Abstract
The aim of the present study was to explore the miRNA-Gene regulatory mechanism in chronic lymphocytic leukemia (CLL), and identify new targets for the therapy of CLL. The miRNA expression dataset GSE62137 and mRNA expression dataset GSE22529 were downloaded from National Center of Biotechnology Information Gene Expression Omnibus database. In CLL samples compared with normal B cell samples, differentially expressed miRNAs (DEMs) were identified via the GEO2R instrument of GEO and differentially expressed genes (DEGs) were obtained via the limma package of R. Functional enrichment analysis of the DEGs was performed via the Database for Annotation, Visualization and Integrated Discovery. The targets of the DEMs were identified based on the miRNAWalk platform. The overlaps between the DEGs and the targets of the DEMs were selected, and the miRNA-Gene regulatory network was constructed based on the overlaps and the corresponding DEMs. A total of 63 DEMs and 504 DEGs were identified in CLL samples compared with normal B cell samples. Eleven enriched functional clusters of the DEGs were obtained. 405 miRNA-Gene regulatory pairs were identified. The miRNA-Gene regulatory pairs contained 351 target genes of the DEMs, including 9 overlaps with the DEGs. A miRNA-Gene regulatory network was constructed. Bioinformatics methods could help us develop a better understanding of the molecular mechanism of CLL. MiRNAs may play a critical role in regulating the process of CLL. They may affect CLL by regulating the processes of immunoreactivity and protein degradation. Genes such as Neurogenic Locus Notch Homolog Protein 2, PR/SET domain 4 and A-kinase anchoring protein 12 may be their regulating targets in CLL.
Keywords: chronic lymphocytic leukemia, gene expression omnibus, MiRNA-gene regulatory network, bioinformatics
Introduction
B-cell chronic lymphocytic leukemia (CLL) accounts for about 25% of all leukemia and is the most common form of lymphoid malignancy in western countries (1). CLL is characterized by the accumulation of the clonal expansion and resistance to apoptosis of immature CD5(+) B cells (2). The disease displays a high heterogeneity in its clinical course (3,4). Staging and prognostication of CLL is performed by two similar clinical staging systems (5,6). However, the clinical staging systems cannot fully reflect the high variability of CLL and predict survival of CLL patients, as well as the response to therapy (7). Molecular aberration plays an important role in the initiation and progression of CLL. Several factors that can predict the clinical course have been identified (8–10). The most frequent deletion of genomic DNA in CLL occurs in chromosome 13q13.4. This deletion occurs in ~50% of CLL cases and is associated with a long interval between diagnosis and the need for treatment (11). One study was reported that an increased DPF3 expression in CLL patients that was linked with a significant increase of STAT5 activation in myeloid lineage cells (granulocytes) and not in neoplastic B cells (12). However, the molecular mechanism of these associations is still largely unknown.
miRNAs are small noncoding RNA molecules that negatively regulate gene expression via degradation or translational repression of their targeted mRNAs (13). miRNAs played important roles in the pathogenesis of various human cancers and ~50% of the known human microRNAs were located at cancer-associated regions of the genome (14–16). In addition, miRNAs played critical roles in the development and oncogenesis of B-cell. They were involved in the pathogenesis of CLL (17). MiRNAs expressions were associated with clinical characteristics of CLL, and they were likely to be served as diagnostic and prognostic biomarkers, as well as potential therapeutic targets in CLL (18,19). Negrini et al revealed that miRNAs whose expression was distinctive of cases with mutated vs. unmutated IGHV genes or cases with 13q, 11q, and 17p deletions and trisomy 12 and miR26a, miR532-3p, miR146-5p, and miR29c* were strongly associated with progression-free survival in CLL (20). However, the specific regulatory mechanism of miRNA in CLL was still needed to further explore.
In the current studies usually analyzed gene expression by high throughput RNA sequenc, though miRNA is also known as important in the regulation of gene expression and cancer formation. In this study, we firstly identified the key miRNA-Gene pairs in CLL.
In this study, differentially expressed genes (DEGs) and miRNAs (DEMs) in CLL samples compared with normal B cell samples were identified via bioinformatics methods. The miRNA-Gene regulatory network in CLL was constructed. A better understanding of the regulatory mechanism of miRNAs in CLL was obtained. Our study may provide references for the diagnosis and therapy of CLL.
Materials and methods
Microarray data
The miRNA expression dataset GSE62137 (21) and mRNA expression dataset GSE22529 (22) were downloaded from National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database. The miRNA dataset GSE62137 contained 44 samples, including 38 CLL cell samples and 6 normal B cell samples. Some of these samples were treated with IL-4. Our study was performed based on the untreated samples (23 CLL cell samples and 3 normal B cell samples cultured with nothing). MiRNA expression profile was detected via the Agilent-021827 Human miRNA Microarray G4470C platform. The mRNA dataset GSE22529 contained 52 samples, including 41 CLL cell samples and 11 normal B cell samples. The microarray data we used was detected based on GPL96 [HG-U133A] Affymetrix Human Genome U133A Array platform.
Identification of differently expressed miRNAs and genes
For the miRNA dataset, DEMs in CLL samples compared with normal B cell samples were identified via the GEO2R application of GEO. The screening threshold was FDR corrected P<0.05 and |log2 (fold-change)|>1. For the mRNA dataset, the raw data with CEL files were background corrected, normalized and log2 transformed using the affy package in R. (23). If multiple probes correspond to one gene, the mean expression value was defined as expression value. DEGs in CLL samples compared with normal B cell samples were identified via the limma package (24) of R. The DEGs were identified according to the criteria of |log2 (fold-change) |>1 and FDR corrected P<0.05.
Functional clustering analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) was a widely used web-based tool for genomic functional annotations (25). In the present study, Gene Ontology (GO) terms enrichment analysis were performed via DAVID. Enrichment score >1 was used as the criteria to identify significant functional clusters.
Screening of target genes of the DEMs
The targets of the DEMs, as well as the miRNA-Gene pairs, were identified based on the miRNAWalk platform. Five databases: miRnada (26), miRDB (27), miRWalk (28), RNA22 (29) and TargetScan (30) were used in this study to predicted the targets of the DEMs. Only the targets that simultaneously appeared in the five databases were screened out.
Construction of the miRNA-Gene regulatory network
The overlaps between the DEGs and the targets of the DEMs were selected. The miRNA-Gene regulatory network was constructed based on the miRNA-Gene pairs, and then visualized via Cytoscape software (version 3.4.0; www.cytoscape.org).
Results
The DEMs and the DEGs
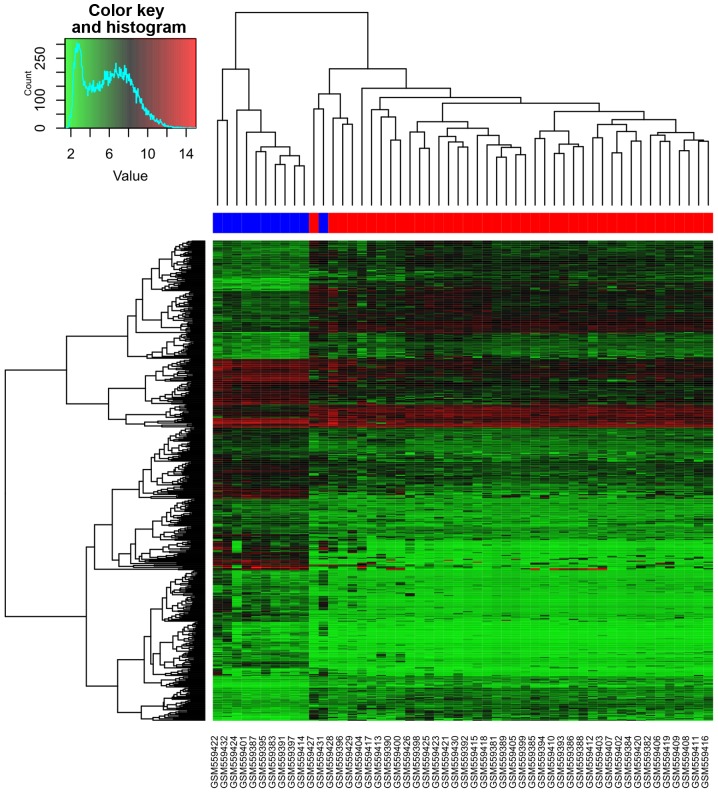
A total of 63 DEMs were identified in CLL samples compared with normal B cell samples, including 51 downregulated ones and 12 upregulated ones. The top 20 DEMs were listed in Table I. For the mRNA dataset, gene expression values of each sample after normalization were shown in Fig. 1. 504 DEGs were identified in CLL samples compared with normal B cell samples, including 316 downregulated ones and 188 upregulated ones. Table I showed the top 20 DEGs. Cluster analysis of CLL samples and normal B cell samples based on the DEGs expression values was shown in Fig. 2. From the heatmap (Fig. 2), we found that the gene expression of 10 normal B cell samples were distinguished from the 41 CLL samples. One normal B cell samples was assigned to CLL samples. Accuracy rate of prediction is 98.07%.
Table I.
The top 20 DEMs in CLL samples compared with normal B cell samples.
| MiRNA ID | P-value | LogFC |
|---|---|---|
| hsa-miR-582-5p | 5.32×10−17 | −8.23 |
| hsa-miR-181a | 8.86×10−04 | −8.56 |
| hsa-miR-132 | 8.86×10−04 | −6.42 |
| hsa-miR-95 | 8.86×10−04 | −4.34 |
| hsa-miR-548c-3p | 8.86×10−04 | −3.65 |
| hsa-miR-181c | 8.86×10−04 | −3.22 |
| hsa-miR-150 | 8.86×10−04 | 1.54 |
| hsa-miR-486-5p | 8.86×10−04 | 5.65 |
| hsa-miR-451 | 8.86×10−04 | 6.43 |
| hsa-miR-144 | 8.86×10−04 | 8.56 |
| hsa-miR-28-5p | 1.81×10−03 | 1.57 |
| hsa-miR-885-3p | 1.42×10−02 | −3.2 |
| hsa-miR-199a-3p | 1.71×10−02 | −5.62 |
| hsa-miR-155 | 1.74×10−02 | 2.00 |
| hsa-miR-126 | 2.22×10−02 | −6.29 |
| hsa-miR-29a | 2.26×10−02 | 1.27 |
| hsa-miR-21 | 2.32×10−02 | 1.42 |
| hsa-miR-202 | 2.47×10−02 | −3.53 |
| hsa-miR-29b | 3.93×10−02 | 1.15 |
| hsa-miR-199a-5p | 3.95×10−02 | −3.42 |
miRNA, microRNA; DEMs, differentially expressed miRNAs; FC, fold-change; CLL, chronic lymphocytic leukemia.
Figure 1.
Gene expression value of each sample in GSE22529 after normalization.
Figure 2.
Cluster analysis of chronic lymphocytic leukemia samples and normal B cell samples based on the differentially expressed genes. Lighter red in the heat-map represents high expression and darker green indicates low expression, black denotes medial expression.
Enriched functional clusters
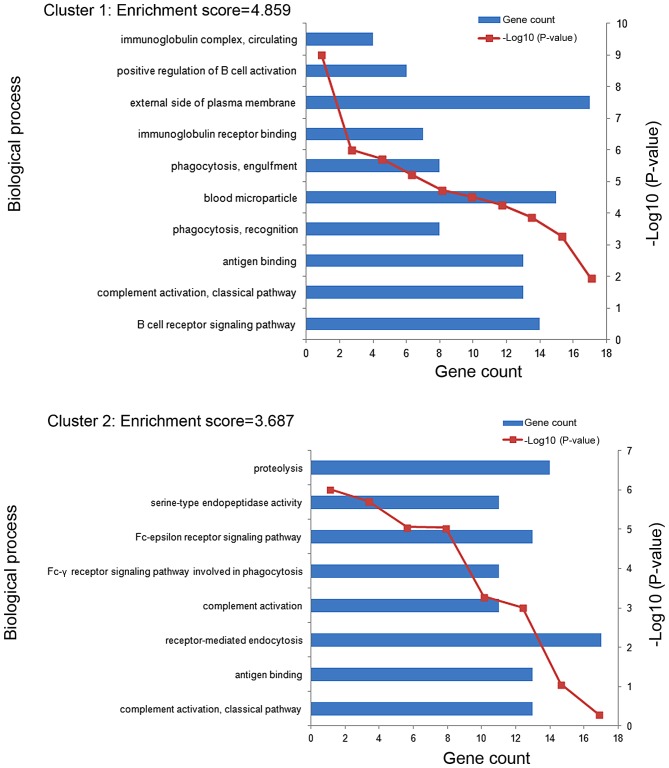
11 enriched functional clusters of the DEGs were obtained. The enriched GO terms in the top 2 clusters (cluster 1 and cluster 2) were shown in Fig. 3. The GO terms of clusters 1 were significantly enriched in the process of immune response, including B cell receptor signaling pathway (GO:0050853), complement activation classical pathway (GO:0006958), antigen binding (GO:0003823) and so on. The GO terms of clusters 2 were mainly enriched in the process of immune recognition receptors includes complement activation classical pathway (GO:0006958), antigen binding (GO:0003823), receptor-mediated endocytosis (GO:0006898) and so on. The most enriched GO term in these 2 clusters involve an immune response.
Figure 3.
The enriched Gene Ontology terms in the top 2 functional clusters (cluster 1 and cluster 2).
The miRNA-gene regulatory network
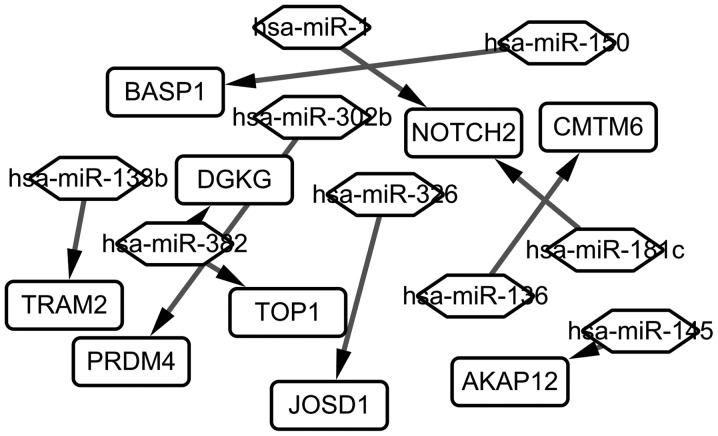
A total of 405 miRNA-Gene regulatory pairs were identified via the miRNAWalk platform. The miRNA-Gene regulatory pairs contained 351 target genes of the DEMs, including 9 overlaps with the DEGs. These overlaps were regulated by 10 DEMs, and then 10 miRNA-Gene regulatory pairs were obtained. Afterwards, a regulatory network between these overlaps and the DEMs were constructed and visualized (Fig. 4). As seen in Fig. 4, 9 target genes differentially expressed (TRAM2, BASP1, PRDM4, DGKG, TOP1, JOSD1, NOTCH2, AKAP12, CNTN6) are regulated by 9 different miRNAs (hsa-miR-138b, hsa-miR-352, hsa-miR-1, hsa-miR-302b, hsa-miR-326, hsa-miR-136, hsa-miR-181c, hsa-miR-145, hsa-miR-150).
Figure 4.
The miRNA-Gene regulatory network in chronic lymphocytic leukemia. The rectangle represents the gene, and the rhombus represents the miRNA. Has, homo sapiens; miRNA, microRNA; Notch2, neurogenic locus notch homolog protein 2; PRDM4, PR/SET domain 4; AKAP12, A-kinase anchoring protein 12; BASP1, brain abundant membrane attached signal protein 1; DGKG, diacylglycerol kinase γ; CMTM6, CKLF like MARVEL transmembrane domain containing 6; TRAM2, translocation associated membrane protein 2; TOP1, topoisomerase (DNA) I; JOSD1, josephin domain containing 1.
Discussion
CLL is the most common form of leukemia in western countries with the incidence of approximately 1 out of 100,000 patients per year (31), but little is known about its initiation and progression (32). MiRNAs play critical roles in B-cell oncogenesis by regulating the expression of many genes, and they are involved in the pathogenesis of CLL. In this study, a miRNA-Gene regulatory network in CLL was constructed via bioinformatics methods, which would help us make a better understanding of the molecular mechanism of CLL.
In the enriched GO terms in cluster 1, immunoglobulin complex, circulating and positive regulation of B cell activation were most closely related to the initiation and progression of CLL. While, in the enriched GO terms in cluster 2, proteolysis and serine-type endopeptidase activity had the most closely relationship with CLL. The mutational status of immunoglobulin heavy-chain variable-region (IgVH) genes in the leukemic cells of CLL was an important prognostic factor in the disease. Approximately 50–70% of patients with CLL had somatic hypermutation in IgVH genes of the leukemic cells (9,33). In addition, immunoglobulin could also be used as a therapeutic agent for CLL, and intravenous immunoglobulin G may alter the response of CLL cells to chemotherapy (34). B cell activation was an important initiation factor of CLL. CLL was characterized by the progressive accumulation of clonal mature B cells in the blood, bone marrow, and secondary lymphoid organs (35). B cell receptor signaling represented one of the central pathways to enhance survival and proliferation in CLL (10). One research showed that differences existed in the activation of B-CLL cells in vivo, and these patterns were correlated with disease activity (36). Although the importance of protein proteolysis in biological functions was frequently unclear, new technologies had started to unravel the critical role of clipping in cellular homeostasis and disease (37,38). Specific histone H2A proteolysis as disruption of the histone code was reported to drive hematopoietic cells in lymphomagenesis and result in lymphoid malignancies (39). Furthermore, matriptase was reported to be highly upregulated in CLL, which promoted cancer cell invasion (40). Serine-type endopeptidase also played critical roles in CLL. A serine endopeptidase, kallikrein B1 (KLKB1) was reported to be overexpressed in CLL, and its expression could be served as a novel molecular biomarker for the diagnosis of CLL (41).
The miRNA-Gene regulatory network contained 9 DEGs; they were BASP1, NOTCH2, CMTM6, DGKG, TRAM2, TOP1, PRDM4, JOSD1 and AKAP12. DAVID is a comprehensive system of biological function annotation information for large-scale genes, so it is too little to perform the DAVID analysis on this 9 overlapping DEGs. According to the European Molecular Biology Laboratory (EMBL) database (http://www.ebi.ac.uk/), we found 9 DEGs were enriched the following GO terms. BASP1, NOTCH2, TOP1, PRDM4, AKAP12 and JOSD1 were included in the biological process of protein binding (GO:0005515) which child terms included antigen binding, immunoglobulin receptor binding. JOSD1 was aslo included in the biological process of proteolysis (GO:0006508). JOSD1, NOTCH2 and AKAP12 wwere included in the biological process of hydrolase activity (GO:0016787), receptor activity (GO:0004872), regulation of protein kinase C signaling (GO:0090036), respectively, which were closely related to complement activation, classical pathway, positive regulation of B cell activation. TRAM2 and DGKG were respectively included in the biological process of integrall component of menbrane (GO:001601), intracellular signal transduction (0035556) which were closely related to signal transduction. To sum up, the GO terms of 9 DEGs were associated with the GO terms of all DEGs. Many of these genes were associated with the pathobiology of CLL. For example, NOTCH2 was a member of the NOTCH gene family. The Notch gene family encodes transmembrane receptors that modulate differentiation, proliferation and apoptotic programs in response to extracellular ligands expressed on neighboring cells (42). Enforced expression of NOTCH1IC in bone marrow stem cells caused T-cell leukemia in mice, indicating a causative role for NOTCH family in T-cell oncogenesis (43). The NOTCH2 oncogene was reported to be overexpressed in B-CLL cells, and was also related to the failure of apoptosis characteristic for this disease. Deregulation of NOTCH2 signaling was involved in the aberrant expression of CD23 in B-CLL (44). The overexpression of CD23 was one of the hallmarks of B-CLL cells (45). PRDM4 was a highly conserved member of the PRDM family (46). One study reported that PRDM4 could control proliferation and differentiation, which played critical roles in tumorigenesis (47). Another study reported that PRDM4 protein mapped to a tumor suppressor locus on human chromosome and could affect the processes of ovarian, gastric, and pancreatic cancers (48). AKAP12, which functioned as a kinase scaffold protein and as a dynamic regulator of the b2-adrenergic receptor complex, was one of the A-kinase anchoring proteins (49). AKAP12 played an important role in regulating cell cycle, cytokinesis cell adhesion, signaling, and oncogenic suppression (50). AKAP12 expression was closely related to tumorigenesis. The downregulation of AKAP12 expression had been reported in human prostate cancers in vivo, suggesting that the inactivation of AKAP12 expression was associated with oncogenesis (51). In gastric cancer, AKAP12 may function as an important negative regulator of the survival pathway (49). In addition, AKAP12 expression could also be served as a predictor for survival in CLL (52).
In conclusion, miRNAs played a critical role in regulating the process of CLL. They could affect CLL by regulating the processes of immunoreaction and protein degradation. Genes such as NOTCH2, PRDM4 and AKAP12 proved be their regulating targets in CLL. These DEGs which were related to CLL, could potentially serve as biomarkers for detection, prognosis, monitoring and predicting therapeutic responses in CLL. However, further studies were still needed to confirm our results.
Table II.
The top 20 DEGs in CLL samples compared with normal B cell samples.
| Gene name | P-value | LogFC |
|---|---|---|
| LMO2 | 2.84×10−19 | −3.95 |
| GLRX | 1.59×10−18 | −3.31 |
| IGHV1-69 | 2.46×10−17 | −5.23 |
| DENND3 | 5.06×10−17 | −1.87 |
| CCDC88A | 5.06×10−17 | 4.07 |
| HOPX | 9.31×10−16 | −2.71 |
| CST3 | 1.10×10−15 | −4.84 |
| IGHV3-48 | 1.10×10−15 | −4.42 |
| PIK3C2B | 1.10×10−15 | −3.27 |
| CEBPD | 1.10×10−15 | −2.47 |
| FPR1 | 1.10×10−15 | −1.86 |
| CD14 | 2.09×10−15 | −5.12 |
| CPVL | 2.09×10−15 | −3.74 |
| IGKV3-20 | 2.09×10−15 | −2.94 |
| CSF1R | 2.09×10−15 | −2.62 |
| SCN3A | 3.03×10−15 | −3.69 |
| SLC2A3 | 6.60×10−15 | −3.21 |
| SORL1 | 1.51×10−14 | −5.08 |
| FOS | 3.61×10−14 | −6.24 |
| SLC2A14 | 3.72×10−14 | −3.32 |
DEGs, differentially expressed genes; FC, fold-change; CLL, chronic lymphocytic leukemia.
Acknowledgements
We would like to thank all the members of our research group for their enthusiastic participation in the present study.
References
- 1.Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 2.Reddy KS. Chronic lymphocytic leukaemia profiled for prognosis using a fluorescence in situ hybridisation panel. Br J Haematol. 2006;132:705–722. doi: 10.1111/j.1365-2141.2005.05919.x. [DOI] [PubMed] [Google Scholar]
- 3.Byrd JC, Stilgenbauer S, Flinn IW. Chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2004:1–183. doi: 10.1182/asheducation-2004.1.163. [DOI] [PubMed] [Google Scholar]
- 4.Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia-what do we need to know? Nat Rev Clin Oncol. 2011;8:38–47. doi: 10.1038/nrclinonc.2010.167. [DOI] [PubMed] [Google Scholar]
- 5.Binet JL, Auquier A, Dighiero G, Chastang C, Piguet H, Goasguen J, Vaugier G, Potron G, Colona P, Oberling F, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48:198–206. doi: 10.1002/1097-0142(19810701)48:1<198::AID-CNCR2820480131>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 6.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46(2):219-234. Blood. 2016;128:2109. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 7.Kay NE, O'Brien SM, Pettitt AR, Stilgenbauer S. The role of prognostic factors in assessing ‘high-risk’ subgroups of patients with chronic lymphocytic leukemia. Leukemia. 2007;21:1885–1891. doi: 10.1038/sj.leu.2404802. [DOI] [PubMed] [Google Scholar]
- 8.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, Neuberg DS, Flinn IW, Rai KR, Byrd JC, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 9.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 10.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 11.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 12.Theodorou M, Speletas M, Mamara A, Papachristopoulou G, Lazou V, Scorilas A, Katsantoni E. Identification of a STAT5 target gene, Dpf3, provides novel insights in chronic lymphocytic leukemia. PLoS One. 2013;8:e76155. doi: 10.1371/journal.pone.0076155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers; Proc Natl Acad Sci USA; 2004; pp. 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 16.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 17.Fernando TR, Rodriguez-Malave NI, Rao DS. MicroRNAs in B cell development and malignancy. J Hematol Oncol. 2012;5:7. doi: 10.1186/1756-8722-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, Rovira C, Naya H, Dighiero G, Cayota A. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 19.Ferracin M, Zagatti B, Rizzotto L, Cavazzini F, Veronese A, Ciccone M, Saccenti E, Lupini L, Grilli A, De Angeli C, et al. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol Cancer. 2010;9:123. doi: 10.1186/1476-4598-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negrini M, Cutrona G, Bassi C, Fabris S, Zagatti B, Colombo M, Ferracin M, D'Abundo L, Saccenti E, Matis S, et al. microRNAome expression in chronic lymphocytic leukemia: Comparison with normal B-cell subsets and correlations with prognostic and clinical parameters. Clin Cancer Res. 2014;20:4141. doi: 10.1158/1078-0432.CCR-13-2497. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Lafuente N, Alcaraz-García MJ, Sebastián-Ruiz S, García-Serna AM, Gómez-Espuch J, Moraleda JM, Minguela A, García-Alonso AM, Parrado A. IL-4 up-regulates MiR-21 and the MiRNAs hosted in the CLCN5 gene in chronic lymphocytic leukemia. PLoS One. 2015;10:e0124936. doi: 10.1371/journal.pone.0124936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez A, Jr, Tschumper RC, Wu X, Shanafelt TD, Eckel-Passow J, Huddleston PM, III, Slager SL, Kay NE, Jelinek DF. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood. 2010;116:2975–2983. doi: 10.1182/blood-2010-02-269878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 24.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 26.Ma N, Gao X. β-actin is predicted as one of the potential targets of miR-145: Choose internal control gene in verification of microRNA target. Carcinogenesis. 2013;34:236. doi: 10.1093/carcin/bgs354. [DOI] [PubMed] [Google Scholar]
- 27.Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-database: Prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Rigoutsos I, Miranda K, Huynh T. rna22: A unified computational framework for discovering miRNA precursors, localizing mature miRNAs, identifying 3′ UTR target-islands and determining the targets of mature-miRNAs. IBM Corporation; Yorktown Heights, NY: 2007. [Google Scholar]
- 30.Edris B. A comparison of the Oligomap and TargetScan algorithms for miRNA target analysis. Stanford, CA: 2011. Publication no. Bmi231. [Google Scholar]
- 31.Pfeil AM, Imfeld P, Pettengell R, Jick SS, Szucs TD, Meier CR, Schwenkglenks M. Trends in incidence and medical resource utilisation in patients with chronic lymphocytic leukaemia: Insights from the UK Clinical Practice Research Datalink (CPRD) Ann Hematol. 2015;94:421–429. doi: 10.1007/s00277-014-2217-7. [DOI] [PubMed] [Google Scholar]
- 32.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer. 2016;16:145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 33.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, Schulman P, Vinciguerra VP, Rai K, Rassenti LZ, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besa EC. Use of intravenous immunoglobulin in chronic lymphocytic leukemia. Am J Med. 1984;76:209–218. doi: 10.1016/0002-9343(84)90344-9. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Y, El-Gamal D, Dubovsky JA, Beckwith KA, Harrington BK, Williams KE, Goettl VM, Jha S, Mo X, Jones JA, et al. Selinexor suppresses downstream effectors of B-cell activation, proliferation and migration in chronic lymphocytic leukemia cells. Leukemia. 2014;28:1158–1163. doi: 10.1038/leu.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tötterman TH, Carlsson M, Funderud S, Simonsson B, Oberg G, Nilsson K. Chronic B-lymphocytic leukemia-expression of B cell activation markers in relation to activity of the disease. Nouv Rev Fr Hematol. 1988;30:279–281. [PubMed] [Google Scholar]
- 37.Doucet A, Butler GS, Rodríguez D, Prudova A, Overall CM. Metadegradomics: Toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Rogers LD, Overall CM. Proteolytic post-translational modification of proteins: Proteomic tools and methodology. Mol Cell Proteomics. 2013;12:3532–3542. doi: 10.1074/mcp.M113.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor KH, Briley A, Wang Z, Cheng J, Shi H, Caldwell CW. Aberrant epigenetic gene regulation in lymphoid malignancies. Semin Hematol. 2013;50:38–47. doi: 10.1053/j.seminhematol.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Gao L, Liu M, Dong N, Jiang Y, Lin CY, Huang M, Wu D, Wu Q. Matriptase is highly upregulated in chronic lymphocytic leukemia and promotes cancer cell invasion. Leukemia. 2013;27:1191–1194. doi: 10.1038/leu.2012.289. [DOI] [PubMed] [Google Scholar]
- 41.Adamopoulos PG, Kontos CK, Papageorgiou SG, Pappa V, Scorilas A. KLKB1 mRNA overexpression: A novel molecular biomarker for the diagnosis of chronic lymphocytic leukemia. Clin Biochem. 2015;48:849–854. doi: 10.1016/j.clinbiochem.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 43.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubmann R, Schwarzmeier JD, Shehata M, Hilgarth M, Duechler M, Dettke M, Berger R. Notch2 is involved in overexperssion of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.V99.10.3742. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Matas M, Rodriguez-Justo M, Morilla R, Catovsky D, Matutes E. Quantitative expression of CD23 and its ligand CD21 in chronic lymphocytic leukemia. Haematologica. 2000;85:1140–1145. [PubMed] [Google Scholar]
- 46.Bogani D, Morgan MA, Nelson AC, Costello I, McGouran JF, Kessler BM, Robertson EJ, Bikoff EK. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol Cell Biol. 2013;33:3936–3950. doi: 10.1128/MCB.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chittka A, Nitarska J, Grazini U, Richardson WD. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J Biol Chem. 2012;287:42995–43006. doi: 10.1074/jbc.M112.392746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang XH, Huang S. PFM1 (PRDM4), a new member of the PR-domain family, maps to a tumor suppressor locus on human chromosome 12q23-q24.1. Genomics. 1999;61:319–325. doi: 10.1006/geno.1999.5967. [DOI] [PubMed] [Google Scholar]
- 49.Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim TY, Kim NK, Bang YJ. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095–7103. doi: 10.1038/sj.onc.1207932. [DOI] [PubMed] [Google Scholar]
- 50.Akakura S, Gelman IH. Pivotal role of AKAP12 in the regulation of cellular adhesion dynamics: Control of cytoskeletal architecture, cell migration and mitogenic signaling. J Signal Transduct. 2012;2012:529179. doi: 10.1155/2012/529179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia W, Unger P, Miller L, Nelson J, Gelman IH. The Src-suppressed C kinase substrate, SSeCKS, is a potential metastasis inhibitor in prostate cancer. Cancer Res. 2001;61:5644–5651. [PubMed] [Google Scholar]
- 52.van't Veer MB, Brooijmans AM, Langerak AW, Verhaaf B, Goudswaard CS, Graveland WJ, van Lom K, Valk PJ. The predictive value of lipoprotein lipase for survival in chronic lymphocytic leukemia. Haematologica. 2006;91:56–63. [PubMed] [Google Scholar]