regions of reduced fluctuation and close approach to the protein. To automate the identification of such events in all 72 cholesterol trajectories, a cutoff distance of 0.63 nm was defined from the first minimum of the radial distribution function of the protein surface-cholesterol center of mass distance, indicated by the horizontal dashed line in Fig. 2. Periods of time during which this distance falls below the cutoff are identified as interaction events.
Figure 2.
Distance to protein surface for a single cholesterol during a 50 μs segment of a 200 μs Martini simulation. Interaction events are visible as periods of close approach and reduced fluctuation. Any distance <0.63 nm (red dashed line) is identified as an interaction event. To see this figure in color, go online.
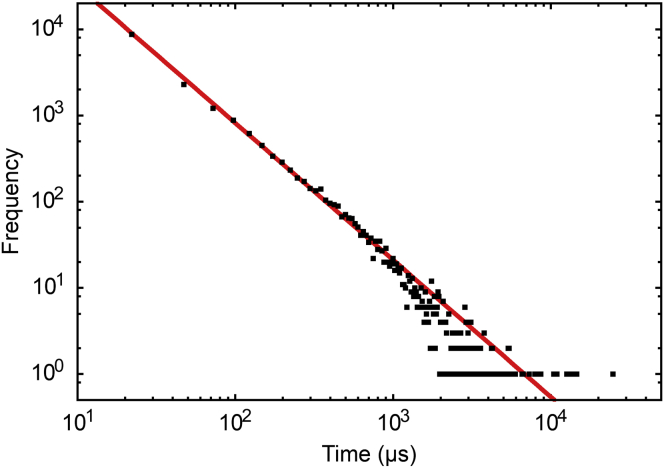
The durations of each event for all cholesterol trajectories are compiled into a histogram, shown in Fig. 3. Individual cholesterol-protein interactions span four orders of magnitude, and are well-described by a power law (t−1.59) up to ∼1 μs, after which the statistical representation of very long events is not sufficient to obtain a reliable fit. The broad, power-law distribution of event durations suggests that the free-energy barriers governing the unbinding kinetics are themselves broadly distributed (although not necessarily as a power law), which in turn implies that cholesterol-protein interactions are very heterogeneous. Stochastic processes of this type may be described by a fractional Klein-Kramers equation (65). Although the power law is similar to a one-dimensional first-passage time distribution (t−3/2), the correspondence is coincidental. The cutoff that defines a cholesterol-protein interaction is very short-ranged, corresponding to direct contact between the cholesterol and protein.
Figure 3.
Histogram of cholesterol interaction event durations. A total of 18,459 events are included, with durations from 10 ns to 25 μs. The line is fit to t−1.59. To see this figure in color, go online.
Scoring residues by binding event duration and contact number identifies putative interaction sites
There are 18,459 interaction events in the histogram in Fig. 3. Most are weak, short-lived, and nonspecific, but some likely involve well-defined cholesterol interaction sites on the protein surface. Locations on the protein that are repeatedly visited by cholesterol are more likely to be such specific sites, especially if cholesterol dwells at these locations for an extended period of time, and if a cholesterol makes multiple contacts with the protein at the same location.
A residue-based scoring function was therefore developed that incorporates both locality and duration of interaction. For each cholesterol, every interaction event defined by the threshold in Fig. 2 is identified. Events with durations <1 μs are discarded. This cutoff in duration is arbitrary, as there is no timescale evident in the distribution in Fig. 3; however, Fig. S3 shows that the ranking of the residues is insensitive to this parameter, and Fig. S4 shows that it is insensitive to the 0.63 nm distance cutoff. A score for each residue is accumulated by summing the number of cholesterol-residue contacts for that residue during the interaction events with duration >1 μs. After summing these scores for each residue, they are normalized by the total trajectory duration and the number of side chain sites in the Martini model for that residue. Although the absolute value of the score is arbitrary, it nonetheless ranks the residues by their propensity to interact with cholesterol.
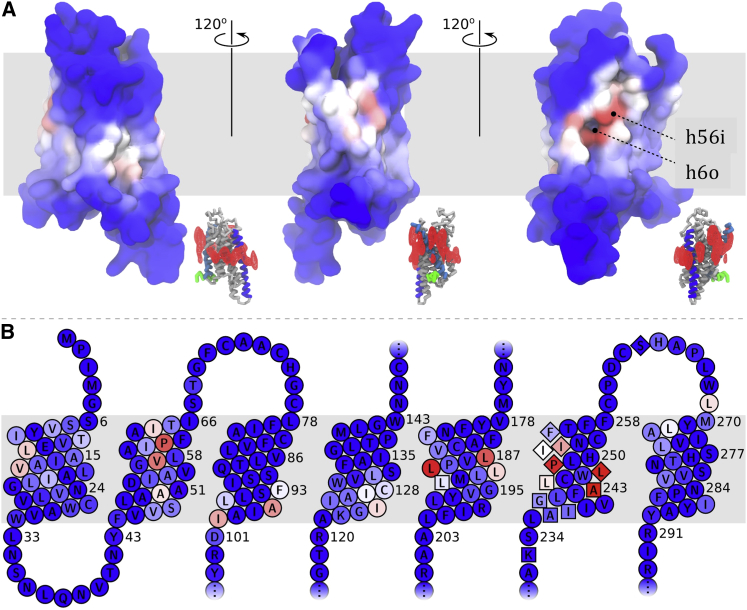
When rendered on top of the protein structure, the residue scores identify several putative cholesterol interaction sites, as shown in Fig. 4. The most highly ranked residues cluster around a deep cleft between helices 5 and 6. These residues span both leaflets, and will be referred to as h6o (helix 6, outer leaflet) and h56i (helices 5 and 6, inner leaflet). Cholesterol is also found at h6o in every high-resolution crystal structure of A2AR bound to the inverse agonist ZM241385, several of which were obtained by different methods and by different research groups (30, 31, 32).
Figure 4.
Cholesterol interaction scores for every residue, rendered on the membrane-facing surface (A) and a snake plot (B). Blue indicates no interaction with cholesterol, red indicates a high probability for cholesterol interaction. Inset of (A) shows the middle row from Fig. 1, to orient the viewer and to facilitate comparison to cholesterol densities. The highest-ranked locations are referred to in the text as h56i and h6o, and appear in the far right image. In (B), the residues of h6o are indicated as diamonds, and h56i as squares. To see this figure in color, go online.
Cholesterol also interacts with highest-ranked sites from Martini analysis in all-atom simulations, revealing additional structural and dynamic details
Cholesterol also interacts with both h6o and h56i in unbiased all-atom simulations. Two simulations of the receptor, one bound to ZM241385 and one apo, each 6 μs in duration, were obtained in a dipalmitoyl phosphatidyl choline/dioleoyl phosphatidyl choline/cholesterol mixture. The choice of receptor states was motivated by recent experimental work indicating a difference in lipid-protein interactions upon ligand binding (N. Malmstadt, personal communication). A single cholesterol remains tightly bound to h56i throughout the entire ZM241385 trajectory; in the apo trajectory a cholesterol occupies h56i continuously for 2.6 μs. An illustrative configuration in Fig. 5 A from the apo trajectory shows several residues that form the h56i interaction site. Note the presence of a lysine positioned to interact with the hydroxyl, a common feature in other cholesterol interaction motifs, including the CRAC and CCM. Note also that the site is positioned between helices 5 and 6, and therefore definition of the site requires superimposing the sequence on top of the structure; identifying the residues of the interaction site on the basis of sequence alignment alone would be challenging.
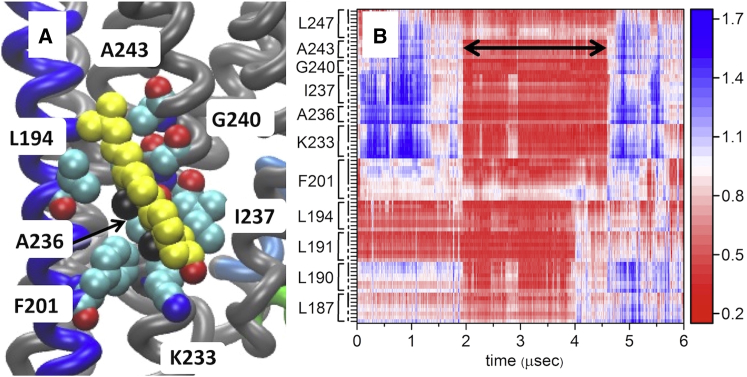
Figure 5.
Snapshot showing disposition of cholesterol with h56i in all-atom simulation (A), and distance between cholesterol and h56i residues during all-atom simulation (B). Cholesterol (yellow) interacts with h56i in all-atom simulations of both ZM241385-bound and apo (shown). Note the position of K233, interacting with the hydroxyl, shown in red. The methyl groups of the β-face of cholesterol are shown in black. (B) shows the distance between each heavy atom of selected side chains and the closest atom of any cholesterol (in nm, scale bar on the right). Although many cholesterols make transient contacts with some residues of h56i, a binding event (black arrow) appears as a sustained, close approach of several structurally contiguous residues. The sequence is indicated at the left, the protein backbone is indicated by the dashed line, and the side chains by the solid vertical lines. To see this figure in color, go online.
A map of the distance of each residue to the nearest atom of any cholesterol (Fig. 5 B) reveals the difference between transient encounters and long-lived interaction events. Interaction events entail the close approach to several residues simultaneously; when this occurs, the interaction is maintained for a significant period of time. An interaction event observed during the apo receptor simulation is used to illustrate the point, as during the ZM241385 trajectory a single cholesterol remains bound throughout the entire simulation (Fig. S5). Fig. 5 B also shows that Leu191 and Leu194 are always in contact with a cholesterol, even when a single cholesterol is not bound to the h56i. These residues were among the most highly scored in the Martini analysis, as shown in Fig. 4 B.
Cholesterol is dynamic even when tightly interacting with the protein, frequently rotating about its long axis. This behavior is evident in a 200 ns portion of the apo receptor interaction event detailed in Fig. 5, and shown in Movie S1. Although this is in contrast with the conventional view of ligand binding, in which the ligand is expected to occupy a well-defined binding pose, small ligands are also observed to be quite dynamic when bound to the orthosteric binding pocket of A2AR (66).
Cholesterol also interacts with h6o in both trajectories. Fig. 6 B shows the distance between the closest of all cholesterol atoms and the residues of h6o during the ZM241385 bound trajectory. Two distinct cholesterol binding events are apparent, indicated by the simultaneous close approach of a cholesterol to all residues of h6o; these events are marked by two sets of black arrows. That these events are interactions with single cholesterols is easily verified by visual inspection, as shown in Fig. 6 A and Movie S2. That these binding events are also dynamic is clear from Movie S2. In contrast, cholesterol only interacts with h6o during the first 250 ns of the apo trajectory (Fig. S6). At other times during the apo trajectory, cholesterol makes transient contacts with some of the residues, but it never binds all the residues of the site simultaneously after the first 250 ns. In particular, it never forms a sustained interaction with hydroxyl of S263. In a third simulation of the receptor bound to the agonist NECA, there is no interaction of cholesterol whatsoever with h6o, as shown in Fig. S7.
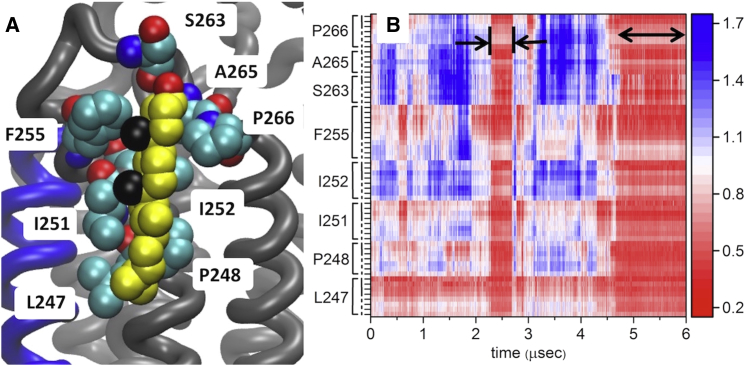
Figure 6.
Snapshot showing disposition of cholesterol with h6o in all atom simulation (A), and distance between cholesterol and h6o residues during all-atom simulation (B). A snapshot of cholesterol interacting with h6o in the all-atom simulation of the receptor bound to ZM241385 is shown in (A). All labeled residues are also observed to interact with cholesterol in several crystal structures of the receptor bound to ZM241385. Note the position of S263, interacting with the hydroxyl, shown in red. The methyl groups of the β-face are shown in black. (B) shows the distance between the nearest cholesterol atom and each heavy atom of the residues (in nm, scale bar on the right) indicated in the (A) over the entire simulation. Two interaction events are indicated with horizontal black arrows, the backbone of the protein is indicated with dashes, and the side chains with solid vertical lines. To see this figure in color, go online.
Although the trajectories of the receptor in three different states are suggestive of a ligand-dependent interaction of cholesterol at h6o, none of the cholesterol-binding events appear to be connected to obvious conformational changes in either the global structure, or the individual helices. This is shown in Figs. S8 and S9, which show overall conformational landscape of the protein, and the root mean square deviation of individual helices during the apo and ZM241385-bound simulations. Comparison of the helix root mean square deviations in particular and the cholesterol-binding events does not reveal any change in helix 5 or 6 upon cholesterol binding or unbinding. Thus, we expect that ligand-dependent preference for cholesterol is “built in” to the initial structure.
The two cholesterol-binding sites are located in the same region of the protein, but in opposite leaflets, with Leu247 located near the bilayer center and positioned to interact with cholesterols in either leaflet. The motif of h56i is similar to other sites reported on GPCRs: it occupies a groove between helices 5 and 6, and has a Lys positioned to interact with the headgroup of cholesterol. In contrast, h6o is not located in a groove between helices, and instead is located mainly on helix 6, and has a serine located on extracellular loop 3. Interacting with the hydroxyl on cholesterol, the serine takes the place of the lysine that is often found on cholesterol-binding motifs.
Discussion
CG simulations of the A2AR with Martini show the local environment of the protein to be enriched in cholesterol, and identify areas on the protein surface that prefer interacting with cholesterol over other lipids. A residue-based scoring function that rewards protein-cholesterol contacts and the duration of interaction identifies two cholesterol interaction sites. The first, h56i, is located between helices 5 and 6 in the inner leaflet, and the second, h6o, is located on helix 6 in the outer leaflet.
A total of 12 μs of unbiased all-atom simulations of the receptor in two different ligation states (apo, and bound to the inverse agonist ZM241385) strongly support the interaction of cholesterol at these locations, with cholesterol interacting at both locations in both trajectories; in one case, a cholesterol remains bound at h56i for the entire 5 μs trajectory. The all-atom data reveal the interactions to be dynamic, with the cholesterol free to rotate about its long axis even when associated closely with the interaction sites. In addition, the interaction of cholesterol at h6o is strikingly similar to several recent high-resolution crystal structures of A2AR bound to ZM241385, in which cholesterol is found at the same location, interacting with the same residues (30, 31, 32). Recently, Guixà-González, et al. (20) proposed that cholesterol may access the orthosteric-binding site from the same location. We do not observe spontaneous entry of cholesterol in 17 μs of AA simulation.
Prior work, both simulation (20, 21, 26, 67, 68) and experimental (10, 14, 18), on cholesterol-GPCR interactions has often focused on a single interaction site, whether it is the CRAC motif or the CCM. This is logical, as these were the only known cholesterol interaction sites, identified through a combination of crystallography, bioinformatic analysis, and functional studies. After earlier, unbiased simulations of cholesterol A2AR interactions (27), our results suggest that cholesterol interaction sites on single GPCRs may be more numerous than previously thought, with a broad distribution of interaction times. This should be taken into account when designing experiments and interpreting cholesterol-dependent functional data, as changing the bulk cholesterol concentration will change the occupancy of all cholesterol interaction sites. Thus, a catalog of all cholesterol interaction sites on the protein would be of great use in designing mutants to test the functional consequences of direct cholesterol interactions.
Although anecdotal, the all-atom data suggest that cholesterol interaction at h6o is favored when the receptor is in the inactive state. Cholesterol is observed to interact at this location in the ZM241385-bound receptor, but not when bound to the agonist NECA. The apo simulation was initiated from the ZM241385 structure by deleting the ligand, and therefore is expected to retain a memory of the inactive state. In this case, cholesterol briefly binds h6o in the beginning of the simulation, but then does not interact during the remainder of the simulation. High-resolution structures of the receptor have cholesterol at this location, and all of these structures are bound to the inverse agonist ZM241385.
A related function of cholesterol interaction sites may be to couple trafficking/localization and function, especially given that there seem to be multiple cholesterol interaction sites on A2AR. These appear to be relatively low affinity interactions, with the consequence that they will not be occupied by cholesterol when the protein is inserted into the membrane of the endoplasmic reticulum, and only bind cholesterol once the protein is located in a higher cholesterol environment, perhaps ensuring that the protein signals after being trafficked to the plasma membrane.
Our results suggest that GPCRs may possess several cholesterol interaction sites, which may interact with cholesterol in a ligand-dependent way. An exhaustive catalog of putative cholesterol interaction sites should be of great value, in tandem with experimental tests of cholesterol-dependent function. Work is ongoing to experimentally test the sites reported here for their effect on cholesterol-dependent signaling.
Author Contributions
E.L. and C.A. designed the research. E.L., C.A., and E.R. wrote the manuscript. C.A., E.R., and L.Y. performed the analysis.
Acknowledgments
We thank Alex Sodt for sharing his Voronoi analysis of lipid solvation shells. The Anton2 machine at National Resource for Biomedical Supercomputing/Pittsburgh Supercomputing Center was generously made available by D.E. Shaw Research.
C.A., E.L., and L.Y. were supported by National Institutes of Health grants P20GM104316 and R01 GM120351. E.R. was supported by the Delaware Space Grant college program, National Aeronautics and Space Administration (NNX15AI19H). Martini simulations were performed on the “Naja” cluster at the University of Delaware, supported by the National Institutes of Health Institutional Development Award program through grant P20GM104316. Anton2 computer time was provided by the National Resource for Biomedical Supercomputing, the Pittsburgh Supercomputing Center, and the Biomedical Technology Research Center for Multiscale Modeling of Biological Systems through grant P41GM103712-S1 from the National Institutes of Health.
Editor: D. Peter Tieleman.
Footnotes
Supporting Materials and Methods, nine figures, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31070-6.
Supporting Material
See Fig. 5 legend for explanation of rendering scheme.
See Fig. 6 legend for explanation of rendering scheme.
References
- 1.Gerl M.J., Sampaio J.L., Simons K. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin X., Lorent J.H., Levental I. Domain stability in biomimetic membranes driven by lipid polyunsaturation. J. Phys. Chem. B. 2016;120:11930–11941. doi: 10.1021/acs.jpcb.6b06815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sezgin E., Levental I., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 5.Epand R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006;45:279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Barrett P.J., Song Y., Sanders C.R. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 8.Baier C.J., Fantini J., Barrantes F.J. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci. Rep. 2011;1:69. doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fantini J., Barrantes F.J. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jafurulla M., Tiwari S., Chattopadhyay A. Identification of cholesterol recognition amino acid consensus (CRAC) motif in G-protein coupled receptors. Biochem. Biophys. Res. Commun. 2011;404:569–573. doi: 10.1016/j.bbrc.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Pucadyil T.J., Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin1A receptors from bovine hippocampus. Biochim. Biophys. Acta. 2004;1663:188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Levitan I., Singh D.K., Rosenhouse-Dantsker A. Cholesterol binding to ion channels. Front. Physiol. 2014;5:65. doi: 10.3389/fphys.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paila Y.D., Chattopadhyay A. The function of G-protein coupled receptors and membrane cholesterol: specific or general interaction? Glycoconj. J. 2009;26:711–720. doi: 10.1007/s10719-008-9218-5. [DOI] [PubMed] [Google Scholar]
- 14.Hanson M.A., Cherezov V., Stevens R.C. A specific cholesterol binding site is established by the 2.8 A structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagerström M.C., Schiöth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 16.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 18.Pydi S.P., Jafurulla M., Chattopadhyay A. Cholesterol modulates bitter taste receptor function. Biochim. Biophys. Acta. 2016;1858:2081–2087. doi: 10.1016/j.bbamem.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez M.G., Mansfield K.S., Malmstadt N. The functional activity of the human serotonin 5-HT1A receptor is controlled by lipid bilayer composition. Biophys. J. 2016;110:2486–2495. doi: 10.1016/j.bpj.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guixà-González R., Albasanz J.L., Selent J. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun. 2017;8:14505. doi: 10.1038/ncomms14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manna M., Niemelä M., Vattulainen I. Mechanism of allosteric regulation of β2-adrenergic receptor by cholesterol. eLife. 2016;5:e18432. doi: 10.7554/eLife.18432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sodt A.J., Venable R.M., Pastor R.W. Nonadditive compositional curvature energetics of lipid bilayers. Phys. Rev. Lett. 2016;117:138104. doi: 10.1103/PhysRevLett.117.138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedger G., Sansom M.S.P. Lipid interaction sites on channels, transporters and receptors: recent insights from molecular dynamics simulations. Biochim. Biophys. Acta. 2016;1858:2390–2400. doi: 10.1016/j.bbamem.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta D., Chattopadhyay A. Identification of cholesterol binding sites in the serotonin1A receptor. J. Phys. Chem. B. 2012;116:12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- 25.Khelashvili G., Grossfield A., Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins. 2009;76:403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitman M.C., Grossfield A., Feller S.E. Role of cholesterol and polyunsaturated chains in lipid-protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J. Am. Chem. Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 27.Lee J.Y., Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J. Am. Chem. Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cang X., Du Y., Jiang H. Mapping the functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B. 2013;117:1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 29.Neale C., Herce H.D., García A.E. Can specific protein-lipid interactions stabilize an active state of the beta 2 adrenergic receptor? Biophys. J. 2015;109:1652–1662. doi: 10.1016/j.bpj.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batyuk A., Galli L., Cherezov V. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci. Adv. 2016;2:e1600292. doi: 10.1126/sciadv.1600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W., Chun E., Stevens R.C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segala E., Guo D., Cooke R.M. Controlling the dissociation of ligands from the adenosine A2A receptor through modulation of salt bridge strength. J. Med. Chem. 2016;59:6470–6479. doi: 10.1021/acs.jmedchem.6b00653. [DOI] [PubMed] [Google Scholar]
- 33.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson K.A. Crystal structures of the A2A adenosine receptor and their use in medicinal chemistry. In Silico Pharmacol. 2013;1:22. doi: 10.1186/2193-9616-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlsson J., Yoo L., Jacobson K.A. Structure-based discovery of A2A adenosine receptor ligands. J. Med. Chem. 2010;53:3748–3755. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Malley M.A., Helgeson M.E., Robinson A.S. The morphology and composition of cholesterol-rich micellar nanostructures determine transmembrane protein (GPCR) activity. Biophys. J. 2011;100:L11–L13. doi: 10.1016/j.bpj.2010.12.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnarez C., Marrink S.J., Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci. Rep. 2013;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wassenaar T.A., Ingólfsson H.I., Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 39.Periole X., Cavalli M., Ceruso M.A. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 2009;5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 40.Melo M.N., Ingólfsson H.I., Marrink S.J. Parameters for Martini sterols and hopanoids based on a virtual-site description. J. Chem. Phys. 2015;143:243152. doi: 10.1063/1.4937783. [DOI] [PubMed] [Google Scholar]
- 41.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 42.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 43.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 44.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 45.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 46.Doré A.S., Robertson N., Marshall F.H. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 48.Wu E.L., Cheng X., Im W. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 50.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanommeslaeghe K., MacKerell A.D., Jr. Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 56.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 57.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 58.Lippert R.A., Predescu C., Shaw D.E. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J. Chem. Phys. 2013;139:164106. doi: 10.1063/1.4825247. [DOI] [PubMed] [Google Scholar]
- 59.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 60.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 61.Shan Y., Klepeis J.L., Shaw D.E. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005;122:54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 62.Kräutler V., van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 63.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 64.Beaven A.H., Maer A.M., Im W. Gramicidin A channel formation induces local lipid redistribution I: experiment and simulation. Biophys. J. 2017;112:1185–1197. doi: 10.1016/j.bpj.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metzler R., Klafter J. From a generalized chapman−kolmogorov equation to the fractional klein−kramers equation. J. Phys. Chem. B. 2000;104:3851–3857. [Google Scholar]
- 66.Lee J.Y., Lyman E. Agonist dynamics and conformational selection during microsecond simulations of the A(2A) adenosine receptor. Biophys. J. 2012;102:2114–2120. doi: 10.1016/j.bpj.2012.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J.Y., Patel R., Lyman E. Ligand-dependent cholesterol interactions with the human A(2A) adenosine receptor. Chem. Phys. Lipids. 2013;169:39–45. doi: 10.1016/j.chemphyslip.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyman E., Higgs C., Voth G.A. A role for a specific cholesterol interaction in stabilizing the Apo configuration of the human A(2A) adenosine receptor. Structure. 2009;17:1660–1668. doi: 10.1016/j.str.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See Fig. 5 legend for explanation of rendering scheme.
See Fig. 6 legend for explanation of rendering scheme.