Abstract
Abstract: Xylooligosaccharides (XOS) have gained increased interest as prebiotics during the last years. XOS and arabinoxylooligosaccharides (AXOS) can be produced from major fractions of biomass including agricultural by-products and other low cost raw materials. Endo-xylanases are key enzymes for the production of (A)XOS from xylan. As the xylan structure is broadly diverse due to different substi-tutions, diverse endo-xylanases have evolved for its degradation. In this review structural and functional aspects are discussed, focusing on the potential applications of endo-xylanases in the production of dif-ferently substituted (A)XOS as emerging prebiotics, as well as their implication in the processing of the raw materials. Endo-xylanases are found in at least eight different glycoside hydrolase families (GH), and can either have a retaining or an inverting catalytic mechanism. To date, it is mainly retaining endo-xylanases that are used in applications to produce (A)XOS. Enzymes from these GH-families (mainly GH10 and GH11, and the more recently investigated GH30) are taken as prototypes to discuss substrate preferences and main products obtained. Finally, the need of new and accessory enzymes (new specifici-ties from new families or sources) to increase the yield of different types of (A)XOS is discussed, along with in vitro tests of produced oligosaccharides and production of enzymes in GRAS organisms to fa-cilitate use in functional food manufacturing.
Keywords: Xylanase, glycoside hydrolase, XOS, AXOS, arabinooligosaccharides, GH10, GH11, GH30, subsites
1. Introduction
Biomass utilization is a topic of wide interest today, due to shrinking fossil resources combined with future needs in the fields of energy, chemicals and food production [1]. Polysaccharides are the major components in plant biomass, and serve either as storage for reserve food (e.g. starch) or contribute to mechanical strength (e.g. cellulose and hemicelluloses).
Hemicelluloses form a matrix between the cellulose microfibrils and lignin polymers. Most hemicelluloses are heteropolysaccharides, and are (despite additional substituents) often collectively named based on the most common sugar units in the polymer (e.g. xylans, galactoglucomannans, xyloglucans, etc.). The most common type of hemicellulose are the xylans, which in terrestrial plants have a backbone built of β-1,4-linked xylose units with diverse substituent decorations that are both dependent on the source and the tissue in the plant where they occur [2, 3]. In marine environments xylans with a backbone of β-1,3-linked xylose units are also found [4-6].
The enzymes that hydrolyze the backbone of the differently substituted xylans are termed xylanases. These enzymes can be either endo-acting (hydrolyzing bonds in the interior of xylan polymers) or exo-acting enzymes (hydrolyzing xylan from either the reducing or non-reducing end), and can act on β-1,4- (or β-1,3-) linked xylose units. It is the endo-acting xylanases that are the focus of this review.
Endo-1,4-β-xylanases (EC 3.2.1.8) that randomly cleave the β-1,4-linked backbone of xylans are most common and most commonly investigated. In addition, some enzymes are known that either require substituents linked to the xylose residues for activity such as the glucuronoarabinoxylan endo-1,4-β-xylanases (EC 3.2.1.136), or accept a substituent (then sometimes classified with double enzyme commission numbers). Yet another group of endo-xylanases cleaves β-1,3-linked xylose units (EC3.2.1.32). Enzymes characterized as specific endo-1,3-β-xylanases (not cleaving β-1,4-linked xylan) are still uncommon. Following the sequence-based classification in the carbohydrate active enzymes database CAZy (www.cazy.org) the characterized candidates are in principle found in glycoside hydrolase family 26 (GH26, containing six enzymes from marine organisms and genes [Genbank codes]: Alcaligenes sp. [BAB88993.1], Brevundimonas vesicularis [AIX87981.1], Pseudomonas sp. [BAB79289.1], Thermotoga neapolitana [ACM22793.1], and two strains of Vibro [BAD51934.1, BAA94698.1]). In the same database, see Schedule 1 for classification terminology, endo-1,4-β-xylanases with characterized specificity EC 3.2.1.8, are to date classified under eight GH-families: GH5, GH8, GH10, GH11, GH30, GH43, GH51 and GH98.
Schedule 1. Terminology for Glycoside hydrolase classification. Adapted from www.cazy.org.
Among the GH families, enzymes classified as endo-1,4-β-xylanases dominate GH10 and GH11, and are together with enzymes specifically acting on glucuronoxylan (EC 3.2.1.136), comprising a subfamily of GH30 (classified as subfamily 8). Classification under EC 3.2.1.8 is either relatively less common in other GH-families, or comprises subfamilies with few characterized candidate enzymes (Table 1).
Table 1.
Glycoside hydrolase (GH) families including enzymes with EC 3.2.1.8 (endo-1,4-β-xylanase) specificity and/or other specificity for endo-activity on substituted xylans (from www.cazy.org, March 2016).
|
Clan
(overall fold) |
Family
(number of characterized enzymes) |
Mechanism |
Subfamily
(number of characterized enzymes) |
EC number | Number of characterized endo-xylanases | Number of structure determined endo-xylanases |
|---|---|---|---|---|---|---|
| GH-A ((αβ)8) |
GH5 (538) |
retaining | subfamily 1-53 | |||
| subfamily 4 (95) |
EC 3.2.1.8/3.2.1.4 (cellulase) | 4(bifunctional) | 1 | |||
| subfamily 21 (4) |
EC 3.2.1.8 | 4 | 0 | |||
| subfamily 25 (11) |
1(bifunctional) | 1 | ||||
| subfamily 34 (1) |
EC 3.2.1.- (arabinoxylan specific) | 1 | 1 | |||
| GH10 (342) |
retaining | - | EC 3.2.1.8 | >300 | 38 | |
| GH30 (34) |
retaining | subfamily 1-8 | ||||
| subfamily 8 (15) |
enzymes acting on xylan/glucuronoxylan | |||||
| EC 3.2.1.8 | 10 | 1 | ||||
| EC 3.2.1.136 (glucuronoarabinoxylan endo-1,4-β-xylanase) | 2 | 1 | ||||
| EC 3.2.1.8/3.2.1.136 | 3(bifunctional) | 2 | ||||
| GH51 (76) |
retaining | - | GH-family of predominantly arabinofuranosidases EC 3.2.1.8/3.2.1.4 |
1(bifunctional) | 0 | |
| GH-C (β-jelly roll) |
GH11 (270) |
retaining | - | EC 3.2.1.8 | >250 | 29 |
| GH-F (5-fold β-propeller) |
GH43 (139) |
inverting | subfamily 1-37 | GH family of mainly exo-acting enzymes | ||
| subfamily16 | EC 3.2.1.8 | 3 | 0 | |||
| EC 3.2.1.8/3.2.1.55 (arabinofuranosidase) | 2(bifunctional) | 0 | ||||
| subfamily29 | EC 3.2.1.8 | 1 | 0 | |||
| GH-M (α/α)6 |
GH8 (71) |
inverting | - | multi-specific family: | ||
| EC 3.2.1.8 | 8 | 1 | ||||
| EC 3.2.1.156 (oligosaccharide reducing-end xylanase) | 4 | 1 | ||||
| No clan | GH98 (5) |
inverting | - | small GH family with galactosidase and xylanase activities EC 3.2.1.8 |
1 |
0 |
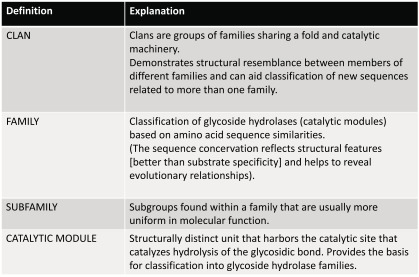
Enzymes in GH26, are together with GH5, GH10, GH11, GH30 and GH51 retaining enzymes (Fig. 1), making the double displacement mechanism dominating for both endo-1,3- and endo-1,4-β-xylanases. Inverting endo-1,4-β-xylanases are found in GH8, GH43 and GH98, limiting the number of characterized enzymes with this activity and mechanism to relatively few characterized candidates (to date, March 2016, eight enzymes in GH8, six in GH43 and one in GH98 are listed in CAZy in the respective family (Table 1)).
Fig. (1).
Representation of the chemical mechanisms for endo-xylanases based on the general mechanism of glycoside hydrolases [7]. (A) Inverting. The reaction occurs via single displacement where one carboxylic group is the general base and the other one is the general acid, and these residues are situated around 10.5 Å from each other. (B) Retaining. The reaction occurs via double displacement where one carboxylic group acts as general acid/base, and the other one as nucleophile forming a covalent intermediate. In this case, both carboxylic groups are situated around 5.5 Å from each other. In both mechanisms the transition states adopt distorted conformations due the oxocarbenium-ion-like structure, with boat (2,5B in GH11) and half-chair (4H3 in GH10) conformations being most energetically favored [8].
The distribution of the endo-xylanases is also reflected in their use as catalytic aids in applications, and thus far enzyme candidates from GH10 and GH11 are most commonly used. A number of applications have recently been reviewed [9, 10], highlighting the use of xylanases as processing aids, for example in saccharification (prior to fermentation for production of e.g. lactic acid or ethanol, or for xylitol production). Other established applications involve use as additives in feed (where xylanases are added to improve digestibility) and food (where xylanases are used as ingredients in dough to improve bread quality) [10]. Production of xylooligosaccharides (XOS) using xylanases as processing aids has received increased interest, as a consequence of the establishment of XOS as putative prebiotics [9, 11] (see schedule 2 for definitions). Today, galacto- (GOS) and fructo-oligosaccharides (FOS) are established prebiotics, and have a significant market with major application sectors in food and beverages, dietary supplements and animal feed. With increasing interest and an increasing number of oligosaccharide compounds claimed as prebiotics (e.g. XOS and arabinoxylooligosaccharides, AXOS), the global oligosaccharide production is expected to grow to 450.000 tons in 2018 (www.dairymark.com/gos.html).
Schedule 2. Definitions of prebiotics, probiotics and synbiotics [12].
Xylans are (as described below) dependent on origin decorated with different substituents, and the oligosaccharides obtained after selective enzymatic hydrolysis are thus also differently substituted. It has for example been shown that non-substituted XOS stimulate a broader group of intestinal bacteria associated with health and well-being than arabinose-substituted AXOS, which instead stimulate strains of bifidobacteria more selectively [13-15]. In this review we overview enzymes used for both AXOS and XOS production (together abbreviated (A)XOS), and highlight substrate interactions of structure-determined enzymes used for this purpose. Knowledge in this field is expected to increase possibilities to choose enzyme candidates or mixtures that will improve the overall yield of (A)XOS or obtain specifically substituted oligosaccharides upon enzymatic hydrolysis of xylan from various sources.
2. Xylan structures and substituents
Xylans are found universally in annuals (often isolated from cereal grains), hardwoods, to a lesser extent in softwoods and finally also in seaweeds in marine environments (Table 2). Most xylans have a varying degree of short side-chains (Fig. 2), although a few examples of linear, nonsubstituted xylan exist, including a fraction isolated from esparto grass (Stipa tenacissima) and a xylan isolated from tobacco stalks (Nicotiana tabacum) [2].
Table 2.
General properties of xylans from different sources.
| Xylan | Source | Back-bone linkage | DP | Major functional groups | References |
|---|---|---|---|---|---|
|
O-acetyl-4-O-methyl- glucuronoxylan |
Hardwood | β-1,4 | 150-200 | Acetyl-groups [5-7 residues/10 xylose residues (linked at C2 or C3)] 4-O-methylglucuronic acid [1 residue/10 xylose residues (linked at C2)] |
[2, 17, 19, 25] |
| Arabino-4-O-methyl- glucuronoxylan |
Softwood | β-1,4 | 70-130 | 4-O-methylglucuronic acid [2 residues/10 xylose residues (linked at C2)] α-l-arabinofuranose [1 residues/10 xylose residues, linked by 1,3-linkages (at C3) to the main chain.] |
[2, 17, 19, 25] |
| Arabinoxylans | Grasses | β-1,4 | >70 |
l-arabinofuranose linked by 1,3-linkages (at C2 or C3) to the main chain 4-methyl-D-glucuronic acid and smaller proportions of D-glucuronic acid Acetyl-groups ferulic or coumaric acid (small amounts, esterified to C5) |
[2, 17, 21, 22] |
| Mixed linkage xylan | red algae | β-1,4, β-1,3 | 25-80 | Methyl-groups (sulfation) |
[4, 6, 26] |
| Xylan | green algae | β-1,3 | approx. 40 | Methyl-groups | [4, 5, 26] |
Fig. (2).
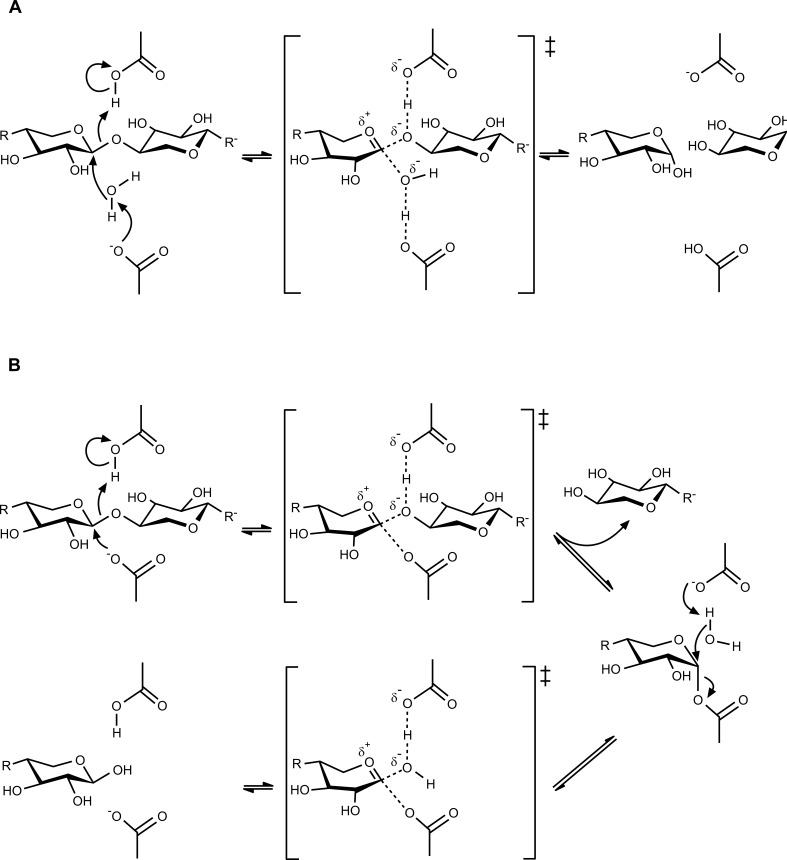
Hypothetical substituted xylan (1) and some examples of the wide variety of potential oligosaccharides produced by the activity of endo-β-xylanases: (2) arabinoxylobiose (A3X), (3) double substituted arabinoxylotriose (XA2+3X), (4) 4-O-methylglucuronoxylotriose (XU4m2X), (5) feruloylated arabinoxylotriose (XA5f3X), (6) xylobiose (X2), (7) xylotriose (X3) and (8) xylotetraose (X4). One-letter code system (in brackets) for oligosaccharides used here is according with the proposed by Fauré [18].
In vascular plants the backbone consists of β-d-xylopyranose units, which are linked by β-1,4-bonds. A common side group is 4-methyl-α-d-glucuronic acid (uronic acid), linked at C2 to some of the xylopyranose residues in the main chain, resulting in acidic xylan. The absence of a primary alcohol functional group diminishes the capacity of xylan chains to join with each other and with other polysaccharides. In contrast, there is evidence for close association, including covalent bonds, between xylan and lignin [16, 17].
In hardwood the predominant hemicellulose is O-acetyl-4-O-methylglucurono-β-d-xylan (sometimes called acetylglucuronoxylan) and the xylan content varies within the range 15-35% of the dry wood [9, 17, 19, 20].
Softwoods have lower xylan content, and the xylans generally have a lower degree of polymerization (DP) [19]. In this material, l-arabinofuranose groups and uronic acids are linked by 1,3-linkages (at C3) to the main chain in softwood xylan. The uronic acids in softwood xylan seem to be localized rather than randomly distributed along the xylan chain [2].
The heteroxylans from Graminae (grasses including cereals) have the same backbone as wood xylans but are generally more branched. It has also become clear that the xylan structure varies in different parts of the plant and during different ages [2, 17, 21]. The side-chains contain large proportions of l-arabinose residues, which may occur in combination with other sugars (e.g. xylose and galactose), or in combination with ferulic or coumaric acid (esterified to C5) [2, 19]. Recently Rumpagaporn and co-workers [22] performed a detailed analysis of linkage features of different cereal arabinoxylans connected to fermentation rate by fecal microbiota, showing that the major structural factors that related to slow fermentation were the frequency of branching and type of linkage of these branched substituents.
Finally, xylans are not only found in terrestrial plants, but are also present in marine resources, such as seaweeds (Table 2). Unique for these xylans is the presence of 1,3-linkages in the backbone, and relatively few substituents.
It is difficult to fully describe all possible covalent linkages in xylan. They do not only vary as a consequence of the plant origin, but some bonds are also selectively broken during isolation processes [17, 23, 24]. Acetyl, feruloyl and p-coumaroyl substituents are alkaline labile (ester linkages are saponified). The chain length can also be decreased by isolation procedures involving heat and pressure [17].
3. Enzymatic xylooligosaccharides production from plant xylans
The production of XOS from plant materials can be divided into two general stages: xylan extraction (sometimes called pre-treatment), and hydrolysis where the latter can be either enzymatic or thermochemical [27]. Xylan extraction is often performed by using alkaline solutions (1 to 2% w/v) of KOH, NaOH, Ca(OH)2, [28] or NH3 [29] in a moderate temperature during 2 to 3 hours, with subsequent neutralization. Hot water extraction is another relatively common method, frequently performed under high pressure in an autoclave [30] or in a special pressurized chamber (e.g. steam explosion). The choice of method, efficiency in extraction, and need of additional steps in the extraction procedure depends on the type of raw material (methodologies and yields are for example reviewed by Otieno and Ahring, [27]), and a number of combinations of different steps are possible. For example, in cereal bran extraction using hot water, an added pretreatment step involving addition of amylases for starch removal could be necessary to improve purity [30]. In some cases, proteases have also been added in the processing to remove excess of gluten. The sensitivity of the xylan-substituents to the processing conditions should be considered, as it is well-known that e.g. feruloyl and acetyl groups, naturally present in xylan, are sensitive to high temperature and alkaline conditions. Prolonged time of heat treatment at high temperature could thus produce (undesirable) side products such as furfural and 5-hydroxymethylfurfural that potentially could inhibit the enzymatic hydrolysis to oligosaccharides.
To date it is predominantly bacterial and fungal enzymes from GH10 and GH11 that are used in trials to produce different types of XOS (Table 3). Bacterial endo-β-xylanases are frequently recombinantly produced, and the genes have mainly been obtained from Gram-positive bacteria classified under the phyla Firmicutes and Actinobacteria. The enzymes derived from Firmicutes commonly include candidates from the classes Bacilli (Bacillus aerophilus, B. halodurans, B. mojavensis, B. licheniformis, B. subtilis, Geobacillus thermodenitrificans, and G. thermoleovorans), and Clostridia (Clostridium thermocellum). Candidates from Actinobacteria are mainly derived from the genus Streptomyces (Streptomyces rameus, S. halstedii, S. matensis, S. olivaceovirides) (Table 3). Less commonly, enzymes have been retrieved from bacterial species of other phylogenetic origin, such as the Gram-negative thermophile Rhodothermus marinus, from which a thermostable GH10 xylanase have been cloned, produced and used in AXOS and XOS production from hardwood and cereal xylans [14, 30, 31].
Table 3.
Xylanases used in the production of xylooligosaccharides as potential prebiotics listed together with recent publications using the respective enzyme.
| Source | Enzyme |
Structure
PDB1 |
Xylan source | Main products2 | Temperature (°C) | pH | Time course (h) | References | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH10 bacterial endo-xylanases | ||||||||||||||||||||||
| Bacillus halodurans S7 | XynA | 2UWF | Wheat straw | X2, X3 | 60 | 9 | 24 | [37] | ||||||||||||||
| Bacillus subtilis | Sunzymes, Sunhy Biology Co. Ltd, China | Wheat bran | X2 - X7 | 50 | 5 | 16 | [38] | |||||||||||||||
| Clostridium thermocellum ATCC 27405 | XynZ | Sugarcane bagasse | X2 - X4 | 50 | 6.4 | 5 | [39] | |||||||||||||||
| Geobacillus thermodenitrificans TSAA1 | rXyl-gtd | Wheat, oat spelts, birchwood, beech wood, wheat bran, corn cobs and sugarcaneb agasse | X2 - X4 | 70 | 9 | 2 | [40] | |||||||||||||||
| Geobacillus thermoleovorans | (xyl-gt) | Wood and agro-residue | X2 - X5 | 80 | 8.5 | 12 | [41] | |||||||||||||||
|
Rhodothermus marinus DSM 4252 |
RmXyn10A | Hardwood and cereal xylans | X2 - X5 | 70 | 7.5 | 24 | [14] | |||||||||||||||
| Streptomyces halstedii JM8 | Xys1S | 1NQ6 | Beechwood | X2 | 55 | 5 | 30 | [42] | ||||||||||||||
| Streptomyces olivaceovirides E-86 | Crude FYNX |
1XYF | Birchwood, oat spelt, and beechwood | X2, X3 | 55 | 6.3 and 5.8 | 24 | [43] | ||||||||||||||
| GH11 and GH30 bacterial endo-xylanases | ||||||||||||||||||||||
| Bacillus mojavensis UEB-FK | XynB | Garlic straw | X2, X3 | 60 | 4 | 24 | [44] | |||||||||||||||
| Bacillus licheniformis | BlxA | Birchwood | X2 - X5 | 40 | 6 | 24 | [45] | |||||||||||||||
| Bacillus subtilis 1A1 | rXynA | 1XXN | Birchwood | X2 - X4 | 50 | 5.5 | 24 | [46] | ||||||||||||||
| Bacillus subtilis | XynA | Sugarcane bagasse | X3 - X5 | 50 | 6 | 48 | [47] | |||||||||||||||
| Bacillus subtilis MR44 | Crude Xyn30C, (GH30) Xyn11A (GH11) | 3GTN (Xyn30C) | Sweet sorghum | U-XOS with DP X11-X12 | 37 | 48 | [33] | |||||||||||||||
| Streptomyces matensis DW67 | Purified | Birchwood | X2, X3 | 50 | 7 | 24 | [48] | |||||||||||||||
| Thermobifida fusca NTU22 | Crude product | 3ZSE | Oat-spelt, corncob, bagasse, wheat bran, and peanut shell | X2 | 60 | 7 | 10 | [49] | ||||||||||||||
| Non classified bacterial endo-xylanases | ||||||||||||||||||||||
| Bacillus aerophilus KGJ2 | Extracted | Corncob | X2 - X4 | 70 | 4 | 48 | [50] | |||||||||||||||
| Bacillus mojavensis A21 | Xyn1 Xyn2 |
Corncob | X2, X3 | 50 | 8 | 24 | [51] | |||||||||||||||
| Bacillus subtilis KCX006 | Crude xylanase | Sugarcane bagasse | X2, X3 and substituted XOS | 50 | 5.5 | 30 | [52] | |||||||||||||||
| Streptomyces rameus L2001 | Purified | Birchwood and oat spelt | X2, X3 | 50 | 7 | 2 | [53] | |||||||||||||||
| GH10 fungal xylanases | ||||||||||||||||||||||
| Penicillium funiculosum | XynD | Wheat | X2 | 70 | 5 | [54] | ||||||||||||||||
| Pseudozyma hubeiensis NCIM 3574 | Purified PhX33 (GH103) and PhX20 (GH113) |
Oat spelt | X3 - X7 | 60 | 5.4 | [36] | ||||||||||||||||
| Thermoascus aurantiacus | Purified | 1TUX | Corncob | FeXOS | 50 | 5 | 1 | [55] | ||||||||||||||
| Trichoderma sp. K9301 | Purified EX1 (GH10) and EX2 (GH11) |
Beechwood, birchwood, and oat-spelt | X2 - X5 X2 - X6 |
50 | 24 | [34] | ||||||||||||||||
| GH11 and GH30 fungal xylanases | ||||||||||||||||||||||
| Penicillium occitanis Pol6 | PoXyn3 | Corncobs | X2, X3 | 50 | 3 | 8 | [56] | |||||||||||||||
| Penicillum funiculosum ATCC 62998 | XynC | Sugarcane bagasse | X2 - X6 | 50 | 5 | 2 | [57] | |||||||||||||||
| Sporotrichum thermophile ATCC 34628 | Purified | Birchwood, beechwood, and oat spelt | X2, X3, U4m2XX, XU4m2XX |
50 | 4 and 5.5 | 20 | [58] | |||||||||||||||
| Thermomyces lanuginosus IMI 84400, IMI 96213 and Erwinia chrysanthemi | Secreted XynA (GH11) and XynA (GH30)5 |
1YNA4 (GH11) 1NOF (GH30)5 |
Beechwood glucuronoxylan | Branched oligosaccharides, including XU4m2XX | 30 | 5 | 16 | [59] | ||||||||||||||
| Source | Enzyme |
Structure PDB1 |
Xylan source | Main products2 | Temperature (°C) | pH | Time course (h) | References | ||||||||||||||
| Thermomyces lanuginosus 1YNA | Pentopan Mono BG, Novozymes, Denmark |
Rye bran | X2 - X4 and AXOS | 70 | 7.4 | 15 | [31] | |||||||||||||||
|
Trichoderma reesei RUT-C30 and Thermotoga maritima |
Hybrid Xyn2-A2 (PTXC2) | 1XYO | Oat spelt, birchwood and beechwood | X3 | 50 | 5 | 6 days | [60] | ||||||||||||||
| Non classified fungal endo-xylanases | ||||||||||||||||||||||
| Aspergillus foetidus MTCC 4898 | Purified | Birchwood, wheat straw and rice straw | X2 - X5 | 45 | 5.3 | 2 -24 | [61] | |||||||||||||||
| Aspergillus foetidus MTCC 4898 | Partially purified | Corncob | X2 - X5 | 45 | 5.3 | 2 - 24 | [62] | |||||||||||||||
| Aspergillus niger | AB Enzymes, Germany | Tobacco stalk | X2 - X4 | 40 | 5.5 | 24 | [63] | |||||||||||||||
|
Aspergillus niger and Trichoderma longibrachiatum |
AB Enzymes, Germany and Danisco, Finland |
Tobacco stalk, cotton stalk, sunflower stalk and wheat straw | X2 >X3 >X4 >X5 > X6 | 40 and 50 | 5.5 and 4.6 | 24 | [64] | |||||||||||||||
| Aspergillus oryzae MTCC 5154 | Secreted | Corncob | X2 - X7 | 50 | 5.4 | 42 | [65] | |||||||||||||||
| Aspergillus versicolor | Secreted | Birchwood and oat spelt | X2 - X6 | 60 | 5 | 10 | [66] | |||||||||||||||
| Paecilomyces themophila J18 | Purified | Corncobs | X2, X3 | 70 | 7 | 2.5 | [67] | |||||||||||||||
|
Pleurotus sp. BCCB068 Pleurotus tailandia |
Secreted | Oat spelt | Not identified separately | 50 | 5 | 40 days | [68] | |||||||||||||||
| Pichia stipitis | Secreted | Triploid Populas tomentosa | X2 - X4 | 50 | 5 | 24 | [69] | |||||||||||||||
| Pichia stipitis | Secreted | Sugarcane bagasse | X2 - X4 | 50 | 5.4 | 20 | [70] | |||||||||||||||
| Talaromyces thermophilus Stolk | Purified | Wheat bran | X2, X3 | 50 | 6 | [71] | ||||||||||||||||
| Thermoascus aurantiacus ATCC 204492 | Secreted | Sugarcane bagasse | X2 - X5 | 50 | 7 - 8 | 3 to 36 | [72] | |||||||||||||||
1 If several structures are available, the first deposited structure is given. 2 One-letter code system for oligosaccharides used here is according to the system proposed by Fauré [18]. 3 Families deduced from the partial protein-sequences published in the referenced article. 4 Structure 1YNA belongs to another strain of Thermomyces lanuginosus (SSBP/ATCC 46882/ DSM10635), but is based on sequence data identical to the one used in the referenced article. 5 Formerly classified in family GH5, currently in GH30.
From fungal sources, the enzymes are most commonly originating from different strains of Aspergillus (Aspergillus foetidus, A. niger, A. oryzae, A. versicolor) and produced in the native organism (Table 3). A range of species from other fungal genera has also been used to obtain xylanases, using a combination of recombinant and native organism production systems (Paecilomyces themophila, Penicillium occitanis, P. funiculosum, Pleurotus tailandia, Pichia stipites, Pseudozyma hubeiensis, Sporotrichum thermophile, Talaromyces thermophilus, Thermoascus aurantiacus, Thermomyces lanuginosus, Trichoderma reesei, and T. longibrachiatum) (Table 3).
Recently, a few publications have also indicated use of GH30 candidates for this purpose [32, 33]. These enzyme candidates have thus far originated from bacterial and fungal sources (Table 3).
The most common XOS produced by enzymatic hydrolysis are xylobiose (X2), xylotriose (X3) and xylotetraose (X4) (Table 3, Fig. 2). The pattern of oligosaccharides obtained is to some extent reported to be GH-family dependent. This was for instance shown in a study of Trichoderma sp. xylanases reporting that GH10 xylanases had higher activity on small substrate molecules producing preferably XOS with lower DP (X2-X5), while xylanases from family GH11 were more active on larger xylan substrates and produced XOS with higher DP (X2-X6) [34]. The higher accessibility of GH11 into larger (insoluble) substrate molecules, could to some extent be attributed to differences in size, as the smaller (generally single domain) GH11 enzymes have better accessibility into the complex structures of xylan, than the bigger (in many cases multidomain [35]) GH10 enzymes. However, two xylanases from the yeast Pseudozyma hubeiensis, representing GH10 and GH11, respectively, were reported as non-producers of xylose (X) and X2, and both with only X3 to X7 as main products [36]. This shows that the pattern of XOS is not only family, but also enzyme dependent. Despite hydrolyzing the same chemical bond, the specificity towards heteroxylan differs as a consequence of differences in subsite interactions in the catalytic site (Described in more detail under “Structures of xylanases from GH 10, 11 and 30 (subfamily 8) and their active site interactions”, below). In line with this, the activity of a single enzyme is of course dependent on the heteroxylan structure. For instance, the total yield of XOS (X2 – X5) in enzymatic birchwood xylan hydrolysis was 20% (w/w) compared to 3.3% hydrolysis of rye flour, after 4 hours, using a thermostable endo-xylanase of Rhodothermus marinus [14]. However, in rye flower, only known fragment types were taken into account, leaving out an important fraction of less characterized branched AXOS and XOS (Table 3). In addition to this, processing conditions (e.g. temperature, pH, and reaction time), enzyme stability and cycles of reuse (in the case of immobilized enzymes) all affect the yield of AXOS and XOS.
4. Structures of xylanases from GH 10, 11 and 30 (subfamily 8) and their active site interactions
All products formed by a xylanase from a given substrate are results of the substrate specificity which in turn is determined by the structure of the xylanase. The spatial arrangement of the amino acid residues in the active site determines which substrates can form a complex with the xylanase and subsequently be hydrolyzed. Hydrogen bonds and hydrophobic interactions between the xylanase and the substrate favor the binding, yielding a lower binding energy. Each sugar residue in the active site binds into a subsite which is defined by its amino acid interactions to the sugar. The cleavage point divides the sub-sites into glycone and aglycone subsites where sugar residues in the non-reducing and reducing end of the substrate bind into the glycone and aglycone subsites, respectively. The number of glycone and aglycone subsites, the binding energy of the individual sub-sites and the space for substituents are factors influencing the specificity of the xylanase. For the production of short XOS and AXOS, relatively low binding energy in the proximal sub-sites, and in the latter case, space for arabinose substituents, are required.
The reasons for the obtained distribution of XOS from the trials using the xylanases from families 10, 11 and 30 (subfamily 8), summarized in Table 3, are investigated after a deeper study of the available structural data from the respective family (Fig. 3). As a basis for our discussion, we use the nomenclature recommended for glycoside hydrolase classification as summarized in Schedule 1, and the subsite nomenclature according to Davies et al. [73].
Fig. (3).
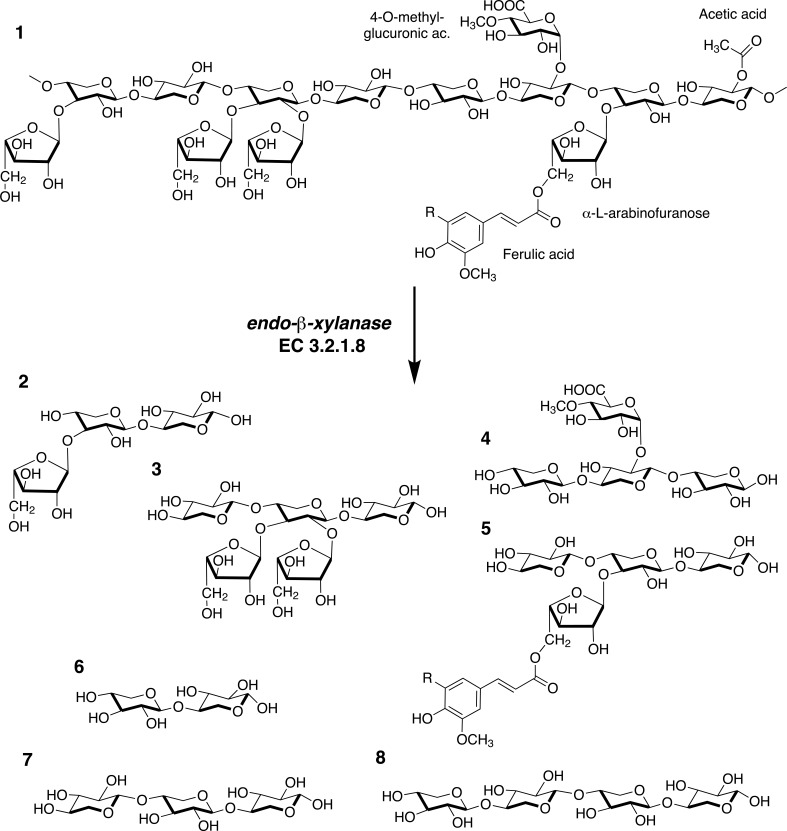
Representative xylanases. To the left, side view and to the right, top view. (A) Xys1Δ from Streptomyces halstedii, PDB 1NQ6 [74], illustrating the typical TIM-barrel of GH10 xylanases. The α-helices and β strands of the (α/β)8 fold are colored in orange and green respectively. (B) XynA from Thermomyces lanuginosus, PDB 1YNA [75], illustrating the typical β-jelly roll fold of GH11 xylanases. The α-helix and β-sheets A and B are colored in purple, yellow and red, respectively. (C) EcXynA from Erwinia chrysanthemi, PDB 1NOF [76], illustrating the typical TIM-barrel with the side β-structure of GH30 subfamily 8 xylanases. The β-strands of the β-structure are colored in blue and the α-helices and β-strands of the (α/β)8 fold are colored in orange and green respectively. (The color version of the figure is available in the electronic copy of the article).
5. GH10 endo-xylanases
5.1. Overall Structure
Members of glycoside hydrolase family 10 belong to the clan A which shares the TIM barrel (α/β)8 fold, consisting of alternating α-helices and β-strands. The eight β-strands form a parallel β-sheet with cylindrical shape in the hydrophobic core of the enzyme while the eight α-helices form the outer surface of the enzyme (Fig. 3A). One face of the enzyme typically has a larger radius than the other, 45 Å compared to 30 Å [77]. The active site is situated at the wider face with the acid/base and nucleophile at the C-terminal end of β-strand 4 and 7, respectively.
5.2. Specificity
GH10 xylanases hydrolyze internal β-1,4-xylosidic linkages in heteroxylan but can tolerate glucose in the active site to some extent. Under low efficiency, even cellooligosaccharides can be hydrolyzed [77]. According to Beaugrand and coworkers, GH10 xylanases have a low capacity to act on insoluble xylan [78], while substitutions to the xylose backbone are not a big hindrance (Table 4) [78, 79]. Arabinose substitutions have been observed in crystal structures in subsites -2, +1, +2 and +3 [80, 81]. However, only substitutions in subsite -2 are shown for several GH10 xylanases. Substitutions of methylglucuronic acid is accepted in subsite -3 and for some GH10 xylanases also in subsite +1 [82]. Tolerance to glucose units as well as to β-1,3 linked carbohydrates in the aglycone sites suggest that the glycone sites are more specific [77]. Soaking of XOS with a DP up to five into the crystal structures of the GH10 xylanase from Penicillium simplicissimum revealed strong binding in the glycone subsites compared to the aglycone sites [83]. X2 and X3 bound into subsite -2 to -1 and -3 to -1 respectively. Longer substrates resulted in cleavage after the third xylose units from the non-reducing end. The leaving groups, X and X2, did not clearly bind into the aglycone subsites. GH10 xylanases show high activity on short XOS indicating a small substrate binding site.
Table 4.
Substituents allowed in the subsites of GH family 10, 11 and 30 subfamily 8. Araf: arabinose substituent, MeGlcA: methylglucuronic acid substituent, P: permitted, B: banned, N: necessary. P/B: not conserved, permitted in some, banned in others.
| GH family | Substituent | Glycone subsites | Aglycone subsites | ||||
|---|---|---|---|---|---|---|---|
| -3 | -2 | -1 | +1 | +2 | +3 | ||
| 10 | Araf | P | B | P/B | P/B | P/B | |
| MeGlcA | P | B | B | P/B | |||
| 11 | Araf | P | P/B | B | B | P | |
| MeGlcA | P/B | P/B | B | B | P | ||
| 30_81 | MeGlcA | N | B | ||||
1Only typical GH30_8 xylanases with glucuronoxylanase activity are considered. These enzymes show no or very low activity on arabinoxylan.
5.3. Active Site
The glycone subsites of the active sites of GH10 xylanases are well conserved (Fig. 4). In subsite -1, four conserved residues are involved in hydrogen bonding with the substrate; the nucleophile and an Asn to O2 and a Lys and a His to O3 [83-85]. Subsite -2 is also well conserved regarding hydrogen bonding; a Trp to O2, an Asn to O3 and a Lys to the endocyclic oxygen. An additional hydrogen bond from Glu is present in many GH10 xylanases. Lack of this hydrogen bond in CjXyn10C from Cellvibrio japonicus reduced the affinity for XOS but not for xylan [84]. Existence of a -3 subsite varies within the family, when it exists, a Tyr performs stacking interaction with the xylose unit which gives rise to a strong binding affinity [84].
Fig. (4).
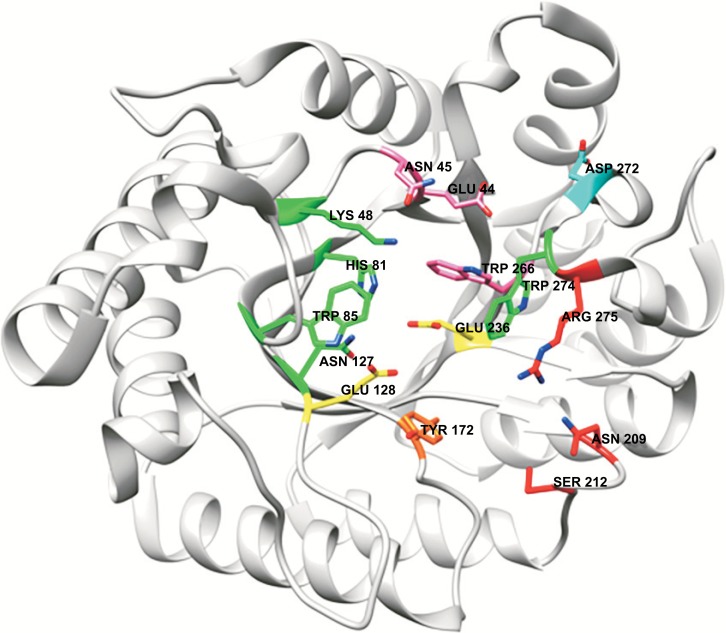
A typical active site of GH10 xylanases, here illustrated by SoXyn10A from Streptomyces olivaceoviridis E-86, PDB entry 1V6X [80]. The amino acids are colored in accordance with subsite-numbering; -2: pink, -1: green, +1: orange and +2: red. The catalytic residues are colored in yellow. Lys48 is also a part in subsite -2 (pink) and Trp266 is also a part of subsite -1 (green). (The color version of the figure is available in the electronic copy of the article).
The amino acids making up the aglycone subsites are less conserved, which can explain differences in hydrolysis products among different members of the family [81, 86]. These subsites are more dominated by aromatic residues favoring hydrophobic interaction rather than hydrogen bonding [87]. Hydrophobic interaction from at least one aromatic residue is conserved in the +1 subsite, but in some cases the xylose unit is sandwiched between two aromatic residues [81, 83, 84, 88, 89]. The O2 and O3 of the xylose unit bound in subsite -1 are completely hidden and substitution in subsite -1 is not possible [90]. The ability to hydrolyze glucose derived substrates was found in Cex from Cellulomonas fimi in subsites -1 and -2 where a Trp and a Gln, respectively, was rearranged to avoid steric hindrance of the extra hydroxylmethyl group of glucose [91]. For Xyn10A from Streptomyces lividans, the same rearrangement is necessary for binding of glucose units but results in a higher energy penalty, which could explain the lower activity on glucose derived substrates for this enzyme [92].
6. GH11 endo-xylanases
6.1. Overall Structure
Glycoside hydrolase family 11 consists so far only of xylanases and the structure is more conserved compared to GH10 [77]. The family is characterized by a small molecular mass, < 30 kDa. GH11 belongs to clan C which has a β-jelly roll fold. The structure contains two antiparallel β-sheets and one α-helix. The β-sheets A and B have the shape of fingers, palm and thumb of a right hand where the fingers and the thumb are slightly bent (Fig. 3B) [93]. The larger and twisted β-sheet B makes up the thumb and the inside of the hand whereas the smaller β-sheet A makes up the outer part of the fingers. The α-helix is situated beneath β-sheet B, close to the thumb.
6.2. Specificity
GH11 enzymes show high activity towards heteroxylan with a backbone of xylose units, while glucose in the backbone is not permitted. Cellulases from GH12 are also members of Clan C but do not comprise the thumb loop. These cellulases generally allow both glucose and xylose in the active site. In GH11, the thumb narrows the binding cleft and the additional hydroxylmethyl group of glucose does not fit. Also, the acid/base catalytic residues are perpendicular in the two families, where the position in GH11 prevents glucose from binding due to steric hindrance and loss of hydrogen bond to O6 of the glucose unit [93] (Paës et al., 2012). GH11 enzymes prefer longer substrates and show no activity on XOS with a DP > 3. The affinity for XOS increases from DP3 up to DP5 [93]. GH11 enzymes do not tolerate any decorations in subsite -1 or +1 due to the narrow binding cleft. However, substitution with arabinofuranosyl residues is in many GH11 xylanases possible in subsite -3, -2, and +2, as well as 4-O-methylglucuronic acid substitution in subsites -3 and +2 (Table 4) [93]. Linkages of the type β-1,3 is a larger obstacle for GH11 xylanases compared to enzymes from GH10 [77, 79]. The restrictions of substitutions generally result in a lower yield of total degradation of substituted xylan compared to GH10 xylanases which tolerate a higher degree of substitutions in the active site [77, 79]. However, probably due to the smaller size of GH11 xylanases, these are more efficient in degrading insoluble xylan compared to GH10 xylanases [78].
6.3. Active Site
The active site of GH11 xylanases is situated in a long, narrow and deep cleft in the palm of the hand. The nucleophile and catalytic acid/base are situated in the middle of the cleft. The cleft can be 30-35 Å long suggesting five or six subsites. Previously, biochemical data have suggested up to seven subsites for some GH11 xylanases due to an increase in activity on XOS with increasing DP up to seven [94, 95]. However, five subsites, -2 to +3, is common for most GH11 xylanases which have been characterized based on structural studies of conserved interactions (Fig. 5) [93]. Subsite -1 provides two hydrogen bonds from Arg and Pro of the thumb loop and subsite -2 provides one hydrogen bond from a Tyr and stacking interaction with a Trp. Further, two Tyr-residues in the +1 and +2 subsites are well conserved and important for both subsite binding and activity [93, 96]. At least three more conserved aromatic residues are involved in substrate binding in subsite +2 and +3 [97]. Mutation of an important stacking interaction in the +3 subsite of BsXynA from Bacillus subtilis decreased the activity of the enzyme suggesting that the +3 subsite is important for activity due to unfavorable binding energies of the closer aglycone subsites. This finding would explain the low activity on XOS with a DP < 3 [97, 98].
Fig. (5).
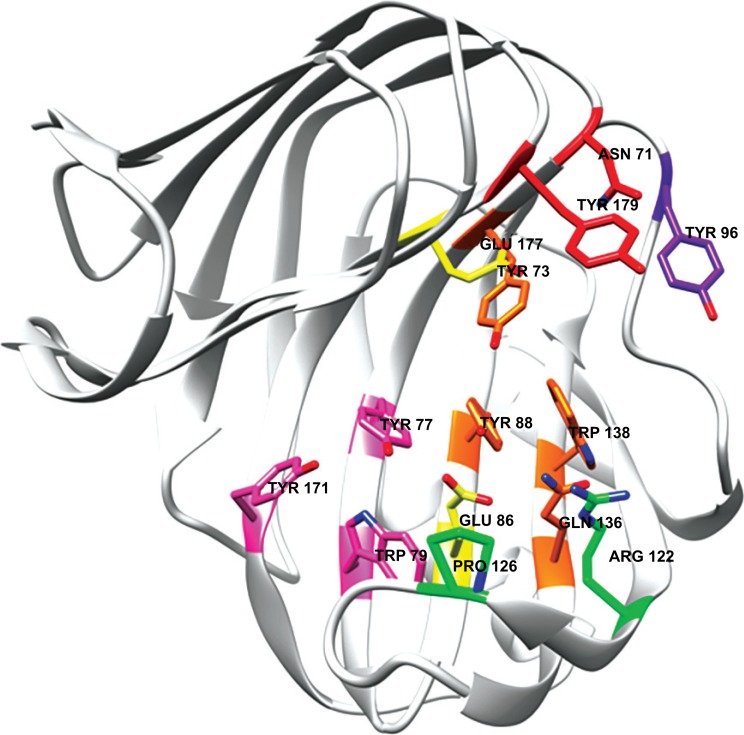
A typical active site of GH11 xylanases, here illustrated by Xyn11A from Trichoderma reesei RUT-C30, PDB 1XYO [98]. The amino acids are colored in accordance with subsite-numbering; -2: pink, -1: green, +1: orange, +2: red, and +3: purple. The catalytic residues are colored in yellow. (The color version of the figure is available in the electronic copy of the article).
6.4. Thumb Loop
The thumb loop is well conserved and the most flexible part of GH11 xylanases, and adopts several conformations [99]. During substrate binding and product release the loop moves up to 3.6 Å shifting between an open and a closed conformation. Modelling of the enzymes reveal observations of more stable conformations with even larger changes, up to 10 Å, during substrate binding, hydrolysis and product release. In most GH11 xylanases, the thumb loop consists of eleven residues, the residues in the tip being Pro, Ser, Ile, any amino acid and Gly. During substrate binding, the Pro and Ser interact with the substrate in the narrowest part of the active site, subsite -2 and -1. It is possible that the loop aids in product release by a catapult mechanism where Ile on the tip of the thumb is positioned behind the product and pushes it away from the enzyme [99].
7. GH30 subfamily 8 endo-xylanases
7.1. Overall Structure
The first xylanases assigned to GH30 were previously members of GH5 but were reclassified as phylogenetic analysis showed that they were more similar to GH30 [100]. GH5 and GH30 are, like GH10, members of Clan A which share the TIM-barrel (α/β8) fold. GH30 enzymes distinguish from GH5 by having the catalytic module fused with a β-structure consisting of a 9-stranded aligned β-sandwich structure (Fig. 3C). Both the N- and C-terminal of the catalytic module is connected with the side β-structure. The hinge-region connecting to the side β-sequence is well conserved in GH30 enzymes. Another well conserved region is a continuous region starting from α-helix 3 and ending at the catalytic acid/base on β-strand 4. These features are not shared with GH5 enzymes which also differ from GH30 by a cap-like structure in the N-terminus at the non-catalytic side of the barrel. GH30 is divided into 8 subfamilies, mainly separated based on the arrangement of the secondary structure forming the side β-structure. Xylanase activity is mainly found in subfamily 8, the only exception being two characterized but not structure determined xylanases in subfamily 7 (www.cazy.org) [101, 102]. Today (March 2016), four characterized GH30 xylanases are structure determined: BsXynC from Bacillus subtilis [103, 104], EcXynA from Erwinia chrysanthemi [76, 105], CpXyn30A from Clostridium papyrosolvens [106] and Xyn30D from Paenibacillus barcinonensis [107] as well as the not characterized Xyn30A from Clostridium thermocellum (PDB structures 4CKQ, 4UQ9 and 4UQA to AUQE).
7.2. Specificity
BsXynC and EcXynA were the two first xylanases to be placed in GH30 and present the common features of GH30_8 xylanases. They differ from xylanases in family 10 and 11 by having a high selectivity for glucuronoxylan and xylooligosaccharides substituted with glucuronic acid (GlcA) or methylglucuronic acid (MeGlcA) via an α-1,2 linkage (Table 4). These enzymes show very low selectivity for unsubstituted xylan, arabinoxylan and XOS. Thus, the two enzymes are assigned to EC 3.2.1.136 (glucuronoarabinoxylan endo-1,4-β-xylanase, with low activity in EC3.2.1.8). Xyn30D from Paenibacillus barcinonensis is only assigned to EC 3.2.1.136 and is exclusively active on glucuronoxylan [107]. Another structure determined GH30_8 enzyme, CpXyn30A from Clostridium papyrosolvens, is however showing opposite selectivity, and is only assigned to EC 3.2.1.8 [106]. This xylanase shows moderate activity on glucuronoxylan while it is active on arabinoxylan and XOS.
7.3. Active Site
BsXynC and EcXynA have similar overall subsite interactions characterized by very strong interactions between the substituent group in subsite -2 and weaker interaction to the xylose residues (Fig. 6). At least five hydrogen bonds and also ionic interactions with the substituent have been observed, whereas only stacking interactions and one or two hydrogen bonds are formed with each xylose residue in subsite -3, -2 and -1. Several of the amino acids interacting with the substituent in subsite -2 are situated on the β8-α8 and β7-α7 loops. Stacking interactions seem to be dominating in the aglycone subsites. The xylan chain is observed to be bent, compared to its regular 3-fold configuration, while bound in the active site. Both the stacking interactions in the aglycone subsites and the very strong interaction to the charged substituent in subsite -2 are responsible for the bending that facilitates hydrolysis. Due to the weak interactions with the xylose units comprising the xylan backbone in the glycone subsites, a linear chain of xylan will not get the correct conformation for hydrolysis. An uncharged substituent, like an arabinofuranosyl residue, will also not generate strong enough interactions for hydrolysis to occur. Thus, these xylanases generally show low activity on arabinoxylan and unsubstituted xylan.
Fig. (6).
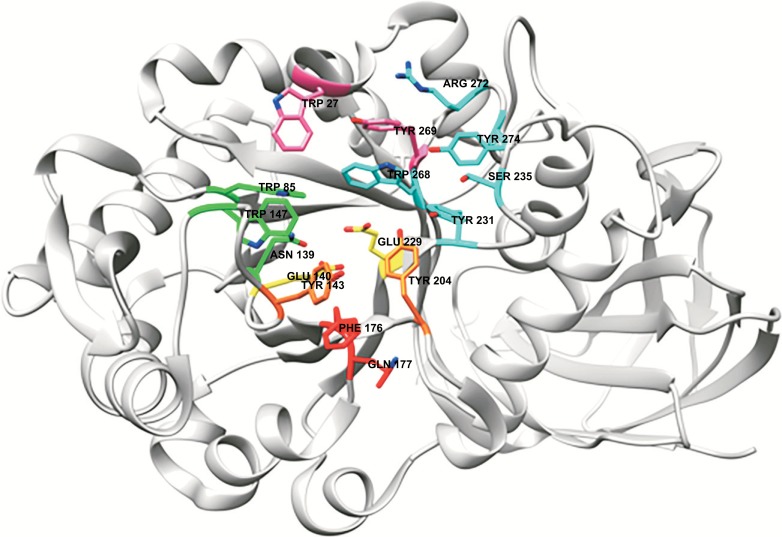
A typical active site of a GH30_8 xylanase with glucuronoxylanase activity, here illustrated by BcXyn30C from Bacillus subtilis, PDB 3KLO [104]. The amino acids are colored in accordance with subsite-numbering; -2: pink, -2 substitution: cyan, -1: green, +1: orange and +2: red. The catalytic residues are colored in yellow. Tyr204 is also a part of subsite +1 (orange), Trp268 is also a part of subsite -1 (green) and Arg272 as well as Trp27 are possibly part of subsite -3. (The color version of the figure is available in the electronic copy of the article).
However, the more recently structure determined CpXyn10A does not share these features and shows several structural differences [106]. The altered specificity of this enzyme is believed to be a result of residue changes in the β8-α8 loop in CpXyn10A (lacking several of the important amino acid interactions described in the other GH30_8 xylanases) and the β7-α7 loop also display differences resulting in loss of interactions to a glucuronic acid substituent.
7.4. β-Structure
The side β-structure of BsXynC binds glucuronic acid substituted xylooligosaccharides in a similar manner as the catalytic site with strong interaction to the substituent, suggesting that this structure has a substrate recognition function similar to CBMs. A binding site in the β-structure of EcXynA has also been proposed. However, the binding site of the β-structure proposed for EcXynA is not conserved within GH30_8 [107]. Interestingly, BsXynC shows the highest overall identity to Xyn30D but when comparing only the β-structure CpXyn30A shows the highest identity to Xyn30D despite the great difference in specificity of the two xylanases [107]. Both CpXynA and Xyn30D are bimodular containing a carbohydrate binding module.
8. Stability adaptations of xylanases used in (A)XOS production
Stability of xylanases is another factor of importance, for the process of converting xylan to (A)XOS. Adaptations to different conditions such as high and low temperature, acidic and alkaline environments are found in many xylanases and are of special interest for industrial applications such as (A)XOS productions.
High temperatures for instance, reduce the viscosity of the reaction-mix facilitating the enzyme-substrate interactions, particularly when the xylan substrates have high molecular weight and low solubility [1]. Therefore, thermostable enzymes are advantageous. Use of thermostable enzymes at suboptimal temperatures also increases the half-life allowing long processing that favor a more complete hydrolysis and higher product yields [14, 37].
Several thermostable GH11 xylanases used in XOS production (Table 3) have been structure determined, e.g. the recombinant rXynA from Bacillus subtilis strain 168 (1A1) [108], TfGH11 from Thermobifida fusca [109] and XynA from Thermomyces lanuginosus [75]. These three enzymes have a higher ratio of hydrophobic residues, especially Tyr and Trp, compared to mesophilic GH11 xylanases. These residues are believed to keep the integrity of the protein by strengthening of the hydrophobic core. The study of rXynA revealed that the fingers and palm of the β-jelly fold are rigid structures and that an increased optimal temperature depends on temperature dependent conformational changes in the thumb region. In TfGH11, non-conserved water mediating hydrogen bonds in the glycone binding site was observed and believed to increase the internal hydrophobicity by fewer solvent-exposed residues. For T. lanuginosus XynA, thermostability was explained by a non-conserved disulphide bridge and an increase in the number of ion-pair interactions compared to mesophilic GH11 xylanases.
In GH10, Xyn10A from Thermoascus aurantiacus is a thermostable xylanase, keeping maximum activity for eight hours at 70 °C [110]. Special features found in the structure of the xylanase, believed to account for the thermostability include: i) efficient packaging of the hydrophobic core, ii) stabilization of helices and iii) absence of long flexible loops [111, 112]. A proline in α-helix 6 was found to introduce an extra hydrogen bond between α-helix 6 and 7 and ten salt bridges were found in the xylanase, one which connected the bottom of β-strand 4 and 7 containing the catalytic residues. Six short helices were also observed which are believed to aid shortening of the connecting loops.
Enzymes working at different pHs can also be advantageous for processing. An example operating in a relatively broad pH spectrum is the GH11 xylanase Xyn11A from Trichoderma reesei RUT-30 which is active from pH 4 to 7.5 [113]. By solving the structure at two different pH, the optimum pH of 5.0 and pH 6.5, two different conformations of the protein were revealed involving several residues in the active site [113, 114]. Especially the catalytic acid/base which shows a large conformational change, and differences in hydrogen bonding which alters its pKa at the two conditions, is believed to increase the activity at higher pH [98].
The GH10 xylanase Xyn10A from Bacillus halodurans is an alkaliphilic xylanase with pH optimum above 9 [115]. It contains a higher percentage of Arg and His and lower percentage of alkali labile Asn residues in its amino acid composition. This type of composition has been pointed out to cause the tolerance to high pH. Other proposed features in the structure promoting high pH tolerance include the formation of a water shield from an excess of negative charges on the surface, hydrophobic and acidic residues in the active site, an acidic surface and a deep active site.
Apart from structural features, enzyme immobilization is another way to increase the stability. For instance, multipoint covalent immobilization of xylanase in glyoxyl-agarose derivative resulted in stable preparations of a xylanase from Aspergillus versicolor. The stabilizing factor was 240 compared to the soluble enzyme at 60 °C, and the XOS yield was 13 and 18% for soluble enzyme and the glyoxyl derivative, respectively [66]. Immobilization of a xylanase from Streptomyces halstedii resulted in an increase of 10 °C of the optimal temperature for xylan hydrolysis [42]. Immobilization can also increase pH stability as for a Penicillum occitanis xylanase immobilized in chitosan with glutaraldehyde by covalent coupling [56]. Another advantage of immobilization is of course the possibility to reuse the immobilized enzymes.
9. Further development of enzymatic processing for production of different XOS
9.1. Combinations of Xylanases with Accessory Enzymes
Since xylans are naturally heterogeneous and contain diverse substituent decorations (Fig. 2), optimal enzymatic hydrolysis may include a combined use of debranching enzymes together with endo-xylanases. The accessory enzymes include α-arabinofuranosidases (EC 3.2.1.55), α-glucuronidases (EC 3.2.1.139) and acetyl xylan esterases (EC 3.1.1.72). The degree of synergy can be determined by dividing the total activity of the combination of enzymes used by the sum of activities of each enzyme separately, where any value higher than one indicates synergy. To date, relatively few studies have been made to evaluate and show synergy between endo-xylanases and accessory enzymes in production of different types of XOS. A few examples are however published, proving possibilities to increase the yield by using combinations of enzymes. One example is the combination of endo-xylanase and arabinofuranosidase. Arabinofuranosidases (Abfs) are a diverse group of enzymes, with α-1,2, α-1,3 and α-1,5 hydrolyzing activities grouped in families GH3, GH43, GH51, GH54 and GH62 (www.cazy.org), and for xylan degradation, α-1,2 and α-1,3 activities are of interest [116]. Synergy in XOS production has for example been reported between a GH11 xylanase (XynC from Penicillium funiculosum) and a GH54 AbfB of Aspergillus niger. In this case, the degree of synergy was higher on insoluble wheat flour arabinoxylan (1.6) than on rye flour arabinoxylan (1.2) [57]. This result shows that combinations with accessory enzymes can be useful together with a GH11 xylanase which preferentially hydrolyzes debranched xylan [82]. Acetyl groups could be another hindrance for hydrolysis of xylans by endo-xylanases and is thought to be an adaptive defense by plants towards endo-xylanases from phytopathogen organisms [102]. The most frequent acetylation positions are 2 and 3, including double acetylation of both position 2 and 3 in the xylopyranosyl residues [117]. Alkaline extraction removes acetyl groups and with this pretreatment, no acetylxylan esterases (AcXEs) are needed for the subsequent action of endo-xylanases. However, hydrothermal pretreatment and dimethylsulfoxide (DMSO) extraction results in partially acetylated xylan, and for such raw materials AcXEs could be useful. XOS from oat-spelt have also been produced in higher yield using an endo-xylanase in presence of a thermostable AcXE from Thermobifida fusca, showing a synergetic action [118].
To the best of our knowledge, no synergetic studies between endo-xylanases and glucuronidases for production of XOS are yet reported. Glucuronidases acting on glucuronoxylan are grouped into GH67 and GH115. There is no evidence that GH67 α-glucuronidases can use high molecular mass glucuronoxylan. They only remove MeGlcA or GlcA from the non-reducing end of fragments of glucuronoxylan. In contrast, some members of the family GH155, such as α-(4-O-methyl)-glucuronidase from Pichia stipis CBS 6054, can hydrolyze MeGlcA side chains from terminal or internal positions in glucuronoxylan [119], and could be interesting to explore for synergistic studies with endo-xylanases from GH10 or GH11.
Studies on synergy between enzymes for XOS production should thus consider several important factors: selection of raw material, type of pretreatment, substrate analysis and the selection of the main and accessory enzymes [120]. In most of the studies on synergy for the production of XOS done to date, two enzymes were used. Due to the complexity of the different xylan sources, a higher number of enzymes and combinations could be investigated with the aim to increase the overall yield of XOS. On the other hand, excessive debranching would give mostly linear XOS, while certain branched xylooligosaccharides, such as AXOS, ferouyl-XOS, and glucuronyl-xylooligosaccharides, can have attractive biological effects. Selected combinations of main and accessory enzymes should hence also focus on the possibilities to produce new xylooligo-derivatives.
9.2. New Enzymes Acting on Heteroxylan Substrates
Development of new XOS mainly depends of the use of a variety of enzyme activities. So far, the use of endo-xylanases for XOS production is concentrated to candidates from families GH10 and GH11, and to much lesser extent enzymes from GH30. However, increased possibilities to obtain XOS could reside in the use of xylanases from additional families where these activities occur (Table 1), including e.g. GH8 and GH5. In GH5 more than one subfamily (yet with few characterized candidates) are reported to act on xylan, including one arabinoxylan-specific endo-β-1,4-xylanase (EC 3.2.1.-) from Ruminiclostridium thermocellum which hydrolyzes arabinoxylans with O3 of arabinose linked to the reducing end xylose, but not unsubstituted xylans [121]. In family GH8 different enzyme candidates are also available, such as the cold-adapted xylanase from the psychrophilic Antarctic bacterium Pseudoalteromonas haloplanktis, which has higher activity at lower temperature than mesophilic counterparts, with increased flexibility, enhancing accommodation and transformation of the substrates at low energy costs [122].
9.3. In Vitro Studies of XOS Utilization and Need of Enzyme Production in GRAS Hosts
One of the main applications of XOS is in functional food manufacture, due their prebiotic effects. XOS are not digestible by human enzymes, but stimulate selectively the growth of beneficial gastrointestinal flora, such as lactic acid bacteria and bifidobacteria. In vitro assays have shown that X2 and X3 promote the growth of Bifidobacterium adolescentis, B. infantis, B. longum, B. bifidum (reviewed by Vázquez et al. [123]) and lactic acid bacteria such as Lactobacillus brevis [37] including new potential probiotic strains of Weissella [124, 125] while short AXOS are more selective for Bifidobacterium strains [14, 31]. Other biological effects tested in vitro are antimicrobial effects of acidic xylooligosaccharides, such as aldotetrauronic and aldopentauronic acids [58], and antioxidant activities of XOS from sugarcane bagasse [70].
Manufacture of XOS for food requires the use of enzymes that either are produced in GRAS (generally recognized as safe) organisms, or purified after production combined with testing of the purified enzyme to assure that levels of e.g. endotoxins are below a certain level. Production in a GRAS organism is generally a preferred alternative, considering both safety and costs. The GRAS organism can be either the native organism or a host for recombinant production. One option for use of native xylanase production is to use enzyme-secreting GRAS organisms, such as different fungal species of Aspergillus (e.g. used in the examples in Table 3). The other option is production of recombinant xylanases in GRAS hosts, such as the yeasts Pichia and Saccharomyces, the fungus Aspergillus or bacterial GRAS hosts, such as Bacillus or Corynebacterium.
Conclusions and future perspectives
Endo-xylanases play a key role in the production of high value XOS from agricultural by-products and other low cost raw material. The structural diversity of XOS produced depends of the type of substrate, pretreatment and structure/function of enzyme or enzymes utilized. Most of the endo-xylanases utilized to date belong to GH10 and GH11, GH10 being more tolerant to utilize decorated xylan as substrate, while GH11 enzymes are more restrictive in binding xylan with side groups in the active site. The smaller size of GH11 enzymes can however facilitate their penetration into insoluble substrates. Stability adaptations of endo-xylanases to e.g. high temperatures are also important for efficient processing, and as stable enzymes are found, and strategies for stabilization are identified, this is expected to facilitate use of enzymatic processing for production of XOS. The increased knowledge on the diversity specificity at a structural level is expected to lead to both improvement of the yields of XOS (by combining enzymes), as well as diversification of the types of XOS produced (using specific enzymes). The expansion of new XOS structure-derivatives (e.g. using new candidate enzymes from GH5, GH8 and GH30) is an attractive field that may lead to production of oligosaccharides with biological activities that potentially benefits human/animal health. Combination of accessory enzymes, such as arabinofuranosidases, glucuronidases and acetyl esterases, with endo-xylanases is expected to lead to increased yields (due to synergism), but may also result in new types of XOS derivatives.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge financial support from the Swedish Research Councils (VR and Formas), the Lund University Anti-Diabetic Food Centre (a VINNOVA VINN Excellence Center), and the EU FP7 program SEABIOTECH.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Linares-Pasten J.A., Andersson M., Nordberg Karlsson E. Thermostable glycoside hydrolases in biorefinery technologies. Curr. Biotechnol. 2014;3:26–44. [Google Scholar]
- 2.Stephen A.M. Other plant polysaccharides. In: Aspinall G.O., editor. The polysaccharides. London: Academic Press Inc.; 1983. [Google Scholar]
- 3.Saha B.C. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 2003;30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 4.Aspinall G.O. Classification of polysaccharides. In: Aspinall G.O., editor. The polysaccharides. London: Academic Press Inc.; 1983. [Google Scholar]
- 5.Konishi T., Nakata I., Miyagi Y., Tako M. Extraction of β-1,3 xylan from green seaweed, Caulerpa lentillifera. J. Appl. Glycosci. 2012;59:161–163. [Google Scholar]
- 6.Viana A.G., Noseda M.D., Gonçalves A.G., Duarte M.E., Yokoya N., Matulewicz M.C., Cerezo A.S. β-d-(1→4), β-d-(1→3) ‘mixed linkage’ xylans from red seaweeds of the order Nemaliales and Palmariales. Carbohydr. Res. 2011;346:1023–1028. doi: 10.1016/j.carres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Rye C.S., Withers S.G. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 2000;4:573–580. doi: 10.1016/s1367-5931(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 8.Iglesias-Fernandez J., Raich L., Ardevol A., Rovira C. The complete conformational free energy landscape of β-xylose reveals a two-fold catalytic itinerary for β-xylanases. Chem. Sci. (Camb.) 2015;6:1167–1177. doi: 10.1039/c4sc02240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deutschmann R., Dekker R.F. From plant biomass to bio-based chemicals: Latest developments in xylan research. Biotechnol. Adv. 2012;30:1627–1640. doi: 10.1016/j.biotechadv.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Juturu V., Wu J.C. Microbial xylanases: engineering, production and industrial applications. Biotechnol. Adv. 2012;30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Broekaert W.F., Courtin C.M., Verbeke K., van De Wiele T., Verstraete W., Delcour J.A. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit. Rev. Food Sci. Nutr. 2011;51:178–194. doi: 10.1080/10408390903044768. [DOI] [PubMed] [Google Scholar]
- 12.Roberfroid M.B., Bornet F., Bouley C., Cummings J.H. Colonic microflora: nutrition and health. Summary and conclusions of an international life sciences institute (ilsi) [europe] workshop held in Barcelona, Spain. Nutr. Rev. 1995;53:127–130. doi: 10.1111/j.1753-4887.1995.tb01535.x. [DOI] [PubMed] [Google Scholar]
- 13.FAO (Food and Agriculture Organisation of the United Nations).Biotechnologies for Agricultural Development.; Proceedings of the FAO International Technical Conference on "Agricultural Biotechnologies in Developing Countries: Options and Opportunities in Crops, Forestry, Livestock, Fisheries and Agroindustry to Face the Challenges of Food Insecurity and Climate Change" (ABCD - 10); 2011. [Google Scholar]
- 14.Falck P., Precha-Atsawanan S., Grey C., Immerzeel P., Stålbrand H., Adlercreutz P., Nordberg Karlsson E. Xylooligosaccharides from hardwood and cereal xylans produced by a thermostable xylanase as carbon sources for Lactobacillus brevis and Bifidobacterium adolescentis. J. Agric. Food Chem. 2013;61:7333–7340. doi: 10.1021/jf401249g. [DOI] [PubMed] [Google Scholar]
- 15.Pastell H., Westermann P., Meyer A.S., Tuomainen P., Tenkanen M. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J. Agric. Food Chem. 2009;57:8598–8606. doi: 10.1021/jf901397b. [DOI] [PubMed] [Google Scholar]
- 16.Lawoko M., Henriksson G., Gellerstedt G. Structural differences between the lignin-carbohydrate complexes present in wood and in chemical pulps. Biomacromol. 2005;6:3467–3473. doi: 10.1021/bm058014q. [DOI] [PubMed] [Google Scholar]
- 17.Puls J., Schuseil J. Chemistry of hemicelluloses: Relationship between hemicellulose structure and enzymes required for hydrolysis. In: Coughlan M., Hazlewoog G., editors. Hemicellulose and Hemicellulases. London, Chapel Hill: Portland Press Ltd.; 1993. [Google Scholar]
- 18.Fauré R., Courtin C.M., Delcour J.A., Dumon C., Faulds C.B., Fincher G.B., Fort S., Fry S.C., Halila S., Kabel M.A., Pouvreau L., Quemener B., Rivet A., Saulnier L., Schols H.A., Driguez H., O’Donohue M.J. A brief and informationally rich naming system for oligosaccharide motifs of heteroxylans found in plant cell walls. Aust. J. Chem. 2009;62:533–537. [Google Scholar]
- 19.Coughlan M.P., Tuohy M.G., Filho E.X., Puls J., Claeyssens M., Vrsanská M., Hughes M.M. Enzymological aspects of microbial hemicellulases with emphasis on fungal systems. In: Coughlan M.P., Hazlewood G.P., editors. Hemicellulose and hemicellulases. London, Chapel Hill: Portland Press Ltd.; 1993. [Google Scholar]
- 20.Mikkonen K.S., Tenkanen M. Sustainable food-packaging materials based on future biorefinery products: Xylans and mannans. Trends Food Sci. Technol. 2012;28:90–102. [Google Scholar]
- 21.Wilkie K.C. The Hemicelluloses of Grasses and Cereals. In: Tipson R.S., Derek H., editors. Adv. Carbohydr. Chem. Biochem. Academic Press; 1979. [Google Scholar]
- 22.Rumpagaporn P., Reuhs B.L., Kaur A., Patterson J.A. Structural features of soluble cereal arabinoxylan fibers associated with a slow rate of in vitro fermentation by human fecal microbiota. Carbohydr. Polym. 2015;130:191–197. doi: 10.1016/j.carbpol.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 23.Puls J. Chemistry and biochemistry of hemicelluloses: Relationship between hemicellulose structure and enzymes required for hydrolysis. Macromol. Symp. 1997;120:183–196. [Google Scholar]
- 24.Wong K.K., Tan L.U., Saddler J.N. Multiplicity of β-1,4-xylanase in microorganisms: Functions and applications. Microbiol. Rev. 1988;52:305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gírio F.M., Fonseca C., Carvalheiro F., Duarte L.C., Marques S., Ukasik B-R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010;101:4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- 26.Painter T.J. Algal polysaccharides. In: Aspinall G.O., editor. The polysaccharides. London: Academic Press Inc.; 1983. [Google Scholar]
- 27.Otieno D.O., Ahring B.K. The potential for oligosaccharide production from the hemicellulose fraction of biomasses through pretreatment processes: Xylooligosaccharides (XOS), arabinooligosaccharides (AOS), and mannooligosaccharides (MOS). Carbohydr. Res. 2012;360:84–92. doi: 10.1016/j.carres.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Chanliaud E., Saulnier L., Thibault J.F. Alkaline extraction and characterisation of heteroxylans from maize bran. J. Cereal Sci. 1995;21:195–203. [Google Scholar]
- 29.Zhu Y., Kim T.H., Lee Y.Y., Chen R., Elander R.T. Enzymatic Production of Xylooligosaccharides From Corn Stover and Corn Cobs Treated With Aqueous Ammonia. In: Mcmillan J.D., Adney W.S., Mielenz J.R., Klasson K.T., editors. Twenty-Seventh Symposium on Biotechnology for Fuels and Chemicals. Totowa, NJ: Humana Press; 2006. [DOI] [PubMed] [Google Scholar]
- 30.Immerzeel P., Falck P., Galbe M., Adlercreutz P., Nordberg Karlsson E., Stålbrand H. Extraction of water-soluble xylan from wheat bran and utilization of enzymatically produced xylooligosaccharides by Lactobacillus, Bifidobacterium and Weissella spp. Lebensm. Wiss. Technol. 2014;56:321–327. [Google Scholar]
- 31.Falck P., Aronsson A., Grey C., Stålbrand H., Nordberg Karlsson E., Adlercreutz P. Production of arabinoxylan-oligosaccharide mixtures of varying composition from rye bran by a combination of process conditions and type of xylanase. Bioresour. Technol. 2014;174:118–125. doi: 10.1016/j.biortech.2014.09.139. [DOI] [PubMed] [Google Scholar]
- 32.Padilha I.Q., Valenzuela Mayorga S.V., Grisi T.C., Díaz Lucea P., De Araujo D.A., Pastor Blasco F.I. A glucuronoxylan-specific xylanase from a new Paenibacillus favisporus strain isolated from tropical soil of Brazil. Int. Microbiol. 2014;17:175–184. doi: 10.2436/20.1501.01.220. [DOI] [PubMed] [Google Scholar]
- 33.Wei L., Rhee M.S., Preston J.F. Production of acidic xylooligosaccharides from methylglucuronoarabinoxylans by Bacillus subtilis strain MR44. J. Chem. Technol. Biotechnol. 2016;91(7):2056–2062. [Google Scholar]
- 34.Chen L-L., Zhang M., Zhang D-H., Chen X-L., Sun C-Y., Zhou B-C., Zhang Y-Z. Purification and enzymatic characterization of two β-endoxylanases from Trichoderma sp. K9301 and their actions in xylooligosaccharide production. Bioresour. Technol. 2009;100:5230–5236. doi: 10.1016/j.biortech.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 35.Abou-Hachem M., Olsson F., Nordberg Karlsson E. Probing the stability of the modular family 10 xylanase from Rhodothermus marinus. Extremophiles. 2003;7:483–491. doi: 10.1007/s00792-003-0348-1. [DOI] [PubMed] [Google Scholar]
- 36.Adsul M.G., Bastawde K.B., Gokhale D.V. Biochemical characterization of two xylanases from yeast Pseudozyma hubeiensis producing only xylooligosaccharides. Bioresour. Technol. 2009;100:6488–6495. doi: 10.1016/j.biortech.2009.07.064. [DOI] [PubMed] [Google Scholar]
- 37.Faryar R., Linares-Pastén J.A., Immerzeel P., Mamo G., Andersson M., Stålbrand H., Mattiasson B., Nordberg Karlsson E. Production of prebiotic xylooligosaccharides from alkaline extracted wheat straw using the K80R-variant of a thermostable alkali-tolerant xylanase. Food Bioprod. Process. 2015;93:1–10. [Google Scholar]
- 38.Wang J., Sun B., Cao Y., Tian Y., Wang C. Enzymatic preparation of wheat bran xylooligosaccharides and their stability during pasteurization and autoclave sterilization at low pH. Carbohydr. Polym. 2009;77:816–821. [Google Scholar]
- 39.Mandelli F., Brenelli L.B., Almeida R.F., Goldbeck R., Wolf L.D., Hoffmam Z.B., Ruller R., Rocha G.J., Mercadante A.Z., Squina F.M. Simultaneous production of xylooligosaccharides and antioxidant compounds from sugarcane bagasse via enzymatic hydrolysis. Ind. Crops Prod. 2014;52:770–775. [Google Scholar]
- 40.Verma D., Anand A., Satyanarayana T. Thermostable and alkalistable endoxylanase of the extremely thermophilic bacterium Geobacillus thermodenitrificans TSAA1: Cloning, expression, characteristics and its applicability in generating xylooligosaccharides and fermentable sugars. Appl. Biochem. Biotechnol. 2013;170:119–130. doi: 10.1007/s12010-013-0174-6. [DOI] [PubMed] [Google Scholar]
- 41.Verma D., Satyanarayana T. Cloning, expression and applicability of thermo-alkali-stable xylanase of Geobacillus thermoleovorans in generating xylooligosaccharides from agro-residues. Bioresour. Technol. 2012;107:333–338. doi: 10.1016/j.biortech.2011.12.055. [DOI] [PubMed] [Google Scholar]
- 42.Aragon C.C., Mateo C., Ruiz-Matute A.I., Corzo N., Fernandez-Lorente G., Sevillano L., Díaz M., Monti R., Santamaría R.I., Guisan J.M. Production of xylo-oligosaccharides by immobilized-stabilized derivatives of endo-xylanase from Streptomyces halstedii. Process Biochem. 2013;48:478–483. a. [Google Scholar]
- 43.Ai Z., Jiang Z., Li L., Deng W., Kusakabe I., Li H. Immobilization of Streptomyces olivaceoviridis E-86 xylanase on Eudragit S-100 for xylo-oligosaccharide production. Process Biochem. 2005;40:2707–2714. [Google Scholar]
- 44.Kallel F., Driss D., Bouaziz F., Neifer M., Ghorbel R., Ellouz Chaabouni S. Production of xylooligosaccharides from garlic straw xylan by purified xylanase from Bacillus mojavensis UEB-FK and their in vitro evaluation as prebiotics. Food Bioprod. Process. 2015;94:536–546. [Google Scholar]
- 45.Liu M-Q., Liu G-F. Expression of recombinant Bacillus licheniformis xylanase A in Pichia pastoris and xylooligo-saccharides released from xylans by it. Protein Expr. Purif. 2008;57:101–107. doi: 10.1016/j.pep.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 46.Milessi T.S., Kopp W., Rojas M.J., Manrich A., Baptista-Neto A., Tardioli P.W., Giordano R.C., Fernandez-Lafuente R., Guisan J.M., Giordano R.L. Immobilization and stabilization of an endoxylanase from Bacillus subtilis (XynA) for xylooligosaccharides (XOs) production. Catal. Today. 2015;259:130–139. [Google Scholar]
- 47.Bragatto J., Segato F., Squina F.M. Production of xylooligo-saccharides (XOS) from delignified sugarcane bagasse by peroxide-HAc process using recombinant xylanase from Bacillus subtilis. Ind. Crops Prod. 2013;51:123–129. [Google Scholar]
- 48.Yan Q., Hao S., Jiang Z., Zhai Q., Chen W. Properties of a xylanase from Streptomyces matensis being suitable for xylooligosaccharides production. J. Mol. Catal., B Enzym. 2009;58:72–77. [Google Scholar]
- 49.Yang C-H., Yang S-F., Liu W-H. Production of xylooligosaccharides from xylans by extracellular xylanases from Thermobifida fusca. J. Agric. Food Chem. 2007;55:3955–3959. doi: 10.1021/jf0635964. [DOI] [PubMed] [Google Scholar]
- 50.Gowdhaman D., Ponnusami V. Production and optimization of xylooligosaccharides from corncob by Bacillus aerophilus KGJ2 xylanase and its antioxidant potential. Int. J. Biol. Macromol. 2015;79:595–600. doi: 10.1016/j.ijbiomac.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 51.Haddar A., Driss D., Frikha F., Ellouz-Chaabouni S., Nasri M. Alkaline xylanases from Bacillus mojavensis A21: Production and generation of xylooligosaccharides. Int. J. Biol. Macromol. 2012;51:647–656. doi: 10.1016/j.ijbiomac.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 52.Reddy S.S., Krishnan C. Production of high-pure xylooligo-saccharides from sugarcane bagasse using crude β-xylosidase-free xylanase of Bacillus subtilis KCX006 and their bifidogenic function. Lebensm. Wiss. Technol. 2016;65:237–245. [Google Scholar]
- 53.Li X., Li E., Zhu Y., Teng C., Sun B., Song H., Yang R. A typical endo-xylanase from Streptomyces rameus L2001 and its unique characteristics in xylooligosaccharide production. Carbohydr. Res. 2012;359:30–36. doi: 10.1016/j.carres.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Lafond M., Tauzin A., Desseaux V., Bonnin E., Ajandouz E-H., Giardina T. GH10 xylanase D from Penicillium funiculosum: biochemical studies and xylooligosaccharide production. Microb. Cell Fact. 2011;10:20. doi: 10.1186/1475-2859-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katapodis P., Christakopoulos P. Enzymic production of feruloyl xylo-oligosaccharides from corn cobs by a family 10 xylanase from Thermoascus aurantiacus. Lebensm. Wiss. Technol. 2008;41:1239–1243. [Google Scholar]
- 56.Driss D., Zouari-Ellouzi S., Chaari F., Kallel F., Ghazala I., Bouaziz F., Ghorbel R., Chaabouni S.E. Production and in vitro evaluation of xylooligosaccharides generated from corncobs using immobilized Penicillium occitanis xylanase. J. Mol. Catal., B Enzym. 2014;102:146–153. [Google Scholar]
- 57.Gonçalves T.A., Damásio A.R., Segato F., Alvarez T.M., Bragatto J., Brenelli L.B., Citadini A.P., Murakami M.T., Ruller R., Paes Leme A.F., Prade R.A., Squina F.M. Functional characterization and synergic action of fungal xylanase and arabinofuranosidase for production of xylooligosaccharides. Bioresour. Technol. 2012;119:293–299. doi: 10.1016/j.biortech.2012.05.062. [DOI] [PubMed] [Google Scholar]
- 58.Christakopoulos P., Katapodis P., Kalogeris E., Kekos D., Macris B.J., Stamatis H., Skaltsa H. Antimicrobial activity of acidic xylo-oligosaccharides produced by family 10 and 11 endoxylanases. Int. J. Biol. Macromol. 2003;31:171–175. doi: 10.1016/s0141-8130(02)00079-x. [DOI] [PubMed] [Google Scholar]
- 59.Puchart V., Biely P. Simultaneous production of endo-β-1,4-xylanase and branched xylooligosaccharides by Thermomyces lanuginosus. J. Biotechnol. 2008;137:34–43. doi: 10.1016/j.jbiotec.2008.07.1789. [DOI] [PubMed] [Google Scholar]
- 60.Jun H., Bing Y., Keying Z., Daiwen C. Functional characteri-zation of a recombinant thermostable xylanase from Pichia pastoris: A hybrid enzyme being suitable for xylooligosaccharides production. Biochem. Eng. J. 2009;48:87–92. [Google Scholar]
- 61.Chapla D., Dholakiya S., Madamwar D., Shah A. Characterization of purified fungal endoxylanase and its application for production of value added food ingredient from agroresidues. Food Bioprod. Process. 2013;91:682–692. [Google Scholar]
- 62.Chapla D., Pandit P., Shah A. Production of xylooligosaccharides from corncob xylan by fungal xylanase and their utilization by probiotics. Bioresour. Technol. 2012;115:215–221. doi: 10.1016/j.biortech.2011.10.083. [DOI] [PubMed] [Google Scholar]
- 63.Akpinar O., Erdogan K., Bakir U., Yilmaz L. Comparison of acid and enzymatic hydrolysis of tobacco stalk xylan for preparation of xylooligosaccharides. Lebensm. Wiss. Technol. 2010;43:119–125. [Google Scholar]
- 64.Akpinar O., Erdogan K., Bostanci S. Enzymatic production of Xylooligosaccharide from selected agricultural wastes. Food Bioprod. Process. 2009;87:145–151. [Google Scholar]
- 65.Aachary A.A., Prapulla S.G. Value addition to corncob: Production and characterization of xylooligosaccharides from alkali pretreated lignin-saccharide complex using Aspergillus oryzae MTCC 5154. Bioresour. Technol. 2009;100:991–995. doi: 10.1016/j.biortech.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 66.Aragon C.C., Santos A.F., Ruiz-Matute A.I., Corzo N., Guisan J.M., Monti R., Mateo C. Continuous production of xylooligosaccharides in a packed bed reactor with immobilized–stabilized biocatalysts of xylanase from Aspergillus versicolor. J. Mol. Catal., B Enzym. 2013;98:8–14. b. [Google Scholar]
- 67.Teng C., Yan Q., Jiang Z., Fan G., Shi B. Production of xylooligosaccharides from the steam explosion liquor of corncobs coupled with enzymatic hydrolysis using a thermostable xylanase. Bioresour. Technol. 2010;101:7679–7682. doi: 10.1016/j.biortech.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 68.De Menezes C.R., Silva Í.S., Pavarina É.C., Guímaro Dias E.F., Guímaro Dias F., Grossman M.J., Durrant L.R. Production of xylooligosaccharides from enzymatic hydrolysis of xylan by the white-rot fungi Pleurotus. Int. Biodet. Biodeg. 2009;63:673–678. [Google Scholar]
- 69.Yang H., Wang K., Song X., Xu F. Production of xylooligo-saccharides by xylanase from Pichia stipitis based on xylan preparation from triploid Populas tomentosa. Bioresour. Technol. 2011;102:7171–7176. doi: 10.1016/j.biortech.2011.03.110. [DOI] [PubMed] [Google Scholar]
- 70.Bian J., Peng F., Peng X-P., Peng P., Xu F., Sun R-C. Structural features and antioxidant activity of xylooligosaccharides enzymatically produced from sugarcane bagasse. Bioresour. Technol. 2013;127:236–241. doi: 10.1016/j.biortech.2012.09.112. [DOI] [PubMed] [Google Scholar]
- 71.Maalej-Achouri I., Guerfali M., Gargouri A., Belghith H. Production of xylo-oligosaccharides from agro-industrial residues using immobilized Talaromyces thermophilus xylanase. J. Mol. Catal., B Enzym. 2009;59:145–152. [Google Scholar]
- 72.Brienzo M., Carvalho W., Milagres A.M. Xylooligosaccharides production from alkali-pretreated sugarcane bagasse using xylanases from Thermoascus aurantiacus. Appl. Biochem. Biotechnol. 2010;162:1195–1205. doi: 10.1007/s12010-009-8892-5. [DOI] [PubMed] [Google Scholar]
- 73.Davies G.J., Wilson K.S., Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Canals A., Vega M.C., Gomis-Ruth F.X., Diaz M., Santamaria R.I., Coll M. Structure of xylanase Xys1Δ from Streptomyces halstedii. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1447–1453. doi: 10.1107/s0907444903012629. [DOI] [PubMed] [Google Scholar]
- 75.Gruber K., Klintschar G., Hayn M., Schlacher A., Steiner W., Kratky C. Thermophilic xylanase from Thermomyces lanuginosus: High-Resolution X-ray structure and modeling studies. Biochemistry. 1998;37:13475–13485. doi: 10.1021/bi980864l. [DOI] [PubMed] [Google Scholar]
- 76.Larson S.B., Day J., Barba De La Rosa A.P., Keen N.T., Mcpherson A. First crystallographic structure of a xylanase from glycoside hydrolase family 5: Implications for catalysis. Biochemistry. 2003;42:8411–8422. doi: 10.1021/bi034144c. [DOI] [PubMed] [Google Scholar]
- 77.Collins T., Gerday C., Feller G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005;29:3–23. doi: 10.1016/j.femsre.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 78.Beaugrand J., Chambat G., Wong V.W., Goubet F., Rémond C., Paës G., Benamrouche S., Debeire P., O’Donohue M., Chabbert B. Impact and efficiency of GH10 and GH11 thermostable endoxylanases on wheat bran and alkali-extractable arabinoxylans. Carbohydr. Res. 2004;339:2529–2540. doi: 10.1016/j.carres.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 79.Biely P., Vršanská M., Tenkanen M., Kluepfel D. Endo-β-1,4-xylanase families: differences in catalytic properties. J. Biotechnol. 1997;57:151–166. doi: 10.1016/s0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- 80.Fujimoto Z., Kaneko S., Kuno A., Kobayashi H., Kusakabe I., Mizuno H. Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86. J. Biol. Chem. 2004;279:9606–9614. doi: 10.1074/jbc.M312293200. [DOI] [PubMed] [Google Scholar]
- 81.Pell G., Taylor E.J., Gloster T.M., Turkenburg J.P., Fontes C.M., Ferreira L.M., Nagy T., Clark S.J., Davies G.J., Gilbert H.J. The mechanisms by which family 10 glycoside hydrolases bind decorated substrates. J. Biol. Chem. 2004;279:9597–9605. doi: 10.1074/jbc.M312278200. b. [DOI] [PubMed] [Google Scholar]
- 82.Pollet A., Delcour J.A., Courtin C.M. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit. Rev. Biotechnol. 2010;30:176–191. doi: 10.3109/07388551003645599. a. [DOI] [PubMed] [Google Scholar]
- 83.Schmidt A., Gübitz G.M., Kratky C. Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant. Biochemistry. 1999;38:2403–2412. doi: 10.1021/bi982108l. [DOI] [PubMed] [Google Scholar]
- 84.Pell G., Szabo L., Charnock S.J., Xie H., Gloster T.M., Davies G.J., Gilbert H.J. Structural and biochemical analysis of Cellvibrio japonicus xylanase 10C: How variation in substrate-binding cleft influences the catalytic profile of family GH-10 xylanases. J. Biol. Chem. 2004;279:11777–11788. doi: 10.1074/jbc.M311947200. a. [DOI] [PubMed] [Google Scholar]
- 85.White A., Tull D., Johns K., Withers S.G., Rose D.R. Crystallographic observation of a covalent catalytic intermediate in a β-glycosidase. Nat. Struct. Mol. Biol. 1996;3:149–154. doi: 10.1038/nsb0296-149. [DOI] [PubMed] [Google Scholar]
- 86.Harris G.W., Jenkins J.A., Connerton I., Cummings N., Lo Leggio L., Scott M., Hazlewood G.P., Laurie J.I., Gilbert H.J., Pickersgill R.W. Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites. Structure. 1994;2:1107–1116. doi: 10.1016/s0969-2126(94)00112-x. [DOI] [PubMed] [Google Scholar]
- 87.Zolotnitsky G., Cogan U., Adir N., Solomon V., Shoham G., Shoham Y. Mapping glycoside hydrolase substrate subsites by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA. 2004;101:11275–11280. doi: 10.1073/pnas.0404311101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lo Leggio L., Jenkins J., Harris G.W., Pickersgill R.W. X-ray crystallographic study of xylopentaose binding to Pseudomonas fluorescens xylanase A. Proteins. 2000;41:362–373. doi: 10.1002/1097-0134(20001115)41:3<362::aid-prot80>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 89.Lo Leggio L., Kalogiannis S., Eckert K., Teixeira S.C., Bhat M.K., Andrei C., Pickersgill R.W., Larsen S. Substrate specificity and subsite mobility in T. aurantiacus xylanase 10A. FEBS Lett. 2001;509:303–308. doi: 10.1016/s0014-5793(01)03177-5. [DOI] [PubMed] [Google Scholar]
- 90.Fujimoto Z., Kuno A., Kaneko S., Yoshida S., Kobayashi H., Kusakabe I., Mizuno H. Crystal structure of Streptomyces olivaceoviridis E-86 β-xylanase containing xylan-binding domain. J. Mol. Biol. 2000;300:575–585. doi: 10.1006/jmbi.2000.3877. [DOI] [PubMed] [Google Scholar]
- 91.Notenboom V., Birsan C., Warren R.A., Withers S.G., Rose D.R. Exploring the cellulose/xylan specificity of the β-1,4-glycanase cex from cellulomonas fimi through crystallography and mutation. Biochemistry. 1998;37:4751–4758. doi: 10.1021/bi9729211. [DOI] [PubMed] [Google Scholar]
- 92.Ducros V., Charnock S.J., Derewenda U., Derewenda Z.S., Dauter Z., Dupont C., Shareck F., Morosoli R., Kluepfel D., Davies G.J. Substrate specificity in glycoside hydrolase family 10: Structural and kinetic analysis of the Streptomyces lividans xylanase 10A. J. Biol. Chem. 2000;275:23020–23026. doi: 10.1074/jbc.275.30.23020. [DOI] [PubMed] [Google Scholar]
- 93.Paës G., Berrin J-G., Beaugrand J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv. 2012;30:564–592. doi: 10.1016/j.biotechadv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 94.Bray M.R., Clarke A.J. Action pattern of xylo-oligosaccharide hydrolysis by Schizophyllum commune xylanase A. Eur. J. Biochem. 1992;204:191–196. doi: 10.1111/j.1432-1033.1992.tb16623.x. [DOI] [PubMed] [Google Scholar]
- 95.Vršanská M., Gorbacheva I.V., Krátký Z., Biely P. Reaction pathways of substrate degradation by an acidic endo-1,4-β-xylanase of Aspergillus niger. Biochim. Biophys. Acta. 1982;704:114–122. doi: 10.1016/0167-4838(82)90138-8. [DOI] [PubMed] [Google Scholar]
- 96.Wan Q., Zhang Q., Hamilton-Brehm S., Weiss K., Mustya-kimov M., Coates L., Langan P., Graham D., Kovalevsky A. X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism. Acta Crystallogr. D Biol. Crystallogr. 2014;70:11–23. doi: 10.1107/S1399004713023626. [DOI] [PubMed] [Google Scholar]
- 97.Pollet A., Lagaert S., Eneyskaya E., Kulminskaya A., Delcour J.A., Courtin C.M. Mutagenesis and subsite mapping underpin the importance for substrate specificity of the aglycon subsites of glycoside hydrolase family 11 xylanases. Biochim. Biophys. Acta. 1804;2010b:977–985. doi: 10.1016/j.bbapap.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 98.Törrönen A., Rouvinen J. Structural Comparison of two major endo-1,4-xylanases from Trichoderma reesei. Biochemistry. 1995;34:847–856. doi: 10.1021/bi00003a019. [DOI] [PubMed] [Google Scholar]
- 99.Pollet A., Vandermarliere E., Lammertyn J., Strelkov S.V., Delcour J.A., Courtin C.M. Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases. Proteins. 2009;77:395–403. doi: 10.1002/prot.22445. [DOI] [PubMed] [Google Scholar]
- 100.St John F.J., González J.M., Pozharski E. Consolidation of glycosyl hydrolase family 30: A dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett. 2010;584:4435–4441. doi: 10.1016/j.febslet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 101.Luo H., Yang J., Li J., Shi P., Huang H., Bai Y., Fan Y., Yao B. Molecular cloning and characterization of the novel acidic xylanase XYLD from Bispora sp. MEY-1 that is homologous to family 30 glycosyl hydrolases. Appl. Microbiol. Biotechnol. 2010;86:1829–1839. doi: 10.1007/s00253-009-2410-0. [DOI] [PubMed] [Google Scholar]
- 102.Tenkanen M., Vršanská M., Siika-Aho M., Wong D.W., Puchart V., Penttilä M., Saloheimo M., Biely P. Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity. FEBS J. 2013;280:285–301. doi: 10.1111/febs.12069. [DOI] [PubMed] [Google Scholar]
- 103.St John F.J., Godwin D.K., Preston J.F., Pozharski E., Hurlbert J.C. Crystallization and crystallographic analysis of Bacillus subtilis xylanase C. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009;65:499–503. doi: 10.1107/S1744309109013098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.St John F.J., Hurlbert J.C., Rice J.D., Preston J.F., Pozharski E. Ligand Bound Structures of a Glycosyl Hydrolase Family 30 Glucuronoxylan Xylanohydrolase. J. Mol. Biol. 2011;407:92–109. doi: 10.1016/j.jmb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 105.Urbániková L., Vršanská M., Mørkeberg Krogh K.B., Hoff T., Biely P. Structural basis for substrate recognition by Erwinia chrysanthemi GH30 glucuronoxylanase. FEBS J. 2011;278:2105–2116. [Google Scholar]
- 106.St John F.J., Dietrich D., Crooks C., Pozharski E., Gonzalez J.M., Bales E., Smith K., Hurlbert J.C. A novel member of glycoside hydrolase family 30 subfamily 8 with altered substrate specificity. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2950–2958. doi: 10.1107/S1399004714019531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sainz-Polo M.A., Valenzuela S.V., González B., Pastor F.I., Sanz-Aparicio J. Structural Analysis of glucuronoxylan-specific Xyn30D and its attached CBM35 domain gives insights into the role of modularity in specificity. J. Biol. Chem. 2014;289:31088–31101. doi: 10.1074/jbc.M114.597732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murakami M.T., Arni R.K., Vieira D.S., Degrève L., Ruller R., Ward R.J. Correlation of temperature induced conformation change with optimum catalytic activity in the recombinant G/11 xylanase A from Bacillus subtilis strain 168 (1A1). FEBS Lett. 2005;579:6505–6510. doi: 10.1016/j.febslet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 109.Lammerts Van Bueren A., Otani S., Friis E.P., Wilson K.S., Davies G.J. Three-dimensional structure of a thermophilic family GH11 xylanase from Thermobifida fusca. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:141–144. doi: 10.1107/S1744309111049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khandke K.M., Vithayathil P.J., Murthy S.K. Purification of xylanase, β-glucosidase, endocellulase, and exocellulase from a thermophilic fungus, Thermoascus aurantiacus. Arch. Biochem. Biophys. 1989;274:491–500. doi: 10.1016/0003-9861(89)90462-1. [DOI] [PubMed] [Google Scholar]
- 111.Lo Leggio L., Kalogiannis S., Bhat M.K., Pickersgill R.W. High resolution structure and sequence of T. aurantiacus Xylanase I: Implications for the evolution of thermostability in family 10 xylanases and enzymes with βα-barrel architecture. Proteins. 1999;36:295–306. [PubMed] [Google Scholar]
- 112.Natesh R., Bhanumoorthy P., Vithayathil P.J., Sekar K., Ramakumar S., Viswamitra M.A. Crystal structure at 1.8 Å resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus. J. Mol. Biol. 1999;288:999–1012. doi: 10.1006/jmbi.1999.2727. [DOI] [PubMed] [Google Scholar]
- 113.Tenkanen M., Puls J., Poutanen K. Two major xylanases of Trichoderma reesei. Enzyme Microb. Technol. 1992;14:566–574. [Google Scholar]
- 114.Törrönen A., Harkki A., Rouvinen J. Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO J. 1994;13:2493–2501. doi: 10.1002/j.1460-2075.1994.tb06536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mamo G., Thunnissen M., Hatti-Kaul R., Mattiasson B. An alkaline active xylanase: Insights into mechanisms of high pH catalytic adaptation. Biochimie. 2009;91:1187–1196. doi: 10.1016/j.biochi.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 116.Lagaert S., Pollet A., Courtin C.M., Volckaert G. β-Xylosidases and α-L-arabinofuranosidases: accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014;32:316–332. doi: 10.1016/j.biotechadv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 117.Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 2012;30:1575–1588. doi: 10.1016/j.biotechadv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 118.Huang Y-C., Chen G-H., Chen Y-F., Chen W-L., Yang C-H. Heterologous expression of thermostable acetylxylan esterase gene from Thermobifida fusca and its synergistic action with xylanase for the production of xylooligosaccharides. Biochem. Biophys. Res. Commun. 2010;400:718–723. doi: 10.1016/j.bbrc.2010.08.136. [DOI] [PubMed] [Google Scholar]
- 119.Ryabova O., Vršanská M., Kaneko S., Van Zyl W.H., Biely P. A novel family of hemicellulolytic α-glucuronidase. FEBS Lett. 2009;583:1457–1462. doi: 10.1016/j.febslet.2009.03.057. [DOI] [PubMed] [Google Scholar]
- 120.Van Dyk J.S., Pletschke B.I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes - Factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 2012;30:1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 121.Correia M.A., Mazumder K., Bras J.L., Firbank S.J., Zhu Y., Lewis R.J., York W.S., Fontes C.M., Gilbert H.J. Structure and function of an arabinoxylan-specific xylanase. J. Biol. Chem. 2011;286:22510–22520. doi: 10.1074/jbc.M110.217315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Petegem F., Collins T., Meuwis M.A., Gerday C., Feller G., Van Beeumen J. The structure of a cold-adapted family 8 xylanase at 1.3 Å resolution. Structural adaptations to cold and investgation of the active site. J. Biol. Chem. 2003;278:7531–7539. doi: 10.1074/jbc.M206862200. [DOI] [PubMed] [Google Scholar]
- 123.Vázquez M.J., Alonso J.L., Domínguez H., Parajó J.C. Xylooligosaccharides: manufacture and applications. Trends Food Sci. Technol. 2000;11:387–393. [Google Scholar]
- 124.Falck P., Linares-Pastén J.A., Adlercreutz P., Nordberg Karlsson E. Characterization of a family 43 β-xylosidase from the xylooligosaccharide utilizing putative probiotic Weissella sp. strain 92. Glycobiology. 2016;26:193–202. doi: 10.1093/glycob/cwv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patel A., Falck P., Shah N., Immerzeel P., Adlercreutz P., Stålbrand H., Prajapati J., Holst O., Nordberg Karlsson E. Evidence for xylooligosaccharide utilization in Weissella strains isolated from Indian fermented foods and vegetables. FEMS Microbiol. Lett. 2013;346:20–28. doi: 10.1111/1574-6968.12191. [DOI] [PubMed] [Google Scholar]