Abstract
A sixth base, 5-hydroxymethylcytosine (5hmC), is formed by the oxidation of 5-methylcytosine (5mC) via the catalysis of the ten-eleven translocation (TET) protein family in cells. Expression levels of 5hmC are frequently depleted during carcinogenesis. However, the detailed mechanisms underlying the depletion of 5hmC expression in gastric cancer cells remains unclear, and further research is required. The present study examined the expression levels of 5mC and 5hmC and the expression levels of TET1 and TET2 in gastric cancer tissues using immunohistochemistry. The results revealed that 5hmC expression levels were markedly lower in gastric cancer tissues compared with corresponding adjacent normal tissues. Furthermore, a decrease in 5hmC expression levels was associated with a decrease in TET1 protein expression levels in gastric cancer tissues. The ectopic expression level of TET1 may increase the 5hmC expression level in gastric cancer cells. In addition, the results revealed that TET1 protein expression was markedly different in regards to subcellular localization, and mislocalization was significantly associated with the depletion of 5hmC expression levels in gastric cancer. Together, the results of the present study indicated that TET1 dysfunction reduces 5hmC expression levels, and this phenomenon may serve a crucial role in gastric cancer progression.
Keywords: DNA methylation, hydroxylmethylation, gastric cancer, demethylation
Introduction
Epigenetic regulation includes histone modification and DNA methylation, which are involved in the regulation of cell growth and development in mammals (1,2). The methylation of cytosine occurs via DNA methyltransferases (DNMTs), which use S-adenosylmethionine as the donor for the methyl group. In mammalian cells, DNMT genes are classified into de novo (DNMT3A and DNMT3B) and maintenance (DNMT1) methyltransferases; these genes serve various functions in setting the methylation maps of the mammalian genome (3). During carcinogenesis, the DNA methylation level gradually decreases in the DNA repetitive region, leading to genomic DNA instability (4–7). In general, high methylation of a gene promoter is associated with gene silencing. Numerous tumor suppressors have been identified with a hypermethylated promoter that suppresses their transcription potential in multiple types of human cancer (8–11). Therefore, establishing and maintaining DNA methylation status is essential in human types of cancer.
Previous studies have reported that a sixth base, 5-hydroxymethylcytosine (5hmC), is present in a number of tissues, including the muscle, lung, kidney and heart, and is highly expressed in the brain and embryonic cells (2,12–15). Furthermore, 5hmC is generated by the oxidation of 5-methylcytosine (5mC) by ten-eleven translocation protein (TET), and 5hmC serves critical roles in various tissues (2,13,15–22). Furthermore, 5hmC has been suggested to serve a crucial role in gene regulation via the demethylation process (2,13,15–22). The two following theories suggest the involvement of 5hmC in the DNA demethylation process: i) Passive DNA demethylation, where DNMT protein does not recognize the 5hmC-rich region, thus preventing maintenance methylation during DNA replication; ii) active DNA demethylation, where the 5hmC-rich region is recognized by DNA glycosylase protein, which converts 5hmC to cytosine (2,13,15–22). Previous studies have reported that hyperhydroxymethylation of the promoter region of metalloproteinase and homeobox A9 genes increases the expression levels of these genes (3,23). Therefore, high 5hmC expression levels in the promoter region may activate gene transcription via the promotion of DNA demethylation (2,24).
Although 5mC is the upstream substrate for generating 5hmC via the catalysis of TET proteins, the expression level and distribution of 5hmC is not associated with that of 5mC in human types of cancer (25). Therefore, global DNA hypomethylation in human types of cancer only partially explain the low 5hmC expression levels in human tumors. Multiple previous studies have indicated that the dysfunction of isocitrate dehydrogenase [NAPD(+))1/2, activation-induced deaminase and TET genes are associated with aberrant expression of 5hmC in human cancer cells (16,26–28). However, the mechanism underlying the depletion of 5hmC expression levels in gastric cancer cells remains unclear. The present study investigated the status of DNA modification and TET1 and TET2 protein expression levels in gastric cancer tissue, and provided another potential explanation for 5hmC expression level depletion in gastric cancer.
Materials and methods
Clinical samples and DNA/RNA extraction
A total of 16 gastric cancer and corresponding adjacent normal tissue samples were collected from patients with gastric cancer who underwent surgery at the Department of Surgery, Kaohsiung Veterans General Hospital (Kaohsiung, Taiwan) between July 2013 and August 2014. They did not receive any treatment prior to surgery. These patients included 10 males and 6 females, whose age ranged between 55 and 70 years old. The study protocol was independently reviewed and approved by the Institutional Review Board of Kaohsiung Veterans General Hospital (approval no. VGHKS12-CT3-10). The methods performed were in accordance with approved guidelines, and written informed consent was obtained from all patients prior to enrolment in the present study.
Immunohistochemical (IHC) analysis and scoring
Human gastric carcinoma tissue microarrays, including adjacent normal tissues (cat. no. IMH-341) and tumors (cat. no. IMH-316) from 59 patients with gastric cancer were obtained from Imgenex; Novus Biologicals, LLC (Littleton, CO, USA). IHC analyses were performed using the Novolink Max Polymer Detection System (Leica Microsystems, Ltd., Milton Keynes, UK). The tissue slides were sectioned at a thickness of 5 µm, deparaffinized in xylene and rehydrated in 100, 95 and 75% ethanol for 3 min each. Antigen retrieval was performed by immersing the slides in Tris-EDTA (10 mM; pH 9.0) for 10 min at 125°C in a pressure boiler. Endogenous peroxidase activity was blocked by incubating the slides for 30 min with 3% hydrogen peroxide in methanol. The slides were then blocked with protein block buffer (0.4% casein in PBS, with stabilizers, surfactant and 0.2% bronidox L as a preservative). Following blocking at room temperature (RT) for 30 min, primary antibodies were immediately applied and the slides were incubated overnight at 4°C in a wet chamber. The primary antibodies used were mouse monoclonal anti-5mC (1:500; cat. no. ab10805; Abcam, Cambridge, UK); rabbit polyclonal anti-5hmC (1:1,000; cat. no. 39769; Active Motif, Carlsbad, CA, USA), rabbit polyclonal anti-TET1 (1:150; cat. no. TA309902; OriGene Technologies, Inc., Rockville, MD, USA) and goat polyclonal anti-TET2 (1:50; cat. no. OAEB00839; AVIVA Systems Biology, San Diego, CA, USA) in primary antibody diluent (ScyTek Laboratories, Logan, UT, USA). Following washing with PBS, the slides were incubated with a rabbit anti-mouse poly-horseradish peroxidase (HRP)-immunoglobulin (Ig)G (cat. no. RE7159; Leica Microsystems, Ltd., Milton Keynes, UK) and goat anti-rabbit poly-HRP-IgG (cat. no. RE7161; Leica Microsystems, Ltd., Milton Keynes, UK) for 10 min at RT. Finally, the color was developed using a 0.03% diaminobenzidine solution (ScyTek Laboratories) for 2 min at room temperature. Subsequently the tissue sections were counterstained with hematoxylin for 10 min at room temperature.
The expression scores of individual candidates for nuclear or cytoplasmic staining were determined on the basis of staining intensity (0, no signal; 1, mild; 2, moderate; 3, strong). The proportion of positively stained tumor cells (scored as 0–100%) was evaluated in the whole field of each core. The score of individual candidates was calculated using the following formula: Intensity × percentage of positively stained cells.
Gene expression data
The microarray data of TET1 and TET2 in 311 gastric cancer tissues and 57 adjacent normal tissues were obtained from the Gene Expression Across Normal and Tumor Tissue (GENT) database (http://medicalgenome.kribb.re.kr/GENT/search/search.php) (29).
Western blot analysis
The tissue samples were lysed using a lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.02% sodium azide, 1 µg/ml aproteinin and 1 mM PMSF) at 4°C for 30 min. The lysates were collected and centrifuged at 16,000 × g at 4°C for 10 min to remove cell debris. Protein assays were performed using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) based on the Bradford dye-binding procedure (30). Protein samples (60 µg) were separated by 8–10% SDS-PAGE. The separated proteins were then electrotransferred onto nitrocellulose membranes (GE Healthcare, Chicago, IL, USA). Following blocking overnight at 4°C using 0.1% of Tween in PBS supplemented with 5% skimmed milk, the membranes were incubated with mouse monoclonal anti-TET1 (1:500; cat. no. GTX627420; GeneTex Inc., Irvine, CA, USA), mouse monoclonal anti-green fluorescent protein (GFP; 1:2,000; cat. no. sc-9996; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse monoclonal anti-actin (1:3,000; cat. no. MAB1501; EMD Millipore, Billerica, MA, USA) for 1 h in PBS-Tween supplemented with 5% skimmed milk at room temperature. The membranes were then incubated with HRP-conjugated goat-anti-rabbit-IgG (1:5,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or goat-anti-mouse IgG secondary antibodies (1:5,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at RT. Following washing three times with PBS-Tween, immunoreactive bands were detected using an enhanced chemiluminescence kit (cat. no. K-12045-D50; Advansta, Inc., Menlo Park, CA, USA).
Gastric cancer cell lines and transfection assay
Human gastric cancer AGS cells were obtained from American Type Culture Collection (Manassas, VA, USA), and cultured in Dulbecco's modified Eagle's medium (Biological Industries Israel Beit-Haemek Ltd., Kibbutz Beit-Haemek, Israel) supplemented with 10% fetal bovine serum (GE Healthcare Life Sciences, Logan, UT, USA) and 1% penicillin/streptomycin (Sigma-Aldrich; Merck KGaA). In the present study, transfection assay was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
DNA dot-blot assay
DNA samples were added to the denaturation buffer (0.4 mM NaOH and 10 mM EDTA) and denatured at 100°C for 10 min. Furthermore, the samples were chilled on ice for 5 min and applied on a positive-charged nylon membrane (Roche, Basel, Switzerland). The membrane was UV cross-linked and dried for 1 h at 70°C. The membranes were probed with rabbit polyclonal anti-5hmC (1:5,000; cat. no. 39769; Active Motif, Carlsbad, CA, USA) at 4°C overnight. The membranes were then incubated with HRP-conjugated goat anti-rabbit IgG (1:5,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) for 1 h at RT. The target bands were visualized using WesternBright enhanced chemiluminescence reagent (Advansta, Inc., Menlo Park, CA, USA) and the results of the immunoreactions were analyzed with a BioSpectrum® 500 Imaging System (Ultra-Violet Products Ltd., Cambridge, UK). The membranes were stained with methylene blue (Sigma-Aldrich; Merck KGaA) as a loading control.
Predict nuclear export signals (NESs) by bioinformatic approach
A useful web server (NetNES 1.1) for predicting potential leucine-rich NESs within protein sequences is available at http://www.cbs.dtu.dk/services/NetNES/ (31). The TET1 protein sequences (NP_085128.2) were downloaded from The National Center for Biotechnology Information databases (NCBI; https://www.ncbi.nlm.nih.gov/). TET1 protein sequences in FASTA format was uploaded to NetNES 1.1 website. According to the characteristics and the homology of the NESs (Nuclear export signals), NetNES 1.1 could predict putative NESs within TET1 protein.
In vitro DNA demethylation assay
pEGFP plasmids (Clontech, Mountain View, CA, USA) were completely methylated in vitro using M.SssI methylase enzymes (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Subsequently, methylated reporter vectors were co-transfected with TET1-FL or TET1-delNLS into AGS gastric cancer cells. Following transfection for 24–48 h, demethylation activity in cells was examined by western blotting as aforementioned.
Statistical analysis
The expression levels of 5mC and 5hmC in paired gastric cancer were analyzed using a paired Student's t-test. The expression levels of TET1 and TET2 from the TCGA database and the association between various localizations of TET1 protein and 5hmC expression levels in human gastric cancer was analyzed using Student's t-tests. The TET1 activity assays were performed in triplicate. The intensity of GFP was quantified using ImageJ Software 1.45s (National Institutes of Health, Bethesda, MD, USA) and presented graphically. The data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference and P<0.01 was considered to indicate a highly significant difference.
Results
5hmC expression level depletion in gastric cancer cells
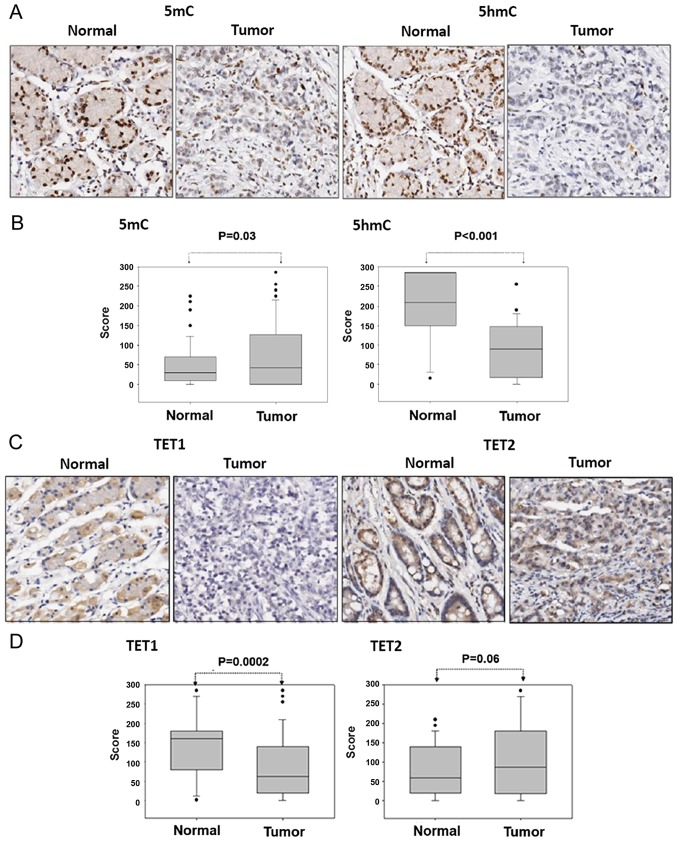
The IHC staining revealed that compared with the corresponding adjacent normal tissues, the expression level of 5mC was slightly increased and that of 5hmC was significantly depleted in gastric cancer tissues (5mC, P=0.03 and 5hmC, P<0.001; Fig. 1A and B). These results indicated that 5hmC expression level depletion was not due to low 5mC expression levels in gastric cancer tissues. The potential mechanism underlying 5hmC expression level depletion may therefore be the dysfunction of metabolic enzymes involved in the DNA demethylation signaling pathway.
Figure 1.
Expression levels of 5mC, 5hmC, TET1 and TET2 were analyzed using IHC in gastric cancer tissue arrays. A representative case is presented in this figure. (A) The IHC assay analyzed the 5mC and 5hmC expression levels in a patient with gastric cancer (magnification, ×100), and (B) the relative 5mC and 5hmC expression levels were scored between gastric cancer and adjacent normal tissues from 58 patients. (C) The IHC assay revealed the expression levels of TET1 and TET2 in a patient with gastric cancer (magnification, ×100). (D) The relative expression levels of TET1 and TET2 were scored between the gastric cancer tissues of 58 patients and their corresponding adjacent normal tissues. IHC, immunohistochemistry; TET, ten-eleven translocation; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine.
Expression levels of TET1/2 in gastric cancer cells
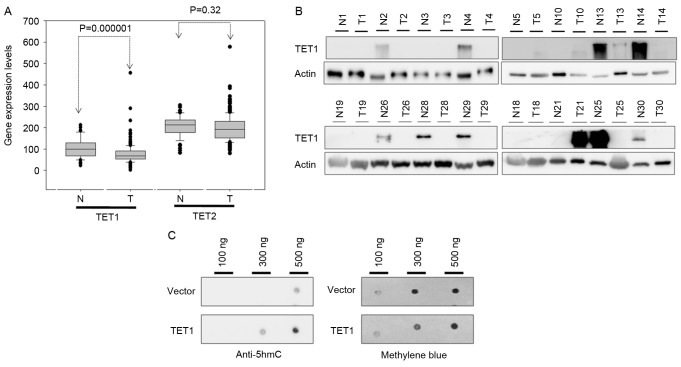
The present study investigated the expression levels of TET1 and TET2 in a gastric cancer tissue array (n=59 cases) using IHC. TET1 protein expression was markedly decreased and TET2 protein expression was slightly increased in the gastric cancer tissues compared with the adjacent normal tissues (TET1, P=0.0002 and TET2, P=0.06; Fig. 1C and D). This result was further confirmed by mRNA and protein expression level analysis using the GENT database and western blotting, respectively. By analyzing the GENT database, the present study revealed that the transcriptional expression levels of TET1 were substantially decreased in the gastric cancer tissues compared with normal adjacent tissues (P<0.001; Fig. 2A). This result was consistent with that of IHC analyses. Furthermore, as presented in Fig. 2B, compared with the corresponding adjacent normal tissues, the protein expression levels of TET1 were frequently decreased in the tumor tissues (in 8 out of 16 cases). The transfection of TET1 into AGS cells for 24 h resulted in an increase in 5hmC expression levels (Fig. 2C). These results indicated that the decrease in 5hmC expression levels in gastric cancer may be induced by low TET1 protein expression levels.
Figure 2.
Expression levels of TET1 and TET2 in gastric cancer. (A) Expression levels of TET1 and TET2 were analyzed in 311 gastric cancer and 57 adjacent normal tissues from the Gene Expression Across Normal and Tumor Tissue database. (B) Expression levels of TET1 protein were decreased in gastric cancer tissues obtained from 16 patients with gastric cancer compared with the normal adjacent tissues. (C) TET1 overexpression increased the global 5hmC expression levels in the genomic DNA of AGS cells. The loading control was visualized using methylene blue staining. N, normal; T, tumor; TET, ten-eleven translocation; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine.
Nuclear exclusion of TET1 is associated with 5hmC expression level depletion in gastric cancer
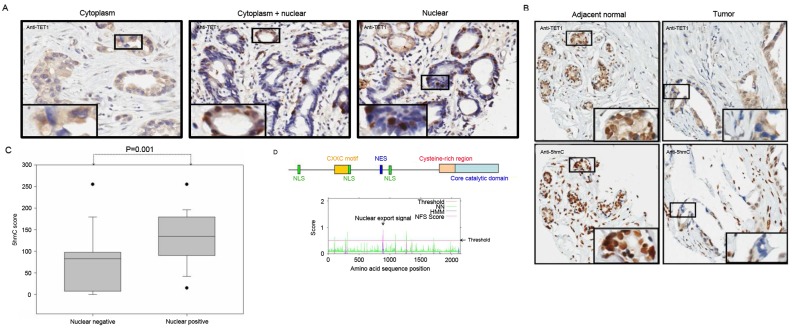
The present study not only observed the depletion of TET1 protein expression levels in gastric cancer tissues, but also observed marked differences in its subcellular localization. As presented in Fig. 3A, TET1 protein was detected in three subcellular localizations in the gastric cancer tissues: Cytoplasm (left panel); cytoplasm and nucleus (middle panel); nuclear accumulation (right panel). Furthermore, the present study detected TET1 expression level in gastric cancer tissue samples, except in 3 samples where no TET1 protein expression was observed. Of the 59 tumor samples, the protein expression level was restricted to the cytoplasm without nuclear staining in 36 (61%) samples, in the cytoplasm and nucleus in 9 (15.3%) samples and predominantly in the nucleus in 11 (18.6%) samples. Furthermore, as presented in Fig. 3B, TET1 nuclear localization was associated with 5hmC expression levels. The comparison of the 5hmC expression levels with TET1 cellular localization revealed that TET1 nuclear staining had a significant association with high 5hmC expression levels (P=0.001) in gastric cancer (Fig. 3C). These results implied that TET1 protein may shuttle between the nucleus and the cytoplasm. Using bioinformatics, the present study identified three nuclear localization signals (NLS; NLS1:aa20-50, NLS2:aa620-653 and NLS3:1158-1162) and one nuclear export signal (NES; aa877-889) in the N-terminal region of TET1 protein (Fig. 3D). However, this result requires further confirmation by future studies.
Figure 3.
Various localizations of TET1 protein are associated with 5hmC status in human gastric cancer. (A) TET1 protein was exclusively expressed in the cytoplasm (left panel), in the cytoplasm and nucleus (middle panel) and predominantly in the nucleus (right panel) (magnification, ×100). (B) The results of immunohistochemistry revealed that nuclear exclusion of TET1 protein was associated with 5hmC depletion in gastric cancer tissues (magnification, ×100). (C) Nucleic TET1 protein expression was significantly associated with high 5hmC expression level in gastric cancer. (D) A schematic display of the structure of TET1 gene. An NES signal was identified in TET1 (aa877-889) using the ENS prediction tool (http://www.cbs.dtu.dk/services/NetNES/). TET, ten-eleven translocation; 5hmC, 5-hydroxymethylcytosine; NES, nuclear export signal; NLS, nuclear localization signals; NN, Neural Network; HMM, Hidden Markov Model.
TET-1 localization may contribute to the active DNA demethylation signaling pathway
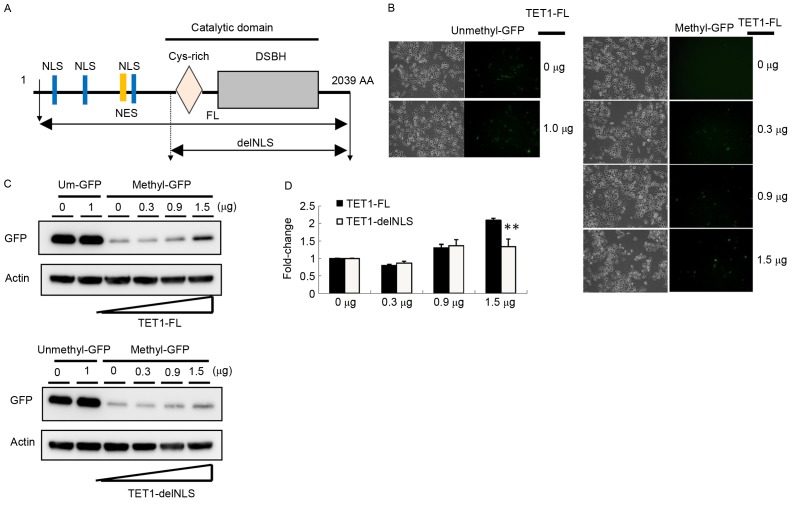
TET1 nuclear exclusion was associated with 5hmC depletion in gastric cancer tissues according to the IHC data, so the present study evaluated whether different cellular localizations of TET1 protein affected its demethylation activity by constructing two TET1 expression vectors, TET1-FL and TET1-delNLS. TET1 contains a critical catalytic domain and three NLSs, whereas TET1-delNLS contained only a catalytic domain (Fig. 4A). A reporter assay was used to assess DNA demethylation activity. The GFP plasmid was methylated in vitro with CpG methyltransferase (M.SssI). Furthermore, the completely methylated pEGFP plasmids were co-transfected into AGS cells with TET1-FL or TET1-delNLS. As presented in Fig. 4B, GFP expression was completely silenced in AGS cells transfected with the methylated GFP expression vector. Following co-transfection with the TET1-FL expression vector for 24 h, the number of GFP-positive cells markedly increased (Fig. 4B). By performing western blot analysis, the present study observed that the GFP expression level increased 2.2-fold following transfection with 1.5 µg TET1 (Fig. 4C and D). Compared with cells transfected with TET1-delNLS, the GFP expression level was slightly increased in cells transfected with 1.5 µg of TET1-delNLS (~1.3-fold). These results indicated that the nuclear localization of TET1 may serve a crucial role in the modulation of gene expression, particularly in gastric cancer.
Figure 4.
TET1 is involved in active DNA demethylation pathway in AGS cells. (A) Schematic diagrams of TET1 expression constructs for TET1-full length and TET1-delNLS. AGS cells were co-transfected with methylated GFP or unmethylated GFP, and TET1-FL or TET1-delNLS. Following transfection for 24 h, the GFP expression was detected by (B) fluorescence microscopy (magnification, ×100) and (C) western blotting. (D) The intensity of GFP was quantified using ImageJ software and was represented graphically. **P<0.01. Actin was used as an internal control. TET, ten-eleven translocation; GFP, green fluorescent protein; NES, nuclear export signal; NLS, nuclear localization signals; TET1-FL, full length of TET1 genes; TET1-delNLS, NLS domain of TET1 was deleted.
Discussion
Previous studies have reported that 5hmC expression levels are significantly lower in gastric cancer tissues compared with in adjacent normal tissues (32–34). In addition, Yang et al (33) reported that a low 5hmC expression level is an independent factor for poor prognosis in patients with gastric cancer. They also reported that a low 5hmC expression level was associated with the majority of recorded clinicopathological characteristics, including the tumor size, stage, lymph node metastasis and overall survival rate. These results indicated that the mechanism underlying 5hmC depletion served a crucial role in gastric cancer progression. The results of the present study revealed that the potential mechanism underlying 5hmC expression level depletion in gastric cancer cells may be a low TET1 protein expression level. Furthermore, Yang et al (33) reported a strong association between the decrease in 5hmC expression levels and decrease in TET1 mRNA expression levels; however, this link was not observed for TET2 and TET3 proteins. Frycz et al (35) demonstrated that TET1 transcript and protein expression levels were associated with the metastasis stage in patients with gastric cancer. The results of the present study revealed that TET1 transcript and protein expression levels were significantly decreased in gastric cancer tissues compared with adjacent normal tissues. In addition, the present study observed a marked difference in subcellular localization. The localization of TET1 protein expression level in the nucleus was associated with 5hmC expression levels and contributed to the active demethylation process. Our previous study reported a similar phenomenon and revealed that the cytoplasmic mislocalization of TET1 reduced the expression level of 5hmC in breast cancer (25). A low 5hmC expression level is an effective independent prognostic biomarker for breast ductal carcinoma, particularly in patients with an endocrine receptor/progesterone receptor-negative subtype (25). Similarly, Müller et al (36) reported that a decrease in 5hmC expression levels is attributable to the nuclear exclusion of TET1 from the nuclei of glioma cells. Using the bioinformatics approach, the present study identified three NLSs and one NES in the N-terminal region of TET1 protein (Fig. 3D). This result suggested that TET1 protein shuttles between the nucleus and the cytoplasm. In conclusion, these results revealed that the nuclear-to-cytoplasm shuttling of TET proteins in tumor cells may be a critical event contributing to human cancer progression.
Acknowledgements
The present study was supported by the Kaohsiung Veterans General Hospital (grant no. VGHKS-105-016), the Zuoying Branch of Kaohsiung Armed Forces General Hospital (grant no. ZBH-103-27) and Yen Tjing Ling Medical Foundation (grant nos. CI-102-13 and CI-103-18).
References
- 1.Esteller M. Cancer epigenetics for the 21st Century: What's next? Genes Cancer. 2011;2:604–606. doi: 10.1177/1947601911423096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dahl C, Grønbæk K, Guldberg P. Advances in DNA methylation: 5-hydroxymethylcytosine revisited. Clin Chim Acta. 2011;412:831–836. doi: 10.1016/j.cca.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis; Proc Natl Acad Sci USA; 2013; pp. 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim B, Kim JH, Kim M, Kim SY. Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J Gastroenterol. 2016;22:1190–1201. doi: 10.3748/wjg.v22.i3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto H, Imai K. Microsatellite instability: An update. Arch Toxicol. 2015;89:899–921. doi: 10.1007/s00204-015-1474-0. [DOI] [PubMed] [Google Scholar]
- 6.Kim KJ, Lee TH, Cho NY, Yang HK, Kim WH, Kang GH. Differential clinicopathologic features in microsatellite-unstable gastric cancers with and without MLH1 methylation. Hum Pathol. 2013;44:1055–1064. doi: 10.1016/j.humpath.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Ling ZQ, Tanaka A, Li P, Nakayama T, Fujiyama Y, Hattori T, Sugihara H. Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 2010;297:244–251. doi: 10.1016/j.canlet.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao HW, Fang WL, Lin WC. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;49:969–980. doi: 10.1002/gcc.20804. [DOI] [PubMed] [Google Scholar]
- 9.Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–1197. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, Kao HW, Fang WL, Huang KH, Chan WC, Lin WC. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129:2600–2610. doi: 10.1002/ijc.25919. [DOI] [PubMed] [Google Scholar]
- 11.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 18.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 19.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai KW, Li GC, Chen CH, Yeh MH, Huang JS, Tseng HH, Fu TY, Liou HH, Pan HW, Huang SF, et al. Reduction of global 5-hydroxymethylcytosine is a poor prognostic factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res Treat. 2015;153:219–234. doi: 10.1007/s10549-015-3525-x. [DOI] [PubMed] [Google Scholar]
- 26.Kinney SR, Pradhan S. Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer. Adv Exp Med Biol. 2013;754:57–79. doi: 10.1007/978-1-4419-9967-2_3. [DOI] [PubMed] [Google Scholar]
- 27.Pérez C, Martínez-Calle N, Martín-Subero JI, Segura V, Delabesse E, Fernandez-Mercado M, Garate L, Alvarez S, Rifon J, Varea S, et al. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS One. 2012;7:e31605. doi: 10.1371/journal.pone.0031605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: Gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duhamel RC, Meezan E, Brendel K. The addition of SDS to the Bradford dye-binding protein assay, a modification with increased sensitivity to collagen. J Biochem Biophys Methods. 1981;5:67–74. doi: 10.1016/0165-022X(81)90007-5. [DOI] [PubMed] [Google Scholar]
- 31.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 32.Park JL, Kim HJ, Seo EH, Kwon OH, Lim B, Kim M, Kim SY, Song KS, Kang GH, Kim HJ, et al. Decrease of 5hmC in gastric cancers is associated with TET1 silencing due to with DNA methylation and bivalent histone marks at TET1 CpG island 3′-shore. Oncotarget. 2015;6:37647–37662. doi: 10.18632/oncotarget.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Q, Wu K, Ji M, Jin W, He N, Shi B, Hou P. Decreased 5-hydroxymethylcytosine (5-hmC) is an independent poor prognostic factor in gastric cancer patients. J Biomed Nanotechnol. 2013;9:1607–1616. doi: 10.1166/jbn.2013.1713. [DOI] [PubMed] [Google Scholar]
- 34.Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670–676. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frycz BA, Murawa D, Borejsza-Wysocki M, Marciniak R, Murawa P, Drews M, Kołodziejczak A, Tomela K, Jagodziński PP. Decreased expression of ten-eleven translocation 1 protein is associated with some clinicopathological features in gastric cancer. Biomed Pharmacother. 2014;68:209–212. doi: 10.1016/j.biopha.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Müller T, Gessi M, Waha A, Isselstein LJ, Luxen D, Freihoff D, Freihoff J, Becker A, Simon M, Hammes J, et al. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol. 2012;181:675–683. doi: 10.1016/j.ajpath.2012.04.017. [DOI] [PubMed] [Google Scholar]