Abstract
Background
Parathyroid tumors are mostly considered monoclonal neoplasms, the rationale for focused parathyroidectomy (PTX) in primary hyperparathyroidism (PHPT). We reported that flow sorting parathyroid tumor cells and methylation-sensitive PCR (me-PCR) of polymorphic HUMARA and PGK alleles in DNA reveals that up to 35% of parathyroid tumors are polyclonal. We sought to confirm these findings and assess for clinical relevance.
Methods
Parathyroid tumors from 286 female PHPT patients were analyzed for clonal status. Tumor clonal status was compared with clinical variables and operative findings. Statistical analysis was performed and significance was established at P< 0.05.
Results
176 (62%) patients were informative for HUMARA and/or PGK. Assignment of clonal status was made in 119 (68%) tumors, of which 64 (54%) were monoclonal and 55 (46%) were polyclonal. Comparison of tumor clonal status to clinical variables in patients with complete operative data (N=82) showed that while clinical features were the same between tumor types, patients with polyclonal tumors more often had multiple gland disease (RR 4.066, CI 1.016 – 16.26; p= 0.039) potentially missed at unilateral neck exploration.
Conclusions
This work confirms that PHPT is often the result of polyclonal tumors, and that parathyroid tumor clonal status may be associated with multiple gland disease.
Introduction
Parathyroid adenoma originating from a single parathyroid gland is the most common cause of nonfamilial primary hyperparathyroidism (PHPT).(1) Less commonly, PHPT patients have primary chief cell hyperplasia or multiple adenomas as the cause of their disease. These processes of parathyroid neoplasia cannot be predicted on clinical grounds and can be difficult to distinguish on pathologic examination. Their importance lies in their relationship with multiple gland disease and its impact on approach to parathyroidectomy (PTX) and results of surgery. Removal of single adenoma by either a focused (i.e. unilateral) or bilateral exploration and PTX is likely curative; however, cure of PHPT due to multiple gland hyperplasia can be less reliable following surgery.(2)
The somatic mutation theory of cancer holds that a finite set of somatic mutations in DNA result in the transformation of cells and their progression to malignancy.(3) According to this framework, parathyroid adenomas in nonfamilial PHPT are predicted to be monoclonal expansions of a single transformed parathyroid cell, whereas hyperplasias may be the result of poly- or oligo-clonal expansions of multiple cells due to exogenous stimuli. Tumor clonal status may then be viewed as a potential surrogate for both underlying etiology and type of parathyroid neoplasia. The topic of parathyroid tumor clonal status has been the subject of several studies with mixed and controversial results.(4–7) In particular, the finding of parathyroid tumor polyclonal status by several investigators has been questioned due to methodologic approach (e.g. use of microdissection to remove polyclonal stroma) and the assays used to assign tumor clonal status.
We previously conducted a study of parathyroid tumors from patients with nonfamilial PHPT due to single gland disease in which cells isolated from these tumors were dispersed and flow sorted to yield purified populations of oxyphil and chief cells. These isolated cells were analyzed both functionally and genetically, and our results showed that a significant proportion (9/14, 36%) of apparent adenomas were in fact polyclonal.(8) Now we have examined an expanded cohort of 119 patients and have found that a significant proportion (up to 46%) of patients have polyclonal parathyroid tumors as the cause of their disease and that these patients are otherwise indistinguishable based on clinical and biochemical criteria. In addition, among 82 patients well-characterized in terms of demographic, biochemical, operative, and pathologic data we found that polyclonal tumor status is associated with the presence of multiple gland disease that may be missed with unilateral exploration. Our findings indicate that the etiology of PHPT is heterogeneous and that underlying parathyroid tumor clonal status may be important to disease outcome following PTX.
Methods
Primary parathyroid tumors and clinical data were obtained from consenting PHPT patients under IRB – approved protocols at The University of Maryland Baltimore (UMB) (N = 135, 2012 – 2016) and Duke University (N=151, 2001 – 1012). De-identified tumor samples were transferred from Duke to UMB under a materials transfer agreement between the two institutions. Tumor samples were collected from resected tumors in the operating room and immediately placed in liquid nitrogen. Duplicate samples were fixed in buffered formalin, embedded, sectioned, stained with hematoxylin and eosin, and examined to ensure parathyroid tumor identity and cellularity. Parathyroid tumor samples were kept at −80°C until use. Peripheral blood lymphocytes (PBLs) were isolated from patient-matched whole blood using RBC Lysis Buffer (Biolegend, CA).
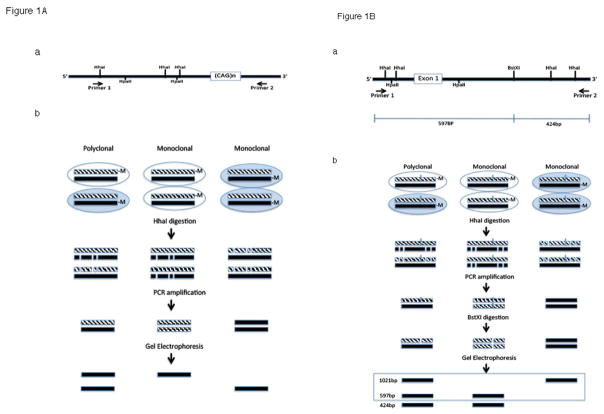
Genomic DNA extraction from peripheral blood lymphocytes and parathyroid adenoma tissue was performed using DNeasy Blood & Tissue Kit (Qiagen). Clonal status at the HUMARA locus was determined via restriction enzyme (HhaI) digestion and PCR amplification following the procedure described by Allen et al.(9). HUMARA primer sequences were: primer 1, 5′-TCCAGAATCTGTTCCAGAGCGTGC -3′; and primer 2, 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′. The PGK clonality assay was modified based on the method described by Gilliland et al.(10). The PGK primer sequences were: primer 1, 5′-TGTGGGGCGGTAGTGTGGGCCCTGTTCCTG-3′; and primer 2, 5′-AACCGTGTTGGCAAGTGACTAGAGATCCAC-3′. A schematic diagram depicting the me-PCR assays for HUMARA and PGK is shown in Figure 1.
Figure 1.
A. The HUMARA me-PCR – based method of clonal analysis with map of the region amplified in the first exon of the HUMARA gene on X chromosome that includes the variably methylated HhaI restriction site B. The PGK PCR-based method of clonal analysis with map of PGK gene in the vicinity of the variably methylated HhaI or HpaII sites and the BstXI polymorphism.
De-identified clinical and research data were maintained in separate password-protected databases linked by a study code. Clonal status from de-identified tumor samples and clinicopatholofic data were analyzed for significant association using SPSS software (International Business Machines, Armonk, NY). Significance was established at p<0.05.
Results
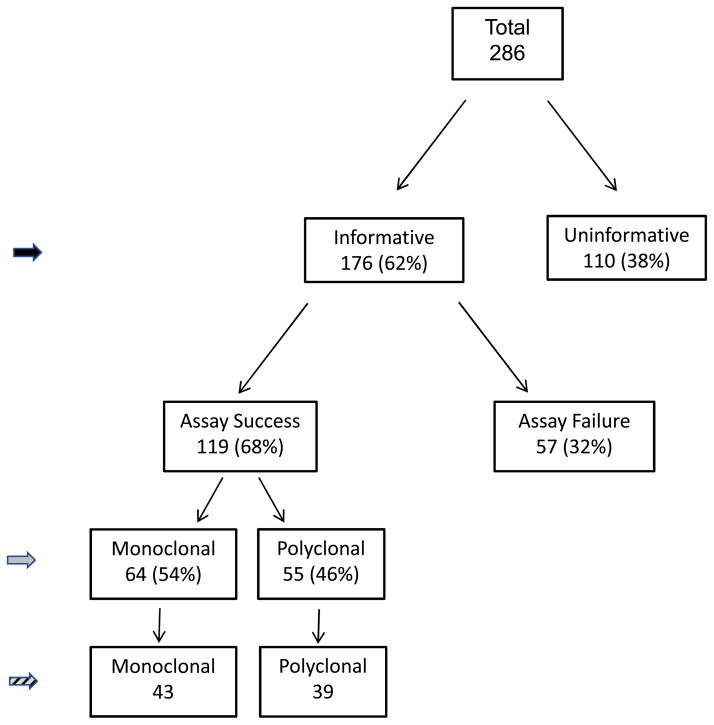
PBL samples from 286 PHPT patients were analyzed for clonal status by me-PCR at both the HUMARA and PGK loci. Of these patients, 176 (62%) were informative for either or both assays in PBL (Figure 2, solid black arrowhead). Definitive assignment of parathyroid tumor clonal status in these informative patients was made in 119 patients based on clear identification of appropriate size PCR bands in undigested and HhaI digested DNA from parathyroid tumor samples (Figure 2, solid grey arrowhead). There were 57 patient tumor samples that yielded ambiguous clonal results on repeated testing and were deemed an assay failure. Reasons behind the unclear results included insufficient tumor tissue for repeat assay, failure of PCR reactions in tumor DNA, inability to resolve appropriate sized bands on the gel, or failure of control (undigested) tumor DNA to show bands as expected from the results of the corresponding patient’s PBL sample. Of the 119 patients with clearly assigned clonal status, 82 patients had complete clinical and operative records available for review (Figure 2, hatched arrowhead).
Figure 2.
Flow diagram of PHPT patients studied. Solid grey arrowhead indicates patients examined to determine the prevalence of monoclonal and polyclonal tumors in the study cohort and the clinical and biochemical features of patients with monoclonal and polyclonal tumors (summarized in table 1). Hatched arrowhead indicates patients examined to determine surgical and pathologic results in patients with monoclonal and polyclonal tumors (summarized in table 2).
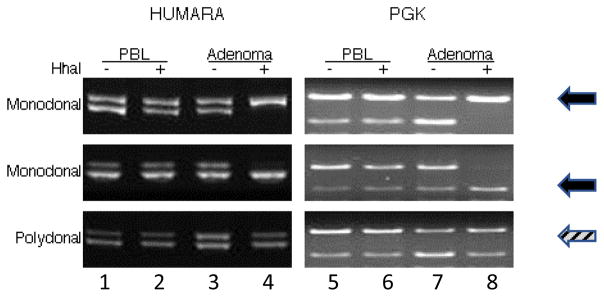
Representative me-PCR gels from two monoclonal and one polyclonal tumors are shown in Figure 3. Amplification of alleles in control DNA from PBL and undigested tumor DNA with primers specific for HUMARA (Figure 3, lanes 1–3) yielded two bands of approximately 280 bp, differing in size by the presence of (CAG)N repeats. Amplification of monoclonal tumor DNA digested with HhaI (Figure 3, lane 4) resulted in loss of one unmethylated HUMARA allele from the single clone yielding a single band (solid arrows), whereas digestion of polyclonal tumor DNA with HhaI resulted in loss of one or the other unmethylated alleles from multiple clones resulting in two bands (hatched arrows). Similarly, amplification of alleles in control DNA from PBL and undigested tumor DNA with primers specific for PGK followed by BstXI digestion of PCR products yielded two bands of 1021 and 597 bp (Figure 3, lanes 5–7). Amplification of monoclonal tumor DNA digested with HhaI (Figure 3, lane 8) resulted in loss of either unmethylated PGK allele with or without the BstXI restriction site from a single tumor clone resulting in a single band of either 1021 bp or 597 bp (solid arrows). In contrast, polyclonal tumor DNA digested with HhaI resulted in loss of one or the other unmethylated alleles with and without the BstXI restriction site from multiple clones resulting in both bands (hatched arrows).
Figure 3.
Clonal status of parathyroid tumors. Me-PCR assays at HUMARA (lanes 1–4) and PGK (lanes 5–8) loci from 3 representative parathyroid tumors are shown. Lanes 1,2 and 5,6 = control DNA from peripheral blood lymphocytes (PBL); Lanes 3,4 and 7,8 = DNA from parathyroid adenomas. Lanes 3 and 7 = undigested adenoma DNA. Lanes 4 and 8 = HhaI digested adenoma DNA. HhaI = methylation sensitive restriction enzyme used to digest input DNA.
Comparison of clinical and laboratory data of 119 patients with unambiguously determined tumor clonal status indicates that these patients are indistinguishable in terms of age, race, BMI, biochemical and imaging results at presentation (Table 1). Symptom data was not available for the Duke patients; however, among 62 UMB patients with preoperatively recorded symptoms, there was no difference in the presence or type of symptoms reported between patients with monoclonal and polyclonal tumors. Patients with monoclonal tumors showed a statistically nonsignificant but nonetheless notable tendency toward reduced bone density (76% vs. 54% with osteoporosis/osteopenia) and positive sestamibi scans (66% vs 50% positive). The frequency of polyclonal tumors was the same in the patients from UMB (29/63, 46%) and Duke (26/56, 46%).
Table 1.
Characteristics of PHPT patients (N = 119)
| Feature | Monoclonal | Polyclonal | P value |
|---|---|---|---|
| Clonal status | 64 (54%) | 55 (46%) | |
| Age (mean, range) | 59, 24 – 85 | 51, 20 – 77 | 0.277 |
| Race | |||
| White | 37 | 27 | 0.385 |
| Black | 9 | 13 | |
| Other/Unknown | 18 | 15 | |
| BMI (Mean, N= 55) | 27 | 30 | 0.211 |
| Presentation | |||
| Asymptomatic | 5 (15%) | 7 (25%) | |
| Symptomatic | 29 (85%) | 21 (75%) | 0.307 |
| Bone density | |||
| OPO/OPE | 20 (76%) | 12 (54%) | 0.101 |
| Normal | 6 (24%) | 10 (46%) | |
| Pre-op serum Ca+2 (mean, mg/dl; N=97) | 10.9 | 11.2 | 0.225 |
| Pre-op Intact PTH (mean, pg/ml; N=97) | 139 | 179 | 0.988 |
| 25-OH Vit D (mean, IU/L; N=66) | 30 | 33 | 0.632 |
| Pre-op serum Cr | 0.88 | 0.84 | 0.567 |
| Pre-op localization (MIBI) | |||
| Positive | 12 (66%) | 10 (50%) | 0.251 |
| Negative | 6 (34%) | 10 (50%) | |
BMI, body mass index; OPO, osteoporosis; OPE osteopenia
Analysis of data from the subset of 82 patients with unambiguous tumor clonal status and complete operative and pathologic information revealed that these patients were not different in terms of operative approach taken, intra-operative PTH kinetics, postoperative calcium, and the frequency of persistent disease (table 2). There was a notable, but statistically non-significant trend toward higher average postoperative PTH levels in patients with polyclonal versus monoclonal tumors (70 pgm/ml vs. 57 pg/ml, respectively). There was no difference in the index parathyroid tumor weight (single adenoma or largest tumor in the case of multiple gland disease) or pathologic diagnosis of hyperplasia or adenoma in polyclonal vs. monoclonal tumors. However, patients with polyclonal tumors were more likely to have multiple gland disease (P = 0.033, O.R. 4.1 (CI 1.02 – 16.3) vs. monoclonal). Altogether, these results demonstrate that patients with monoclonal and polyclonal tumors are indistinguishable based on routine clinical assessment; however, patients with polyclonal tumors were significantly more likely to have multiple gland disease.
Table 2.
Operative results of PHPT patients by tumor clonal status (N = 82)
| Feature | Monoclonal | Polyclonal | P value |
|---|---|---|---|
| Clonal Status | 43 (52%) | 39 (48%) | |
| Operative approach | 0.556 | ||
| UNE | 16 (37%) | 17 (43%) | |
| BNE | 27 (63%) | 22 (57%) | |
| % ioPTH drop | 67 | 69 | 0.787 |
| ioPTH slope (B2 – PTH10) | −14 | −16 | 0.691 |
| Final ioPTH level (mean &range, pg/ml) | 40 | 43 | 0.886 |
| Glands abnormal | |||
| Single | 40 (93%) | 30 (77%) | |
| Multiple | 3 (7%) | 9 (23%) | 0.039 |
| Gland weight** (mean, mg) | 830 | 873 | 0.738 |
| Persistent PHPT (%) | 1 (2%) | 2 (5%) | 0.355 |
| Post-op serum Ca+2 (mean, mg/dl;N=49) | 9.4 | 9.5 | 0.659 |
| Post-op Intact PTH (mean, pg/ml;N=45) | 57 | 70 | 0.927 |
UNE, unilateral neck exploration; BNE, bilateral neck exploration.
B2, baseline 2 defined as PTH level drawn prior to removal of the index parathyroid tumor; PTH10, intra-operative PTH drawn 10 minutes after removal of the index gland; slope = B2-PTH10/10
weight of gland (index tumor) used to determine clonal status
Examination of operative results parsed by operative approach are shown in Table 3. We observed that patients with polyclonal tumors who had unilateral neck exploration were less likely to have multiple abnormal glands identified and removed than those patients who had bilateral exploration (1/16 vs 8/14, P = 0.025) suggesting that patients with polyclonal tumors may have abnormal glands missed at unilateral neck exploration. No such difference was observed in patients with monoclonal glands who had unilateral exploration or in patients with either tumor type who had bilateral exploration.
Table 3.
Parathyroid tumor clonal status and pathology at discovered at surgery
| Clonality | N | Operative Findings @ UE | Operative Findings @ BNE | ||
|---|---|---|---|---|---|
|
| |||||
| SGD | MGD | SGD | MGD | ||
|
|
|||||
| Monoclonal | 43 | 16 | 0 | 24 | 3 |
| Polyclonal | 39 | 16 | 1 | 14 | 8* |
| Total | 82 | 32 | 1 | 38 | 11 |
UE = unilateral exploration; BNE = bilateral neck exploration; SGD = single gland disease; MGD = multiple gland disease
P = 0.025 for MGD identified at BNE vs. UE
Discussion
In this study, we report the largest analysis of parathyroid tumor clonal status to date, showing that a significant proportion of patient with PHPT have polyclonal rather than monoclonal tumors. These findings support results of smaller series that question the notion that most parathyroid tumors are monoclonal in origin (Table 4). (4–8, 11) Our results also show that patients with polyclonal tumors are at greater risk for having multiple gland disease, and these glands may go undiscovered at unilateral neck exploration. Importantly, patients with polyclonal tumors are presently not discernable by standard clinical criteria justifying future investigation for the cause and markers of polyclonal parathyroid neoplasia.
Table 4.
Select studies of clonal status of parathyroid tumors in PHPT
| Author | Year | N | HPT Type | Method | Ratio M:P (%P) |
|---|---|---|---|---|---|
| Fiaklow | 1977 | 3* | Primary | G-6-PD | 0:4 (0) |
| Arnold | 1988 | 8 | Primary | HPRT RFLP | 6:2 (25) |
| Friedman | 1989 | 34 | Primary | Ch11 LOH | 9:25 (73) |
| Shan | 1997 | 8 | Primary | HUMARA me-PCR | 7:1 (12) |
| Sanjuan | 1998 | 13** | Primary | HUMARA me-PCR | 2:11 (85) |
| Shi | 2014 | 14 | Primary | HUMARA/PGK me-PCR | 9:5 (36) |
| Present study | 2017 | 119*** | Primary | HUMARA/PGK me-PCR | 64:55 (46) |
4 tumors from 3 patients;
8 patients with adenomas + 5 patients with hyperplasia;
57 pts with “unclear” clonal status result; (G-6-PDH = glucose-6 phosphate dehydrogenase; HPRT RFLP = hypoxanthine phosphoribosyltransferase restriction-fragment-length-polymorphism; Ch11 LOH = chromosome 11 loss of heterozygosity; HUMARA me-PCR = Human androgen receptor gene methylation-sensitive PCR; PGK me-PCR = Phosphoglycerate kinase gene methylation-sensitive PCR
The method of assigning tumor clonal status in this study is worthy of close scrutiny. Me-PCR has been employed in numerous studies of tumor clonality, including parathyroid and is considered a reliable method of determining clonal status in endocrine neoplasia.(6, 12) While this assay method could be subject to inaccuracy due to somatic tumor DNA methylation, this seems unlikely. Most recently, Corrado et al. reported a HUMARA me-PCR analysis of clonal status in two tumors from patients with neonatal severe hyperparathyroidism, finding that both tumors were polyclonal expansions.(13) They employed a second, independent method, array comparative genomic hybridization, to examine gain/loss of chromosomal material and found concordance between the two methods in terms of parathyroid tumor clonal status. Future studies likely will need to employ sequence based approaches as secondary methods to confirm me-PCR assignment of tumor clonal status.(14)
Strengths of our study include the large number of tumors available for analysis, the use of multiple assays to assign tumor clonal status, and the careful procurement and processing of samples to ensure that contaminating stroma did not yield spurious assignment of polyclonal tumor status. The most significant limitation of the study is that the me–PCR HUMARA and PGK clonality assays are based on the biology of X-inactivation and therefore can be applied only to female patients. To minimize potential assignment error, we included only the 119 patients with informative alleles in PBL who had unambiguous clonality assay results in undigested and digested tumor DNA to determine of the frequency of clonal status in the patients of the study. While it is possible that this filter could introduce bias to the study and overestimate the frequency of polyclonal disease, this seems unlikely since there is no biologic reason why technical failure or ambiguity of results based on size difference of the maternal and paternal HUMARA and PGK alleles would occur differentially in patients with single or multiple gland disease, and so is unlikely to change the conclusion. Future studies using sequence – based fragment analysis of HUMARA alleles may improve upon ambiguous assignment by the gel-based method.(13) The retrospective nature of the analysis and availability of complete surgical and outcome data for 82 of the 119 tumors examined likely makes the study underpowered to detect small, but clinically meaningful clinical differences between tumor types. Finally, the absence of long-term biochemical and symptom follow-up for these patients (median FU 10 months) does not permit us to determine whether tumor clonal status matters to disease recurrence over time. A prospective, multicenter validation study is in development to address this important question.
Despite the limitations of this study, it is reasonable to conclude from our data that all parathyroid tumors are not genetically the same (i.e. monoclonal) and that parathyroid tumors may result from differing underlying etiologies, including a secondary stimulus leading to hyperplasia, polyclonal expansion and apparent adenoma formation. Whether primary chief cell hyperplasia described histologically and associated with multiple gland disease and poorer surgical outcomes is in fact polyclonal and reactive to an exogenous stimulus (e.g. chronic vitamin D deficiency) is an interesting question that will require future study.(2, 15) Regardless, if future work in expanded populations of PHPT confirm that parathyroid tumors clonal status is important to outcome, then it will be critically important to determine clonal status before surgery. Development of an actionable blood biomarker test to accurately predict either a monoclonal tumor amenable to directed PTX, or a polyclonal tumor better approached by BNE, or treatment of the driving stimulus (e.g. vitamin D deficiency) could become of great importance.
This work represents the first assessment of the frequency and clinical significance of parathyroid tumor clonal status in PHPT. These results confirm our published findings that PHPT is often the result of polyclonal parathyroid tumors, a conclusion that refutes the idea that all parathyroid tumors are monoclonal. While biochemical and demographic features of patients with both types of parathyroid tumors are similar, patients with polyclonal tumors more often have multiple gland disease that may be missed at unilateral exploration. Our results support the notion that PHPT is a heterogeneous disorder and reinforces the importance of developing a better understanding of underlying parathyroid tumor biology as a basis of improving treatment of PHPT.
Acknowledgments
Supported by National Institutes of Health Grant R01DK088188 (to J.A.O./J.K.)
Footnotes
Author contribution: J.A.O. and Y.S. designed the project, analyzed data and wrote the manuscript; Y.S. performed the experiments. P.A. performed data analysis, and co-wrote the manuscript. S.W. and J.F. collected clinical data; J.K., S.W., and N.B. edited the manuscript.
Presented at the annual meeting of The American Association of Endocrine Surgeons held in Orlando FL on April 1–4, 2017.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeLellis RA. Parathyroid tumors and related disorders. Mod Pathol. 2011;24(Suppl 2):S78–93. doi: 10.1038/modpathol.2010.132. [DOI] [PubMed] [Google Scholar]
- 2.Wallfelt C, Ljunghall S, Bergstrom R, Rastad J, Akerstrom G. Clinical characteristics and surgical treatment of sporadic primary hyperparathyroidism with emphasis on chief cell hyperplasia. Surgery. 1990;107(1):13–9. [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Fialkow PJ, Jackson CE, Block MA, Greenawald KA. Multicellular origin of parathyroid “adenomas”. N Engl J Med. 1977;297(13):696–8. doi: 10.1056/NEJM197709292971304. [DOI] [PubMed] [Google Scholar]
- 5.Arnold A, Staunton CE, Kim HG, Gaz RD, Kronenberg HM. Monoclonality and abnormal parathyroid hormone genes in parathyroid adenomas. N Engl J Med. 1988;318(11):658–62. doi: 10.1056/NEJM198803173181102. [DOI] [PubMed] [Google Scholar]
- 6.Sanjuan X, Bryant BR, Sobel ME, Merino MI. Clonality Analysis of Benign Parathyroid Lesions by Human Androgen Receptor (HUMARA) Gene Assay. Endocr Pathol. 1998;9(1):293–300. doi: 10.1007/BF02739689. [DOI] [PubMed] [Google Scholar]
- 7.Friedman E, Sakaguchi K, Bale AE, Falchetti A, Streeten E, Zimering MB, et al. Clonality of parathyroid tumors in familial multiple endocrine neoplasia type 1. N Engl J Med. 1989;321(4):213–8. doi: 10.1056/NEJM198907273210402. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Hogue J, Dixit D, Koh J, Olson JA., Jr Functional and genetic studies of isolated cells from parathyroid tumors reveal the complex pathogenesis of parathyroid neoplasia. Proc Natl Acad Sci U S A. 2014;111(8):3092–7. doi: 10.1073/pnas.1319742111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51(6):1229–39. [PMC free article] [PubMed] [Google Scholar]
- 10.Gilliland DG, Blanchard KL, Levy J, Perrin S, Bunn HF. Clonality in myeloproliferative disorders: analysis by means of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1991;88(15):6848–52. doi: 10.1073/pnas.88.15.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shan L, Nakamura M, Nakamura Y, Inoue D, Morimoto S, Yokoi T, et al. Comparative analysis of clonality and pathology in primary and secondary hyperparathyroidism. Virchows Arch. 1997;430(3):247–51. doi: 10.1007/BF01324809. [DOI] [PubMed] [Google Scholar]
- 12.Shattuck TM, Westra WH, Ladenson PW, Arnold A. Independent clonal origins of distinct tumor foci in multifocal papillary thyroid carcinoma. N Engl J Med. 2005;352(23):2406–12. doi: 10.1056/NEJMoa044190. [DOI] [PubMed] [Google Scholar]
- 13.Corrado KR, Andrade SC, Bellizzi J, D’Souza-Li L, Arnold A. Polyclonality of Parathyroid Tumors in Neonatal Severe Hyperparathyroidism. J Bone Miner Res. 2015;30(10):1797–802. doi: 10.1002/jbmr.2516. [DOI] [PubMed] [Google Scholar]
- 14.Prandi D, Baca SC, Romanel A, Barbieri CE, Mosquera JM, Fontugne J, et al. Unraveling the clonal hierarchy of somatic genomic aberrations. Genome Biol. 2014;15(8):439. doi: 10.1186/s13059-014-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castleman B, Cope O. Primary parathyroid hypertrophy and hyperplasia; a review of 11 cases at the Massachusetts General Hospital. Bull Hosp Joint Dis. 1951;12(2):368–78. [PubMed] [Google Scholar]