Abstract
Human papillomaviruses (HPVs) are double stranded circular DNA viruses that infect cutaneous and mucosal epithelial cells. Almost 99% of cervical cancer has a HPV infection. The early oncoproteins E6 and E7 are important in this cellular transformation process. Epigenetic mechanisms have long been known to result in decisive alterations in DNA, leading to alterations in DNA-protein interactions, alterations in chromatin structure and compaction and significant alterations in gene expression. The enzymes responsible for these epigenetic modifications are DNA methyl transferases (DNMTs), histone acetylases and deacetylases. Epigenetics has an important role in cancer development by modifying the cellular micro environment. In this review, the authors discuss the role of HPV oncoproteins E6 and E7 in modulating the epigenetic mechanisms inside the host cell. The oncoproteins induce the expression of DNMTs which lead to aberrant DNA methylations and disruption of the normal epigenetic processes. The E7 oncoprotein may additionally directly bind and induce methyl transferase activity of the enzyme. These modulations lead to altered gene expression levels, particularly the genes involved in apoptosis, cell cycle and cell adhesion. In addition, the present review discusses how epigenetic mechanisms may be targeted for possible therapeutic interventions for HPV mediated cervical cancer.
Keywords: human papillomavirus, E6, E7, cervical, cancer, epigenetics, DNA methyl transferase, histone de-acetylase
1. Introduction
According to World Cancer Report cervical cancer or cancer of the uterine cervix is both the second-most common cause of cancer and the fourth-most common cause of death from cancer in women. It has also been reported that in the year 2012, an estimated 528,000 cases has resulted 266,000 deaths worldwide. It has been observed that the most common causative agent of cervical cancer is by a virus called human papilloma virus, or HPV. The progression of cervical carcinoma occurs from normal to low grade squamous intra-epithelial lesion (LSIL) then to high grade squamous intra-epithelial lesion (HSIL) to carcinoma in situ (CIS) and finally to metastatic cancer (1). Oncogenic human papillomaviruses (HPVs), mostly HPV types-16, 18, 31, 33 are related with invasive cervical cancer lesions. HPV has its viral genome present in the 72-capsomere capsid. The genome is divided into three regions which include-the long control region (LCR) without coding potential; the region of early proteins (E protein) (E1-E7), and the region of late proteins (L1 and L2) (2) LCR, early region and late region regulates DNA replication, oncogenesis, cellular transformation and viral capsule formation respectively. There are over 200 different varieties of HPV (3). The E proteins of HPV are used to determine the risk factor involved in cervical cancer. HPV-6, 11, 42, 43 and 44 are low risk types as they are found in benign tumours; HPV-16, 18, 31, 33, 35, 51, 52 and 58 are high risk types as they are found in both in benign tumors and invasive cancers. The E6 and E7 proteins of the oncogenic HPV interacts with cellular proliferation and apoptotic mechanisms at well-defined targets, the primary targets are p53 and retinoblastoma (Rb) tumour suppressor proteins, respectively (4,5). The E1 and E2 proteins mediate viral replication and act to recruit cellular replication factors to viral replication origins (6).
2. Epigenetic control of cervical cancer
A change in genomic DNA and chromatin resulting in alteration of gene expression which is somatically heritable but does not involve DNA sequence changes is defined as an epigenetic phenomenon. A post-replicative DNA modification that occurs on the 5′-position of cytosine rings located in CpG dinucleotides is termed as DNA methylation. It is the most studied epigenetic change which influence gene expression (7). The reasons for epigenetic changes may be a regular or natural occurrence but can also be influenced by several factors including age, the environment, lifestyle, and disease state. Recently it has been established that in cervical cancer epigenetic changes play a major role in its development (8–11). DNA methylation may be useful as a marker for early detection or predicting the risk of cancer precursors. The existence of HPV alone is not sufficient to cause cervical cancer; presence of DNA methylation in cervical cancer and precancerous lesions support a role for this phenomenon in cervical cancer development (12). This is mainly because there is obscurity in the molecular processes which is the main cause of futility in HPV-mediated cervical cancer. During all the stages of cervical cancer a numerous epigenetic alterations are observed which include DNA hypomethylation, hypermethylation of tumour suppressor genes and histone modifications (12). DNA methylation is done by an enzyme known as DNA methyl transferase (DNMTs) which regulates the gene expression in prokaryotes, eukaryotes and viruses (1). It is a reversible reaction catalyzed by DNMTs which conserve the prototype of methylation during each cellular division. The two de novo methyl transferases are DNMT3a and DNMT3b. DNMTs add a methyl group onto carbon 5 of cytosine residues adjacent to guanine residues (5′-CpG-3′), which mainly occurs in CpG islands. CpG islands are generally found in the promoter regions of protein-coding genes, and expression is silenced upon their methylation. Non-coding genes, such as microRNAs (miRNAs), are also susceptible to regulation by methylation (13). The epigenome could be targeted by environmental factors such as dietary methyl donors. CpG methylation of Type 1 HPV was observed in early transfection studies (14). A study on cell transfection with HPV-16 in vitro methylated genomes proved that DNA is repressed transcriptionally (15). Some of the genes regulated by methylaion are also involved in cervical cancer which acts as transcription factors, viz p53, UTF1, paired box 1 (PAX1) and TWIST1 (16–19).
During early cancer development the DNA modifications which are generally observed are global DNA hypomethylation in repetitive regions and hypermethylation in CpG island regions of tumour-suppressor gene promoters. Cervical cancer is correlated with the silencing of tumour-suppressor genes through DNA hypermethylation. However, silencing of tumour suppressor miRNAs through hypermethylation of CpG islands in their promoter regions has also been a reason for carcinogenesis (20,21). Wang et al, in the year 2013 had reported about the role of DNMT1 in cervical cancer (22). From in vitro and in vivo studies performed it was established that low levels of serum folate and high expression of DNMT1 protein or mRNA were extensively related with cervical carcinogenesis. Integration of high risk-HPV DNA like HPV-16 and −18 into the host's genome is an essential step in cervical carcinogenesis (23). The HPV-16, 18 DNA gets methylated after it integrates into the human genome (24).
A miRNA is a small non-coding (containing about 22 nucleotides) RNA generally found in plants, animals and some viruses. Its main purpose is RNA silencing and post-transcriptional regulation of gene expression (25,26). In cervical cancer high risk HPV (HR-HPV) plays a role in the deregulation of miRNA gene methylation. In patients and cervical cancer cell lines (HeLa, SiHa, CaSki and C33A) it has been observed that silencing of tumour-suppressor miRNAs through aberrant promoter methylation favours cervical carcinogenesis. It has been proposed that HR-HPV can lead to modifications in the methylation pattern of miRNA promoters. After cervical cancer cell lines were subjected to treatment with hypomethylating agents, decreased methylation levels were found for certain miRNAs, which resulted in their increased expression and associated decreased expression of their target genes It is possible that HR-HPV genotypes are involved in the methylation processes of miRNAs in cervical cancer (27–30).
Basal transcription machinery and accessibility of transcription factors are regulated by post-translational modification of histones bound to promoter regions which control gene expression. Acetylation and methylation as well as phosphorylation, ubiquitination, and sumoylation are different types of modifications included (6). The expression of tumour suppressor genes, oncogenes and growth factors are regulated by many histone-modifying enzymes such as the histone deacetylases (HDACs), sirtuins, and histone acetyltransferases (HATs). Thus, dysregulation and mutation of expression of histone modifiers leads to epigenetic control of gene expression (31,32).
3. Role of E6/E7 oncoproteins
The most important oncogenes of HPV are E6 and E7 genes which encode oncoproteins that cause transformation of the host cell. Transcription of E6 and E7 oncogenes has always been observed in cervical carcinomas, being the first indication of a main role of these viral genes in HPV mediated cervical cancer (33). Oncogenic properties of E6 and E7, as well as their effects on p53 and Rb protein (pRb), have provided the general basis for further investigations of the role of HPV in carcinogenesis in the HPV-infected cervix. The oncogenic protein E6 and E7 of HPV initiate dysregulation of cellular proliferation and apoptotic machinery by targeting tumour suppressor proteins such as p53 and Rb (34,35).
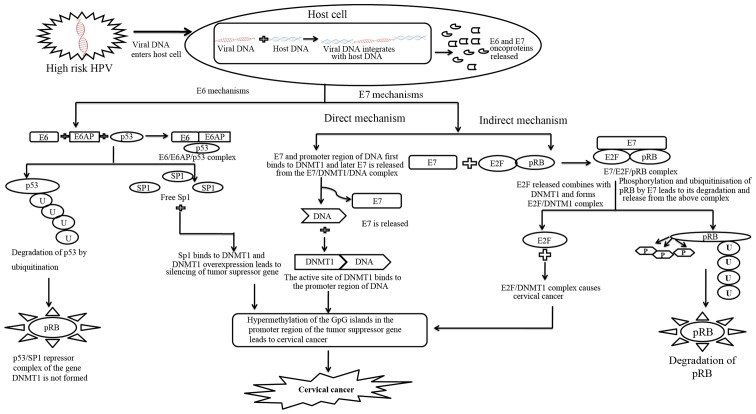
There are 160 amino acids present in the HPV E6 oncoprotein (36). For maintaining cellular transformation E6 protein from HPV is adequate (37–40). The function of tumour suppressor protein p53 is repressed by the action of E6 through the ubiquitin pathway and with the help of a cellular protein known as E6-associated protein (E6AP) (41–43). Genes which help in cell cycle arrest by repairing DNA damage or by activating apoptotic pathways are stimulated by p53 levels (44). Binding of the E7 oncoprotein on pRB provides a complementary function. Binding releases transcription factor E2F that activates expression of genes that stimulate DNA synthesis in the cell. Cellular E3 ubiquitin ligase E6AP is targeted by HR-HPV E6 oncoprotein which transfer ubiquitin peptides from E6AP to p53 and marks p53 for degradation by 26S proteasome pathway. On the other hand, low risk and cutaneous epithelia-infecting HPV E6 oncoproteins are unable to target the cellular p53 protein for proteasome mediated degradation (45,46). The mechanism of E6 mediated gene silencing through DNMT1 has been illustrated in Fig. 1. The mechanism involves degradation of p53 and release of specificity protein 1 (Sp1) transcription activator, which binds to the promoter of DNMT1 and upregulates the gene expression. The higher amount of DNMT1 leads to hypermethylation of DNA.
Figure 1.
The figure shows a common mechanism by which E6 and E7 might be working to induce hypermethylation of genes. The common target of both the oncoproteins is DNMT1 which gets activated by E6/E7 through different pathways. Unlike E6 which follows an indirect mechanism for DNMT1 activation through SP1, E7 follows both direct and indirect mechanism for DNMT1 activation. In both the indirect mechanisms by E6 and E7, there is a proteasomal degradation of two key tumour suppressor proteins i.e., p53 and pRb. The transcription factors associated with p53 and pRb; Sp1 and E2F respectively bind to the DNMT1 promoter to induce overexpression of DNMT1. HPV, human papillomavirus; Sp1, specificity protein 1; pRb, Rb protein; DNMT, DNA methyl transferase; microRNA.
The HPV E7 oncoprotein is about 100 amino acids long (47). The HPV E7 oncoprotein binds to the pRb and disrupts its function by preventing the binding of pRb to E2F transcription factor (48). The E7 oncoprotein was also observed in recent studies to modulate DNA methylation mechanism to control pathways of cellular propagation. It could bring about epigenetic changes through Rb family of tumour suppressor proteins. A study showed that DNA methyl transferase DNMT1 could be linked by HPV-16 E7 both in vitro and in vivo to kindle its enzymatic actions. The mechanism of E7 mediated gene silencing has been described in Fig. 1. This involves direct and indirect pathways involving DNMT1 in both the cases. E7 can directly bind to DNMT1 and induce gene silencing by hypermethylation. E7 can form a tight complex with Rb resulting in release of E2F which goes and binds to DNMT1 causing hypermethylation of CpG islands. In case of cervical neoplasia CpG islands in the promoter region of tumour suppressor genes are frequently inactivated by hyper-methylation (49). The [3H]-S-adenosyl-methionine incorporation assay provided the first evidence for global DNA hypo-methylation in cervical cancerous tissues. As the grade and stage of cervical neoplasia gradually progressed, the extent of hypo-methylation with 3H-methyl group incorporation also increased (48).
Several studied reported that HPV interferes with the cellular DNA methylation machinery; in order to disguise itself or as part of its viral cycle (50–52). During HPV 16 infection, cellular DNA undergoes epigenetic alterations induced by the E6 and E7 oncoproteins (51,53,54). Hence DNA methylation might serve as a defence mechanism of host cell to silence viral DNA (50,55). HPV encoded E6 and E7 interacts with p53 and E2F, inducing the expression of DNMT1 resulting in aberrant methylation. In HPV mediated carcinogenesis, E6/E7 expression is also regulated by E2 protein and other cellular transcription factors (NF1, AP1, KRF1, Oct1, Sp1, and YY1). In HR-HPV E6 oncoproteins increase the expression and activity of DNMT1 by degrading p53 (54). It has been reported that knockdown of E6 is associated with an increase in p53 and a decrease in DNMT1 expression in cervical cancer cell lines SiHa and CaSki (51,54). p53 binds to the Sp1 and chromatin-remodeling proteins, and this resulting complex further binds to the promoter region of DNMT1. This resulting complex prevents Sp1 from activating the transcription of DNMT1 (56).
In addition DNMT1 expression is modulated by E7 in two distinct ways. In the indirect mechanism E7 binds to pRb and releases the transcription factor E2F. As E2F is present at the transcription start site of DNMT1 the release of E2F activates the promoter of DNMT1. In the direct mechanism E7 binds directly to DNMT1 (53). This binding causes conformational changes in DNMT1 exposing the active site and binding of E7/DNMT1 to DNA. A stable DNMT1/DNA complex is formed after E7 dissociates from the complex (51). This results to methylation of the genome due to silencing of the tumour suppressor gene and hence causes cervical cancer.
4. Modulation of gene expression
As stated earlier the oncoproteins of HPV E6 and E7 are responsible for the carcinogenicity of the virus. The promoter, enhancer and silencer sequences in the HPV LCR controls the expression of E6 and E7 (57). Hypomethylation of the promoter region is considered to be an important part of activation of gene in the early stage of cancer which is also a very frequent affair. A study was conducted recently to compare normal samples with cervical cancer samples which reported that cervical cancer is related with progressive hypomethylation (48). It has been established by various studies that in cervical cancer both host and HPV genome are hypomethylated. In cervical cancer not only the HPV genome undergoes hypomethylation but Rap guanine nucleotide exchange factor 1 (RAPGEF1), long interspersed elements (LINE1) and Cancer Antigen Gene (CAGE) promoter are also hypomethylated (58–60).
Like hypomethylation, hypermethylation is also an early and frequent event in cervical cancer. It is mainly related with the CpG island and associated with promoter region which leads to gene silencing in cervical carcinogenesis (20). Badal et al, 2003 illustrated that hypermethylation of HPV genome suppresses the abnormal growth of normal tissue to malignant tissue where as hypomethylation promotes carcinogenesis (61). Research on epigenetic for the past few decades in cervical cancer has established the fact that methylation of DNA can be used for early recognition, analysis and maturity of unique curative approaches along with its pathogenic role (62). Signalling pathways such as cell cycle, cell to cell adhesion, Apoptotic pathway and cell signalling pathways are effected by hypermethylation of HPV in cervical cancer (20). Therefore, the epigenetic pathways that are active in the host cell also cause modification of the viral genome.
In both squamous cell carcinoma (SCC) and adenocarcinoma, the frequently methylated genes are cell adhesion molecule 1 (CADM1), cadherin 1 (CDH1), death-associated protein kinase 1 (DAPK1), EPB41L3, FAM19A4, myelin and lymphocyte (MAL), PAX1, PR domain containing 14 (PRDM14) and telomerase reverse transcriptase (TERT) (63). Recently a wide profiling of methylation in cervical intraepithelial neoplasia (CIN) lesions and cervical cancer has been conducted which requires proper justification and further studies. Tumour suppressive activities in cervical cancer has been observed in these subset of genes-CADM1, WNT signalling pathway inhibitor 3 (DKK3), MAL, secreted frizzled-related protein 2 (SFRP2) and C13ORF18 (also known as KIAA0226L). Other than these findings methylation events and their related biological relevance in cervical cancer remains indefinable (64).
A combined mechanism involving both E6 and E7 acting together on DNMT1 has been shown in Fig. 1. This mechanism is most likely to be occurring in vivo when both E6 and E7 are over expressed. Thus, a combined mechanistic action of both E6 and E7 results in hypermethylation of many tumour suppressor genes leading to unregulated cell proliferation. It might be possible that through indirect mechanisms of E6/E7, there is an over expression of DNMT1, which is bound directly by E7 to cause hypermethylation of the gene promoters. Therefore, the indirect mechanism is likely to precede the direct pathway.
Modulation of genes involved in cell cycle
For cell cycle functioning of cyclin dependent kinases (CDKs) its proper expression is essential (65). In cervical carcinogenesis, CDK gene is not included in HPV induced cervical cancer but the expression of this gene is observed to be abnormal. P16INK4a is a tumour suppressor protein which acts as an inhibitor of CDK4 and CDK6, which initiate the phosphorylation of the Rb tumour suppressor protein, Rb. E7 oncoprotein plays an important role in disrupting the normal cell cycle when it binds to pRB and resulting in activation of p16INK4a which causes cell death by arresting cells in G1 phase. Thus, p16INK4a probable physiological role is in the implementation of irreversible growth arrest termed cellular senescence (66). The normal cell cycle pathway is disrupted when the oncoprotein E7 of HPV binds to pRB and disrupts its function. Hypermethylation results in downregulation of E7 leading to activation of p16INK4A (1). In the year 2011, Huang et al reported that hypermethylation was present in 17.6% of CIN I, 42.1% of CIN II, 55.0% of CIN III, and 65.0% of invasive cancers (67). Additionally, this can be used as a biomarker to foresee the development of low grade dysplasia and reappearance of high grade intraepithelial neoplasias (68). In HPV induced cervical cancer the downregulation of Cyclin A1 expression is observed due to its methylation. Other than these above mentioned proteins, fragile histidine triad (FHIT) is another protein related to cell cycle regulation and apoptosis. In HPV related cervical cancer this protein (FHIT) is inactivated. Promoter hypermethylation is the reason for silencing of the gene rather than genetic alterations (69).
Modulation of genes involved in cell adhesion
A transmembrane glycoprotein called E-cadherin acts as a mediator for calcium-dependent interactions between adjacent epithelial cells. In cervical cancer, previous data showed that the expression of E-cadherin is reduced at both the mRNA and protein levels. Loss of membranous E-cadherin expression was detected in in three HPV mediated cervical cancer cell lines (HeLa, SiHa, C33A) due to hypermethylation of CpG islands in its promoter region. These data suggested that E-cadherin might be an important factor in cervical cancer (70). Later it was reported that 52.6% of gene expression of CDH1 gene is downregulated due to promoter methylation in HPV positive cervical cancer samples (71). Another study conducted on a tumor suppressor gene TSLC1 gene and reported that it is silenced in cervical cancer due to methylation of the promoter region. Hypermethylation of TSLC1 results in downregulation of its expression which was reported in 82 and 91% of cervical cancer cell lines (11). It was also stated that there was 89% epigenetic change in case of invasive cervical cancers whereas only 26% in case of CIN III and none in normal tissues (72). Cancer invasion or metastasis could lead to loss of expression of CADM1 which is same as the TSLC1 gene which is associated with epithelial cell adhesion. A study reported that HR-HPV is associated with the frequency and density of methylation in cervical cancer. Expression of CADM1 is reduced in the methylated regions of high grade CIN and SCC (73). In another study related to chemokine signalling axes stromal derived factor-1 (SDF-1)α/CXCR4, demonstrated its expression to be downregulated at both transcript and protein level in cervical cancer cell line (C33A a HPV positive cervical cancer cell line) and primary tumors due to promoter hypermethylation (74).
Modulation of genes involved in apoptotic pathway
Apoptosis or programmed cell death occurs in multi-cellular organisms. It is a highly synchronized and controlled process which is essential in an organism's lifecycle. For cancer to develop in an organism the cells should be resistant to apoptosis, chemotherapy and radiotherapy. Recognising and identifying the genes which are related to apoptosis might help scientist in early development of new therapeutic approaches to cure cervical cancer.
Methylation of decoy receptor (DcR1 and DcR2) were found in HPV related cervical cancer patients which resulted in silencing and inhibition of apoptosis (75). Recent study showed that PRDM14 is downregulated in cervical cancer HPV positive cell lines due to promoter hypermethylation and its abnormal levels resulted in apoptosis (76). In many tumours it is observed that transcriptional silencing and inactivation of tumour suppressor genes is due to the hypermethylation of CpG islands. It was reported that p73 and p53 genes were hypermethylated among the 50 CpG islands of p21, p27, p57, p53, p73 and Rb1 genes in HPV positive cervical cancer samples as compared to control samples (17). DAPK is a pro-apoptotic serine/threonine kinase which plays a major role in metastasis and tumour pathogenesis (77). This gene is located in the chromosome number 9q34.1. In cervical cancer this gene was reported to be inactivated due to hypermethylation of the promoter region (78,79).
Modulation of genes involved in cell signalling
The retinoic acid receptor β2 (RARβ2) belongs to the nuclear superfamily and is a type of retinoic acid which is regulated by tumour suppressor genes. Re-expression of this gene was observed due to methylation of the promoter region in the cervical cancer cell lines SiHa and HeLa when administered with natural compounds curcumin and genistein (80). Thus, it can be stated that RARβ2 might be used as a biomarker for epigenetic therapy by targeting natural compounds and is related to HPV positive cervical cancer cell lines. The RAS association domain family 1 (RASSF1) is a tumour suppressor gene. When it is inactivated the gene could be correlated with the hypermethylation of its CpG-island promoter region. RASSF1 gene is mainly of two types-RASSF1A and RASSF1C, which are transcribed from separate promoters (81). Cell death is activated during transient over-expression when human scaffold protein CNK1, a c-Raf1 binding protein necessary for Ras-induced Raf activation, binds the human tumor suppressor RASSF1A and is stably reintroduced into human tumor cell lines. RASSF1 gene is not only related to cervical cancer but is related to different other cancer like colon cancer. Adenomatous polyposis coli (APC) is a tumor suppressor protein complex; cell overgrowth is prevented by regulating the genes that encourage cell division by controlling beta-catenin which in turn is controlled by APC. Over-expression of β-catenin targets, including cyclin D, cMyc, ephrins, and caspases are caused due to inactivation or loss of APC expression. Certain proteins such as microtubules, the components for Wnt/Wg pathway-catenin and axin, EB1 and IQGAP1 which are cytoskeletal regulators and the Rac guanine-nucleotide-exchange factor (GEF) Asef1 have their binding site in APC which is a multi-domain protein. Mutation cluster region is the region where ~60% of cancer-linked APC mutations occur. For the tumour suppressor activity of APC interaction with β-catenin or microtubules is considered to be necessary (82). An in vitro study documented that in HPV induced cervical carcinoma cell line HeLa 60% of APC was hypermethylated as compared to 13% in SCCs cell line SiHa (83). When cervical carcinoma cell line HeLa was de-methylated with the drug hydralazine it resulted in re-expression of methylation (84). Thus, methylation is a potential epigenetic marker for cervical adenocarcinoma. From the above mentioned studies it has been observed that epigenetic alterations such as hypermethylation or de-methylation of the promoter region of a gene can act as a marker to predict cervical cancer in the early stage of its onset and cure it accordingly. In 30% of the tumour in cervical cancer the genes have been silenced due to methylation (85,86).
5. Epigenetic biomarkers in clinical applications
A molecule which secreted by a tumour or a specific response of the body due to the presence of cancer or a process which indicates the presence of cancer clinically is known as a cancer biomarker. The types of biomarkers used to detect cancer are- genetic, epigenetic, proteomic glycomic or imaging biomarker. Recently to predict, evaluate and treat the outcomes of cancer, biomarkers have been identified to conduct this process. The biomarkers which show response to anticancer therapy in SCC are- checkpoint with forkhead and ring finger (CHFR); Werner DNA helicases (WRN); hypoxia inducible factor-1α subunit (HIF-1α), which is associated with hypoxic response; epidermal growth factor receptor (EGFR), which may be a molecular target; and cyclooxygenase-2 (COX-2), which predicts radiation sensitivity (87).
However, it is thought that viral gene transcription is caused due to methylation of viral. The common methylating genes for samples with invasive SCC and adenocarcinoma have the following biomarkers-CADM1, CDH1, DAPK1, erythrocyte membrane protein band 4.1-like 3 (EPB41L3) family with sequence similarity 19, chemokine (CC motif)-like, member A4 (FAM19A4) myelin and lymphocyte protein (MAL), PAX1, PR domain-containing 14 (PRDM14), and TERT (63). In case of cervical cancer frequently methylated genes can serve as molecular markers for the detection of neoplastic cells in body fluids such as urine and plasma. It can be served as non-invasive early diagnostic tool to detect cervical cancer. Different stages of cervical carcinogenesis can be represented by different methylation pattern of tumour suppressor genes in serum as well as in tissue samples (88,89). Single epigenetic biomarkers are not adequately sensitive to predict oncogenic progression, thus panels of candidate genes can improve sensitivity. The panel of gene mainly includes genes having high CpG methylation rate such as DAPK1, CDH1, IGSF4, CALCA, which were supplemented with genes of lower methylation rate such as RARβ2, or APC (10,90,91). The Table I shows different classes of genes along with their nature of DNA methylation and how the gene expression gets modulated giving rise to cervical cancer phenotype. A combination of some of these genes could be useful biomarkers for cervical cancer. Aberrant DNA methylation pattern in urine can be compared with exfoliated cervical cells but sensitivity is lower (92).
Table I.
The table shows the different types of genes whose expression is controlled through epigenetic methylation of their promoter DNA. All the genes shown in the table are hypermethylated which leads to reduced gene expression. The genes belong to apoptosis, cell adhesion, cell growth, cell cycle pathways. The table also shows that hypermethylation of these genes is important for the development of cervical cancer and progression to the malignant, invasive cancer. Some of these genes could be used as a biomarker for cervical cancer prognosis.
| Gene | Biological function | Epigenetic status | Included in HPV induced cervical cancer | Modulation of gene after epigenetic alterations | (Refs.) |
|---|---|---|---|---|---|
| MGMT | DNA repair | Hypermethylated | Yes | MGMT is methylated at higher rate in high-grade CIN and in invasive to cytotoxic agents play an important role in cervical cancers. | (9,113) |
| RARβ2 | Vitamin response | Hypermethylated | Yes | Tumor suppressor gene is silenced due to methylation. Drugs like genistein and curcumin reverse hypermethylation of the gene along with apoptosis of the cervical cancer cell lines. | (80) |
| RASSF1A | Ras signaling | Hypermethylated | No | The CpG promoter region of the tumor suppressor gene is hypermethylated and the activity of RASSF1A is silenced when it is attached with CNK1. | (81) |
| p16INK4a, FHIT, CDK | Cell cycle | Hypermethylated | p16INK4a gene here is related to all types of cancer cells and FHIT and CDK gene here are related to HPV induced cervical cancer. | Expression of CDK is observed to be abnormal. P16INK4a is a tumour suppressor protein which during cancer helps in cell arrest during G1 phase of cell cycle. Due to methylation, the expression is reversed. Cyclin A1 expression is downregulated and promoter region of FHIT gene is hypermethylated leading to silencing of the gene. | (114–116) |
| CADM1 | Cell adherence and invasion | Hypermethylated | Yes | Due to promoter methylation expression of CADM1 is reduced in the methylated regions of high grade CIN and SCC. | (73) |
| PRDM14 (PRD1-BF1 and RIZ), DcR1, DcR2, DAPK1, p73, p53 | Apoptosis | Hypermethylated | Yes | PRDM14 is downregulated in cervical cancer due to promoter hypermethylation. Methylation of decoy receptor DcR1 and DcR2) (in cervical cancer resulted in silencing resulted in silencing and inhibition of apoptosis. In cervical cancer DAPK is inactivated due to hypermethylation of the promoter region. p73 and p53 genes are hypermethylated among the 50 CpG island of the p21, p27, p57, p53, p73 and Rb1 genes in cervical cancer as compared samples to normal. | (17,76–79,117) |
| APC, E-cadherin | Wnt signaling pathway | Hypermethylated | Yes | The APC is a tumor suppressor protein which is repeatedly silenced due to hypermethylation of the promoter region in human cervical cancer. The main reason for hypermethylation of E-cadherin gene is the difficulty of it's expression during cervical cancer. | (72,82–84) |
| SYK | Tyrosine kinase signaling cascade functions is transmitting extracellular signals through the cell membrane to the cytoplasm and then to the nucleus, where gene expression is modified. | Hypermethylated | Yes | Promoter region of the gene is hypermethylated leading to some tyrosine kinases to become constitutively active, a nonstop functional state that may contribute to initiation or progression of cancer of the cervix. | (86) |
| CDH1, TSLC1, CXCR4 | Cell adhesion | Hypermethylated | Yes | Hypermethylation of promoter region results in downregulation of CDH1 gene expression in cervical cancer. TSLC1 gene is silenced/downregulated in cervical cancer due to methylation of the promoter region. The property of cell adhesion of the gene CXCR4 is lost due to epigenetic silencing. | (11,70,71,73) |
| ADCYAP1 | Involved in cell growth, proliferation, differentiation, stimulation of adenylate cyclase and subsequently increases the cAMP level in target cells. Aditionally functions as a hypophysiotropic hormone, neurotransmitter and neuromodulator. | Hypermethylated | Yes | Multiple CpG sites in the ADCYAP1 promoter are highly methylated in CIN III and invasive carcinoma cells as well as seven cervical cancer cell lines. Hypermethylation in the ADCYAP1 promoter is responsible for the transcriptional silencing of the ADCYAP1 gene in cervical cancer cells. | (118) |
| ZNF582 | Transcription factor, biological processes related to the DNA damage response, proliferation, cell cycle control, and neoplastic transformation | Hypermethylated | Yes | Gene is silenced due to hypermethylation. | (119) |
| TIMP2/TIMP3 | Tissue inhibitor which inhibit the activity of MMPs | Hypermethylated | Yes | Increased depth of invasion of tumor due to promoter methylation of TIMP2 favours the development of primary cervical tumors. | (120,121) |
| ER1 | Steroid hormone receptor | Hypermethylated | Yes | Hypermethylation or loss of ER1 leads to downregulation of the gene hence causes invasive cervical cancer. | (108,122) |
HPV, human papillomavirus; MGMT, O6-methylguanine DNA methyltransferase; CIN, cervical intraepithelial neoplasia; RARβ2, retinoic acid receptor β2; RASSF1, RAS association domain family 1; FHIT, fragile histidine triad; CDK, cyclin dependent kinases; CADM1, cell adhesion molecule 1; SCC, squamous cell carcinoma; DcR, decoy receptor; DAPK, death-associated protein kinase; Rb, retinoblastoma; APC, adenomatous polyposis coli; SYK, spleen tyrosine kinase; CDH1, cadherin 1; ADCYAP1, adenylate cyclase activating polypeptide 1; ZNF582, zinc finger protein 582; MMP, matrix metalloproteinase; ER1, estrogen receptor 1.
6. Role of histone modifications in HPV-induced cervical cancer
Recently there have been few studies in cervical cancer which are related to histone modification and alteration. Although analysis of this relation between cervical cancer and histone modification is at its beginning stage, some records regarding modification of the histones in cervical cancer samples may help in the identification of this cancer. The molecular level of diagnosis of chromatin modification has still not been fully established.
It was reported that in cervical cancer the expression levels of HDACs increases and due to continuous hypomethylation of DNA Sequence, tumor cells lose monoacethylated and trimethylated histone H4 form (93). In the same year it was also published that silencing of a DNA protein O6-methylguanine DNA methyltransferase (MGMT) which is important for genome stability was caused due to histone modification in cervical cancer (94). Furthermore it was reported that histone deacetylation caused repression of Wnt signalling pathway antagonist dickkopf-related protein 1 (DKK-1) in cervical cancer cell lines (95) by promoter CpG hypermethylation and histone deacetylation. It is generally observed that in HPV infected cervical cancer lesions and warts are formed. These deformities are mainly related to lack of oxygen present in tissues because of amplified expression of pro-angiogenic factors through the activity of oxygen sensitive HIF-1α. It was stated in another study that the transcriptional activity of HIF-1 is enhanced by the oncoprotein E7 of HPV while the suppressive action of p53 gene is neutralized by E6. Thus, they concluded that E7 oncoprotein in HPV mediated cervical cancer can activate the function of a key transcription factor mediating hypoxic response by displacing histone deacetylases HDAC1, HDAC4, and HDAC7 from HIF-1α (96). Histone modification and DNA mehtylation caused silencing of proliferation repressor protein osteo-protegerin (OPG) and RARβ2 in another study (97,98).
The main reason reported for the cause of progression in cervical cancer from CIN I to CIN II and CIN III has been reported to be phosphorylated and acetylated forms of histone H3 (99). For gene transcription, balance between HDACs and HATs plays a very important function which should be maintained in normal cells so that uncontrolled cell death and proliferation is under control (100). But in case of HPV mediated cervical cancer the E6 and E7 oncoprotein disrupts this balance between HDACs and HATS and thus uncontrollable cell growth and cell proliferation of cancer cell occurs.
7. Clinical applications of epigenetics in cancer therapy
Epigenetic therapy is a new and fast growing area of medicines with the use of drugs to cure epigenetic defects. This kind of therapy is considered to be very potential as it can easily reverse the genetic defects and prevent diseases with the help of medical involvement (101). Methylation of the gene promoter is targeted for cancer therapy by reactivation of tumour suppressor genes (102). It has been established through in vitro and in vivo clinical models that inhibition of DNA methylation exerts anti-tumour effects; restore gene expression and inhibit hypermethylation. HDACs is a potential candidate for cancer treatment. Several experimental models demonstrated that HDAC inhibition leads to induced apoptosis, or growth arrest, tumour cell differentiation and to sensitize cells to chemotherapy or radiation therapy (103,104). The molecular interplay between DNA hypermethylation and histone deacetylation for modulation of gene expression and silencing of different cancer related gene predicts synergy of gene expression as well as for anti-tumour activity (105). Therefore, a combination of DNA demethylating agent with HDAC inhibitor can be a potential target for cancer treatment.
Two main classes of epigenetic drugs, methylation inhibitors and HDACs inhibitors, are currently in clinical trials for cancer treatment. These drugs target the epigenome in a non-cell specific way. They act on most or all tumour types as deacetylation machinery or as deregulators of methylation. Hydralazine, a peripheral vasodilatator drug is a stable DNA methylation inhibitor having negligible toxicity. This drug has been widely used to treat hypertension. Hydralazine treatment can induce demethylation of promoter sequences, reactivation of mRNA transcription and protein synthesis of tumour suppressor genes in in vivo as well as in vitro clinical models (106). A randomized phase III trial, conducted by Coronel et al, compared chemotherapy alone with epigenetic therapy with hydralazine and the deacylation agent valproate plus cisplatin and topotecan for treatment of advanced cervical cancer (107). The outcome of the study suggested that patients treated with combination therapy had longer progression free survival than did patients treated with chemotherapy alone. Another widely used anti-epileptic drug, Valproic acid (VPA) is an effective inhibitor of HDACs. VPA has potent anti-tumour effects by modulating multiple cellular pathways including cell cycle arrest, apoptosis, angiogenesis, metastasis, differentiation and senescence in a variety of in vitro and in vivo systems (108). It leads to growth inhibition in HeLa, SiHa and CaSki cells of cervical epithelial origin. This anti-tumour effect of VPA in cervical cancer is mediated by hyperacetylation of p53 protein which protects it from degradation by E6 (109). In addition, Apicidin, a cyclic peptide HDAC downregulate DNMT1 gene expression in HeLa cells (110). In a recent study conducted by Chen et al, in the year 2015 observed there were methylation changes in cervical cancer cell line S3 compared to control cell line SiHa. When S3 cell line was treated with a demethylating agent it reversed the resistance and restored the sensitivity of the cell line to cisplatin, taxol and oxaliplatin drugs. Hence, it was concluded that methylation is caused due to resistance towards drug. This property of the cervical cancer cell line could act as a biomarker and also a therapeutic target to detect drug resistance in cervical cancer (111). Epigenetic therapy has certain limitations due to the fact that both DNA methylation and HDAC inhibitors may activate oncogenes because of non specific targeting which can lead to tumour progression. Furthermore, once cured epigenetic states may revert back to its previous state due to the reversible nature of DNA (112). More investigations are needed for specific targeting of the drugs as well as to maintain the cured epigenetic state and prevent reversal.
8. Future perspectives
The crucial role of epigenetics in development of cervical cancer is well established. Apart from the drugs that target epigenetic machinery (HDAC & DNMT1), new approaches need to be developed to target cancer cells specifically. Using anti-HDAC inhibitors can have a bystander effect on normal, healthy cells causing undue stress to them. Other than drugs, liposome-based cell permeable peptides targeting E6/E7 could be used. These peptides would bind to E6/E7 and prevent their interaction with other cellular components including the epigenetic machinery. Blocking E6/E7 would be the most effective approach as this will have a cascading effect on all the cellular pathways that are modulated by E6/E7. This approach requires more studies and testing to optimize the amount of peptide to be delivered inside the infected cell that would be sufficient to block E6/E7. These peptides can be periodically injected at the site of cancer and the patient can be monitored over time for cancer regression.
This review primarily focuses on modulation of DNA methylation by HPV E6 and E7 oncoproteins in cervical cancer. There are several reviews on DNA methylation and cancers but this review focuses on the key mechanisms that are likely to operate during HPV E6 and E7 mediated modulation of gene expression. The review discusses a wide variety of genes involved in diverse cellular functions that are affected in cervical cancer. The review also discusses various implications in clinical applications of epigenetic based therapy and what classes of drugs would be suitable for epigenetic therapy. Cervical cancer remains one of the lethal cancers of woman worldwide and thus, there is a continuous need to identify non-invasive biomarkers for early detection, prognosis, selection of the treatment, prediction of treatment response, and development of novel therapeutic strategies. Epigenetic based therapy is a new approach toward early prognosis and hence treatment of this lethal cancer. Understanding the role of these epigenetic alterations in cervical carcinogenesis will contribute to developing effective strategies in detecting and treating this disease.
Acknowledgements
Funding from DST-SERB, Govt. of India is duly acknowledeged.
Glossary
Abbreviations
- HPV
human papillomavirus
- LSIL
low grade squamous intra-epithelial lesion
- HSIL
high grade squamous intra-epithelial lesion
- CIS
carcinoma in situ
- LCR
long control region
- E protein
early protein
- Rb
retinoblastoma
- CIN
cervical intraepithelial neoplasia
- DNMT
DNA methyl transferase
- microRNA
miRNA
- E6AP
E6 associated protein
- HR-HPV
high risk HPV
- Sp1
specificity protein 1
- RAPGEF1
Rap guanine nucleotide exchange factor 1
- LINE1
long interspersed element
- CAGE
cancer antigen gene
- CDK
cyclin dependent kinases
- FHIT
fragile histidine triad
- CDH1
cadherin 1
- CADM1
cell adhesion molecule 1
- SDF-1
stromal derived factor-1
- SCC
squamous cell carcinoma
- DcR
decoy receptor
- DAPK
death-associated protein kinase
- RARβ2
retinoic acid receptor β2
- RASSF1
RAS association domain family 1
- APC
adenomatous polyposis coli
- HDAC
histone deacetylase
- HAT
histone acetyltransferases
- VPA
valproic acid
- MGMT
O6-methylguanine DNA methyltransferase
- DKK-1
dickkopf-related protein 1
- HIF-1α
hypoxia inducible factor-1α
- OPG
osteo-protegerin
References
- 1.Bhat S, Kabekkodu SP, Noronha A, Satyamoorthy K. Biological implications and therapeutic significance of DNA methylation regulated genes in cervical cancer. Biochimie. 2016;121:298–311. doi: 10.1016/j.biochi.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Panatto D, Amicizia D, Bragazzi NL, Rizzitelli E, Tramalloni D, Valle I, Gasparini R. Chapter Eight-Human Papillomavirus Vaccine: State of the Art and Future Perspectives. Adv Protein Chemistry Structural Biol. 2015;101:231–322. doi: 10.1016/bs.apcsb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 3.IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Pharmaceuticals: A Review of human carcinogens. (IARC Monographs).IARC monographs on the evaluation of carcinogenic risks to humans. 2012;100A:1–401. [Google Scholar]
- 4.D'Andrilli G. Advances in cervical cancer and ongoing clinical trials. In: Giordano A, Macaluso M, editors. Gynecological Cancers. Springer International Publishing; Cham: 2016. pp. 51–64. [Google Scholar]
- 5.Zhao S. Specific Type Epigenetic Changes in Cervical Cancers. In: Verma M, editor. Cancer Epigenetics, Methods in Molecular Biology (Methods and Protocols) Vol. 1238. Humana Press; New York, NY: 2015. pp. 733–749. [DOI] [PubMed] [Google Scholar]
- 6.Langsfeld E, Laimins LA. Human papillomaviruses: Research priorities for the next decade. Trends Cancer. 2016;2:234–240. doi: 10.1016/j.trecan.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu H, Shu M, He L, Yu X, Liu X, Lu Y, Chen Y, Miao X, Chen X. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br J Cancer. 2017;116:658–668. doi: 10.1038/bjc.2016.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sova P, Feng Q, Geiss G, Wood T, Strauss R, Rudolf V, Lieber A, Kiviat N. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 9.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: Its relationship to clinical outcome. Mol Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Q, Balasubramanian A, Hawes SE, Toure P, Sow PS, Dem A, Dembele B, Critchlow CW, Xi L, Lu H, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji318. [DOI] [PubMed] [Google Scholar]
- 11.Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, Snijders PJ. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 12.Lu Q, Ma D, Zhao S. DNA methylation changes in cervical cancers. Methods Mol Biol. 2012;863:155–176. doi: 10.1007/978-1-61779-612-8_9. [DOI] [PubMed] [Google Scholar]
- 13.Trang P, Weidhaas JB, Slack FJ. MicroRNAs and cancer. In: Coleman WB, Tsongalis GJ, editors. The Molecular Basis of Human Cancer. 2nd. Springer; New York, New York, NY: 2017. pp. 277–286. [DOI] [Google Scholar]
- 14.Leung TW, Liu SS, Leung RC, Chu MM, Cheung AN, Ngan HY. HPV 16 E2 binding sites 1 and 2 become more methylated than E2 binding site 4 during cervical carcinogenesis. J Med Virol. 2015;87:1022–1033. doi: 10.1002/jmv.24129. [DOI] [PubMed] [Google Scholar]
- 15.Doeberitz MVK, Vinokurova S. US Patent 12/740,986. Compounds and methods associated with differential methylation of human papilloma virus genomes in epithelial cells. 2015 Filed October 31, 2008; issued September 29.
- 16.Kan YY, Liou YL, Wang HJ, Chen CY, Sung LC, Chang CF, Liao CI. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer. 2014;24:928–934. doi: 10.1097/IGC.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 17.Jha AK, Nikbakht M, Jain V, Sehgal A, Capalash N, Kaur J. Promoter hypermethylation of p73 and p53 genes in cervical cancer patients among north Indian population. Mol Biol Rep. 2012;39:9145–9157. doi: 10.1007/s11033-012-1787-5. [DOI] [PubMed] [Google Scholar]
- 18.Guenin S, Mouallif M, Deplus R, Lampe X, Krusy N, Calonne E, Delbecque K, Kridelka F, Fuks F, Ennaji MM, Delvenne P. Aberrant promoter methylation and expression of UTF1 during cervical carcinogenesis. PLoS One. 2012;7:e42704. doi: 10.1371/journal.pone.0042704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Missaoui N, Hmissa S, Trabelsi A, Traoré C, Mokni M, Dante R, Frappart L. Promoter hypermethylation of CDH13, DAPK1 and TWIST1 genes in precancerous and cancerous lesions of the uterine cervix. Pathol Res Pract. 2011;207:37–42. doi: 10.1016/j.prp.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Yang HJ. Aberrant DNA methylation in cervical carcinogenesis. Chin J Cancer. 2013;32:42–48. doi: 10.5732/cjc.012.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Wang JT, Ding L, Jiang SW, Hao J, Zhao WM, Zhou Q, Yang ZK, Zhang L. Folate deficiency and aberrant expression of DNA methyltransferase 1 were associated with cervical cancerization. Curr Pharm Des. 2014;20:1639–1646. doi: 10.2174/13816128113199990543. [DOI] [PubMed] [Google Scholar]
- 23.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Jackson R, Rosa BA, Lameiras S, Cuninghame S, Bernard J, Floriano WB, Lambert PF, Nicolas A, Zehbe I. Functional variants of human papillomavirus type 16 demonstrate host genome integration and transcriptional alterations corresponding to their unique cancer epidemiology. BMC Genomics. 2016;17:851. doi: 10.1186/s12864-016-3379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 27.Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botezatu A, Goia-Rusanu CD, Iancu IV, Huica I, Plesa A, Socolov D, Ungureanu C, Anton G. Quantitative analysis of the relationship between microRNA-124a, −34b and −203 gene methylation and cervical oncogenesis. Mol Med Rep. 2011;4:121–128. doi: 10.3892/mmr.2010.394. [DOI] [PubMed] [Google Scholar]
- 29.Yao T, Rao Q, Liu L, Zheng C, Xie Q, Liang J, Lin Z. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in cervical cancer. Virol J. 2013;10:175. doi: 10.1186/1743-422X-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilting SM, Verlaat W, Jaspers A, Makazaji NA, Agami R, Meijer CJ, Snijders PJ, Steenbergen RD. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics. 2013;8:220–228. doi: 10.4161/epi.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29:238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan SA, Reddy D, Gupta S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment? World J Biol Chem. 2015;6:333–345. doi: 10.4331/wjbc.v6.i4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghittoni R, Accardi R, Chiocca S, Tommasino M. The role of human papillomaviruses in carcinogenesis. Ecancermedicalscience. 2015;9:526. doi: 10.3332/ecancer.2015.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruttkay-Nedecky B, Jimenez AM Jimenez, Nejdl L, Chudobova D, Gumulec J, Masarik M, Adam V, Kizek R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins (Review) Int J Oncol. 2013;43:1754–1762. doi: 10.3892/ijo.2013.2105. [DOI] [PubMed] [Google Scholar]
- 35.Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen AA, Gheit T, Franceschi S, Tommasino M, Clifford GM, IARC HPV Variant Study Group Human papillomavirus 18 genetic variation and cervical cancer risk worldwide. J Virol. 2015;89:10680–10687. doi: 10.1128/JVI.01747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans W, Filippova M, Aragon R, Filippov V, Reeves ME, Duerksen-Hughes P. Abstract 1828: Proteomic analysis of the effect of E6 star expression on cellular pathways in HPV positive SiHa and HPV negative C33A cervical carcinoma cells. Cancer Res. 2015;75:1828–1828. doi: 10.1158/1538-7445.AM2015-1828. [DOI] [Google Scholar]
- 38.Illiano E, Demurtas OC, Massa S, Di Bonito P, Consalvi V, Chiaraluce R, Zanotto C, De Giuli Morghen C, Radaelli A, Venuti A, Franconi R. Production of functional, stable, unmutated recombinant human papillomavirus E6 oncoprotein: Implications for HPV-tumor diagnosis and therapy. J Transl Med. 2016;14:224. doi: 10.1186/s12967-016-0978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duensing S, Münger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- 40.Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 42.Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells; Proc Natl Acad Sci USA; 2001; pp. 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mantovani F, Banks L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene. 2001;20:7874–7887. doi: 10.1038/sj.onc.1204869. [DOI] [PubMed] [Google Scholar]
- 44.Paek AL, Liu JC, Loewer A, Forrester WC, Lahav G. Cell-to-cell variation in p53 dynamics leads to fractional killing. Cell. 2016;165:631–642. doi: 10.1016/j.cell.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yim EK, Park JS. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319–324. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganguly N, Parihar SP. Human papilloma virus E6 and E7 oncoproteins as risk factors for tumorigenesis. J Biosci. 2009;34:113–123. doi: 10.1007/s12038-009-0013-7. [DOI] [PubMed] [Google Scholar]
- 47.Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Yin FF, Wang N, Bi XN, Yu X, Xu XH, Wang YL, Zhao CQ, Luo B, Wang YK. Serine/threonine kinases 31(STK31) may be a novel cellular target gene for the HPV16 oncogene E7 with potential as a DNA hypomethylation biomarker in cervical cancer. Virol J. 2016;13:60. doi: 10.1186/s12985-016-0515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dueñas-González A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer. 2005;4:38. doi: 10.1186/1476-4598-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whiteside MA, Siegel EM, Unger ER. Human papillomavirus and molecular considerations for cancer risk. Cancer. 2008;113(Suppl 10):S2981–S2994. doi: 10.1002/cncr.23750. [DOI] [PubMed] [Google Scholar]
- 51.Leonard SM, Wei W, Collins SI, Pereira M, Diyaf A, Constandinou-Williams C, Young LS, Roberts S, Woodman CB. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 52.Jiang P, Yue Y. Human papillomavirus oncoproteins and apoptosis (Review) Exp Ther Med. 2014;7:3–7. doi: 10.3892/etm.2013.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Au Yeung CL, Tsang WP, Tsang TY, Co NN, Yau PL, Kwok TT. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 55.Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS, Chen CY, Lu YY, Tang YA, Yang YC, Yang PC, Wang YC. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 2010;70:5807–5817. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 57.Cai Q, Lv L, Shao Q, Li X, Dian A. Human papillomavirus early proteins and apoptosis. Arch Gynecol Obstet. 2013;287:541–548. doi: 10.1007/s00404-012-2665-z. [DOI] [PubMed] [Google Scholar]
- 58.Banzai C, Nishino K, Quan J, Yoshihara K, Sekine M, Yahata T, Tanaka K. Gynecological Cancer Registry of Niigata: Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. Int J Clin Oncol. 2014;19:127–132. doi: 10.1007/s10147-013-0530-0. [DOI] [PubMed] [Google Scholar]
- 59.Samuelsson J, Alonso S, Ruiz-Larroya T, Cheung TH, Wong YF, Perucho M. Frequent somatic demethylation of RAPGEF1/C3G intronic sequences in gastrointestinal and gynecological cancer. Int J Oncol. 2011;38:1575–1577. doi: 10.3892/ijo.2011.972. [DOI] [PubMed] [Google Scholar]
- 60.Shuangshoti S, Hourpai N, Pumsuk U, Mutirangura A. Line-1 hypomethylation in multistage carcinogenesis of the uterine cervix. Asian Pac J Cancer Prev. 2007;8:307–309. [PubMed] [Google Scholar]
- 61.Badal V, Chuang LS, Tan EH, Badal S, Villa LL, Wheeler CM, Li BF, Bernard HU. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: Genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: Practical considerations and applications. Biochimie. 2012;94:2314–2337. doi: 10.1016/j.biochi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 63.Mersakova S, Nachajova M, Szepe P, Kasajova PS, Halasova E. DNA methylation and detection of cervical cancer and precancerous lesions using molecular methods. Tumor Biol. 2016;37:23–27. doi: 10.1007/s13277-015-4197-1. [DOI] [PubMed] [Google Scholar]
- 64.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 65.Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83–105. doi: 10.1016/j.pharmthera.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherr CJ, Bartek J. Cell cycle-targeted cancer therapies S phase: The DNA synthesis phase of the cell cycle. Annual Review of Cancer Biology. 2017;1:41–57. doi: 10.1146/annurev-cancerbio-040716-075628. [DOI] [Google Scholar]
- 67.Huang LW, Pan HS, Lin YH, Seow KM, Chen HJ, Hwang JL. P16 methylation is an early event in cervical carcinogenesis. Int J Gynecol Cancer. 2011;21:452–456. doi: 10.1097/IGC.0b013e31821091ea. [DOI] [PubMed] [Google Scholar]
- 68.Terra AP, Murta EF, Maluf PJ, Caballero OL, Brait M, Adad SJ. Aberrant promoter methylation can be useful as a marker of recurrent disease in patients with cervical intraepithelial neoplasia grade III. Tumori. 2007;93:572–579. doi: 10.1177/030089160709300610. [DOI] [PubMed] [Google Scholar]
- 69.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14:R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen CL, Liu SS, Ip SM, Wong LC, Ng TY, Ngan HY. E-cadherin expression is silenced by DNA methylation in cervical cancer cell lines and tumours. Eur J Cancer. 2003;39:517–523. doi: 10.1016/S0959-8049(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 71.Jeong DH, Youm MY, Kim YN, Lee KB, Sung MS, Yoon HK, Kim KT. Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: Correlation with clinicopathologic characteristics. Int J Gynecol Cancer. 2006;16:1234–1240. doi: 10.1111/j.1525-1438.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 72.Shivapurkar N, Sherman ME, Stastny V, Echebiri C, Rader JS, Nayar R, Bonfiglio TA, Gazdar AF, Wang SS. Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecol Oncol. 2007;107:549–553. doi: 10.1016/j.ygyno.2007.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Overmeer RM, Henken FE, Snijders PJ, Claassen-Kramer D, Berkhof J, Helmerhorst TJ, Heideman DA, Wilting SM, Murakami Y, Ito A, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008;215:388–397. doi: 10.1002/path.2367. [DOI] [PubMed] [Google Scholar]
- 74.Yadav SS, Prasad SB, Das M, Kumari S, Pandey LK, Singh S, Pradhan S, Narayan G. Epigenetic silencing of CXCR4 promotes loss of cell adhesion in cervical cancer. Biomed Res Int. 2014;2014:581403. doi: 10.1155/2014/581403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narayan G, Xie D, Ishdorj G, Scotto L, Mansukhani M, Pothuri B, Wright JD, Kaufmann AM, Schneider A, Arias-Pulido H, Murty VV. Epigenetic inactivation of TRAIL decoy receptors at 8p12-21.3 commonly deleted region confers sensitivity to Apo2L/trail-cisplatin combination therapy in cervical cancer. Genes Chromosomes Cancer. 2016;55:177–189. doi: 10.1002/gcc.22325. [DOI] [PubMed] [Google Scholar]
- 76.Snellenberg S, Cillessen SA, Van Criekinge W, Bosch L, Meijer CJ, Snijders PJ, Steenbergen RD. Methylation-mediated repression of PRDM14 contributes to apoptosis evasion in HPV-positive cancers. Carcinogenesis. 2014;35:2611–2618. doi: 10.1093/carcin/bgu197. [DOI] [PubMed] [Google Scholar]
- 77.Michie AM, McCaig AM, Nakagawa R, Vukovic M. Death-associated protein kinase (DAPK) and signal transduction: Regulation in cancer. FEBS J. 2010;277:74–80. doi: 10.1111/j.1742-4658.2009.07414.x. [DOI] [PubMed] [Google Scholar]
- 78.Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol Oncol. 2010;116:99–104. doi: 10.1016/j.ygyno.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 79.Yang N, Nijhuis ER, Volders HH, Eijsink JJ, Lendvai A, Zhang B, Hollema H, Schuuring E, Wisman GB, van der Zee AG. Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell Oncol. 2010;32:131–143. doi: 10.3233/CLO-2009-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jha AK, Nikbakht M, Parashar G, Shrivastava A, Capalash N, Kaur J. Reversal of hypermethylation and reactivation of the RARβ2 gene by natural compounds in cervical cancer cell lines. Folia Biol (Praha) 2010;56:195–200. [PubMed] [Google Scholar]
- 81.Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A, Mollahan P, Pfeifer GP, Avruch J, Seed B. The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A-induced cell death. J Biol Chem. 2004;279:29247–29254. doi: 10.1074/jbc.M401699200. [DOI] [PubMed] [Google Scholar]
- 82.Aoki K, Taketo MM. Adenomatous polyposis coli (APC): A multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- 83.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 84.Song Y, Zhang C. Hydralazine inhibits human cervical cancer cell growth in vitro in association with APC demethylation and re-expression. Cancer Chemother Pharmacol. 2009;63:605–613. doi: 10.1007/s00280-008-0773-z. [DOI] [PubMed] [Google Scholar]
- 85.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 86.Jiménez-Wences H, Peralta-Zaragoza O, Fernández-Tilapa G. Human papilloma virus, DNA methylation and microRNA expression in cervical cancer (Review) Oncol Rep. 2014;31:2467–2476. doi: 10.3892/or.2014.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iida M, Banno K, Yanokura M, Nakamura K, Adachi M, Nogami Y, Umene K, Masuda K, Kisu I, Iwata T, et al. Candidate biomarkers for cervical cancer treatment: Potential for clinical practice (Review) Mol Clin Oncol. 2014;2:647–655. doi: 10.3892/mco.2014.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dakubo GD. Cancer Biomarkers in Body Fluids. 1st. Springer International Publishing; Cham: 2016. Methylated DNA as cancer biomarkers in circulation; pp. 103–123. [DOI] [Google Scholar]
- 89.Huang RL, Su PH, Liao YP, Wu TI, Hsu YT, Lin WY, Wang HC, Weng YC, Ou YC, Huang TH, Lai HC. Integrated Epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017;23:263–272. doi: 10.1158/1078-0432.CCR-16-0863. [DOI] [PubMed] [Google Scholar]
- 90.Kahn SL, Ronnett BM, Gravitt PE, Gustafson KS. Quantitative methylation-specific PCR for the detection of aberrant DNA methylation in liquid-based Pap tests. Cancer. 2008;114:57–64. doi: 10.1002/cncr.23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wisman GB, Nijhuis ER, Hoque MO, Reesink-Peters N, Koning AJ, Volders HH, Buikema HJ, Boezen HM, Hollema H, Schuuring E, et al. Assessment of gene promoter hypermethylation for detection of cervical neoplasia. Int J Cancer. 2006;119:1908–1914. doi: 10.1002/ijc.22060. [DOI] [PubMed] [Google Scholar]
- 92.Feng Q, Hawes SE, Stern JE, Dem A, Sow PS, Dembele B, Toure P, Sova P, Laird PW, Kiviat NB. Promoter hypermethylation of tumor suppressor genes in urine from patients with cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2007;16:1178–1184. doi: 10.1158/1055-9965.EPI-06-0694. [DOI] [PubMed] [Google Scholar]
- 93.Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M, Raju GC, Hooi SC. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 94.Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther. 2005;4:61–69. [PubMed] [Google Scholar]
- 95.Lee J, Yoon YS, Chung JH. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecol Oncol. 2008;109:270–274. doi: 10.1016/j.ygyno.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 96.Bodily JM, Mehta KP, Laimins LA. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer Res. 2011;71:1187–1195. doi: 10.1158/0008-5472.CAN-10-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu TY, Kao CF, Lin CT, Huang DY, Chiu CY, Huang YS, Wu HC. DNA methylation and histone modification regulate silencing of OPG during tumor progression. J Cell Biochem. 2009;108:315–325. doi: 10.1002/jcb.22256. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Z, Joh K, Yatsuki H, Zhao W, Soejima H, Higashimoto K, Noguchi M, Yokoyama M, Iwasaka T, Mukai T. Retinoic acid receptor beta 2 is epigenetically silenced either by DNA methylation or repressive histone modifications at the promoter in cervical cancer cells. Cancer Lett. 2007;247:318–327. doi: 10.1016/j.canlet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 99.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 100.Plesa A, Iancu IV, Botezatu A, Huica I, Stoian M, Anton G. The involvement of epigenetic mechanisms in HPV-induced cervical cancer. Rajkumar R, editor. 9Human Papillomavirus – Research in a Global Perspective. 2016 doi: 10.5772/62833. InTech. [DOI] [Google Scholar]
- 101.Easwaran H, Tsai HC, Baylin SB. Cancer Epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de la Cruz-Hernández E, Pérez-Cárdenas E, Contreras-Paredes A, Cantú D, Mohar A, Lizano M, Dueñas-González A. The effects of DNA methylation and histone deacetylase inhibitors on human papillomavirus early gene expression in cervical cancer, an in vitro and clinical study. Virol J. 2007;4:18. doi: 10.1186/1743-422X-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 106.Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: From pathogenesis to therapy. Tumor Biol. 2014;35:5083–5093. doi: 10.1007/s13277-014-1737-z. [DOI] [PubMed] [Google Scholar]
- 107.Coronel J, Cetina L, Pacheco I, Trejo-Becerril C, González-Fierro A, de la Cruz-Hernandez E, Perez-Cardenas E, Taja-Chayeb L, Arias-Bofill D, Candelaria M, et al. A double-blind, placebo-controlled, randomized phase III trial of chemotherapy plus epigenetic therapy with hydralazine valproate for advanced cervical cancer. Preliminary results. Med Oncol. 2011;28(Suppl 1):S540–S546. doi: 10.1007/s12032-010-9700-3. [DOI] [PubMed] [Google Scholar]
- 108.Zambrano P, Segura-Pacheco B, Perez-Cardenas E, Cetina L, Revilla-Vazquez A, Taja-Chayeb L, Chavez-Blanco A, Angeles E, Cabrera G, Sandoval K, et al. A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer. 2005;5:44. doi: 10.1186/1471-2407-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.You JS, Kang JK, Lee EK, Lee JC, Lee SH, Jeon YJ, Koh DH, Ahn SH, Seo DW, Lee HY, et al. Histone deacetylase inhibitor apicidin downregulates DNA methyltransferase 1 expression and induces repressive histone modifications via recruitment of corepressor complex to promoter region in human cervix cancer cells. Oncogene. 2008;27:1376–1386. doi: 10.1038/sj.onc.1210776. [DOI] [PubMed] [Google Scholar]
- 110.Kelly WK, O'Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen CC, Lee KD, Pai MY, Chu PY, Hsu CC, Chiu CC, Chen LT, Chang JY, Hsiao SH, Leu YW. Changes in DNA methylation are associated with the development of drug resistance in cervical cancer cells. Cancer Cell Int. 2015;15:98. doi: 10.1186/s12935-015-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–1166. [PubMed] [Google Scholar]
- 113.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clin Cancer Res. 2001;7:584–589. [PubMed] [Google Scholar]
- 114.Kim YT, Zhao M. Aberrant cell cycle regulation in cervical carcinoma. Yonsei Med J. 2005;46:597–613. doi: 10.3349/ymj.2005.46.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohtani N, Yamakoshi K, Takahashi A, Hara E. The p16INK4a-RB pathway: Molecular link between cellular senescence and tumor suppression. J Med Invest. 2004;51:146–153. doi: 10.2152/jmi.51.146. [DOI] [PubMed] [Google Scholar]
- 116.Ki KD, Lee SK, Tong SY, Lee JM, Song DH, Chi SG. Role of 5′-CpG island hypermethylation of the FHIT gene in cervical carcinoma. J Gynecol Oncol. 2008;19:117–122. doi: 10.3802/jgo.2008.19.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Özören N, El-Deiry WS. Cell surface death receptor signaling in normal and cancer cells. Semin Cancer Biol. 2003;13:135–147. doi: 10.1016/S1044-579X(02)00131-1. [DOI] [PubMed] [Google Scholar]
- 118.Jung S, Yi L, Jeong D, Kim J, An S, Oh TJ, Kim CH, Kim CJ, Yang Y, Kim KI, et al. The role of ADCYAP1, adenylate cyclase activating polypeptide 1, as a methylation biomarker for the early detection of cervical cancer. Oncol Rep. 2011;25:245–252. [PubMed] [Google Scholar]
- 119.Huang RL, Chang CC, Su PH, Chen YC, Liao YP, Wang HC, Yo YT, Chao TK, Huang HC, Lin CY, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One. 2012;7:e41060. doi: 10.1371/journal.pone.0041060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ivanova T, Vinokurova S, Petrenko A, Eshilev E, Solovyova N, Kisseljov F, Kisseljova N. Frequent hypermethylation of 5′flanking region of TIMP-2 gene in cervical cancer. Int J Cancer. 2004;108:882–886. doi: 10.1002/ijc.11652. [DOI] [PubMed] [Google Scholar]
- 121.Widschwendter A, Müller HM, Fiegl H, Ivarsson L, Wiedemair A, Müller-Holzner E, Goebel G, Marth C, Widschwendter M. DNA methylation in serum and tumors of cervical cancer patients. Clin Cancer Res. 2004;10:565–571. doi: 10.1158/1078-0432.CCR-0825-03. [DOI] [PubMed] [Google Scholar]
- 122.Zhai Y, Bommer GT, Feng Y, Wiese AB, Fearon ER, Cho KR. Loss of estrogen receptor 1 enhances cervical cancer invasion. Am J Pathol. 2010;177:884–895. doi: 10.2353/ajpath.2010.091166. [DOI] [PMC free article] [PubMed] [Google Scholar]