ABSTRACT
In Gram-negative bacteria, lipopolysaccharide (LPS) contributes to the robust permeability barrier of the outer membrane (OM), preventing the entry of toxic molecules, such as detergents and antibiotics. LPS is transported from the inner membrane (IM) to the OM by the Lpt multiprotein machinery. Defects in LPS transport compromise LPS assembly at the OM and result in increased antibiotic sensitivity. LptA is a key component of the Lpt machine that interacts with the IM protein LptC and chaperones LPS through the periplasm. We report here the construction of lptA41, a quadruple mutant in four conserved amino acids potentially involved in LPS or LptC binding. Although viable, the mutant displays increased sensitivity to several antibiotics (bacitracin, rifampin, and novobiocin) and the detergent SDS, suggesting that lptA41 affects LPS transport. Indeed, lptA41 is defective in Lpt complex assembly, and its lipid A carries modifications diagnostic of LPS transport defects. We also selected and characterized two phenotypic bacitracin-resistant suppressors of lptA41. One mutant, in which only bacitracin sensitivity is suppressed, harbors a small in-frame deletion in mlaA, which codes for an OM lipoprotein involved in maintaining OM asymmetry by reducing accumulation of phospholipids in the outer leaflet. The other mutant, in which bacitracin, rifampin, and SDS sensitivity is suppressed, harbors an additional amino acid substitution in LptA41 and a nonsense mutation in opgH, encoding a glycosyltransferase involved in periplasmic membrane-derived oligosaccharide synthesis. Characterization of the suppressor mutants highlights different strategies adopted by the cell to overcome OM defects caused by impaired LPS transport.
IMPORTANCE Lipopolysaccharide (LPS) is the major constituent of the outer membrane (OM) of most Gram-negative bacteria, forming a barrier against antibiotics. LPS is synthesized at the inner membrane (IM), transported across the periplasm, and assembled at the OM by the multiprotein Lpt complex. LptA is the periplasmic component of the Lpt complex, which bridges IM and OM and ferries LPS across the periplasm. How the cell coordinates the processes involved in OM biogenesis is not completely understood. We generated a mutant partially defective in lptA that exhibited increased sensitivity to antibiotics and selected for suppressors of the mutant. The analysis of two independent suppressors revealed different strategies adopted by the cell to overcome defects in LPS biogenesis.
KEYWORDS: lipopolysaccharide transport, MlaA, OpgH, outer membrane biogenesis, suppressor analysis
INTRODUCTION
Gram-negative bacteria are characterized by a cell envelope composed of two concentric membranes, the inner membrane (IM) and the outer membrane (OM), separated by a hydrophilic compartment, the periplasm, in which the murein wall is embedded (1, 2). The two membranes have different compositions and permeability properties. While the IM is a typical phospholipid (PL) bilayer, the OM is an asymmetric bilayer with PLs and lipopolysaccharides (LPS) in the inner and outer leaflet, respectively. LPS is a negatively charged amphipathic molecule composed of three covalently linked moieties: lipid A, a core oligosaccharide, and a long polysaccharide called O antigen (Fig. 1A) (3). In the presence of divalent cations, LPS molecules pack tightly together to form a layer that reduces OM fluidity and permeability, which is selectively controlled by dedicated OM proteins (4). LPS organization can be disrupted by defects in OM component assembly (5), in mutants producing LPS with truncated sugar chains (6), or by exposure to antimicrobial peptides and chelating agents, such as EDTA, which displace divalent cations between LPS molecules (4). Perturbation of LPS organization on the cell surface induces PL migration from the inner to the outer leaflet of the OM, generating locally symmetrical bilayer rafts that are more permeable to hydrophobic molecules (4). Cells have evolved systems to monitor the integrity of the OM and to respond to disruption of OM asymmetry either by removing PLs from the outer leaflet or by modifying LPS. Outer membrane phospholipase A (OMPLA), encoded by pldA, is an OM phospholipase that degrades PLs that have accumulated in the outer leaflet of the OM (7). PagP is an OM acyltransferase that transfers a palmitate acyl chain from PLs in the outer leaflet to lipid A (8) and to phosphatidylglycerol (9). A third system, the Mla (maintenance of OM lipid asymmetry) complex, prevents PL accumulation in the outer leaflet of the OM. The Mla system comprises at least six proteins distributed across the cell envelope. MlaA (formerly VacJ) is a predicted OM lipoprotein, MlaC is a periplasmic protein, and MlaFEDB form a putative ABC transporter at the IM (10, 11). Mutations in the Mla system are not lethal but lead to PL accumulation in the outer leaflet of the OM and result in increased sensitivity to sodium dodecyl sulfate (SDS) (10). Recently, MlaA has been found to interact specifically with the OM β-barrel OmpC (12). Notably, cells lacking OmpC accumulate PLs in the outer leaflet of the OM in stationary phase, thus implicating OmpC in the maintenance of lipid asymmetry and suggesting OmpC as an additional OM component of the Mla system (12). The mechanism of Mla complex-mediated retrograde PL transport is not yet completely clear, and nothing is known about the anterograde trafficking of PLs from the IM to the OM. Interestingly, recent work has revealed that two additional proteins with unknown functions, belonging to the same mammalian cell entry (MCE) protein family as MlaD, form transenvelope structures in the periplasm (13). This suggests that much remains to be understood about lipid trafficking between membranes in Gram-negative bacteria and highlights the relevance of multiprotein complexes in OM biogenesis.
FIG 1.
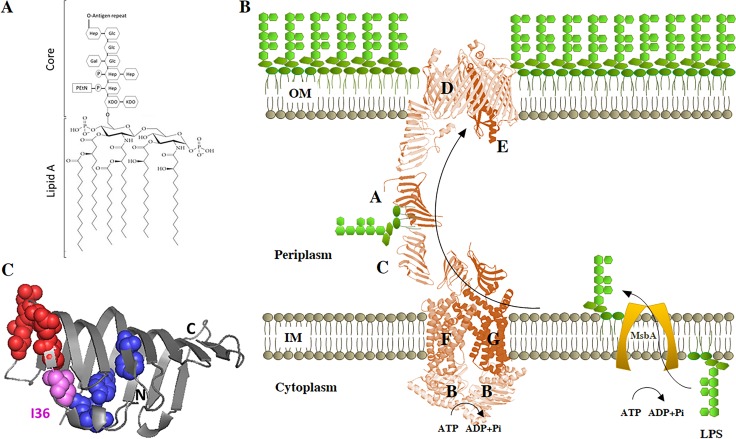
LPS transport system. (A) Chemical structure of LPS. The lipid A and core structures depicted correspond to those of E. coli K-12. The O antigen, if present, is highly variable. Gal, d-galactose; Glc, d-glucose; Hep, l-glycero-d-manno-heptose; KDO, 2-keto-3-deoxy-octonic acid; EtN, ethanolamine; P, phosphate. (B) Transport of LPS from IM to OM. After flipping over the IM by MsbA protein, LPS is transported across the periplasm and assembled at the cell surface by the Lpt machinery. (C) Graphic representation of the crystal structure of E. coli LptA (Protein Data Bank [PDB] 2r19, chain B). Residues involved in LptC interaction according to reference 55 (I38, E39, S40, Q42, and Q43) are represented by red spheres; residues involved in LPS interaction according to reference 21 (T32, F95, Y114, and L116) are represented by blue spheres. Residue I36 is involved in both LPS and LptC interactions and is represented in pink.
The asymmetric distribution of LPS in the OM is generated by the Lpt system, a molecular machine that transports and assembles LPS exclusively in the outer leaflet of the membrane. In Escherichia coli, the Lpt machinery is composed of seven essential proteins (LptABCDEFG) that span the entire envelope (14–18) (Fig. 1B). This multiprotein machinery is organized into two subassemblies (19, 20). At the IM, the ABC transporter LptB2FG, associated with the bitopic protein LptC, energizes the system (21). At the OM, the β-barrel LptD protein and the lipoprotein LptE constitute the OM translocon, characterized by a peculiar plug-and-barrel architecture (22–26), which is responsible for the final stages of LPS assembly at the cell surface. LptA, which connects the IM LptB2FGC and the OM LptDE subcomplexes, receives the LPS from LptC in an energy-dependent process and is thought to deliver it to the LptDE complex for insertion in the outer leaflet of the OM (21). The crystal structures of LptA from E. coli and of the LptH homologue from Pseudomonas aeruginosa have been solved (27, 28). LptA and LptH display very similar β-jellyroll folds, despite very low sequence similarity. Notably, a very similar β-jellyroll architecture is also present in the periplasmic region of LptC, in the periplasmic domains of LptF and LptG, and in the N-terminal domain of LptD proteins from several organisms (25, 29–31). It thus appears that the presence of such an “Lpt fold” is crucial for the assembly of a functional Lpt machine in which the C terminus of LptC interacts with the N terminus of LptA and the C terminus of LptA interacts with the N terminus of LptD (32, 33). These proteins therefore constitute the protein bridge that connects the IM and the OM and build up a hydrophobic groove able to accommodate the lipid moiety of LPS during its journey across the periplasm (19).
LptA is a key component of the protein bridge; nevertheless, it is still not well understood how the protein interacts with its LptC and LptD partners and with the LPS ligand. LptA undergoes oligomerization in vitro (27, 34, 35), yet it is not known whether it functions as an oligomer in the Lpt machine or how many LptA molecules form the protein bridge that connects the IM and OM. Photo-cross-linking experiments have highlighted a few amino acids implicated in LptA-LptC or LptA-LptA (H37) and LptA-LptD (V163) interactions (32) and several residues implicated in LptA-LPS binding (I36, F95, T32, Y114, and L116) (21). However, replacements of none of these amino acids have been shown to impair LptA function in vivo (21, 27). Only two inactive E. coli LptA mutants, Q111P (36) and G138R (28), which have not been extensively characterized, have been reported so far.
In an effort to extend the LptA structure-function relationship, we report here the characterization of a viable quadruple lptA mutant that is impaired in the assembly of the Lpt machinery, although it retains the ability to bind LPS. Although viable, the quadruple lptA mutant disrupts OM asymmetry and severely impairs the OM permeability barrier. Suppressor mutants that partially reverse the OM permeability defects provide insights into the strategies adopted by cells to restore OM functionality.
RESULTS
Generation of a quadruple lptA mutant.
Several conserved amino acid residues located at the N-terminal rim of the LptA protein, namely, I36, I38, R76, and K83 (Fig. 2A), have been previously postulated to be important for its functionality (27). Indeed, I36 has been implicated in LPS binding (21) and LptA oligomerization (35). Three of the residues (I36, I38, and R76) had been previously mutated individually to both D and E without an appreciable phenotype, as site-specific single mutations in any of these residues do not impair LptA function (27).
FIG 2.
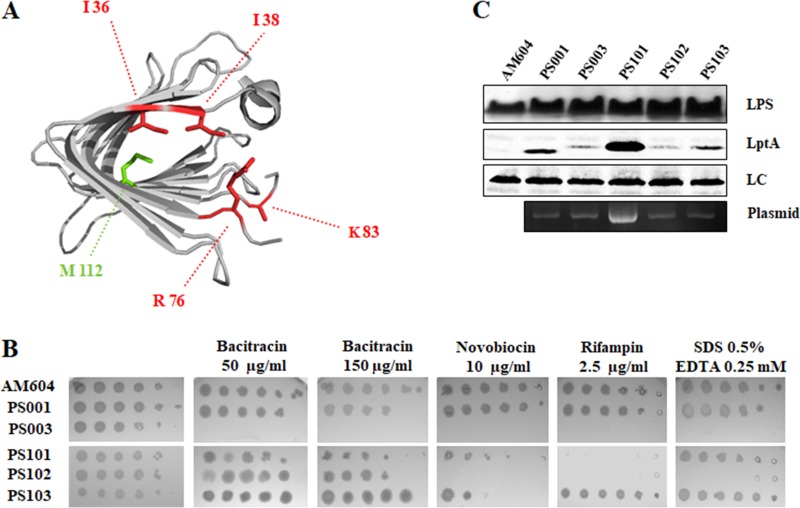
LptA41 mutations and lptA41 suppressor phenotypes. (A) Ribbon diagram of E. coli LptA. Residues mutated by site-directed mutagenesis (I36, I38, R76, and K83) are indicated in red. The intragenic suppressor (lptA42 quintuple-mutant allele) encodes an additional amino acid change at the position indicated in green (M112). (B) OM permeability assay of the lptA41 strain and suppressor mutants. Serial 10-fold dilutions of stationary-phase cultures of AM604 (wild-type reference strain); PS001 (wild-type lptA control strain ectopically expressing lptAB); PS003 (ectopically expressing lptA41 lptB); and PS101, PS102, and PS103 (PS003 derivative suppressor mutants) were replicated on LD (for AM604) or LD-ampicillin agar plates supplemented with bacitracin (50 μg/ml), novobiocin (10 μg/ml), and rifampin (2.5 μg/ml), as indicated. (C) Cell lysates from AM604, PS001, PS003, PS101, PS102, and PS103 were analyzed by Western blotting with anti-LptA and anti-LPS antibodies as described in Materials and Methods. Culture samples with equal OD600 values were processed and loaded into each lane. The different electrophoretic mobility of LptA41 relative to LptA can be explained by the different net charge of the mutant protein. A nonspecific band was used as a loading control (LC).
We thus tested whether multiple mutations (in the lptA41 allele, encoding the following amino acid substitutions: I36A, I38A, R76D, and K83D) could impair LptA functionality. Since both lptA and lptB are essential genes, we introduced pWSK29-LptA41 LptB, a low-copy-number plasmid expressing the lptA41-lptB operon, into FL907, a conditional expression mutant in which the lptAB operon is inducible by arabinose (16). We observed that this strain could grow both in the presence and in the absence of arabinose, and thus the lptA41 allele is viable.
Partial impairment of LPS transport, albeit nonlethal, may be associated with increased OM permeability to hydrophobic toxic compounds and detergents (2). To test whether lptA41 is a partial loss-of-function allele, plasmid pWSK29-LptA41 LptB was introduced into the reference wild-type strain AM604, and the chromosomal lptAB operon was replaced with the ΔlptAB::kan allele, generating the lptA41 mutant strain PS003, in which deletion of the lptAB operon is complemented. The OM permeability of PS003 was probed by testing its sensitivity to a panel of hydrophobic (novobiocin [Nov] and rifampin [Rif]) and hydrophilic (bacitracin [Bct]) antibiotics. As a control, the isogenic strain PS001, in which the ΔlptAB::kan allele is complemented by plasmid pWSK29-LptA LptB expressing wild-type LptA, was used. As shown in Fig. 2B, the lptA41 mutant displayed increased sensitivity to the antibiotics tested compared to the isogenic strain expressing wild-type lptA, suggesting that lptA41 is a partial-loss-of-function allele of lptA.
The lptA41 allele induces modifications in lipid A.
Conditions that disrupt OM asymmetry, such as defective LPS biogenesis (15, 24) or LPS release from the OM upon EDTA treatment, result in translocation of PLs to the outer leaflet of the OM (37). Bacteria respond to such stress conditions by activating enzymes that modify the basic lipid A structure (38). We thus analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) the composition of lipid A extracted from the ectopically complemented lptA41 (PS003) and lptA (PS001) isogenic strains to detect lipid A modifications as a marker of LPS transport defects (39); the parental reference strain AM604 (wild type), untreated and treated with EDTA, was used as a control.
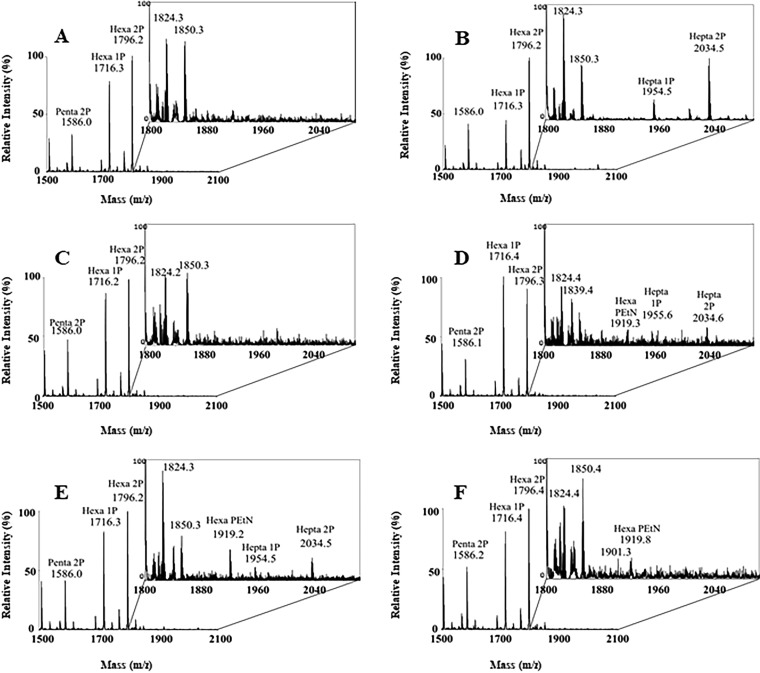
As shown in Fig. 3, untreated AM604 and PS001 (both expressing wild-type LptA) cells (Fig. 3A and C, respectively) did not produce detectable peaks corresponding to modified lipid A species. On the other hand, hepta-acylated lipid A mono- and bis-phosphate species were detected both in the AM604 EDTA-treated and in PS003 (lptA41-complemented) cells (Fig. 3B and D, respectively). Interestingly in the latter sample, a peak corresponding to phosphoethanolamine (PEtN)-modified lipid A species was also detected. This modification is catalyzed by the EptA enzyme, which decorates the phosphate in position 1 by adding a PEtN moiety and is known to be induced in response to OM perturbation by antimicrobial peptides (38, 39). These data suggest that the lptA41 allele, although complementing the ΔlptA mutant for viability, leads to the translocation of PLs in the outer leaflet of the OM, a diagnostic trait of LPS transport defects.
FIG 3.
Lipid A modifications in lptA41 suppressor strains. MALDI-TOF profiles of lipid A isolated from wild-type strain AM604 (A and B) treated (B) or not treated (A) with EDTA, PS001 (lptA+) (C), PS003 (lptA41) (D), and suppressor strains PS102 (mlaA102) (E) and PS103 (lptA42/opgH103) (F). The percent intensity relative to the most abundant species for each peak is reported. The inset plots show 10-fold-magnified views of the region around m/z 1,800 to 2,050. Relevant lipid A species are indicated. For details, see Materials and Methods.
The LptA41 mutant fails to assemble the Lpt complex.
We then assessed whether the defect associated with lptA41 could depend on lower levels or stability of LptA protein in the cell. As shown in Fig. 2C, the steady-state level of LptA41 was about 3-fold lower than that of the wild-type LptA, as revealed by densitometric analysis of bands in Western blots with anti-LptA antibodies, suggesting lower stability of the quadruple-mutant protein. It should be noted, however, that the LptA41 protein abundance in the complemented ΔlptA mutant was greater than that of the wild-type LptA expressed from the chromosome (undetectable under our experimental conditions) (Fig. 2C). Thus, the defects caused by the lptA41 allele cannot be solely attributed to lower abundance of the mutant protein but must involve other functional aspects.
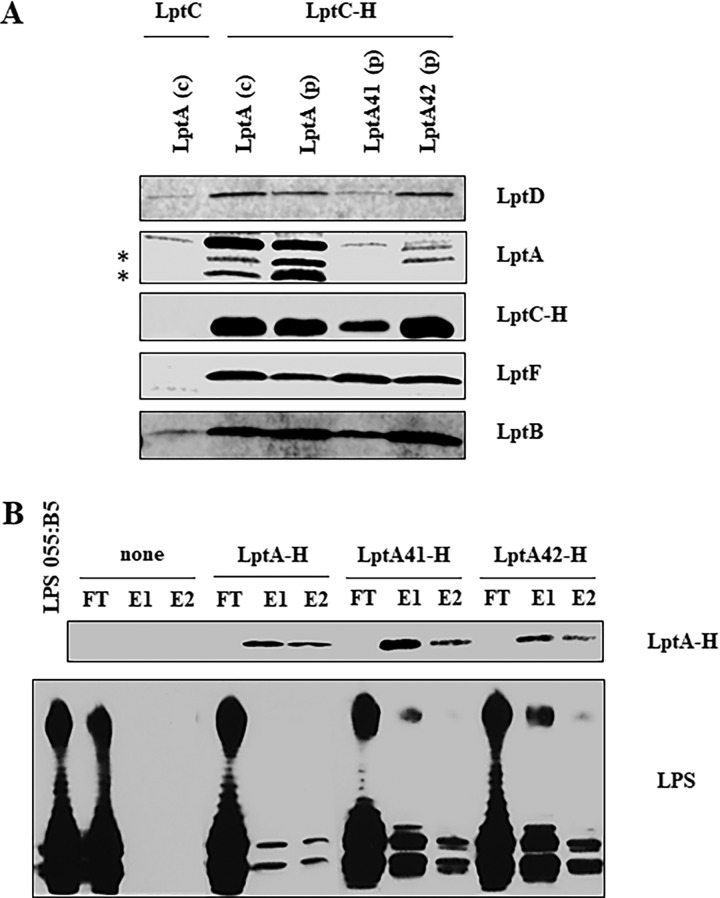
To determine whether LptA41 affects Lpt complex assembly, we performed affinity purification experiments with solubilized membranes of strains AM604, PS001, and PS003 ectopically expressing C-terminally His-tagged LptC (LptC-H) from plasmid pGS108. As a negative control, affinity purification was carried out from solubilized membranes of strain AM604 ectopically expressing untagged LptC from plasmid pGS103. Affinity-purified samples were then analyzed by SDS-PAGE immunoblotting with a panel of specific antibodies. As shown in Fig. 4A, in the LptA41-complemented strain PS003, LptC-H copurified LptF and LptB as in the positive controls, AM604 and PS001, suggesting that the IM LptB2FGC subcomplex was properly assembled. On the other hand, neither the LptA41 mutant protein nor LptD was enriched in affinity-purified membranes from PS003 (Fig. 4A), suggesting that LptA41 cannot interact with LptC and, therefore, that the OM LptDE complex cannot be properly recruited into the Lpt complex. It should be noted that the amount of LptC-H purified from membranes of the PS003 mutant was slightly lower than that purified from the control strains expressing wild-type LptA. However, the amount of LptC-H did not affect the amount of the inner membrane component LptF in the affinity-purified membranes, which appeared to be similar in all the strains. This suggests that the plasmid-encoded LptC-H protein is expressed in excess relative to the other components of the Lpt complex, and thus, its level cannot account for the lack of LptA41 copurification in the PS003 mutant.
FIG 4.
Lpt complex assembly and LPS binding by LptA mutants. (A) Assembly of wild-type and mutant LptA proteins into the Lpt complex. Total membranes were collected from strains AM604 expressing chromosomal LptA [LptA (c)], PS001 ectopically expressing LptA [LptA (p)], PS003 ectopically expressing LptA41 [LptA41 (p)], PS111 ectopically expressing LptA42 [LptA42 (p)] harboring pGS108 expressing His-tagged LptC (LptC-H), or pGS103 expressing the nontagged LptC (LptC) as a negative control. Samples were solubilized with DDM and affinity purified using a Talon metal affinity resin. The proteins were then fractionated by SDS-PAGE, and immunoblotting was performed with anti-LptD, anti-LptF, anti-LptB, and anti-His (to detect LptC-H) antibodies. For LptA detection, samples were analyzed by Tricine SDS-PAGE, and immunoblotting was performed using anti-LptA antibody. The asterisks indicate degradation products of LptA. (B) Binding of wild-type and mutant LptA proteins to LPS. The abilities of His-tagged LptA, LptA41, and LptA42 to bind purified LPS were assessed by their coelution from Ni-NTA chromatography resin. LPS and the purified His-tagged proteins were incubated and affinity purified on Ni-NTA resin as described in Materials and Methods. As a negative control, LPS was incubated without any added protein (none). FT, flowthrough; E1 and E2, elutions. To monitor LPS-LptA complex formation, equal volumes of the collected chromatographic fractions were analyzed by denaturing gel electrophoresis. LptA-H protein was detected by SDS-PAGE and Western blotting with anti-His antibodies; for LPS visualization, samples were analyzed by Tricine SDS-PAGE, and Western blotting was performed with anti-lipid A core antibodies.
On the other hand, the lower production of LptA41 from the lptA41 allele cannot account for the lack of LptA41 copurification by LptC-H in strain PS003, since the steady-state level of chromosomally expressed LptA in AM604 is much lower than those in strains PS003 and PS001 (Fig. 2C), whereas the amount of LptA copurified with LptC-H from AM604 is comparable to that in PS001. These observations suggest that the absence of LptA signal in the lane corresponding to the PS003 sample is evidence of impaired Lpt complex assembly in the lptA41 mutant.
LptA and LptC directly interact with LPS, and some residues involved in this interaction have been identified by photo-cross-linking experiments; one such residue is LptAI36, which is mutated in LptA41 (21). Therefore, we performed an in vitro LPS binding assay using purified C-terminally His-tagged LptA and LptA41 proteins (LptA-H and LptA41-H, respectively) and smooth-type LPS (34, 40). It should be noted that the LptA-H construct is functional, as when coexpressed with LptB, it complemented the lptAB conditional-expression mutant under nonpermissive conditions (data not shown). As shown in Fig. 4B, LptA41 retains the ability to interact with LPS.
Overall, these data suggest that the phenotype associated with the lptA41 allele is due to defects in Lpt complex assembly and not in LPS binding.
Screen for suppressors of the lptA41 increased antibiotic susceptibility phenotype.
Analysis of suppressor mutants is a powerful tool to identify genetic and functional interactions between the gene of interest and other genes in the same or different pathways (41). We therefore selected for spontaneous phenotypic revertants to bacitracin resistance by plating independent cultures of the lptA41 mutant on low-salt LB medium (LD medium) supplemented with 0.15 μg/ml bacitracin, a lethal concentration for the lptA41 strain but not for the wild type.
Fifteen independent bacitracin-resistant mutants, which arose at a frequency of approximately 10−8, were colony purified and tested for sensitivity to bacitracin, novobiocin, and rifampin. None of the mutants fully reverted to the wild-type phenotype (Fig. 2B). Based on their sensitivity profiles, the mutants could be grouped in three classes, and a representative of each class (strains PS101, PS102, and PS103) was chosen for subsequent analysis. PS102 retained sensitivity to novobiocin, rifampin, and SDS-EDTA, suggesting that suppression was acting on a pathway specific for bacitracin rather than alleviating the permeability barrier defect associated with lptA41. In addition to bacitracin, in PS103, resistance to rifampin and SDS-EDTA and partial resistance to novobiocin were restored, whereas PS101 exhibited resistance only to SDS-EDTA and partially to novobiocin.
Sequencing the plasmid-borne lptA genes of the three mutants revealed that PS101 and PS102 harbored the lptA41 allele, whereas in PS103, an additional mutation causing the M112I amino acid substitution was present; the quintuple mutant was designated lptA42.
Analysis of the LptA steady-state level in the suppressor strains showed that in the PS101 mutant the steady-state level of LptA41 was highly increased relative to that of the parental strain, PS003 (lptA41), and this appeared to correlate with a higher copy number of the complementing plasmid (Fig. 2C). In PS102, the LptA level was similar to that in the parental strain, PS003, whereas in PS103, the level of LptA appeared to be intermediate between the wild type and the lptA41 mutant, suggesting that the additional M112I substitution may to some extent stabilize the mutant protein (Fig. 2C).
To identify potential chromosomally encoded suppressors of the increased antibiotic sensitivity, we performed genomic sequencing of the three selected strains. The total number of reads obtained for each strain allowed us to reach more than 360-fold mean coverage for the coding portion of each strain's genome (see Table S1 in the supplemental material). In order to identify single nucleotide variations (SNVs) or insertions/deletions (indels) in the coding sequences (CDS), which could be potential suppressors of the antibiotic sensitivity phenotype, we mapped all the reads obtained for each of the three suppressor strains and their PS003 parental strain against the reference genome of MC4100 (parental strain, AM604). PS003 harbored several variations relative to MC4100, many of which were common to all the suppressor strains and thus may simply represent mutations accumulated in a different laboratory line. Table 1 reports the mutations in the suppressor strains not shared with the parental strain, PS003. In our subsequent analysis, we considered neither the variants identified in lacZ, a gene largely manipulated in MC4100 (42), nor single nucleotide changes in intergenic regions relevant.
TABLE 1.
Mutations in suppressor strains
| Strain | Genomic coordinatea | Mutation | Gene | Product | Gene coordinatec | Amino acid changed |
|---|---|---|---|---|---|---|
| PS101 | 290676 | T→G transversion | Intergenic | NAb | NA | NA |
| 4178702 | G→C transversion | lacZ | β-Galactosidase | 246 | Arg116Gly | |
| 4178861 | G insertion | lacZ | β-Galactosidase | 187 | Ser63fs | |
| 4178862 | A→G transition | lacZ | β-Galactosidase | 186 | None | |
| 4178862 | CGG insertion | lacZ | β-Galactosidase | 183 | Asp62fs | |
| PS102 | 290676 | T→G transversion | Intergenic | NA | NA | NA |
| 1574918 | A insertion | tus | Replication terminus site-binding protein | 577 | Ser193fs | |
| 2348710 | AAGTTG deletion | mlaA | Putative lipoprotein | 120 | ΔAsn41-Phe42 | |
| 2613172 | G→A transition | Intergenic | NA | NA | NA | |
| 4178702 | G→C transversion | lacZ | β-Galactosidase | 346 | Arg116Gly | |
| 4178862 | A→G transition | lacZ | β-Galactosidase | 186 | None | |
| 4178862 | CG insertion | lacZ | β-Galactosidase | 184 | Asp62fs | |
| PS103 | 425601 | A→G transition | rhsD | rhsD element | 358 | Ser120Gly |
| 1015255 | G→A transition | mdoH | Glucosyltransferase | 1245 | Trp415Stop | |
| 3667499 | T→G transversion | Intergenic | NA | NA | NA | |
| 3228885e | G→T transversion | lptA | LPS transport protein | 336 | Met112Ile |
For deletions and insertions, the coordinate indicates the first deleted base and the base after which insertion occurred, respectively.
NA, not applicable.
Coordinates from the first base of the open reading frame (ORF). For deletions and insertions, the coordinate indicates the first deleted amino acid and the amino acid after which insertion occurred, respectively.
fs, frameshift starting at the codon indicated.
This region is actually harbored by the complementing plasmid.
Analysis of genomic-sequencing results from PS101 did not reveal any mutation that could be correlated with the suppression and/or the increased complementing plasmid copy number and/or LptA41 abundance (Fig. 2C). It is possible that the increased copy number of the plasmid, and consequently the higher expression level of LptA41, contributes to the suppression of bacitracin and novobiocin susceptibility. Variations in nonsequenced regions (gaps) may contribute to suppression and/or increased plasmid copy numbers. No further analysis was performed on this suppressor strain.
A mutation in mlaA is implicated in suppression of PS102 bacitracin sensitivity.
Strain PS102 harbors, in addition to mutations in lacZ and in intergenic regions, a nucleotide insertion that causes a frameshift mutation at codon 193 of tus (here named the tus-102 mutation) and a 6-nucleotide deletion in mlaA (here named the mlaA102 allele) that removes amino acids N41 and F42 of the encoded MlaA protein (Table 1).
Tus is a nonessential E. coli protein implicated in replication termination at ter sites (43). MlaA is the OM lipoprotein component of the Mla system, which is thought to maintain OM lipid asymmetry in E. coli (10). Therefore, we sought to determine whether mlaA102 and/or tus-102 mutations could be implicated in the suppression of bacitracin sensitivity observed in strain PS102. We therefore sequenced the chromosomal mlaA and tus genes from five independent members of this class of suppressor mutants and found that three of them harbored the same mlaA102 allele; in another mutant, an IS1 element was inserted downstream of mlaA codon 227, whereas in the last one, no mutation was present in mlaA. The tus genes of these five independent mlaA mutants were sequenced and did not bear any mutation, indicating that tus-102 is not implicated in suppression.
To genetically characterize mlaA102, we transformed wild-type, lptA41, and ΔmlaA strains with plasmids expressing the wild-type and the mutant mlaA alleles from the ptac promoter and assessed the impacts of the different mlaA and lptA alleles, in all possible combinations, on OM permeability using bacitracin and SDS-EDTA as probes.
Bacitracin was used at a concentration (50 μg/ml) that reliably discriminates lptA41 from the suppressors, independently of the concentrations used from the selection and the genetic background. We used SDS-EDTA at concentrations of 0.5% and 0.25 mM, respectively, which are tolerated by wild-type strains. The complementation and suppression tests are summarized in Table 2. Both wild-type and ΔmlaA strains (Table 2, compare rows 1 to 3 with 13 to 15) appear to tolerate SDS-EDTA. In a previous report, an E. coli ΔmlaA mutant was shown to be sensitive to a critical SDS-EDTA concentrations (Sdss) but not to other drugs, including bacitracin (10). Under our experimental conditions, however, both wild-type and ΔmlaA strains did not tolerate higher SDS-EDTA concentrations. The lptA41 strain PS003 showed severe permeability defects, as in addition to bacitracin, it showed increased sensitivity to SDS-EDTA compared to the isogenic wild-type strain (compare rows 1 and 2 with 5 and 6). Ectopic expression of mlaA102 was sufficient to restore resistance to bacitracin in lptA41 mutant strains, both in the haploid (row 20) and in the heterozygous (row 8) states. Interestingly, under both conditions, mlaA102 also conferred sensitivity to SDS, suggesting that mlaA102 is (at least partially) a dominant-negative mutant that suppresses the bacitracin sensitivity of lptA41. As mlaA102 also confers the Sdss phenotype irrespective of the presence of the wild-type mlaA allele (rows 4 and 11), it can be hypothesized that mlaA102 is a gain-of-function allele that negatively affects OM permeability. This is in line with the finding that in the PS102 suppressor strain, the lipid A modification pattern is very similar to that observed in the parental PS003 mutant (Fig. 3), further strengthening the hypothesis that the suppression mechanism does not act through restoration of the OM permeability barrier.
TABLE 2.
Phenotypic suppression pattern of the mlaA102 allele
| Strain | Chromosomal version of: |
Plasmid |
EOPa |
Row | ||||
|---|---|---|---|---|---|---|---|---|
| lptA | mlaA | lptAb | mlaAc | LD | Bct (50 μg/ml) | SDS EDTA | ||
| PS001 | ΔlptA | mlaA+ | lptA+ | NP | + | + | + | 1 |
| − | + | + | + | 2 | ||||
| mlaA+ | + | + | + | 3 | ||||
| mlaA102 | + | + | − | 4 | ||||
| PS003 | ΔlptA | mlaA+ | lptA41 | NP | + | − | − | 5 |
| − | + | − | − | 6 | ||||
| mlaA+ | + | − | − | 7 | ||||
| mlaA102 | + | + | − | 8 | ||||
| PS102 | ΔlptA | mlaA102 | lptA41 | NP | + | + | − | 9 |
| − | + | + | − | 10 | ||||
| mlaA+ | + | + | − | 11 | ||||
| mlaA102 | + | + | − | 12 | ||||
| PS130 | ΔlptA | ΔmlaA | lptA+ | NP | + | + | + | 13 |
| − | + | + | + | 14 | ||||
| mlaA+ | + | + | + | 15 | ||||
| mlaA102 | + | + | − | 16 | ||||
| PS107 | ΔlptA | ΔmlaA | lptA41 | NP | + | − | − | 17 |
| − | + | − | − | 18 | ||||
| mlaA+ | + | − | − | 19 | ||||
| mlaA102 | + | + | − | 20 | ||||
EOP relative to growth on LD agar. +, EOP > 10−2; −, EOP < 10−3.
lpt+, pWSK29-LptA LptB; lptA41, pWSK29-LptA41 LptB.
NP, no plasmid; −, empty vector pGS100; mlaA+, pGS100-MlaA; mlaA102, pGS100-MlaA102.
Bacitracin resistance in an mlaA102 suppressor mutant is associated with hypervesiculation.
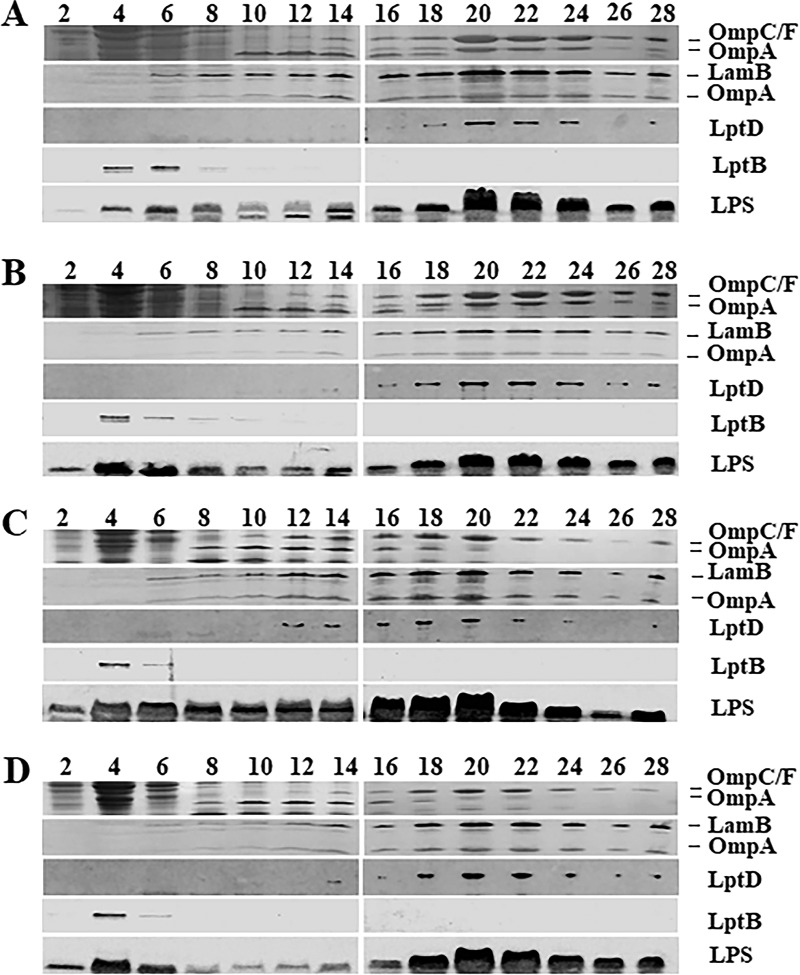
The PS102 mutant could suppress bacitracin sensitivity while not restoring the OM permeability barrier. To gain hints about the mechanism of suppression of bacitracin sensitivity by mlaA102, we analyzed the composition of the OM in the suppressor mutant and in the corresponding isogenic parental strains. The analysis of LPS levels from whole-cell lysates of the PS102 mutant compared with the isogenic lptA+ (PS001) and parental lptA41 (PS003) strains failed to reveal any difference (Fig. 2C). We then analyzed PS102 IM and OM protein profiles. Whole-cell lysates obtained from PS003 and PS102 were fractionated by sedimentation on sucrose density gradients, as described in Materials and Methods. The fractions were assayed by SDS-PAGE, followed by Coomassie staining for OmpC, OmpF, and OmpA porin profiles and by immunoblotting with antibodies against LamB (these antibodies cross-react with OmpA [16]), LptD, LptB, and LPS. The fractionation profile of the isogenic PS001 wild-type strain was used as a reference in this experiment.
As shown in Fig. 5A, in PS001 cells, the OM equilibrated at fractions 20 to 24, as defined by the distribution profile of LPS, porins, and LptD. Conversely, the IM equilibrated at fractions 4 to 6, as judged by the profile of the IM-associated protein LptB. In PS003 mutant cells, the IM and OM equilibrated at the same densities as in the wild-type strain PS001, as witnessed by LptB and porin distributions, respectively. A slightly greater accumulation of LPS in the IM than in the OM fractions was observed in PS003, in agreement with the LPS transport impairment associated with the lptA41 allele (Fig. 5B). Interestingly, in suppressor PS102 cells, the distribution of porins, LPS, and LptD was skewed toward fractions 16 to 20, with densities lower than those of strain PS003 (fractions 20 to 24), indicating that in this mutant, the OM floated at a density lower than that of the parental strain, PS003. Of note, LPS distribution between the IM and the OM was comparable to that in PS003 (Fig. 5C), confirming that LPS transport was not restored in the PS102 mutant. We suggest that the lower OM density in PS102 might result from a lower LPS/PL ratio than in the PS003 OM, associated with mlaA102 mutation.
FIG 5.
Membrane fractionation of wild-type, lptA41, and suppressor strains. Cultures of PS001 (lptA+) (A), PS003 (lptA41) (B), PS102 (mlaA102) (C), and PS103 (lptA42 opgH103) (D) strains were grown to an OD600 of ∼0.6. Crude extracts were fractionated on a sucrose density gradient. Fractions were collected from the top of the gradient. The profiles of the major OM porins (OmpC/F-OmpA) were determined by SDS-PAGE, followed by Coomassie blue staining. LamB-OmpA and LptD profiles were determined as OM markers by immunoblotting using anti-LamB and anti-LptD antibodies, respectively. LptB was detected as an IM marker using anti-LptB antibody. LPS distribution across fractions was determined by Tricine SDS-PAGE and immunoblotting using anti-LPS WN1 222-5 monoclonal antibody. The numbers are fraction numbers.
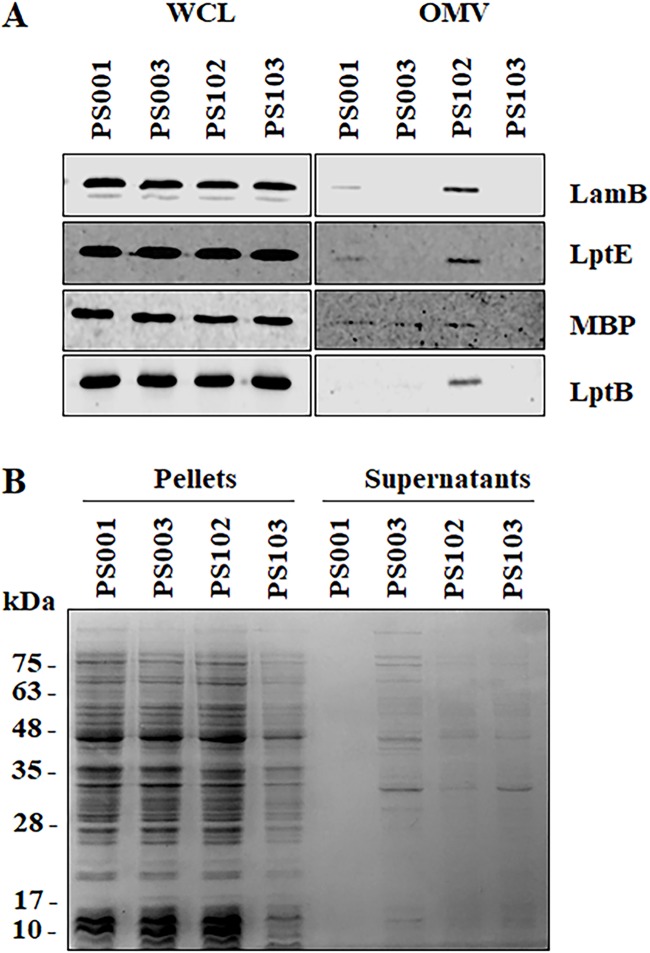
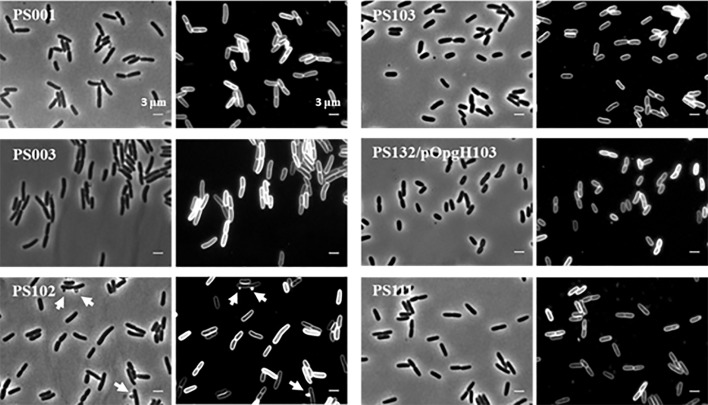
The same mlaA102 allele (referred to as mlaA*) was recently associated with a novel cell death mechanism that occurs under starvation conditions in medium with limited cation concentrations (44). Notably, in cells carrying the mlaA* mutation, PL accumulation at the OM leads to increased OM blebbing at cell division sites (44). As PS102 showed an altered OM (Fig. 3E and 5C), we assessed whether increased outer membrane vesicle (OMV) production was an additional phenotype of the PS102 mutant. To test this hypothesis, we filtered culture supernatants of the PS102 mutant and its parental strain, PS003, to eliminate whole cells and concentrated samples by ultrafiltration to collect OMVs. The ultrafiltered samples were analyzed by SDS-PAGE and immunoblotting using antibodies directed against OM (LamB and LptE) and periplasmic (maltose binding protein [MBP]) proteins, whose levels are indicative of OM vesiculation. As a control, we analyzed the isogenic wild-type strain PS001. As shown in Fig. 6A, we detected the periplasmic MBP in filtered supernatants of PS001, PS003, and PS102. The substantial increase in OM markers only in the supernatant of PS102 suggested that the mutant hypervesiculates compared to the isogenic wild type and its parental strain, PS003. This finding was also confirmed by the presence of OM blebbing in microphotographs of PS102 mutant cells (Fig. 7). Interestingly, the IM-associated LptB protein was also detected in OMVs from PS102, probably reflecting the ability of the protein to colocalize with the OM (14). The presence of MBP alone in PS003 supernatant appeared to be related to leakage of proteins into the growth medium, as assessed by SDS-PAGE and Coomassie blue staining of the corresponding culture supernatants that showed a protein profile identical to that of total cell lysates (Fig. 6B). Leakage of proteins in culture media is a common phenomenon in envelope biogenesis mutants (45).
FIG 6.
OMV isolation and determination of protein release in cell supernatants. (A) Whole-cell lysates (WCL) and OMVs from PS001 (lptA+), PS003 (lptA41), and suppressor strains PS102 (mlaA102) and PS103 (lptA42 opgH103) were prepared, separated by SDS-PAGE, and analyzed by immunoblotting using anti-LamB, anti-LptE, anti-MBP, and anti-LptB antibodies. (B) Pellets containing equal numbers of cells, based on the OD600, and 20-fold-concentrated culture supernatants of strains PS001, PS003, PS102, and PS103 were subjected to SDS-PAGE and stained with Coomassie blue. Molecular mass markers are indicated.
FIG 7.
Morphology of lptA41 and suppressor cells. Shown are phase-contrast and membrane stain (FM5-95) images of PS001 (lptA+), PS003 (lptA41), suppressor strains PS102 (mlaA102) and PS103 (lptA42 opgH103), PS132/pOpgH103 (ΔopgH ectopically expressing OpgH103 mutant protein), and PS111 (lptA42). Cell length measurements are reported in Table 5. The arrows in suppressor strain PS102 indicate OMVs.
An additional amino acid substitution in LptA41 and a missense mutation in opgH contribute to lptA41 suppression in PS103.
In the suppressor strain PS103, resistance to bacitracin, rifampin, and SDS-EDTA and partial resistance to novobiocin were restored. The altered OM permeability profile of PS103 is in line with the finding that only hexa-acylated lipid A decorated with PEtN and not the hepta-acylated lipid A species could be detected in the mutant (Fig. 3F). Genomic sequencing of PS103 revealed that, besides the additional mutation in the plasmid-borne lptA41 allele that generated the lptA42 allele, the strain harbored a W415Stop mutation in opgH (opgH103 allele) (Table 1). opgH (formerly mdoH) encodes an IM glycosyltransferase of 847 amino acids implicated in both the synthesis of osmoregulated periplasmic glucans (OPGs), also known as membrane-derived oligosaccharides (MDOs) (46, 47), and the control of cell size via interaction with FtsZ in a nutrient-dependent manner (48). We thus analyzed the possible contribution of each mutation to the suppressed phenotype exhibited by strain PS103.
As expected, the LptA42 mutant protein retained the ability to copurify LPS in vitro (Fig. 4B). We then tested Lpt complex assembly by performing affinity purification experiments from solubilized membranes of a strain bearing the lptA42 allele and ectopically expressing LptC-H. As shown in Fig. 2C, the steady-state level of LptA42 appeared to be higher than that of LptA41, suggesting that the LptAM112I mutation slightly improved protein stability. Affinity purification from solubilized membranes of PS103 using LptC-H as bait revealed partially restored LptA42-LptC interaction, as judged by the increased amount of an LptA42 degradation product in Western blot analysis using anti-LptA antibodies and by the enrichment of LptD (Fig. 4A). These data suggest that in the PS103 mutant, the Lpt complex could assemble, albeit with weaker affinity than in the wild type (Fig. 4A).
In line with the above-mentioned data is the finding that both the analysis of LPS levels from whole cells and the membrane fractionation profile of PS103 failed to show relevant differences in membrane composition relative to the parental strain, PS003 (Fig. 2C and 5D). We thus analyzed the contributions of the opgH103 and lptA42 alleles, alone or in combination, to the suppression of sensitivity to toxic compounds in a suitable set of strains harboring ectopic and chromosomal lptA and opgH alleles in different combinations. It should be mentioned that E. coli ΔopgH strains have been reported to be sensitive to ≥0.5% SDS (49). However, under our experimental conditions, the ΔopgH mutant was tolerant of 0.5% SDS and 0.25 mM EDTA, whereas, as shown in Table 2, the lptA41 mutant was sensitive.
The data presented in Table 3 indicate that (i) in the PS103 suppressor strain (row 1), tolerance for bacitracin, SDS, and rifampin has been restored, whereas sensitivity to novobiocin was only partially suppressed compared with the parental strain (line 6), and (ii) the lptA42 allele was sufficient to restore bacitracin and SDS tolerance but not rifampin or novobiocin tolerance in a wild-type chromosomal opgH background (lines 10 and 11). On the other hand, lptA42 fully restored rifampin tolerance and partially restored novobiocin tolerance only in the absence of a wild-type opgH allele, irrespective of its chromosomal or plasmid location (compare lines 22, 23, and 25 with 10 to 12 and 24). The merodiploid opgH+ opgH103 heterozygous strain (ectopic opgH103) was only partially tolerant of novobiocin (line 13), indicating that the opgH103 allele is recessive to the wild-type opgH allele.
TABLE 3.
Phenotypic suppression pattern for lptA42 and opgH103 alleles
| Strain | Chromosomal version of: |
Plasmid |
EOPa |
Row | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| lptA | opgH | lptAb | opgHc | LD | Bct (150 μg/ml) | SDS-EDTA | Rif (2.5 μg/ml) | Nov (10 μg/ml) | ||
| PS103 | ΔlptA | opgH103 | lptA42 | NP | + | + | + | + | ± | 1 |
| PS001 | ΔlptA | opgH+ | lptA+ | NP | + | + | + | + | + | 2 |
| − | + | + | + | + | + | 3 | ||||
| opgH+ | + | + | + | + | + | 4 | ||||
| opgH103 | + | + | + | + | + | 5 | ||||
| PS003 | ΔlptA | opgH+ | lptA41 | NP | + | − | − | − | − | 6 |
| − | + | − | − | − | − | 7 | ||||
| opgH+ | + | − | − | − | − | 8 | ||||
| opgH103 | + | − | − | − | − | 9 | ||||
| PS111 | ΔlptA | opgH+ | lptA42 | NP | + | + | + | − | − | 10 |
| − | + | + | + | − | − | 11 | ||||
| opgH+ | + | + | + | − | − | 12 | ||||
| opgH103 | + | + | + | − | ± | 13 | ||||
| PS112 | ΔlptA | ΔopgH | lptA+ | NP | + | + | + | + | + | 14 |
| − | + | + | + | + | + | 15 | ||||
| opgH+ | + | + | + | + | + | 16 | ||||
| opgH103 | + | + | + | + | + | 17 | ||||
| PS109 | ΔlptA | ΔopgH | lptA41 | NP | + | − | − | − | − | 18 |
| − | + | − | − | − | − | 19 | ||||
| opgH+ | + | − | − | − | − | 20 | ||||
| opgH103 | + | − | − | − | − | 21 | ||||
| PS110 | ΔlptA | ΔopgH | lptA42 | NP | + | + | + | + | ± | 22 |
| − | + | + | + | + | ± | 23 | ||||
| opgH+ | + | + | + | − | − | 24 | ||||
| opgH103 | + | + | + | + | ± | 25 | ||||
EOP relative to growth on LD agar. +, EOP > 10−2; ±, EOP between 10−2 and 10−3; −, EOP < 10−3.
lptA+, pWSK29-LptA LptB; lptA41, pWSK29-LptA41 LptB; lptA42, pWSK29-LptA42 LptB.
NP, no plasmid; −, empty vector pGS100; opgH+, pGS100-OpgH; opgH103, pGS100-OpgH103.
The opgH103 allele reduces mutant cell size.
In E. coli, inactivation of opgH leads to increased expression of colanic acid (CA) (50). Consistently, our ΔopgH and opgH103 strains produced mucoid colonies. To test whether colanic acid production could play any role in the suppressed phenotype, we inactivated the wcaJ gene, which codes for the UDP-glucose lipid carrier required for the biosynthesis of CA (51, 52). Deletion of wcaJ in PS103 and in the PS132 ΔopgH lptA42 mutant strain barely affected the antibiotic resistance of the parental strains (Table 4), suggesting that CA overproduction in PS103 is not implicated in suppression of the increased antibiotic susceptibility. Interestingly, inactivation of wcaJ in the ΔopgH mutant expressing the lptA41 allele conferred some amelioration of OM permeability. Overall, these results suggest that suppression of the lptA41 phenotype in PS103 is ensured by the combination of LptA mutation and the production of a truncated OpgH protein and is independent of CA production.
TABLE 4.
Colanic acid production does not affect PS103 suppressor phenotype
| Strainb | Chromosomal version of: |
Plasmid lptAc | EOPa |
Row | |||||
|---|---|---|---|---|---|---|---|---|---|
| opgH | wcaJ | LD | Bct (50 μg/ml) | Bct (150 μg/ml) | Rif (2.5 μg/ml) | Nov (10 μg/ml) | |||
| PS103 | opgH103 | wcaJ+ | lptA42 | + | + | + | + | ± | 1 |
| PS113 | opgH103 | ΔwcaJ | lptA42 | + | + | + | + | ± | 2 |
| PS112 | ΔopgH | wcaJ+ | lptA+ | + | + | + | + | + | 3 |
| PS109 | ΔopgH | wcaJ+ | lptA41 | + | − | − | − | − | 4 |
| PS110 | ΔopgH | wcaJ+ | lptA42 | + | + | + | + | − | 5 |
| PS116 | ΔopgH | ΔwcaJ | lptA+ | + | + | + | + | + | 6 |
| PS114 | ΔopgH | ΔwcaJ | lptA41 | + | ± | ± | ± | − | 7 |
| PS115 | ΔopgH | ΔwcaJ | lptA42 | + | + | + | + | − | 8 |
EOP relative to growth on LD agar. +, EOP > 10−2; ±, EOP between 10−2 and 10−3; −, EOP < 10−3.
All the strains harbor the ΔlptAB::kan chromosomal deletion.
lptA42, pWSK29-LptA42 LptB; lptA+, pWSK29-LptA LptB; lptA41, pWSK29-LptA41 LptB.
OpgH has been implicated in modulating cell size by antagonizing the assembly of FtsZ at the nascent septal site (48). We therefore determined whether the production of a truncated OpgH protein impacted cell size by comparing the average size of PS103 mutant cells with those of the parental PS003, the isogenic wild-type PS001, and the suppressor PS102 cells. As shown in Fig. 7 and Table 5, PS103 mutant cells were 28.8% and 19.1% smaller than those of strains PS003 and PS001, respectively. On the other hand, suppressor PS102 mutant cells were similar in size to the parental PS003 cells, suggesting that the slight decrease in cell size may have a role in the PS103-specific mechanism of lptA41 phenotype suppression.
TABLE 5.
Cell lengths
| Strain | Relevant genotype | Lengtha (μm ± SD) |
|---|---|---|
| PS001 | ΔlptAB/pWSK29lptAB | 3.40 ± 0.52 |
| PS003 | ΔlptAB/pWSK29lptA41lptB | 3.86 ± 0.63 |
| PS102 | ΔlptAB vacJ102/pWSK29lptA41lptB | 3.90 ± 0.67 |
| PS103 | ΔlptAB opgH103/pWSK29lptA42lptB | 2.75 ± 0.57 |
| PS132/pOpgH103 | ΔopgH/pGS100-opgH103 | 2.52 ± 0.51 |
| PS111 | ΔlptAB/pWSK29lptA42lptB | 3.10 ± 0.67 |
Average lengths of cells from 100 cells per strain.
DISCUSSION
LptA is the periplasmic component of the Lpt machine that ferries LPS across the periplasm to the cell surface. LptA connects, supposedly as a head-to-tail dimer/oligomer (27, 34), the inner membrane ABC transporter formed by the LptB2FGC subcomplex with the OM LptDE translocon by contacting the C-terminal region of LptC at the IM and the N-terminal region of LptD at the OM via its N- and C-terminal domains, respectively (19, 20, 32). LptA also binds LPS and is thought to provide a hydrophobic environment for the passage of the amphipathic LPS through the periplasm (21, 40). LptA is the prototype of the peculiar β-jellyroll fold (27), conserved in LptC (29), the N-terminal domain of LptD (25, 31), and the periplasmic domains of LptF and LptG (30) and thought to be relevant for both protein-protein and protein-LPS interactions. In agreement with structural data, photo-cross-linking studies have identified specific residues implicated in LptA-LptC and LptA-LPS interactions (21, 32). Despite the wealth of information available to date, several aspects of the molecular basis of LptA function remain elusive, highlighting the need for structure-function studies. LptA is essential for cell viability (17), and the only nonviable mutants in lptA isolated so far have been Q111P (36) and G138R (28) substitutions; however, it is not known how the above-mentioned mutations affect LptA function.
In order to gain further information about the molecular role of LptA in LPS transport, we generated a mutant LptA protein potentially impaired in interactions with LPS and/or other Lpt proteins by changing four residues (I36, I38, R76, and K83) that lie at the N-terminal rim of the LptA internal cavity and are conserved in representative gammaproteobacteria (27). Furthermore, I36 and I38 are located in a region implicated by photo-cross-linking in LptA-LptC and/or LptA-LptA interaction (32). Such a quadruple mutant (lptA41) complemented both LptA-depleted cells and lptA deletion mutants for viability; thus, modification of the above-mentioned four residues is not sufficient to abolish LPS transport to the OM. LptA41 did not seem to be compromised in LPS binding, whereas it failed to interact with LptC in affinity purification experiments using His-tagged LptC as bait. This suggests that LptA41-LptC interaction is too weak to be detected by affinity purification but is sufficient to form a functional, albeit partially impaired, Lpt complex. It has been previously reported that in vitro LptA-LptC interaction and LptA self-oligomerization are independent of LptA or LptC binding to LPS, as these interactions can occur in the absence of LPS, as assessed by size exclusion chromatography and electron paramagnetic resonance (EPR) and nuclear magnetic resonance (NMR) spectroscopy (53–55). However, it is not possible to exclude the possibility that, in vivo, LPS binding influences LptA assembly within the Lpt complex. To our knowledge, this is the first lptA mutant in which LPS binding to LptA and LptA-LptC interaction can be genetically separated. Notably, mutant Lpt machines missing the LptC component but carrying specific amino acid substitutions of a single residue in the LptF periplasmic domain have been identified (56). This finding suggests that under specific conditions, LptA-LPS binding is independent of LptA-LptC interaction. Clearly, in the mutant Lpt machines missing the LptC component, different protein-protein and protein-LPS interactions must occur to ensure functional LPS transport.
Although E. coli lptA41 is viable, the mutant exhibits increased sensitivity to three antibiotics (the large hydrophilic bacitracin and the hydrophobic rifampin and novobiocin [6]) and to the detergent SDS. Such a sensitivity profile is diagnostic of a defective OM that is more accessible to the detergent, which may disrupt the OM itself, and more permeable to antibiotics, which must diffuse through the OM to reach their targets either in the periplasm (bacitracin) or in the cytoplasm (rifampin and novobiocin). Multiple mechanisms may contribute to the increased permeability of the OM (4, 57). Among these, defective LPS transport may interfere with OM protein assembly and increase the PL concentration in the outer leaflet of the OM, thus making the membrane more permeable to lipophilic molecules, such as rifampin and novobiocin. On the other hand, bacitracin, a hydrophilic molecule too large to diffuse through porins, may enter the periplasm through transient breaching (“cracks”) in the OM bilayer at disordered LPS-PL junctions (58). In the lptA41 mutant, LPS transport may not be as efficient as in the wild type, thus leading to a defective OM with increased LPS-PL junctions. Accordingly, in this mutant, we observed LPS species with hepta-acylated (both mono- and diphosphorylated) lipid A or decorated by PEtN. The former modification, catalyzed by the lipid A palmitoyltransferase PagP, is induced by translocation of PLs to the outer leaflet of the OM (8, 59), where PLs are used as palmitoyl donors, and is consistent with the hypothesis of defective transport. On the other hand, PEtN decoration, promoted by EptA (3, 38) and known to be induced by exposure of cells to mild acid (60) or antimicrobial peptides (61), occurs on the outer surface of the IM (38), where LPS might idle under conditions of inefficient transport (15, 24). Although such lipid A modifications may be part of the homeostatic cell response to diverse envelope stress conditions, they do not appear to be sufficient to fully restore the OM permeability barrier in the lptA41 mutant.
By searching for spontaneous phenotypic revertants to bacitracin resistance (Bctr), we obtained suppressors showing different antibiotic resistance profiles, none of which fully reverted to the resistant phenotype of the lptA wild-type parental strain.
The suppressor mutation of strain PS102 is mlaA102, a two-codon, in-frame deletion in mlaA. In E. coli, MlaA is the OM lipoprotein component of the Mla machinery, which is thought to contribute, together with other systems, to the maintenance of OM lipid asymmetry by removing PLs from its outer leaflet (10). It has been suggested that upon disruption of the Mla system, PL rafts accumulate in the OM outer leaflet (10, 62). Notably, the same mlaA in-frame deletion (named mlaA*) has been recently isolated as responsible for suppressing the bacitracin sensitivity of an lptD mutant defective in LPS assembly at the OM (44). Moreover, the IM MlaD protein increases in abundance in cells where LPS transport is severely impaired (63). These findings further strengthen a functional link between the Mla system and the Lpt machinery.
Disruption of the Mla system does not significantly impair the OM permeability barrier but facilitates access to detergents, as shown by increased sensitivity to a critical SDS (0.5%)-EDTA (1.1 mM) concentration (10). However, under our experimental conditions, the mlaA deletion mutants analyzed in this work tolerate SDS (0.5%)-EDTA (0.25 mM) like their isogenic parental strains (Table 2), whereas neither could grow in the presence of higher SDS-EDTA concentrations. This discrepancy with previously reported data might be ascribed to the genetic differences in laboratory strains and/or laboratory conditions. Interestingly, the mlaA102 allele confers increased sensitivity to SDS but not to bacitracin under both haploid and merodiploid heterozygous conditions, suggesting that mlaA102 is a dominant-negative mutation that impairs the Mla system. These observations fit the role proposed for mlaA* as a gain-of-function allele that transfers PLs from the inner to the outer leaflet of the OM, which is the reverse of mlaA wild-type activity (44). The mlaA* allele has been shown to be associated with cell death in stationary phase occurring after loss of OM material through vesiculation because of an increased LPS level and fatty acid depletion (44). The phenotypes associated with PS102, namely, persistence of lipid A modifications (Fig. 3E), decreased OM density, and altered LPS distribution between the IM and OM (Fig. 5C), indicate that in this suppressor, LPS transport is still impaired and the integrity of the OM is compromised, possibly as a consequence of PL translocation to the outer leaflet of the OM. However, unlike the mlaA* mutant, PS102 showed no increase in LPS level compared with its parental wild-type strain (Fig. 2C) and does not appear to undergo cell death, in agreement with the finding that a decreased LPS level in the OM due to LPS transport impairment suppresses the death phenotype of mlaA* cells (44).
The mlaA102 allele suppresses sensitivity to bacitracin caused by lptA41, whereas it does not restore tolerance for rifampin and novobiocin, suggesting that the suppressor mutation is specific for bacitracin resistance. While the presence of PL rafts justifies the sensitivity of PS102 to the hydrophobic rifampin and novobiocin, it is not immediately evident how the suppressor mutant recovers resistance to the large hydrophilic polypeptide antibiotic bacitracin. It has been shown that accumulation of PLs in the outer leaflet of the OM leads to OMV formation (61). Accordingly, the mlaA102 mutant has been shown to hypervesiculate. In addition to playing important roles in Gram-negative bacterial physiology and pathogenesis, OMV production has been shown to be a strategy to increase resistance to antibacterial molecules by sequestering them from the cellular targets, as in the cases of colistin and polymyxin B (64). We thus propose that in the PS102 suppressor strain hypervesiculation specifically restores Bctr. Bacitracin is a cyclic polypeptide that interferes with generation of undecaprenyl phosphate (C55-P) from its precursor, undecaprenyl pyrophosphate (C55-PP) (65), for the synthesis of cell wall polysaccharides, such as peptidoglycan, LPS O antigen, and teichoic acids (3, 66, 67). In E. coli, Bctr has been shown to occur through several indirect mechanisms, including amplification of the C55-PP phosphatase BacA or mutations impairing the production of colanic acid that otherwise would contribute to further deplete the pool of C55-PP carrier (68), and unlike rifampin and novobiocin, no chromosomal mutations in the antibiotic target can be isolated (69). In this work, we highlight a novel potential mechanism of Bctr involving removal of the antibiotic from its target via hypervesiculation. Interestingly, recent selection for E. coli mutants resistant to a novel expanded-spectrum antibiotic (tigecycline) has allowed the isolation of the same mlaA two-codon deletion present in the mlaA102 allele (70); whether this may involve a hypervesiculation mechanism as well needs to be proved.
In the suppressor strain PS103, bacitracin, rifampin, and SDS tolerances were restored, whereas novobiocin sensitivity was not fully suppressed. The additive effects of an intragenic suppressor and an extragenic suppressor appear to contribute to this phenotype.
The intragenic suppressor (lptA42 allele) encodes an additional amino acid change (M112I) in LptA41. The LptA42 quintuple-mutant protein partially recovered the ability to copurify with LptC, and it appeared to be more stable than LptA41, as judged by the higher steady-state level of the protein. We suggest that this phenotype is due to the reintroduction of an isoleucine residue pointing toward the interior of the cavity of LptA (Fig. 2A). Interestingly, the region around the M112 residue of LptA has been recently reported to undergo a chemical shift perturbation in the presence of LptC and LPS in NMR experiments (55).
The lptA42 allele was sufficient to restore tolerance for bacitracin and SDS but not for rifampin and novobiocin in a wild-type chromosomal opgH background. The partially restored OM asymmetry would prevent the formation of PL rafts, which may favor access to detergents and decrease the number of LPS-PL junctions in the OM and hence permeability to large hydrophilic molecules, such as bacitracin. In keeping with this hypothesis, in the PS103 strain, hepta-acylation of lipid A was not observed (Fig. 3F). Nevertheless, since LPS transport was not restored to wild-type levels, as witnessed by persistence of lipid A decorated by PEtN and LPS distribution between the IM and the OM, the LPS layer would remain sufficiently weakened to allow diffusion of hydrophobic compounds.
The extragenic suppressor associated with PS103 is an amber mutation within the opgH reading frame, which encodes a Glucosyltransferase implicated in the biosynthesis of the so-called OPGs, a class of d-glucose oligosaccharides, heterogeneous in size and structure, found in the periplasm of all proteobacteria (47). OPGs, whose synthesis is induced by low osmolarity (46, 71), have been implicated in several processes, including chemotaxis, virulence, osmoregulation of OM protein expression, synthesis of CA, and resistance to SDS (49, 50, 72, 73), but the underlying mechanisms are poorly understood. Here, we showed that the opgH103 allele does not exert its effect by inducing CA production, even if inhibition of CA production in the ΔopgH strain carrying the lptA41 allele ameliorated OM permeability (Table 4, compare row 4 with row 7).
In addition to its enzymatic role in OPG biosynthesis, OpgH has been implicated in control of cell division as a UDP-glucose-activated inhibitor of FtsZ ring formation in E. coli (48). OpgH is an integral membrane protein that contains 8 transmembrane domains, with the N and C termini located in the cytoplasm (74). OpgH has three cytoplasmic domains encompassing residues 1 to 138 (termed the N domain), 211 to 514 (termed the M domain), and 702 to 848 (termed the C domain) (48). The model of OpgH functioning postulates that UDP-glucose binding to the M domain leads to a conformational change that promotes cell division inhibition mediated by the N domain (48). The opgH103 allele produces a truncated OpgH comprising the N domain and part of the M domain. Deletion of both opgH and the opgH103 nonsense mutation restores resistance to rifampin and, partially, to novobiocin in the lptA42 mutant, thus recapitulating the phenotype of the PS103 suppressor strain. opgH103 also appears to be recessive (strain PS111 with ectopic opgH103 is Rifs, albeit not fully Novs). It thus seems that the lack of OPGs in the periplasm and/or of the full-length OpgH in the IM makes the cell envelope of E. coli lptA42 less permeable to the lipophilic antibiotic rifampin and, to a lesser extent, to novobiocin, which is about 3-fold more lipophilic than rifampin (6). PS103 cells are smaller than the parental PS003 cells, and this phenotype seems to be correlated with the opgH103 allele, since the PS132 ΔopgH strain with ectopic opgH103 shared the same phenotype. Therefore, decreased cell size, and hence membrane surface, might help the cell to cope with impaired LPS transport of the lptA42 allele. Similarly, decreased cell size and growth rate due to alteration of fatty acid biosynthesis have been previously shown to suppress mutations that confer LPS transport defects (75).
In conclusion, in this work, we have identified two different mechanisms that fix, at least partially, the OM permeability barrier compromised in a mutant with defective LPS transport to the cell surface. In the case of the mlaA102 mutant, the suppression appears to be quite specific for the antibiotic used for the selection and relies on the peculiar function acquired by the mlaA102 gain-of-function allele that is associated with hypervesiculation. In the case of the PS103 suppressor, on the other hand, we found a more global mechanism based on the synergistic effect of LptA mutation/stabilization and modulation of the cell size associated with the opgH103 allele.
Overall, this work revealed different strategies adopted by the cell to preserve OM barrier integrity. An understanding of the molecular mechanisms underlying the permeability properties of the OM is fundamental for the design of novel antibacterial drugs and for the improvement of our current arsenal of antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used in this study are listed in Tables 6 and 7, respectively, with a brief outline of their construction by standard techniques. The oligonucleotides used in strain and plasmid construction are listed in Table 8. All the cloned DNA regions obtained by PCR were sequenced to rule out the presence of mutations. Site-directed λRed-mediated mutagenesis of E. coli was performed as described previously (76) and as specified in Table 6. ΔlptAB::kan DNA was obtained by three-step PCR using the external primers AP54-AP267 and, as templates, the kan cassette PCR amplified from pKD4 using AP79-AP80 primers and the two flanking homology regions obtained by PCR amplification of E. coli MG1655 DNA with oligonucleotide pairs AP54-AP268 and AP266-AP267. Transduction with P1 HFT (77) was performed as described previously (78). Unless otherwise stated, bacteria were grown at 30°C in LD (79) or M9 minimal medium supplemented with 0.2% glucose as a carbon source (80) and, when required, 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 100 μg/ml ampicillin, 30 μg/ml chloramphenicol, and 25 μg/ml kanamycin. Solid media were as described above with 1% (wt/vol) agar.
TABLE 6.
E. coli strains
| Strain | Parental strain | Relevant characteristicsa |
Constructionb | Source or reference | |
|---|---|---|---|---|---|
| Chromosomal | Plasmid | ||||
| AM604 | MC4100 | Ara+ | Spontaneous Ara+ revertant | 18 | |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | 87 | |||
| BW25113 | laclq rrnBT14 lacZWJ16 hsdR514 araBADAH33 rhaBADLD78 | 76 | |||
| DH10B | araD139Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR ϕ80dlacZΔM15 endA1 nupG recA1 mcrA(mrr hsdRMS mcrBC) | 88 | |||
| EM001 | JW2343 | ΔmlaA | By FLP-mediated excision of kan cassette | This work | |
| EM004 | BW25113/pACYC184-LptA LptB | ΔlptAB::kan | plac-lptA lptB; Ampr | By site-directed λRed-mediated mutagenesis with ΔlptAB::kan DNA | This work |
| FL907 | AM604 | AM604 ϕ(kan araC araBp-lptA)1 | 16 | ||
| JW2343 | BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔmlaA754::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | 89 | ||
| PS001 | AM604/pWSK29-LptA LptB | ΔlptAB::kan | plac-lptA lptB; Ampr | By P1 HFT*EM004 transduction, selection for Kanr | This work |
| PS003 | AM604/pWSK29-LptA41 LptB | ΔlptAB::kan | plac-lptA41 lptB; Ampr | By P1 HFT*EM004 transduction, selection for Kanr | This work |
| PS101 | PS003 | ΔlptAB::kan | plac-lptA41 lptB; Ampr | Bctr; spontaneous suppressor mutant | This work |
| PS102 | PS003 | mlaA102 ΔlptAB::kan | plac-lptA41 lptB; Ampr | Bctr, spontaneous suppressor mutant | This work |
| PS103 | PS003 | mdoH103 ΔlptAB::kan | plac-lptA42 lptB; Ampr | Bctr, spontaneous suppressor mutant | This work |
| PS107 | PS134/pWSK29-LptA41LptB | ΔmlaA ΔlptAB::kan | plac-lptA41 lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS109 | PS132/pWSK29-LptA41 LptB | ΔopgH ΔlptAB::kan | plac-lptA41 lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS110 | PS132/pWSK29-LptA42 LptB | ΔopgH ΔlptAB::kan | plac-lptA42 lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS111 | AM604/pWSK29-LptA42 LptB | ΔlptAB::kan | plac-lptA42 lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS112 | PS132/pWSK29-LptA LptB | ΔopgH ΔlptAB::kan | plac-lptA lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS113 | PS103 | opgH103 ΔlptAB::kan ΔwcaJ::cat | plac-lptA42 lptB; Ampr | By P1 HFT*PS135 transduction, selection for Catr | This work |
| PS114 | PS109 | ΔopgH ΔlptAB::kan ΔwcaJ::cat | plac-lptA41 lptB; Ampr | By P1 HFT*PS135 transduction, selection for Catr | This work |
| PS115 | PS110 | ΔopgH ΔlptAB::kan ΔwcaJ::cat | plac-lptA42 lptB; Ampr | By P1 HFT*PS135 transduction, selection for Catr | This work |
| PS116 | PS112 | ΔopgH ΔlptAB::kan ΔwcaJ::cat | plac-lptA lptB; Ampr | By P1 HFT*PS135 transduction, selection for Catr | This work |
| PS130 | PS134/pWSK29-LptA LptB | ΔmlaA ΔlptAB::kan | plac-lptA lptB; Ampr | By P1 HFT*PS001 transduction, selection for Kanr | This work |
| PS131 | AM604 | ΔopgH::cat | By site-directed λRed-mediated recombination; primers FG3116/FG3117, template pKD3 | This work | |
| PS132 | PS131 | ΔopgH | By FLP-mediated excision of cat cassette | This work | |
| PS133 | AM604 | ΔmlaA::kan | By P1 HFT*JW2343 transduction (selection for Kanr) | This work | |
| PS134 | PS133 | ΔmlaA | By FLP-mediated excision of kan cassette | This work | |
| PS135 | AM604 | ΔwcaJ::cat | By site-directed λRed-mediated recombination; primers FG3153-FG3154, template pKD3 | This work | |
| XL1-Blue | F− λ− recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZ ΔM15 Tn10(Tetr)] | Agilent Technologies | |||
Amp, ampicillin; Tet, tetracycline.
Kan, kanamycin; Cat, chloramphenicol acetyltransferase.
TABLE 7.
Plasmids
| Plasmid | Parental plasmid/replicon | Relevant characteristicsa | Construction, origin, or reference |
|---|---|---|---|
| pACYC184 | pSC101 ori p15A Camr Tetr | 90 | |
| pACYC184-LptA LptB | pACYC184 | plac-lptA lptB Camr | lptAB genes were excised from plasmid pWSK29-LptA LptB and subcloned into BamHI-SalI sites of pACYC184. |
| pCP20 | bla cat; thermosensitive replication | 76 | |
| pET-LptA41-H | pET21a | pT7-lptA41-His6 | lptA41 allele was PCR amplified with AP182 and AP183 and cloned into BamHI-NotI sites of pET-LptA-H. |
| pET-LptA42-H | pET21a | pT7-lptA42-His6 Ampr | lptA42 was obtained by site-directed mutagenesis with FG3191-FG3192 primers from pET-LptA41-H as the template. |
| pET-LptA-H | pET21a | pT7-lptA-His6 Ampr | 27 |
| pGS100 | pGZ119EH | ptac-TIR cat oriVColD | 91 |
| pGS100-OpgH | pGS100 | ptac-opgH Camr | opgH was PCR amplified with FG3069-FG3070 primers from PS003 genomic DNA and cloned into XbaI-PstI sites of pGS100. |
| pGS100-OpgH103 | pGS100 | ptac-opgH103 Camr | opgH103 was PCR amplified with FG3069-FG3070 primers from PS003 genomic DNA and cloned into XbaI-PstI sites of pGS100. |
| pGS100-MlaA | pGS100 | ptac-mlaA Camr | mlaA was PCR amplified with FG3067-FG3068 primers from PS003 genomic DNA and cloned into XbaI-PstI sites of pGS100. |
| pGS100-MlaA102 | pGS100 | ptac-mlaA102 Camr | mlaA102 was PCR amplified with FG3067-FG3068 primers from PS102 genomic DNA and cloned into XbaI-PstI sites of pGS100. |
| pGS103 | pGS100 | ptac-lptC Camr | 91 |
| pGS108 | pGS100 | ptac-lptC-His6 cat oriVColD | 16 |
| pKD3 | oriRγ Ampr Camr; source of cat cassette | 76 | |
| pKD4 | oriRγ Ampr Kanr; source of kan cassette | 76 | |
| pKD46 | oriR101 repA101ts araC araBp-λ red bla | 76 | |
| pWSK29 | pBSIISK | pSC101 ori f1 ori lacZa Ampr | 92 |
| pWSK29-LptA LptB | pWSK29 | plac-lptA41 lptB Ampr | 93 |
| pWSK29-LptA36 LptB | pWSK29-LptA LptB | plac-lptAI36A I38A lptB Ampr | By site-directed mutagenesis with primers AP184-AP185 |
| pWSK29-LptA37 LptB | pWSK29-LptA36 LptB | plac-lptA36R76D lptB Ampr | By site-directed mutagenesis with primers AP112-AP113 |
| pWSK29-LptA41 LptB | pWSK29-LptA37 LptB | plac-lptA37K83D lptB Ampr | By site-directed mutagenesis with primers AP186-AP187 |
| pWSK29-LptA42 LptB | pWSK29-LptA41 LptB | plac-lptA41M112I lptB Ampr | Spontaneous mutant |
Cam, chloramphenicol.
TABLE 8.
Oligonucleotides
| Name | Sequencea | Notes |
|---|---|---|
| AP54 | cgagaggaattcaccATGAGTAAAGCCAGACGTTGGG | ΔlptAB::kan cassette for EM004 construction by three-step PCR, with AP268; EcoRI |
| AP79 | GTGTAGGCTGGAGCTGCTTCG | Amplification kan cassette from pKD4, with AP80 |
| AP80 | CATATGAATATCCTCCTTAG | Amplification kan cassette from pKD4, with AP79 |
| AP112 | CAAAGTGGTCGTTACCGATCCGGGCGGCGAACAAGG | pWSK29-LptA41 LptB (R76D) construction, with AP113 |
| AP113 | CCTTGTTCGCCGCCCGGATCGGTAACGACCACTTTG | pWSK29-LptA41 LptB (R76D) construction, with AP112 |
| AP182 | ctcgacgcggccgcTATATACCCTTCTTCTGTG | pET-LptA41-H construction, with AP183; NotI |
| AP183 | cgagatggatccATGAAATTCAAAACAAACAAAC | pET-LptA41-H construction, with AP182; BamHI |
| AP184 | CACTGATCAGCCGGCCCACGCTGAATCGGACCAG | pWSK29-LptA41 LptB (I36A and I38A) construction, with AP185 |
| AP185 | CTGGTCCGATTCAGCGTGGGCCGGCTGATCAGTG | pWSK29-LptA41 LptB (I36A and I38A) construction, with AP184 |
| AP186 | GCGGCGAACAAGGTGATGAAGTGATTGACGGC | pWSK29-LptA41 LptB (K83D) construction, with AP187 |
| AP187 | GCCGTCAATCACTTCATCACCTTGTTCGCCGC | pWSK29-LptA41 LptB (K83D) construction, with AP186 |
| AP266 | ctaaggaggatattcatatgGATAGGGTAGAAGTTTGCG | ΔlptAB::kan cassette construction; kan hybrid primer for EM004 construction by three-step PCR, with AP267 |
| AP267 | CTAATGATCAGTCTGGCCTC | ΔlptAB::kan cassette for EM004 construction by three-step PCR, with AP266 |
| AP268 | gaagcagctccagcctacacGATTAAGGCTGAGTTTG | ΔlptAB::kan cassette construction; kan hybrid primer for EM004 construction by three-step PCR, with AP54 |
| AP354 | GTCATGGATGGCAAACTG | mlaA102-SPA::kan cassette construction; SPA-kan hybrid primer for EM002 construction by three-step PCR, with AP403 |
| AP387 | TCCATGGAAAAGAGAAG | Amplification of SPA::kan cassette from CAG60297, with AP388 |
| AP388 | CATATGAATATCCTCCTTAG | Amplification of SPA::kan cassette from CAG60297, with AP387 |
| AP400 | GGTATCGACAACCAAGAACC | mlaA102-SPA::kan cassette construction for EM002 construction by three-step PCR, with AP404 |
| AP403 | catcttctcttttccatggaTTCAGAATCAATATCTTTTAAATC | mlaA102-SPA::kan cassette construction; SPA-kan hybrid primer for EM002 construction by three-step PCR, with AP354 |
| AP404 | ctaaggaggatattcatatgGAAACAAATAAAAAAGGTG | mlaA102-SPA::kan cassette construction; SPA-kan hybrid primer for EM002 construction by three-step PCR, with AP400 |
| FG3067 | cgactagtctagaATGAAGCTTCGCCTGTC | pGS100-MlaA and pGS100-MlaA102 construction, with FG3068; XbaI |
| FG3068 | cgagatctgcagTTATTCAGAATCAATATC | pGS100-MlaA and pGS100-MlaA102 construction, with FG3067; PstI |
| FG3069 | cgactagtctagaATGAATAAGACAACTGAGTAC | pGS100-OpgH and pGS100-OpgH103 construction, with FG3070; XbaI |
| FG3070 | cgagatctgcagTTATTGCGAAGCCGCATC | pGS100-OpgH and pGS100-OpgH103 construction, with FG3069; PstI |
| FG3116 | gtgaaacctggagctaccagttacctgccaatgaataagGTGTAGGCTGGAGCTGCTTC | ΔopgH::cat cassette construction, with FG3117 |
| FG3117 | gtaggcctgataagcgtagcgcatcaggcaactacgttttCATATGAATATCCTCCTTAG | ΔopgH::cat cassette construction, with FG3116 |
| FG3153 | catcgttaatctctatggtgcaacgcttttcagatatcacGTGTAGGCTGGAGCTGCTTC | ΔwcaJ::cat cassette construction, with FG3154 |
| FG3154 | caggaaaacgattttgatatcgaaccagacgctccattcgCATATGAATATCCTCCTTAG | ΔwcaJ::cat cassette construction, with FG3153 |
| FG3191 | GTCACGCTTCCCAGATTCACTACGAACTGGC | pET-LptA42-H construction, with FG3192 |
| FG3192 | GCCAGTTCGTAGTGAATCTGGGAAGCGTGAC | pET-LptA42-H construction, with FG3191 |
Uppercase letters, sequence present in the template; lowercase letters, additional/modified sequence not present in the template; boldface letters, codons mutated by site-directed mutagenesis. Restriction sites are underlined.
Isolation of bacitracin-tolerant phenotypic revertants.
Independent overnight cultures of PS003 were grown from single colonies in 10 ml of LD medium supplemented with ampicillin, pelleted, and individually resuspended in 100 μl of LD before plating onto LD agar with bacitracin at the indicated concentrations. Upon overnight incubation at 37°C, Bctr phenotypic revertants were colony purified in the presence of bacitracin.
Outer membrane permeability assay.
OM sensitivity was evaluated by measuring the efficiency of plating (EOP) on LD agar plates containing bacitracin, rifampin, novobiocin, and SDS-EDTA at noninhibitory concentrations for the reference (wild-type) strain, as indicated. Overnight cultures were grown from single colonies in 5 ml LD supplemented with the antibiotic required for the maintenance of the hosted plasmid. Cultures were serially diluted in LD in microtiter plates, replica plated on the selective LD agar plates, and incubated overnight (or up to 24 h). The EOP was estimated relative to plating on LD agar.
Genomic-DNA sequencing and data analysis.
The library for genomic-DNA sequencing was prepared according to the TruSeq DNA sample preparation protocol (Illumina). Briefly, 1 μg of genomic DNA was sonicated into fragments with a median length of 400 bp; after end repair, indexed adapters were ligated at the DNA fragment ends, and the libraries were quantified by quantitative real-time PCR (qPCR) using Kapa Library Quant kits (Kapa Biosystems). After a short amplification step, the library was sequenced on an Illumina GAIIX sequence analyzer to generate 85-bp paired-end reads. The raw reads were individually mapped to the E. coli MC4100 genome (RefSeq accession number HG738867) using the accurate alignment BWA mem algorithm (81) allowing 1% error. Removal of duplicated reads was performed with SAMtools (82); only high-quality reads having mapping quality score (MAQ) values of >30 were used for the analysis of variant detection. A VCF file containing all the variants for each sample relative to E. coli MC4100 was obtained by using SAMtools and Bcftools (82) and filtered for low-quality variants. SNVs having coverage lower than five high-quality reads were discarded. Predicted indel mutations having coverage lower than six high-quality reads were discarded. Then, the VCF files were analyzed using SNPeff version 4.0 (83), and high-quality SNVs and indels were subsequently annotated to determine their effects and impacts on coding sequences.
Determination of LptA abundance.
LptA abundance was assessed by Western blot analysis. Bacterial cultures were grown overnight at 30°C in LD supplemented with 100 μg/ml of ampicillin. Samples for protein analysis were centrifuged for 5 min at 16,000 × g, and the pellets were resuspended in a volume (in milliliters) of SDS sample buffer equal to 1/24 of the total optical density (OD) of the sample. The samples were boiled for 10 min, and equal volumes (20 μl) were fractionated by SDS-12.5% polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (GE Healthcare), and Western blot analysis was performed as previously described (17). Polyclonal sera raised against LptA (GenScript Corporation) were used as primary antibodies at a dilution of 1:1,000. Goat anti-mouse immunoglobulins (Li-Cor) were used as secondary antibodies at a dilution of 1:7,000, and bands were visualized by an Odyssey Fc imaging system (Li-Cor GmbH). The band analysis tools of Image Studio Lite software version 5.0 (Li-Cor GmbH) were used to perform densitometric analysis of band intensity.
Lipid A analysis by mass spectrometry.
E. coli strains were grown in 1 liter of LD supplemented with 100 μg/ml of ampicillin up to an OD at 600 nm (OD600) of 0.9, yielding approximately 1 g of lyophilized biomass, at 30°C under different conditions, as specified. Cells were harvested by centrifugation at 4°C (5,000 × g; 15 min). The cell pellets were washed once with 20 ml of phosphate-buffered saline (PBS), 0.1 mM, pH 7.4, and lyophilized. Each pellet was treated twice with 10 ml of PCP solution (petroleum ether-chloroform-phenol, 90%; 2:5:8 [vol/vol/vol]) (84). For each strain, the two supernatants were removed at each passage by centrifugation (5,000 rpm; 10 min), pooled, and concentrated in a rotary evaporator to a final volume of approximately 3 ml. Lipooligosaccharide (LOS) precipitation occurred by dropwise addition of water (ca. 0.35 ml). The solid phase was recovered by centrifugation, dissolved in water, dialyzed (cutoff, 12,000 to 14,000 Da) against water for 2 days with five water changes, and freeze-dried. On average, 5 mg of pure LOS was obtained for each strain. LOS (0.5 to 1.5 mg) was dissolved in 0.5 ml of 1% acetic acid (AcOH) in water and heated at 100°C for 2 h to cleave the lipid A moiety that was recovered as a precipitate after centrifugation (5,000 rpm; 30 min) in 20 to 25% yield.
Reflectron MALDI-TOF MS of the lipid A fraction was performed, in negative-ion mode, on a 4800 Proteomic analyzer (Applied Biosystems) supplied with a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser (wavelength, 355 nm, with a <500-ps pulse and a 200-Hz firing rate). External calibration was performed using the ABSciex peptide calibration mixture, and mass accuracy was better than 50 ppm. The matrix solution was prepared by dissolving 2,4,6-trihydroxyacetophenone (THAP) in CH3OH-0.1% trifluoroacetic acid (TFA)-CH3CN (7:2:1 [vol/vol/vol]) at a concentration of 75 mg/ml. A sample-matrix solution mixture (1:1 [vol/vol]) was deposited (1 μl) onto a stainless-steel sample MALDI probe tip and dried at room temperature.
Affinity purification of membrane Lpt complexes.
Membrane Lpt complexes were affinity purified from strains expressing His-tagged LptC from pGS108 plasmids as previously described (23, 33) with a few modifications. Cells were lysed by a single cycle through a cell disruptor (One Shot model; Constant Systems Ltd.) at a pressure of 22,000 lb/in2, and the membranes were collected by ultracentrifugation of the supernatant at 100,000 × g for 2 h. The membranes were extracted at 4°C for 30 min with 5 ml of 50 mM Tris-HCl, pH 7.4, 10% glycerol, 1% dodecyl β-d-maltoside (DDM) (Sigma-Aldrich), and 5 mM MgCl2. The mixture was centrifuged again at 100,000 × g for 1 h, and insoluble material was discarded. The supernatant was incubated with 0.5 ml Talon resin suspension for 20 min at 4°C, and the mixture was then loaded onto a column, washed with 10 ml of 50 mM Tris-HCl, pH 7.4, 10% glycerol, 0.05% DDM, 20 mM imidazole, and eluted with 5 ml of 50 mM Tris-HCl, pH 7.4, 10% glycerol, 0.05% DDM, and 200 mM imidazole. The eluate was concentrated using an ultrafiltration device with a 10,000-Da cutoff (Vivaspin; GE Healthcare) by centrifugation at 7,000 × g to a final volume of 50 μl. Samples were mixed with 2× loading buffer, boiled, separated by SDS-12.5% PAGE (85), electroblotted, and immunodetected using anti-His monoclonal antibodies (1:3,000; Sigma-Aldrich) to detect LptC-H and anti-LptD (1:500) and anti-LptF (1:3,000; GenScript Corporation) and anti-LptB (1:10,000; kindly provided by D. Kahne and N. Ruiz) sera to detect the cognate proteins. For LptA detection, samples were fractionated by 15% Tricine SDS-PAGE and immunodetected using a 1:1,000 dilution of anti-LptA serum (GenScript Corporation). As secondary antibodies, goat anti-rabbit and anti-mouse immunoglobulins (Li-Cor) were used at a dilution of 1:15,000.
Protein expression and purification.
Cultures of E. coli strain BL21(DE3) carrying the pET vector expressing full-length LptA or LptA mutant proteins fused to a C-terminal tag (SGRVEH6) (27) were grown in M9 minimal medium to an OD600 of 0.6 at 30°C. Expression was induced by the addition of 0.5 mM IPTG (Sigma-Aldrich) and further incubation for 16 to 18 h at 20°C. Cells were harvested by centrifugation at 4°C (5,000 × g; 20 min). The cell pellets were resuspended in buffer A (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol), followed by 30 min incubation at 4°C with shaking in the presence of lysozyme (1 mg/ml), DNase (100 μg/ml), 10 mM MgCl2, and 1 mM phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich). The cells were disrupted as described above. Unbroken cells and cell debris were removed by centrifugation at 4°C (39,000 × g; 30 min). The soluble proteins were purified from the supernatant with Ni-nitrilotriacetic acid (Ni-NTA) affinity columns (Qiagen). The columns were washed with 10 column volumes (CV) of 4% buffer B (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 500 mM imidazole, 10% glycerol) in buffer A. Proteins were eluted by a 10%, 20%, 50%, 70%, and 100% buffer B stepwise gradient, at 1 CV per step. Fractions were analyzed by SDS-12.5% PAGE. Pooled fractions containing the purified protein were dialyzed against 20 mM Tris-HCl, pH 7.4, and 300 mM NaCl through cellulose membranes (10,000-Da cutoff; Sigma-Aldrich). The protein concentration was determined with a Coomassie (Bradford) assay kit (Thermo-Pierce), using bovine serum albumin as a standard.
LPS binding assay.
The in vitro LPS binding assay was based on a protocol described previously (40), with minor modifications. Briefly, assays (500 μl) were carried out in buffer C (50 mM sodium phosphate, pH 8, 50 mM NaCl) containing 25 μM purified His-tagged protein and a 5-fold molar excess of purified smooth LPS from E. coli serotype O55:B5 (Sigma-Aldrich; assumed mass, 10,000 Da). The reaction mixtures were incubated at room temperature for 1 h on a rotary shaker to allow the formation of LPS-protein complexes. Ni-NTA resin (200 μl; His-Select nickel affinity gel; Sigma-Aldrich), washed in 1 ml of buffer C, was added to the reaction mixtures and incubated for another hour to allow binding of the LPS-protein complexes. The reaction mixture was centrifuged at 13,000 × g for 1 min, and the supernatant (flowthrough [FT]) was collected. The resin was then washed four times with buffer C, and the protein-LPS complexes were eluted in two steps, with 500 μl of buffer C containing 300 mM imidazole (elution 1 [E1]) or 500 mM imidazole (E2), respectively. To monitor LPS-LptA complex formation, equal volumes (20 μl) of the collected chromatographic fractions were analyzed by denaturing gel electrophoresis. For LPS visualization, samples were fractionated by 18% Tricine SDS-PAGE (86) and immunodetected using a 1:3,000 dilution of the anti-LPS core WN1 222-5 monoclonal antibodies (HyCult Biotechnology b.v.). LptA was analyzed by SDS-12.5% PAGE and immunodetected using a 1:3,000 dilution of the anti-His monoclonal antibodies (Sigma-Aldrich).
Membrane fractionation.
IMs and OMs were separated by isopycnic sucrose gradient centrifugation as described previously (33). Proteins were separated by SDS-12.5% PAGE, followed by Coomassie blue staining to determine the porin profile, or immunoblotted using anti-LamB (a kind gift from T. J. Silhavy). This antibody cross-reacts with OmpA, LptD (GenScript Corporation), and LptB antibodies (kindly provided by N. Ruiz and D. Khane). For the LPS profile, fractions were analyzed by 18% Tricine SDS-PAGE and immunoblotted using anti-LPS WN1 222-5 monoclonal antibodies.
OM vesicle preparation.
OM vesicle isolation was performed as previously described (44). Briefly, for whole-cell lysate control samples, 1 ml of culture was collected and resuspended in a volume (in milliliters) of SDS sample buffer equal to 1/8 of the total OD of the sample. For OMV preparation, a total amount of 16 OD was pelleted (5,000 × g; 10 min; 4°C). Equal volumes of the culture supernatants were filtered through 0.22-μm filters to retain whole cells. OM vesicles were isolated and concentrated from the filtered supernatants by filtration through an Amicon Ultra-4 100K centrifugal filter (Millipore). Samples were resuspended from the 100K filter in 50 μl of SDS sample buffer. Whole-cell lysate and OM vesicle samples were boiled for 10 min, resolved by SDS-12.5% PAGE, and immunoblotted. Monoclonal antibodies against MBP (Thermo Fisher) were used at a dilution of 1:2,000, whereas polyclonal sera against LptE, LptB, and LamB (kindly provided by D. Khane, N. Ruiz, and T. J. Silhavy) were used at dilutions of 1:5,000, 1:10,000, and 1:5,000, respectively. As secondary antibodies, anti-goat, anti-rabbit, and anti-mouse immunoglobulins (Li-Cor) were used at a dilution of 1:15,000.
Determination of protein release in cell supernatants.
Equal amounts of cells were collected for 5 min at 16,000 × g. The pellets were resuspended in SDS sample buffer and boiled for 10 min. Cell-free supernatants were obtained by filtration through a 0.22-μm filter (Millipore), subsequently 20-fold concentrated with trichloroacetic acid, and boiled for 10 min. Protein samples were separated by SDS-12.5% PAGE and stained with Coomassie blue.
Microscopy.
Cells grown to an OD600 of 0.3 were fixed with 1.8% formaldehyde and 0.03% glutaraldehyde in PBS and incubated for 30 min at 30°C, with shaking. Cells were then harvested, washed with PBS, and resuspended in 0.4 ml of PBS. A cell suspension (5 μl) was spotted onto a microscope slide coated with a thin layer of 1% agarose. Images were acquired with a Zeiss Axiovert 200M microscope coupled with an AxioCam Mrm device camera (Zeiss) and with Metamorph imaging software (Universal Imaging). For membrane staining, cells were mounted on a slide coated with 1% agarose supplemented with the membrane dye FM5-95 (ThermoFisher) to a final concentration of 2 μg/ml. Images were analyzed with ImageJ (http://rsb.info.nih.gov/ij/).
Accession number(s).
The genomic data are available in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra), accession number PRJNA345432.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by MIUR-Regione Lombardia, project no. 30190679, Nuovi antibiotici mediante rational design (A. Polissi and G. Dehò); by MIUR PRIN no. 2012WJSX8K, Host-microbe interaction models in mucosal infections—development of novel therapeutic strategies (A. Polissi); and by a Train2Target project grant from the European Union's Horizon 2020 framework program for research and innovation (project no. 721484) (A. Polissi). A generous donation from the Stellalucente Trust is gratefully acknowledged (D. Garozzo).
We are grateful to Shu Sin Chng for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00487-17.
REFERENCES
- 1.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz N, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz NWT, Khane D, Silhavy TJ. 2006. Probing the barrier function of the outer membrane with chemical conditionality. ACS Chem Biol 1:385–395. doi: 10.1021/cb600128v. [DOI] [PubMed] [Google Scholar]
- 6.Young K, Silver LL. 1991. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol 173:3609–3614. doi: 10.1128/jb.173.12.3609-3614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker N. 2000. Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol 35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 8.Bishop RE, Guina T, Trent MS, Miller SI, Raetz CR. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J 19:5071–5080. doi: 10.1093/emboj/19.19.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thong S, Ercan B, Torta F, Fong ZY, Wong HY, Wenk MR, Chng SS. 2016. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5:e19042. doi: 10.7554/eLife.19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong ZS, Woo WF, Chng SS. 2015. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol 98:1133–1146. doi: 10.1111/mmi.13202. [DOI] [PubMed] [Google Scholar]
- 13.Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285. doi: 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chng S-S, Gronenberg LS, Kahne D. 2010. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol 189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperandeo P, Martorana AM, Polissi A. 2017. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim Biophys Acta 1862:1451–1460. doi: 10.1016/j.bbalip.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Okuda S, Freinkman E, Kahne D. 2012. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chimalakonda G, Ruiz N, Chng SS, Garner RA, Kahne D, Silhavy TJ. 2011. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chng SS, Ruiz N, Chimalakonda G, Silhavy TJ, Kahne D. 2010. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc Natl Acad Sci U S A 107:5363–5368. doi: 10.1073/pnas.0912872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freinkman E, Chng SS, Kahne D. 2011. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc Natl Acad Sci U S A 108:2486–2491. doi: 10.1073/pnas.1015617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 26.Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C. 2014. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511:52–56. doi: 10.1038/nature13464. [DOI] [PubMed] [Google Scholar]
- 27.Suits MDL, Sperandeo P, Dehò G, Polissi A, Jia Z. 2008. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein a and mutagenesis analysis. J Mol Biol 380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Bollati M, Villa R, Gourlay LJ, Benedet M, Dehò G, Polissi A, Barbiroli A, Martorana AM, Sperandeo P, Bolognesi M, Nardini M. 2015. Crystal structure of LptH, the periplasmic component of the lipopolysaccharide transport machinery from Pseudomonas aeruginosa. FEBS J 282:1980–1997. doi: 10.1111/febs.13254. [DOI] [PubMed] [Google Scholar]
- 29.Tran AX, Dong C, Whitfield C. 2010. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem 285:33529–33539. doi: 10.1074/jbc.M110.144709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Q, Yang X, Yu S, Shi H, Wang K, Xiao L, Zhu G, Sun C, Li T, Li D, Zhang X, Zhou M, Huang Y. 2017. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat Struct Mol Biol 24:469–474. doi: 10.1038/nsmb.3399. [DOI] [PubMed] [Google Scholar]
- 31.Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, Wojtowicz D, Barnard TJ, Gumbart JC, Buchanan SK. 2016. Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure 24:965–976. doi: 10.1016/j.str.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freinkman E, Okuda S, Ruiz N, Kahne D. 2012. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Deho G, Bolognesi M, Kahne D, Polissi A. 2013. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J Bacteriol 195:1100–1108. doi: 10.1128/JB.02057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santambrogio C, Sperandeo P, Villa R, Sobott F, Polissi A, Grandori R. 2013. LptA assembles into rod-like oligomers involving disorder-to-order transitions. J Am Soc Mass Spectrom 24:1593–1602. doi: 10.1007/s13361-013-0687-9. [DOI] [PubMed] [Google Scholar]
- 35.Merten JA, Schultz KM, Klug CS. 2012. Concentration-dependent oligomerization and oligomeric arrangement of LptA. Protein Sci 21:211–218. doi: 10.1002/pro.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma B, Reynolds CM, Raetz CR. 2008. Periplasmic orientation of nascent lipid A in the inner membrane of an Escherichia coli LptA mutant. Proc Natl Acad Sci U S A 105:13823–13828. doi: 10.1073/pnas.0807028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leive L. 1965. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun 21:290–296. doi: 10.1016/0006-291X(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 38.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J Biol Chem 274:18503–18514. [DOI] [PubMed] [Google Scholar]
- 40.Tran AX, Trent MS, Whitfield C. 2008. The LptA protein of Escherichia coli is a periplasmic lipid A-binding protein involved in the lipopolysaccharide export pathway. J Biol Chem 283:20342–20349. doi: 10.1074/jbc.M802503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prelich G. 1999. Suppression mechanisms. Trends Genet 15:261–266. doi: 10.1016/S0168-9525(99)01749-7. [DOI] [PubMed] [Google Scholar]
- 42.Ferenci T, Zhou Z, Betteridge T, Ren Y, Liu Y, Feng L, Reeves PR, Wang L. 2009. Genomic Sequencing reveals regulatory mutations and recombinational events in the widely used MC4100 lineage of Escherichia coli K-12. J Bacteriol 191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill TM, Tecklenburg ML, Pelletier AJ, Kuempel PL. 1989. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci U S A 86:1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Kennedy EP, Rumley MK. 1988. Osmotic regulation of biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J Bacteriol 170:2457–2461. doi: 10.1128/jb.170.6.2457-2461.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohin JP. 2000. Osmoregulated periplasmic glucans in proteobacteria. FEMS Microbiol Lett 186:11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x. [DOI] [PubMed] [Google Scholar]
- 48.Hill NS, Buske PJ, Shi Y, Levin PA. 2013. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet 9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajagopal S, Eis N, Bhattacharya M, Nickerson KW. 2003. Membrane-derived oligosaccharides (MDOs) are essential for sodium dodecyl sulfate resistance in Escherichia coli. FEMS Microbiol Lett 223:25–31. doi: 10.1016/S0378-1097(03)00323-9. [DOI] [PubMed] [Google Scholar]
- 50.Ebel W, Vaughn GJ, Peters HK III, Trempy JE. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J Bacteriol 179:6858–6861. doi: 10.1128/jb.179.21.6858-6861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumon C, Priem B, Martin SL, Heyraud A, Bosso C, Samain E. 2001. In vivo fucosylation of lacto-N-neotetraose and lacto-N-neohexaose by heterologous expression of Helicobacter pylori alpha-1,3 fucosyltransferase in engineered Escherichia coli. Glycoconj J 18:465–474. doi: 10.1023/A:1016086118274. [DOI] [PubMed] [Google Scholar]
- 52.Patel KB, Toh E, Fernandez XB, Hanuszkiewicz A, Hardy GG, Brun YV, Bernards MA, Valvano MA. 2012. Functional characterization of UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferases of Escherichia coli and Caulobacter crescentus. J Bacteriol 194:2646–2657. doi: 10.1128/JB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowyer A, Baardsnes J, Ajamian E, Zhang L, Cygler M. 2011. Characterization of interactions between LPS transport proteins of the Lpt system. Biochem Biophys Res Commun 404:1093–1098. doi: 10.1016/j.bbrc.2010.12.121. [DOI] [PubMed] [Google Scholar]
- 54.Schultz KM, Lundquist TJ, Klug CS. 2017. Lipopolysaccharide binding to the periplasmic protein LptA. Protein Sci 26:1517–1523. doi: 10.1002/pro.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laguri C, Sperandeo P, Pounot K, Ayala I, Silipo A, Bougault CM, Molinaro A, Polissi A, Simorre JP. 2017. Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci Rep 7:9715. doi: 10.1038/s41598-017-10136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benedet M, Falchi FA, Puccio S, Di Benedetto C, Peano C, Polissi A, Deho G. 2016. The lack of the essential LptC protein in the trans-envelope lipopolysaccharide transport machine is circumvented by suppressor mutations in LptF, an inner membrane component of the Escherichia coli transporter. PLoS One 11:e0161354. doi: 10.1371/journal.pone.0161354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaara M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother 37:2255–2260. doi: 10.1128/AAC.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nikaido H. 2005. Restoring permeability barrier function to outer membrane. Chem Biol 12:507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 59.Jia W, Zoeiby AE, Petruzziello TN, Jayabalasingham B, Seyedirashti S, Bishop RE. 2004. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem 279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 60.Gibbons HS, Kalb SR, Cotter RJ, Raetz CR. 2005. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol Microbiol 55:425–440. doi: 10.1111/j.1365-2958.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 61.Gunn JS, Miller SI. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol 178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM, Fang FC. 2014. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun 82:660–669. doi: 10.1128/IAI.01057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martorana AM, Motta S, Di Silvestre D, Falchi F, Deho G, Mauri P, Sperandeo P, Polissi A. 2014. Dissecting Escherichia coli outer membrane biogenesis using differential proteomics. PloS One 9:e100941. doi: 10.1371/journal.pone.0100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manning AJ, Kuehn MJ. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol 11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson BA, Anker H, Meleney FL. 1945. Bacitracin: a new antibiotic produced by a member of the B. subtilis group. Science 102:376–377. doi: 10.1126/science.102.2650.376. [DOI] [PubMed] [Google Scholar]
- 66.van Heijenoort J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep 18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 67.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pollock TJ, Thorne L, Yamazaki M, Mikolajczak MJ, Armentrout RW. 1994. Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides. J Bacteriol 176:6229–6237. doi: 10.1128/jb.176.20.6229-6237.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cain BD, Norton PJ, Eubanks W, Nick HS, Allen CM. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J Bacteriol 175:3784–3789. doi: 10.1128/jb.175.12.3784-3789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He F, Xu J, Wang J, Chen Q, Hua X, Fu Y, Yu Y. 2016. Decreased susceptibility to tigecycline mediated by a mutation in mlaA in Escherichia coli strains. Antimicrob Agents Chemother 60:7530–7531. doi: 10.1128/AAC.00917-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lacroix JM, Loubens I, Tempete M, Menichi B, Bohin JP. 1991. The mdoA locus of Escherichia coli consists of an operon under osmotic control. Mol Microbiol 5:1745–1753. doi: 10.1111/j.1365-2958.1991.tb01924.x. [DOI] [PubMed] [Google Scholar]
- 72.Fiedler W, Rotering H. 1988. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem 263:14684–14689. [PubMed] [Google Scholar]
- 73.Bontemps-Gallo S, Lacroix JM. 2015. New insights into the biological role of the osmoregulated periplasmic glucans in pathogenic and symbiotic bacteria. Environ Microbiol Rep 7:690–697. doi: 10.1111/1758-2229.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Debarbieux L, Bohin A, Bohin JP. 1997. Topological analysis of the membrane-bound glucosyltransferase, MdoH, required for osmoregulated periplasmic glucan synthesis in Escherichia coli. J Bacteriol 179:6692–6698. doi: 10.1128/jb.179.21.6692-6698.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. 2012. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci U S A 109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wall JD, Harriman PD. 1974. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 59:532–544. doi: 10.1016/0042-6822(74)90463-2. [DOI] [PubMed] [Google Scholar]
- 78.Miller RC., Jr 1972. Asymmetric annealing of an RNA linked DNA molecule isolated during the initiation of bacteriophage T7 DNA replication. Biochem Biophys Res Commun 49:1082–1086. doi: 10.1016/0006-291X(72)90323-3. [DOI] [PubMed] [Google Scholar]
- 79.Ghisotti D, Chiaramonte R, Forti F, Zangrossi S, Sironi G, Deho G. 1992. Genetic analysis of the immunity region of phage-plasmid P4. Mol Microbiol 6:3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- 80.Kunz DA, Chapman PJ. 1981. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWWO) plasmid. J Bacteriol 146:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Baets G, Van Durme J, Reumers J, Maurer-Stroh S, Vanhee P, Dopazo J, Schymkowitz J, Rousseau F. 2012. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res 40:D935–D939. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galanos C, Luderitz O, Westphal O. 1969. A new method for the extraction of R lipopolysaccharides. Eur J Biochem 9:245–249. doi: 10.1111/j.1432-1033.1969.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 85.Laemmli UK, Beguin F, Gujer-Kellenberger G. 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J Mol Biol 47:69–85. doi: 10.1016/0022-2836(70)90402-X. [DOI] [PubMed] [Google Scholar]
- 86.Lesse AJ, Campagnari AA, Bittner WE, Apicella MA. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Immunol Methods 126:109–117. doi: 10.1016/0022-1759(90)90018-Q. [DOI] [PubMed] [Google Scholar]
- 87.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 88.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartolome B, Jubete Y, Martinez E, de la Cruz F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75–78. doi: 10.1016/0378-1119(91)90541-I. [DOI] [PubMed] [Google Scholar]
- 91.Sperandeo P, Pozzi C, Deho G, Polissi A. 2006. Non-essential KDO biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res Microbiol 157:547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 92.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 93.Santambrogio C, Sperandeo P, Barbieri F, Martorana AM, Polissi A, Grandori R. 2015. An induced folding process characterizes the partial-loss of function mutant LptAI36D in its interactions with ligands. Biochim Biophys Acta 1854:1451–1457. doi: 10.1016/j.bbapap.2015.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.