ABSTRACT
A network of genes and at least two peptide signaling molecules tightly control when Streptococcus mutans becomes competent to take up DNA from its environment. Widespread changes in the expression of genes occur when S. mutans is presented with competence signal peptides in vitro, including the increased production of the alternative sigma factor, ComX, which activates late competence genes. Still, the way that gene products that are regulated by competence peptides influence DNA uptake and cellular physiology are not well understood. Here, we developed and employed comprehensive transposon mutagenesis of the S. mutans genome, with a screen to identify mutants that aberrantly expressed comX, coupled with transposon sequencing (Tn-seq) to gain a more thorough understanding of the factors modulating comX expression and progression to the competent state. The screens effectively identified genes known to affect competence, e.g., comR, comS, comD, comE, cipB, clpX, rcrR, and ciaH, but disclosed an additional 20 genes that were not previously competence associated. The competence phenotypes of mutants were characterized, including by fluorescence microscopy to determine at which stage the mutants were impaired for comX activation. Among the novel genes studied were those implicated in cell division, the sensing of cell envelope stress, cell envelope biogenesis, and RNA stability. Our results provide a platform for determining the specific chemical and physical cues that are required for genetic competence in S. mutans, while highlighting the effectiveness of using Tn-seq in S. mutans to discover and study novel biological processes.
IMPORTANCE Streptococcus mutans acquires DNA from its environment by becoming genetically competent, a physiologic state triggered by cell-cell communication using secreted peptides. Competence is important for acquiring novel genetic traits and has a strong influence on the expression of virulence-associated traits of S. mutans. Here, we used transposon mutagenesis and genomic technologies to identify novel genes involved in competence development. In addition to identifying genes previously known to be required for comX expression, 20 additional genes were identified and characterized. The findings create opportunities to diminish the pathogenic potential of S. mutans, while validating technologies that can rapidly advance our understanding of the physiology, biology, and genetics of S. mutans and related pathogens.
KEYWORDS: ComX/SigX, transposon mutagenesis, single-cell behaviors, peptide signals, stress tolerance
INTRODUCTION
Genetic competence, the process by which bacteria are able to internalize DNA, is a trait shared across bacterial phyla (1). The acquisition of the state of natural genetic competence is transient, and under most conditions, the genes encoding products required for DNA uptake are not expressed. The activation of the transcription of these genes occurs in response to specific signals and, even then, only when environmental conditions are permissive. In the Gram-positive streptococci, the competence machinery is typically regulated by externalized peptides, which when present, enhance the production of ComX (also known as SigX), an alternative sigma factor that associates with RNA polymerase and enables the recognition of promoters of late competence genes (2). ComX is essential for competence development and is highly conserved in competent streptococci. However, multiple other networks influence the expression of comX or the levels of ComX or turn off the competence regulon, and these can differ substantially between species (2, 3). Genetic competence regulation in Streptococcus mutans, an etiologic agent of dental caries, is particularly interesting because it relies on two genetic circuits, including one that is integrated with the production of bacteriocins, which appear to be critical for the establishment and persistence of S. mutans in human oral biofilms (2).
S. mutans produces at least two signal peptides that can activate cells to develop competence, namely, competence-stimulating peptide (CSP) and comX-inducing peptide (XIP). The addition of synthetic CSP or XIP to early-exponential-phase cultures of S. mutans can lead to enhanced expression of comX (4). The activation of comX expression can occur directly when XIP, an extracellular 7-amino-acid (aa) peptide derived from its 17-aa precursor, ComS, is internalized and forms a complex with the transcriptional regulator ComR (5). The ComR-XIP complex binds directly upstream of the promoters for comX and comS. In the case of CSP, this 18-aa peptide is derived from ComC by secretion and processing. Externalized CSP interacts with the histidine kinase ComD, which leads to the phosphorylation of the response regulator ComE (ComE∼P). ComE∼P directly activates the expression of a variety of genes encoding bacteriocins and products involved in bacteriocin biogenesis and immunity (6). Although ComE∼P does not directly activate the transcription of comX, the provision of CSP to S. mutans at levels as low as 30 nM can increase the transformation efficiency by as much as 1,000-fold, and higher concentrations of CSP trigger increased comX mRNA and ComX protein levels (4, 7). The mechanism by which CSP activates the competence cascade is poorly understood, but it requires ComRS and may involve the bacteriocin CipB; transcription of cipB is activated by the binding of ComE∼P to the cipB promoter region (6). In addition to the ComRS and CSP-ComDE circuits, many other gene products and environmental inputs can influence comX expression and ComX protein levels, and the effects of these factors differ depending on the signal peptide used and the medium composition. Among the factors that have the greatest influence on S. mutans competence gene expression and the transition to, or exit from, the competent state are (p)ppGpp levels modulated by RelA (8), the products of the rcrRPQ operon (9, 10), the pH of the local environment (11), the carbohydrate source (12), the exposure to oxygen (13), the cell density (14), and the growth in biofilms versus planktonic culture (15). It is also relevant that CSP induces comX expression and competence only in rich (peptide-containing) media, whereas XIP is effective only in chemically defined media lacking peptides (4).
The regulons of ComE∼P, ComR-XIP, and ComX have been explored using microarrays and transcriptome sequencing (RNA-Seq) (16–19). In S. mutans, ComE∼P controls the expression of 4 operons, ComR-XIP controls 2 operons, and ComX affects the expression of 21 operons (16). Collectively, these studies constitute a valuable foundation for understanding how CSP and XIP influence the transcriptome. Still, there are some significant gaps in our understanding of the regulation of genetic competence in S. mutans, including how XIP is processed and externalized, the way in which the ComCDE pathway stimulates the ComRS circuit to activate competence, the mechanisms by which (p)ppGpp exerts its influence on many aspects of competence, and how the ABC transporters and peptides encoded in the rcrRPQ operon so profoundly influence comX-xrpA gene expression and the ability of S. mutans to be transformed. Other significant deficiencies in our knowledge include how tolerance of environmental stress and biofilm formation are integrated with regulators of the competence network and how lytic behaviors are coordinated by inputs from stress tolerance and competence systems. While functional genomics and approaches employing next-generation sequencing (e.g., transcriptomics) are helping to understand the complexities of competence and its integration with the biology of S. mutans, complementary approaches are still needed to close the gaps in knowledge noted above. Interestingly, there have been no classical genetic screens performed with the aim of identifying genes that affect genetic competence in S. mutans or that may integrate competence with other networks that are germane to the establishment, persistence, and virulence of the organism. One reason for this may be that transposon mutagenesis techniques that achieve unbiased saturation of the genome of S. mutans have not been available, in contrast to the systems available for many Gram-negative organisms and for certain Gram-positive bacteria, such as Streptococcus pneumoniae and Bacillus subtilis (20, 21).
Here, we employed classical transposon mutagenesis and screening, coupled with transposon sequencing (Tn-seq), to examine the competence regulon and how it may be integrated with pathways that govern physiologic homeostasis. Virtually all previously known regulators of genetic competence in S. mutans were identified in the screen, as were an additional 20 strains with insertions in coding or noncoding regions of the genome that had not previously been associated with competence. This study also validates Tn-seq as a highly effective addition to the technologies available for genetic studies with S. mutans.
RESULTS
Two screens for genes affecting competence in S. mutans.
To develop a more comprehensive overview of the gene products that may influence comX promoter activity, a transposon insertion library was constructed in an otherwise wild-type genetic background in S. mutans UA159 carrying a fusion of a Streptococcus salivarius lacZ gene to the comX promoter (PcomX-lacZ) (12). Using in vitro transposon mutagenesis (22), a mariner-based transposon library was generated that contained 2.5 × 104 apparently random insertions, which is roughly equivalent to a collection of mutants that carried transposon insertions every 80 bp in the 2 megabase genome of S. mutans UA159. A Southern blot analysis of eight mutants using a probe specific for the aad9 spectinomycin resistance gene in the transposon showed that the mutants had single magellan6 insertions (see Fig. S1 in the supplemental material). Sanger sequencing of PCR products, generated using a transposon-specific primer in conjunction with random primers (arbitrary PCR), from 168 individual PcomX-lacZ defective mutants always gave unique results, providing further evidence that multiple insertions rarely occurred in each mutant (data not shown). The library was screened on tryptone-yeast extract-glucose (TYG) agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), on which the PcomX-lacZ strain formed blue colonies after 40 h of incubation. Importantly, the deletion of the comR gene (ΔcomR mutant), with ComR being required for transcriptional activation of comX, in the PcomX-lacZ strain yielded gray colonies on TYG–X-Gal plates (see Fig. S2). Additionally, cipB expression in colony biofilms was observed using green fluorescent protein (GFP) reporters and confocal laser scanning microscopy, indicating that the CSP-ComDE circuit was active under the conditions tested (Fig. 1A). A total of 233 PcomX-lacZ magellan6 insertion mutants (which we designated mutants aberrant in competence [mac] strains) were isolated on the basis of the formation of colonies that appeared white on TYG–X-Gal plates (see Fig. 1B for an example of the appearance of a colony with reduced lacZ expression). All mac strains were assayed to determine if they displayed phenotypes with reduced transformation efficiencies (Fig. 1C) and decreased sensitivity to an inhibition of growth when CSP was included in the medium. Of the original 233, 168 mac strains met these initial criteria. By the use of PCR and sequencing, the insertion of the transposons in these 168 strains was determined to have occurred in 55 distinct coding and noncoding regions of the genome. Table S1 shows that our screen identified most of the genes that have been shown to influence comX expression previously: comR, comS, rcrR, clpX, ciaH, cipB, comD, and comE. These strains were also resistant to growth inhibition by 2 μM CSP and either displayed very low transformation efficiencies or could not be transformed, except for clpX and ciaH mutants that were resistant to CSP but displayed only modestly diminished transformation efficiencies. That we could discover previously identified genetic competence regulators validated our methodology; thus, we characterized the remaining mutants carrying transposons inserted in the 40 coding and noncoding regions that represented novel regulators of comX expression.
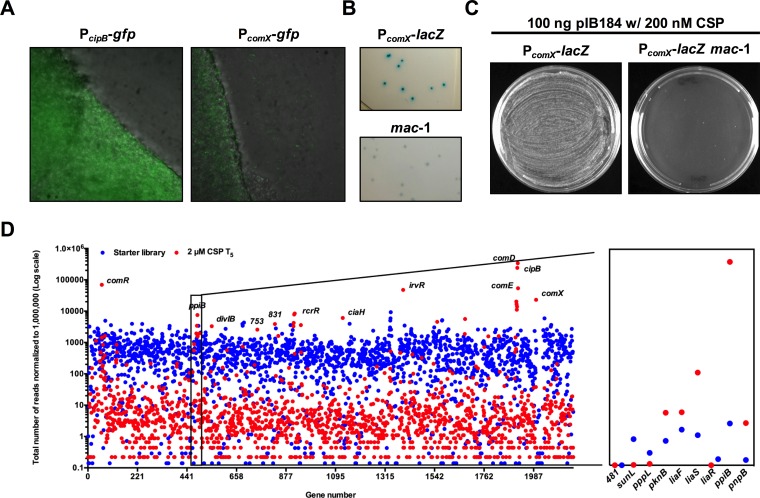
FIG 1.
PcomX-lacZ and Tn-seq screens for novel genes required for optimal comX expression. (A) Photograph of otherwise wild-type S. mutans organisms carrying PcipB-gfp or PcomX-gfp on the surface of TYG agar. Fluorescent strains were spotted (10 μl) on TYG agar and incubated for 24 h before the images were obtained on a fluorescence microscope using a 20× objective. (B) Example of the appearance of wild-type PcomX-lacZ activity (blue colonies) and a transposon mutant (mac-1) with reduced PcomX-lacZ activity (light-blue colonies) on the surface of TYG–X-Gal agar. (C) Screening for transformation defects in transposon strains (mac-1 shown) compared to wild-type S. mutans. Wild-type S. mutans and mac strains were incubated in BHI broth with 200 nM CSP and plasmid pIB184 (Ermr) to monitor DNA uptake after plating 100 μl of the transformation sample on BHI-erythromycin agar. (D) Tn-seq results summarized using a scatterplot that shows changes in the numbers of transposon insertions in S. mutans genes before and after growth in 2 μM CSP. Reads were normalized to 1 million reads per sample. Inset chart displays insertions in genes from SMu.481 to pnpB. The complete list of insertions that were overrepresented following passage in BHI-CSP can be found in Table S2 in the supplemental material.
In addition to the high degree of randomness of insertion of the magellan6 transposon and the ability to rapidly determine insertion sites, another benefit of this transposon library was the ability to perform deep sequencing (Tn-seq) on populations prior to and after growth in selected in vitro or in vivo environments, thereby enabling the enumeration of mutants before and after an imposed challenge to the population. Tn-seq was used here to identify genes that influenced the fitness when cells were exposed to 2 μM CSP. The sequencing of the starting library prior to selection showed that the insertions were randomly distributed across the genome, except for genes that were essential for growth under the conditions used (Fig. 1D) (R. C. Shields, L. Zeng, D. J. Culp, and R. A. Burne, in preparation). The library was inoculated in brain heart infusion (BHI) broth and BHI broth containing 2 μM CSP and passaged 5 times. More specifically, fresh medium was inoculated with 107 cells from the initial library and grown to an optical density at 600 nm (OD600) of 1.0 (∼2 × 109 cells), and then the culture was diluted 1:100 in fresh medium and cultured to an OD600 of 1.0 four more times, corresponding to approximately 30 generations. The hypothesis was that mutants carrying transposon insertions that caused diminished comX expression or that disrupted the connection between the CSP-ComDE circuit and those pathways that elicit CSP-dependent growth inhibition and/or lysis would be present in greater proportions in the library after passage with CSP (6, 16) than in the library grown in BHI broth alone. As with the X-Gal screen, the strains carrying Tn insertions in genes known to affect comX expression were identified as overrepresented after passage with CSP (Fig. 1D; see also Table S2). The strongest positively selected transposon mutants (log2 fold change greater than 0.5) were those with Tn insertions in the cipB (bacteriocin) and comDE operons. Enhanced fitness was also observed for strains with Tn insertions in irvR, comR, comX, rcrR, and ciaH, which are all known to impact competence, as well as in 28 coding regions not previously reported to influence comX expression, competence, or sensitivity to CSP.
Validation of mutations affecting CSP-induced genetic competence.
Across the two screens, 37 coding and noncoding regions were identified that were deemed to warrant further investigation. Three genes, pknB, liaS, and irvR, were previously linked to genetic competence (23–26) but have not been extensively characterized, and so these mutants were also examined. In total, 38 genes and 2 intergenic regions were deleted and replaced with nonpolar kanamycin resistance cassettes. Phenotypic characterization of these defined allelic exchange mutants began with the monitoring of their growth characteristics in the presence of CSP or XIP. At high concentrations of exogenously supplied peptides, both CSP and XIP caused growth arrest and/or lysis of S. mutans UA159, phenotypes that have been shown to be dependent on ComX (25, 27). Mutant strains displayed a wide range of growth behaviors in response to CSP during growth (Fig. S3A). For example, in the presence of CSP, the final OD600 of all mutants was significantly higher than for the wild type (P = 0.0004). There was also variability in the durations of the lag phase and exponential growth rates of the mutants. To begin to interpret the basis for these various behaviors, a phenotypic clustering model was developed that could be used to predict potential functional interrelationships between genes (Fig. 2A; see also Text S1), with the rationale that if growth phenotypes were similar between mutants in an environment, then the gene products that clustered together in the model may participate in a common pathway or function. The model took into consideration the lag phase duration, exponential growth rate, final yield (maximal OD600), and other parameters (Text S1). Using this approach, we discovered putative gene-gene interactions between known genetic competence regulators and the newly discovered genes (Fig. 2A), and previously established interactions were also predicted. For instance, comD and comDE mutants shared growth characteristics with the cipB mutant, consistent with the fact that cipB is directly regulated by phosphorylated ComE (16). In addition, there was a strong connection between liaS and pspC mutant strains. LiaS is a sensor kinase of a two-component system (TCS), and its cognate response regulator, LiaR, binds to the promoter region of the pspC gene in vitro (28). As another example, our clustering model indicated that a ΔcomS mutant shared phenotypic behaviors with ΔciaH, ΔpspC, ΔliaS, and ΔrgpI mutants. In Fig. S3B, we plotted the ratios of the relative growth rates of mutants in BHI broth versus those in BHI broth with 2 μM CSP. The strongest phenotypes were observed for strains with mutations in genes involved in the early stages of the competence cascade, whereas the behaviors of strains carrying mutations in genes for the latter phases of competence showed less profound phenotypes.
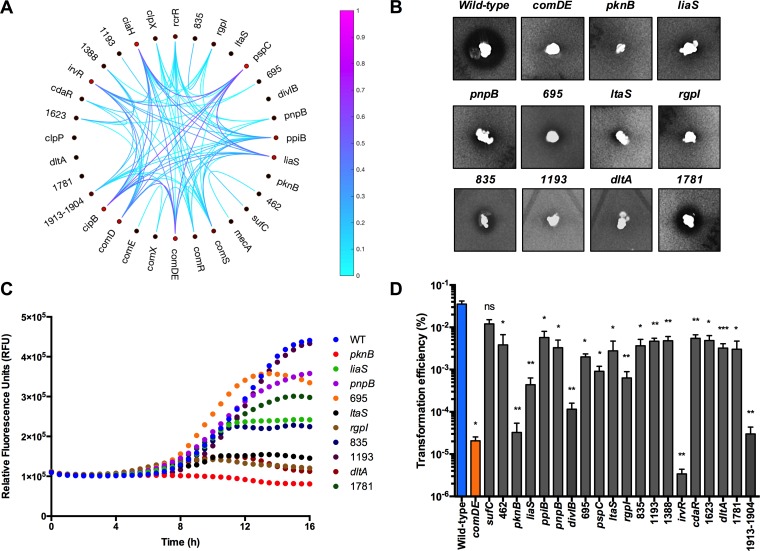
FIG 2.
Discovery of mutant strains with aberrant responses to CSP. (A) Phenotypic clustering of growth characteristics in the presence of 2 μM CSP (see Text S1 in supplemental material for criteria and analytical methods). Links between genes (lines) are based on correlations between phenotypes in the presence of 2 μM CSP. Genes with increased numbers of connections are highlighted by red circles. Additionally, values of linkages (denoted by the scale bar) were determined from the normalized distance matrix discussed in Text S1. (B) Deferred antagonism assay to monitor bacteriocin production by wild-type S. mutans, S. mutans ΔcomDE, and various mutant strains using Streptococcus gordonii DL1 as the indicator strain. Clear zones produced around the stabbed S. mutans colonies indicate lysis of the indicator strain (S. gordonii). (C) PcipB-gfp activity was measured in defined medium (FMC) after the addition of 200 nM CSP. Fluorescence of the PcipB-gfp reporter was measured every 30 min for 16 h using a Synergy microplate reader. Natural PcipB-gfp expression occurs in the wild-type strain during the exponential growth phase. Raw fluorescence readings are plotted without any subtraction for background fluorescence. (D) Transformation efficiencies of wild-type and mutant strains. Cells were grown to an OD600 of 0.1, and 200 nM CSP along with 100 ng pIB184 (Ermr) were added; cells were incubated for 2.5 h and then plated on BHI-erythromycin agar and BHI agar. Bars represent the mean values derived from 3 or 4 biological replicates, with error bars denoting the standard errors. Two-sample t tests were performed to determine statistical significance. *, P < 0.05; **, P < 0.01, ***, P < 0.001; ns, not significant.
The primary function of the CSP-ComDE circuit of S. mutans appears to be to control the production of bacteriocins. However, the expression and activity of bacteriocins, particularly CipB (6), have also been linked to comX expression via ComRS, although the mechanism is undefined. Therefore, we chose to examine cipB expression and bacteriocin production to begin to probe why certain mutants exhibited competence defects. To evaluate bacteriocin production by the panel of mutants identified above, S. mutans UA159 and the deletion replacement mutants were stabbed into BHI agar, and then a soft agar overlay containing either of two sensitive oral commensal microorganisms, Streptococcus gordonii or Streptococcus sanguinis, was poured evenly onto the plates. As deduced from the sizes of the zones of inhibition of growth of the commensals by S. mutans, 11 mutants displayed statistically significant reductions in inhibitory effects against S. gordonii and nine against S. sanguinis (Fig. 2B; see also Fig. S4). In general, the mutant strains exhibited the most significant reductions in inhibition of S. gordonii, as opposed to S. sanguinis. Additional studies will be needed to know if this is related to the unequal impacts on expression of different bacteriocins and/or to the sensitivities of the mutants to antagonistic factors produced by the commensal streptococci. To investigate whether these mutants had reduced bacteriocin gene expression, a PcipB-gfp reporter was introduced into each null strain, the strains were incubated in the presence of CSP (200 nM), and GFP fluorescence measurements were obtained in a microplate reader (Fig. 2C). PcipB-gfp activity was greatly reduced in the ΔpknB, ΔltaS, ΔrgpI, and ΔdltA mutants and moderately reduced in strains carrying deletions of S. mutans 835 (SMu.835) and liaS.
To assess the transformation efficiency for the deletion mutants, we incubated early-exponential-phase cells growing in BHI broth with CSP (200 nM) and 100 ng of plasmid pIB184. For cells treated with CSP, 19 mutants exhibited reduced transformation efficiencies, ranging from 6-fold to over 10,000-fold, compared to those of S. mutans UA159 (Fig. 2D). The deletion mutants with the lowest transformation efficiencies under the conditions tested were ΔpknB, ΔdivIB, ΔirvR, and ΔSMu.1913 to SMu.1904 (Δ1913-1904) mutants.
Identifying genes that are involved in XIP-mediated genetic competence.
A major question that remains unresolved is how the treatment of cells with CSP results in enhanced transformability and induction of comX expression. CSP will not activate competence in a comX or comR mutant, and CipB may play some role in connecting the pathways, but details of how the CSP-ComDE circuit activates the ComRS pathway are lacking. We hypothesized that, among the mutants, there may be isolates that displayed diminished competence because they could no longer couple CSP signaling to the ComRS pathway. Thus, we investigated whether any of the mutants had alterations in XIP-induced genetic competence, which requires the ComR circuit and is most efficient when the comS positive feedback loop is intact (4). We began by monitoring the growth of the mutant strains in the presence of 2 μM XIP in chemically defined medium (FMC). The most substantial changes in the sensitivity to inhibition of growth by XIP were seen with the ΔirvR, ΔltaS, ΔrgpI, and Δ835 deletion mutants, but another 9 strains exhibited an increased final OD600 compared with that of the wild-type strain (Fig. 3A). Certain mutants, namely, ΔsufC, Δ462, ΔpknB, and ΔdltA mutants, grew more poorly than the wild-type strain in FMC alone (see Fig. S5). When cultured in CDM, a chemically defined medium with a composition different from that of FMC, only the Δ462 mutant displayed a noticeable growth defect, although not as substantial as the defect seen in FMC (Fig. S5). The principal albeit not the only difference between the media is that CDM has substantially more phosphate and, as a result, a better buffer capacity than FMC.
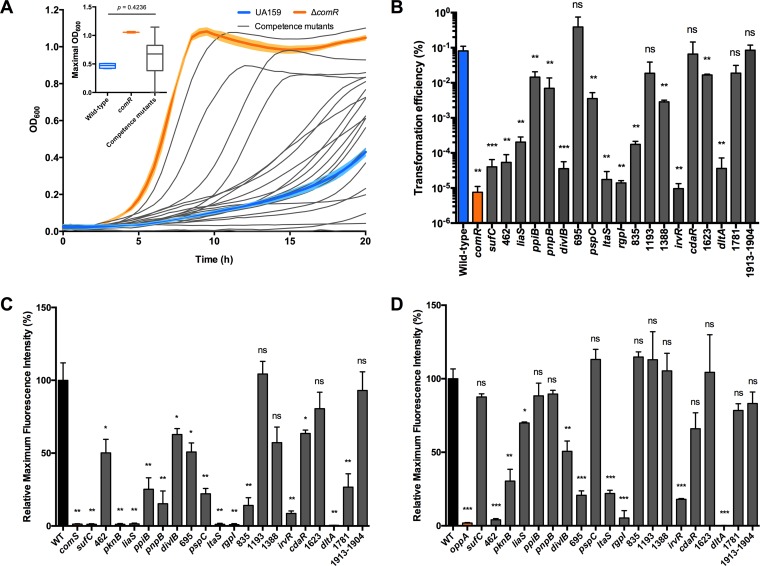
FIG 3.
Newly identified competence mutants also exhibit comRS-dependent phenotypes. (A) Growth of S. mutans UA159, S. mutans ΔcomR, and competence mutant strains in the presence of 2 μM XIP monitored at an OD600. Data points are averages from triplicate samples with the standard errors shown for UA159 and ΔcomR strains. The maximal OD600 achieved by the competence mutants showed considerable variability (inset), with the wild type and a comR mutant as reference strains. (B) Comparison of transformation efficiencies between wild-type S. mutans, S. mutans ΔcomR, and newly identified competence mutants. Strains were grown in FMC using 200 nM XIP to activate transformation and 100 ng of purified pBGS plasmid (Spr) to identify mutants no longer able to take up DNA at the same efficiency as the wild type. (C and D) Strains harboring a PcomX-gfp reporter plasmid were grown in CDM (C) and CDM containing 200 nM exogenous XIP (D). Maximal green fluorescence relative to that of the wild-type strain is shown. Bars indicate means from three biological replicates, with error bars denoting standard errors from the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
When XIP was used to induce competence in S. mutans growing in FMC, 14 of the deletion mutants exhibited reduced transformation efficiencies, ranging from 5-fold to 8,500-fold, compared to that of S. mutans UA159 (Fig. 3B). The mutants that displayed the greatest decreases in transformation efficiencies carried deletions of sufC, SMu.462, divIB, ltaS, rgpI, irvR, and dltA. Of these, only the ΔdivIB and ΔirvR mutants exhibited similar reductions in transformation efficiency when treated with CSP in BHI broth. The total numbers of null mutants with transformation defects in CSP only, XIP only, and in both were 6, 1, and 13, respectively. Thus, mutants were identified that appear to have lost the ability to couple the CSP signal to the induction of competence.
As shown in Fig. 1, comX is naturally expressed when cells are grown as colonies on the surfaces of BHI agar plates, without the addition of CSP or XIP. Interestingly, comX is also induced in CDM without the addition of exogenous XIP; this does not occur in BHI broth or FMC, likely due to the suppressive effects of lower pH (11). The ability of cells to “autoactivate” comX in CDM enabled us to quickly and quantitatively screen reporter strains in a microtiter-based assay, measuring GFP fluorescence and cell growth simultaneously for 16 h. The loss of ltaS, rgpI, irvR, and dltA significantly reduced PcomX-gfp activity with or without the addition of synthetic XIP (Fig. 3). Strong reductions of PcomX-gfp activity in the absence of exogenously supplied XIP were evident in strains carrying deletions of pknB, liaS, ppiB (also named SMu.488 or SMu.1631), pnpB, pspC, SMu.835, or SMu.1781, but the induction of comX could be fully, or at least partially, restored in these mutants by the addition of synthetic XIP (Fig. 3). Interestingly, aberrant expression from PcomX was observed in the Δ462 and Δ695 mutants only when exogenous XIP was added to the CDM, but these two mutants behaved like the wild type in the absence of supplemental XIP (Fig. 3D). The Δ1193, Δ1388, and Δ1913-1904 mutant strains did not behave differently than the wild type in terms of PcomX-gfp activity in CDM. Importantly, these results demonstrate that certain mutations may influence both XIP production and the sensing of exogenous XIP, whereas others could be identified that could not “autoactivate” but were capable of sensing exogenously supplied XIP. Since it is presently not known how XIP arises from ComS, further analysis of these mutants could provide crucial new insights into competence regulation.
Analysis of mutant behaviors at the single-cell level.
The 20 mutants described above displayed growth defects in the presence of CSP and XIP, as well as diminished transformation efficiencies compared with that of the wild-type strain. Of relevance here, in planktonic cultures growing batchwise, the induction of comX expression in S. mutans UA159 by CSP in complex medium is bimodal (i.e., only a subpopulation of cells activates comX transcription), whereas comX transcription is activated unimodally when cells are exposed to XIP in a chemically defined medium lacking peptides (4). Also of note, XIP-dependent activation of comX becomes bimodal in mature biofilms cultivated in vitro (15). Therefore, we sought to determine whether the changes in the growth and competence behaviors of the mutants were due to uniform changes across the population or to changes in the behaviors of a subpopulation of cells. We examined the activation of comX expression at the single-cell level in various mutants in response to CSP or XIP. In the ΔpknB, ΔliaS, ΔppiB, and ΔpnpB mutants, substantial defects in transformation efficiency were observed in BHI broth with CSP (BHI-CSP) and FMC with XIP (FMC-XIP) (Fig. 2D and 3B). Additionally, Tn insertions in these mutants were overrepresented after passage in BHI-CSP (Table S2). The pknB gene encodes a serine/threonine protein kinase, liaS encodes a histidine kinase of a TCS, ppiB encodes a predicted protein with an N-terminal haloacid dehydrogenase (HAD)-like hydrolase domain and a C-terminal peptidyl-prolyl cis-trans isomerase domain, and pnpB is predicted to encode a polynucleotide phosphorylase involved in mRNA turnover and processing. The pspC gene, encoding a putative phage shock domain and a stress-responsive transcriptional regulator domain, was also included in the analysis as it is regulated by LiaR (29). With respect to PcomX-gfp activity, the ΔpknB mutant was noteworthy in that a substantially decreased proportion of the cells induced comX expression when exposed to 200 nM CSP (Fig. 4A) compared with that observed in the wild-type genetic background, and lower proportions of cells in the ΔliaS, ΔppiB, ΔpnpB, and ΔpspC mutant strains were GFP positive than in the wild type (Fig. 4A). Although ΔliaS, ΔppiB, ΔpnpB, and ΔpspC cells expressed comX unimodally when supplied with exogenous XIP, the average fluorescence intensity of these mutants was lower than that in the S. mutans UA159 genetic background (Fig. 4A).
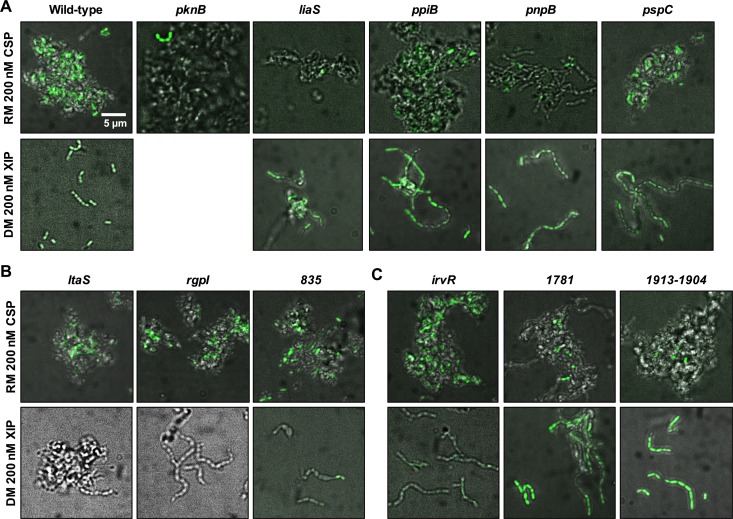
FIG 4.
Diverse comX expression phenotypes of competence-defective mutants revealed by fluorescence microscopy. A selection of mutant strains that displayed CSP and/or XIP growth or transformation phenotypes were explored in greater detail using a PcomX-gfp reporter system and fluorescence microscopy. Strains were grown in rich medium (RM) with 200 nM CSP or defined medium (DM; FMC) with 200 nM XIP and incubated for 2.5 h before images were collected by fluorescence microscopy. Overlays of the green and bright-field channels are shown. Deletion of pknB leads to a significant growth defect in FMC, and so this condition was omitted. Images are representative of three independent experiments and were taken at ×63 magnification.
We also focused on the ltaS, rgpI, and SMu.835 genes. The ltaS gene is predicted to encode a phosphoglycerol transferase involved in envelope biogenesis. The rgpI gene product shares homology with glycosyltransferase 2-like proteins, which are often involved in cell wall biogenesis, and has been shown to affect rhamnose-glucose polysaccharide synthesis by S. mutans (30). Lastly, SMu.835 is annotated as a possible membrane protein that contains low-complexity repetitive sequences. Although the deletion of these genes reduced the transformation efficiency and decreased the sensitivity to growth inhibition relative to those in the wild type in BHI broth containing CSP, more profound phenotypes were noted in cells growing in FMC and treated with XIP (Fig. 2 and 3). In particular, the ΔltaS and ΔrgpI strains were essentially nontransformable in FMC containing XIP. The deletion of these genes led to no visible induction of PcomX-gfp activity when cells were treated with XIP in FMC compared with that in the wild type (Fig. 4B). The mutant lacking SMu.835 still activated PcomX-gfp in response to XIP in FMC, but the level of expression was significantly reduced compared with that of the UA159 genetic background, and, importantly, this strain displayed evidence of bimodality when treated with XIP in FMC (Fig. 4B).
Four additional mutants (ΔdivIB, ΔirvR, Δ1781, and Δ1913-1904 mutants) displayed interesting PcomX-gfp single-cell phenotypes. The irvR gene encodes a LexA-like protein that is required for CSP-induced transformation of S. mutans (24). In accordance with what was previously reported (24), comX expression in the irvR mutant was not adversely affected in cells treated with CSP. However, the loss of IrvR led to decreased PcomX-gfp activity in cells growing in FMC and treated with XIP (Fig. 4C). The function of the SMu.1781 product is unknown, although it shares homology with other uncharacterized proteins associated with RNase G and RNase E (RNase Y in streptococci). Far fewer GFP-positive cells were visualized in the SMu.1781 mutant in the presence of CSP than in the strain with the wild-type genetic background, but the mutant had a response to XIP that mirrored that of the wild type (Fig. 4C). Several transposon insertions leading to decreased PcomX-lacZ activity were identified as insertions in genes and noncoding regions downstream of cipB (SMu.1914c) (Table S1). A strain carrying a deletion of the SMu.1904 to SMu.1913 genes showed greatly reduced proportions of PcomX-gfp-positive cells in the presence of CSP, but the deletion of this cluster of genes had no impact on comX expression when cells were exposed to exogenous XIP (Fig. 4C), providing further evidence that there are gene products that appear essential for coupling the CSP-ComDE circuit with the ComRS-XIP pathway.
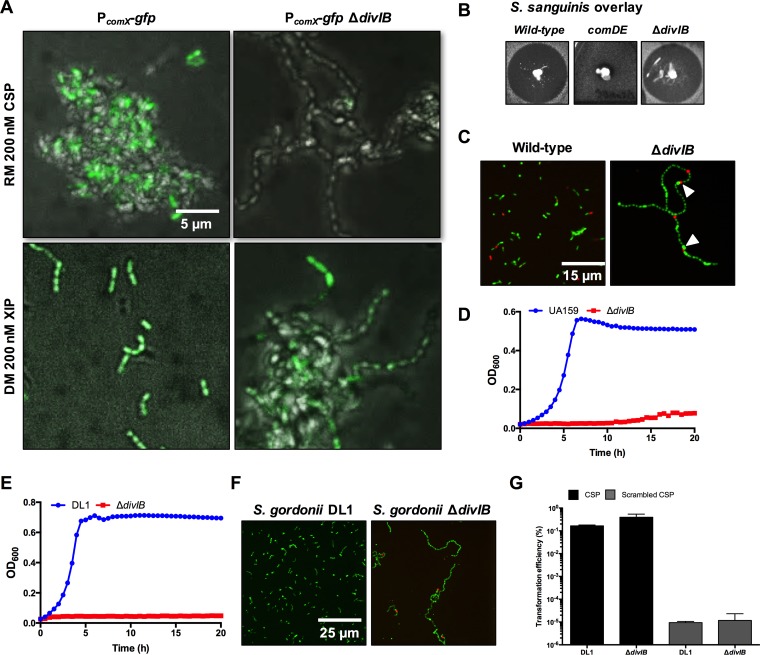
The divIB gene is predicted to encode a protein that participates in the regulation of cell division, and potential divIB homologues have been studied in Streptococcus pneumoniae (31) and Escherichia coli (divIB is designated ftsQ in Gram-negative bacteria) (32). Tn insertions in divIB decreased PcomX-lacZ activity (Table S1) and led to improved growth in BHI broth containing CSP (Table S2). The divIB mutant was virtually nontransformable, either in BHI broth with CSP or in FMC with XIP. Interestingly, the deletion of divIB did not result in improved growth in the presence of XIP in FMC (Fig. 2 and 3). Compared to that in S. mutans UA159, the proportion of cells that exhibited PcomX-gfp activity in the divIB mutant was reduced when cultured in BHI broth containing CSP. Only modest reductions in comX expression were seen in FMC supplemented with XIP, and PcomX-gfp activity remained unimodal (Fig. 5A). Intriguingly, this mutant produces bacteriocins at a level similar to that of the wild-type S. mutans when treated with CSP, indicating the ComDE signaling pathway is fully intact (Fig. 5B). We observed noticeable cell division defects in this mutant, evident from the increased chaining and a subgroup of cells that appeared to bulge (Fig. 5C). As in Bacillus subtilis (33), cells carrying a deletion of divIB displayed thermosensitivity, with no growth evident at 42°C even after 20 h of incubation (Fig. 5D). We sought to compare the above phenotypes with those of Streptococcus gordonii, an organism with a true comCDE system controlling genetic competence (ComE regulates comX expression directly). S. gordonii ΔdivIB shared similar cell division and thermosensitive phenotypes with S. mutans, but divIB deletion had no impact on the transformation efficiency in S. gordonii (Fig. 5E to G).
FIG 5.
DivIB is required for comX activation in S. mutans in response to CSP. (A) PcomX-gfp activity was visualized by fluorescence microscopy when 200 nM exogenous CSP or XIP was added to rich medium (RM; BHI broth) or defined medium (DM; FMC), respectively. Several phenotypes of the S. mutans ΔdivIB strain were also assessed (B to D). (B) Bacteriocin production (using a Streptococcus sanguinis indicator strain) was not altered in the ΔdivIB mutant compared with that in the S. mutans UA159. (C) The ΔdivIB mutant forms longer chains than S. mutans UA159, with occasional cell bulging (arrowheads). Cells were stained with the LIVE/DEAD BacLight bacterial viability kit according to the manufacturer's instructions and visualized with fluorescence microscopy (63× objective). (D) The ΔdivIB mutant (red line) grew poorly in BHI broth at 42°C. Growth was measured using a Bioscreen C automated growth curve analysis system. (E to G) The impacts of a divIB deletion on the following S. gordonii phenotypes were also characterized: growth at 42°C in BHI broth (E), cell chaining using a similar assay to that used for S. mutans (F), and transformation efficiency (G). For the transformation efficiency assay, 200 nM S. gordonii CSP (DVRSNKIRLWWENIFFNKK) or scrambled CSP (same amino acid composition, different sequence [DKRFKWWILKVFNSNEINR]) was added to cultures at an OD600 of 0.1 in BHI broth along with 100 ng of purified pDL278 plasmid (Spr). After incubating for 2.5 h, samples were plated on BHI-Sp agar or BHI agar, and plates were incubated for 2 days and the transformation efficiency was calculated.
DISCUSSION
In this study, we describe the first application of transposon sequencing to S. mutans, employing Tn-seq and a classical genetic screen to identify genes that influence genetic competence. Tn-seq enabled us to assess in a fairly comprehensive manner the relative contribution of gene products to the responses to competence peptides. This systematic approach, confirmed using targeted gene knockouts, has led to the discovery of 20 additional genes that have substantial impacts on the regulation of competence and competence-related phenotypes. We further expanded our analysis by exploring comX expression dynamics at the single-cell level. Since many of the genes that we discovered have not been studied in the context of genetic competence, these investigations contribute in novel and important ways to the overall understanding of cell-cell communication in S. mutans.
Overview of the transposon screens.
The addition of CSP or XIP to growth media causes global changes in gene expression (16, 18, 19). However, the comprehensive mutational analysis performed here indicates that the total number of genes that are required for optimal comX expression is relatively small, approximately 35 genes. By comparison, sporulation in B. subtilis requires >170 of the approximately 4,100 genes in the typical B. subtilis genome (21). The relatively small number of genes affecting optimal comX transcription and ComX levels may reflect the evolutionary pressures that also severely restricted the conditions under which S. mutans is able to activate comX transcription and progress to the competent state. For example, even in the presence of sufficient exogenously supplied CSP or XIP, the activation of comX occurs only within a very narrow pH range; for CSP this is between 6.7 and 7.7 and for XIP the range is 6.3 to 7.9, although the basis for this narrow window is not yet defined (11). Notably, genetic competence in S. mutans appears to be more intimately intertwined with physiologic homeostasis and stress tolerance than in many other bacteria that have the capability to acquire natural genetic competence. In fact, regulators of competence in S. mutans have potent impacts on biofilm formation (34), the stringent response (8, 10), oxidative stress responses (13, 35), and persistence (36). Thus, a long-term goal of this research is to understand how competence and other essential biological processes are coordinated and whether this coordination evolved to enhance the persistence and virulence of S. mutans. The studies presented here have identified new genes in the competence circuit and reveal possible pathways for the integration of the ComRS and ComDE pathways, and many of the genes identified are likely to influence stress tolerance and play essential roles in other homeostatic processes. Future studies will be oriented toward determining the extent to which these newly identified gene products integrate stress and competence. Likewise, analyses of these mutants should clarify how activation of bacteriocin gene expression influences cellular physiology in a way that modifies the development of competence.
While the quality of the transposon library and the results obtained were such that the likelihood was high that we could identify all the genes participating in the processes of interest, there are probably additional gene products and possibly noncoding regions of the genome that were not identified by the mutant screen or by Tn-seq. While genes that impact comX expression on solid surfaces (LacZ screen on agar) and in liquid media in a microaerophilic environment (Tn-seq) were isolated, it is also easy to envision that additional genes could participate in the modulation of comX expression in the presence of higher concentrations of oxygen, on carbohydrate sources other than glucose, or perhaps in response to other known factors that influence genetic competence (12, 13, 35). Tn-seq is somewhat limited in its ability to enable the discovery of small genes or small RNAs, where transposon density may not be sufficiently high to detect during screening. Likewise, genes that encode products with impacts on comX expression that can be fulfilled by other gene products (redundancy) would probably not have been identified in our screens. However, now that this technology has been established in S. mutans, it is possible to begin to probe how screening in different environmental conditions or screening strains carrying certain preexisting mutations might influence the ability to identify other genes that may contribute to competence.
Cell division and cell envelope integrity shape competence decisions.
Interestingly, the majority of the genes that we identified that had no previous links to comX regulation are those that function to control cell division or cell wall metabolism. Single-cell comX-GFP fluorescence microscopy studies facilitated the discovery of genes that perturb the ability of the ComDE TCS system to relay information to the ComRS pathway or genes that act at the ComRS regulatory level. The cell division protein DivIB is a notable example of a gene that specifically reduces the proportion of cells activating comX in response to CSP without having a profound effect on the XIP pathway. The machinery involved in S. mutans cell division is not well studied, but in S. pneumoniae, DivIB has a role in septal peptidoglycan synthesis during the late stages of cell division (31). DivIB interacts with two other cell division proteins, FtsL and DivIC. In S. pneumoniae, DivIB is important for stabilizing FtsL, with both FtsL and DivIC being essential; mutations in ftsL or divIC appear lethal in S. pneumoniae, and we were not able to obtain transformants when we attempted to knockout the ftsL or divIC genes (data not shown) (31). Interestingly, the mutation of divIB in S. gordonii had no impact on transformation efficiency, despite the fact that loss of DivIB in this commensal altered cell morphology and thermosensitivity in a manner similar to that observed in the divIB mutant of S. mutans, reinforcing the idea that factors that control competence development are in some cases species specific. We are presently contrasting other behaviors of the S. mutans and S. gordonii divIB mutants to understand why DivIB is integrated with competence in S. mutans, with one goal being to unmask the molecular link(s) between the ComDE and ComRS networks in S. mutans.
Many of the gene products identified as influencing comX expression participate in cell envelope biogenesis or in monitoring the integrity of the cell envelope. Among these were proteins with somewhat well-defined roles, including signal transduction involved in cell division (PknB) (23), cell envelope stress regulation (LiaS and PspC) (28, 29), and d-alanylation of teichoic acids (DltA) (37, 38). Also identified were genes for a glycosyltransferase enzyme (RgpI) involved in side chain formation during rhamnose-glucose polysaccharide synthesis (30), for a phosphoglycerol transferase (LtaS), and for a protein with a putative peptidoglycan-binding domain (SMu.695). The dltA, rgpI, and ltaS mutants had severe defects in XIP-dependent activation of comX but showed less-severe impairment in their ability to activate comX when provided with CSP; an unusual finding considering that the CSP circuit appears to work through ComR. One possible explanation for these observations is that the mechanism by which these gene products affect XIP signaling is related to the mutants having alterations in the cell envelope that compromise peptide transport and thus internalization of XIP, perhaps by influencing the translocation or membrane insertion of transport proteins (39, 40). However, it is possible that the enzymes that alter cell wall characteristics (DltA, RgpI, LtaS, and SMu.695) have the observed phenotypes because they impact cell membrane-associated sensing and signal transduction systems like LiaS and PknB. Homologues of PknB in other bacterial species sense cell wall precursors required during cell division (41, 42). PknB homologues also regulate the VicRK two-component system (also known as WalRK) by phosphorylating the response regulator VicR (43). VicRK positively regulates cell wall metabolism in S. mutans (44). In agreement with the cross talk between these signaling systems, the deletion of pknB in S. mutans caused severe cell morphology defects, as determined by transmission electron microscopy, and the PknB regulon was shown to overlap that of VicK (23). The liaFSR two-component system of S. mutans also has a regulatory role in cell envelope biogenesis and is important for sensing cell envelope stress (28). Therefore, the monitoring of and responding to changes in the cell envelope by LiaS and PknB could play critical regulatory roles in the reception of competence signal inputs and/or the progression to the competent state. It must also be considered that the mutants simply have a diminished capacity to take up DNA, since much of the machinery required for the active transport of single-stranded DNA (ssDNA) is membrane associated. Such overlaps in the regulatory inputs could have evolved to enable cells to integrate environmental stimuli, including the reductions in pH during carbohydrate metabolism, the production of cationic bacteriocins (known to be regulated by comDE), and the modifications induced by the competence-induced murein hydrolase LytF (regulated by ComX), into the control of the activation of, or the exit from, the competent state.
Novel components of comX regulation.
We also identified genes that have an impact on comX expression with functions that are more difficult to predict, including SMu.1781 and several genes that are in the cipB operon. Mutations in either of these regions greatly diminished comX expression and increased growth rates when exogenous CSP was added to BHI broth. SMu.1781 has a domain that is found in proteins associated with the endoribonuclease enzymes RNase G and RNase E (RNase Y in Gram-positive bacteria). If the predicted function of the product of SMu.1781 is accurate, it may modulate the stability of RNAs in S. mutans. In Gram-positive bacteria, the protein complex responsible for RNA decay (known as the RNA degradosome) has not been well studied, but the core components are RNase Y, RNase J1, RNase J2, and polynucleotide phosphorylase (PNPase [pnpA]), along with enolase, phosphofructokinase, and a DEAD box helicase (45). Notably, a PNPase (pnpB, orthologous to pnpA) and a DEAD-like helicase (SMu.1388) were also identified in our screens and induced significant competence defects when mutated. It is unknown whether SMu.1781, pnpB, and SMu.1388 interact with one another and/or the RNA degradosome in S. mutans. However, RNA stability could be an intriguing avenue for genetic competence research for two reasons. First, it is possible that interactions between glycolytic enzymes or intermediates and the RNA degradation machinery explain the finding that the carbohydrate source can affect comX expression in S. mutans (12, 46). Additionally, RNA stability is a neglected biological process that may be an important aspect of CSP-mediated bimodal comX expression, fine-tuning of ComX levels, or exiting the competent state, perhaps by influencing the stability of comRS or comX transcripts.
Concluding remarks.
This study provides candidate interactions which, with detailed investigations, could link competence and stress pathways with the regulation of cell division, the monitoring of envelope integrity, and signal transduction. By validating the mariner-based approach in S. mutans, we demonstrate that Tn-seq can now be applied as a powerful tool for discovering genes that are critical for other key functions, such as the colonization and persistence of this organism, in the human oral microbiome.
MATERIALS AND METHODS
Strains and growth conditions.
Streptococcus mutans strains were cultured from single colonies in BHI broth (Difco). Unless otherwise stated (here or in Text S1 in the supplemental material), S. mutans was routinely cultured at 37°C in a 5% CO2 aerobic atmosphere. Escherichia coli strains were routinely cultured in LB broth (10 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl) at 37°C with aeration. Antibiotics were added to growth media at the following concentrations: spectinomycin, 1.0 mg/ml for S. mutans and 50 μg/ml for E. coli; kanamycin, 1.0 mg/ml for S. mutans, 500 μg/ml for S. gordonii, and 50 μg/ml for E. coli; ampicillin, 100 μg/ml for E. coli. Lists of strains and plasmids (Table S3) and oligonucleotide primers (Table S4) can be found in the supplemental material.
Transposon library construction.
An in vitro transposon mutagenesis protocol was utilized to randomly mutagenize purified chromosomal DNA from S. mutans UA159 (20). Briefly, MarC9 transposase (47, 48) was purified from E. coli pMalC9 (see Text S1). MarC9, S. mutans genomic DNA, and pMagellan6 plasmid, harboring the mariner Himar1 minitransposon derivative magellan6, were combined and incubated for 1 h at 30°C. After transposon junctions were repaired, the fragments of genomic DNA containing magellan6 insertions were transformed into competent S. mutans. Transposon mutants were selected on BHI agar containing relevant antibiotics after 48 h of incubation. Colonies were removed from the surface of the agar, resuspended in BHI broth, and centrifuged at 3,500 × g for 10 min at 4°C and then resuspended in 5 ml BHI broth with glycerol (25% glycerol) and aliquoted in 250-μl volumes for long-term storage at −80°C.
Plate-based screen for mutants that fail to activate comX transcription.
Glycerol stocks of the library were thawed on ice and then diluted 1:50 in fresh BHI broth. Samples were incubated until the OD600 was equal to 0.1 and then were diluted to 1:105. One hundred-microliter aliquots were spread on TYG–X-Gal agar (2 g/liter glucose, 10 g/liter tryptone, 5 g/liter yeast extract, 3 g/liter K2HPO4, 15 g/liter agarose, 50 μg/ml X-Gal). Plates were incubated for 2 days at 37°C in a 5% CO2 incubator. White colonies were picked and cultured overnight in BHI broth, and glycerol stocks were prepared and stored at −80°C. Strains with apparently low PcomX-lacZ activity were then screened for genetic competence defects, and strains with phenotypes different from that of S. mutans UA159 were sequenced with arbitrary PCR to determine the sites of transposon insertion (for details on the method, see Text S1).
Tn-seq.
An aliquot of the transposon library pool was thawed and diluted 1:100 in 50 ml BHI broth or in 50 ml BHI broth plus 2 μM CSP. Samples were cultured to an OD600 of 1.0 and then transferred to fresh BHI broth or BHI-CSP for a total of 5 passages. The total viable counts before and after incubation were enumerated by plating on BHI agar. After each passage, a 10-ml sample was harvested for genomic DNA preparation. Genomic DNA was extracted using the MasterPure Gram Positive DNA purification kit (Epicentre Biotechnologies) in accordance with the supplier's instructions, with minor modifications (49). Genomic DNA restriction digestion, Illumina adapter ligation, library PCR amplification, and Illumina sequencing were performed as previously described (20, 22). Libraries were sequenced at the NextGen DNA sequencing core facility at the University of Florida on a NextSeq 500 (Illumina) with a 75-bp single-end sequencing read (1 × 50 cycles). See Text S1 for Tn-seq bioinformatics analyses.
Phenotypic assays and clustering analysis.
Text S1 details the criteria and methodologies employed to validate putative genetic competence-regulating genes, including transformation assays, growth studies, and single-cell PcomX-gfp reporter investigations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrew Camilli for providing the transposon sequencing plasmids pMalC9 and pMagellan6. We also thank Sara Palmer for helpful discussions on bioinformatics and Jeong Nam Kim for technical advice.
This study was supported by NIDCR R01 grants DE13239 and DE23339.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00508-17.
REFERENCES
- 1.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 2.Fontaine L, Wahl A, Fléchard M, Mignolet J, Hols P. 2015. Regulation of competence for natural transformation in streptococci. Infect Genet Evol 33:343–360. doi: 10.1016/j.meegid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Martin B, Quentin Y, Fichant G, Claverys J-P. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol 14:339–345. doi: 10.1016/j.tim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Son M, Ahn S-J, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn S-J, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaspar J, Kim JN, Ahn S-J, Burne RA. 2016. An Essential role for (p)ppGpp in the integration of stress tolerance, peptide signaling, and competence development in Streptococcus mutans. Front Microbiol 7:1162. doi: 10.3389/fmicb.2016.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn S-J, Kaspar J, Kim JN, Seaton K, Burne RA. 2014. Discovery of novel peptides regulating competence development in Streptococcus mutans. J Bacteriol 196:3735–3745. doi: 10.1128/JB.01942-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seaton K, Ahn S-J, Sagstetter AM, Burne RA. 2011. A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J Bacteriol 193:862–874. doi: 10.1128/JB.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son M, Ghoreishi D, Ahn S-J, Burne RA, Hagen SJ. 2015. Sharply tuned pH response of genetic competence regulation in Streptococcus mutans: a microfluidic study of environmental sensitivity of comX. Appl Environ Microbiol 81:5622–5631. doi: 10.1128/AEM.01421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moye ZD, Son M, Rosa-Alberty AE, Zeng L, Ahn S-J, Hagen SJ, Burne RA. 2016. Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl Environ Microbiol 82:4821–4834. doi: 10.1128/AEM.01205-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn S-J, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol 189:8519–8527. doi: 10.1128/JB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okinaga T, Xie Z, Niu G, Qi F, Merritt J. 2010. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol Oral Microbiol 25:165–177. doi: 10.1111/j.2041-1014.2010.00574.x. [DOI] [PubMed] [Google Scholar]
- 15.Shields RC, Burne RA. 2016. Growth of Streptococcus mutans in biofilms alters peptide signaling at the sub-population level. Front Microbiol 7:1075. doi: 10.3389/fmicb.2016.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan R, Rukke HV, Høvik H, Åmdal HA, Chen T, Morrison DA, Petersen FC. 2016. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reck M, Tomasch J, Wagner-Döbler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenderska IB, Latos A, Pruitt B, Palmer S, Spatafora G, Senadheera DB, Cvitkovitch DG. 2017. Transcriptional profiling of the oral pathogen Streptococcus mutans in response to competence signaling peptide XIP. mSystems 2:e00102-16. doi: 10.1128/mSystems.00102-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeske AJ, Rodrigues CDA, Brady J, Lim HC, Bernhardt TG, Rudner DZ. 2016. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol 14:e1002341. doi: 10.1371/journal.pbio.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Opijnen T, Camilli A. 2010. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol Chapter 1:Unit1E.3. doi: 10.1002/9780471729259.mc01e03s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banu LD, Conrads G, Rehrauer H, Hussain H, Allan E, van der Ploeg JR. 2010. The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect Immun 78:2209–2220. doi: 10.1128/IAI.01167-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu G, Okinaga T, Zhu L, Banas J, Qi F, Merritt J. 2008. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans. J Bacteriol 190:7268–7274. doi: 10.1128/JB.00967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry JA, Cvitkovitch DG, Levesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299:261–266. doi: 10.1111/j.1574-6968.2009.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szafrański SP, Deng Z-L, Tomasch J, Jarek M, Bhuju S, Rohde M, Sztajer H, Wagner-Döbler I. 2017. Quorum sensing of Streptococcus mutans is activated by Aggregatibacter actinomycetemcomitans and by the periodontal microbiome. BMC Genomics 18:238. doi: 10.1186/s12864-017-3618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336:104–112. doi: 10.1111/j.1574-6968.2012.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suntharalingam P, Senadheera MD, Mair RW, Lévesque CM, Cvitkovitch DG. 2009. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans. J Bacteriol 191:2973–2984. doi: 10.1128/JB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankar M, Mohapatra SS, Biswas S, Biswas I, Rodriguez A, Martinez B. 2015. Gene regulation by the LiaSR two-component system in Streptococcus mutans. PLoS One 10:e0128083. doi: 10.1371/journal.pone.0128083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozaki K, Shibata Y, Yamashita Y, Nakano Y, Tsuda H, Koga T. 2002. A novel mechanism for glucose side-chain formation in rhamnose-glucose polysaccharide synthesis. FEBS Lett 532:159–163. doi: 10.1016/S0014-5793(02)03661-X. [DOI] [PubMed] [Google Scholar]
- 31.Le Gouëllec A, Roux L, Fadda D, Massidda O, Vernet T, Zapun A. 2008. Roles of pneumococcal DivIB in cell division. J Bacteriol 190:4501–4511. doi: 10.1128/JB.00376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JC, Weiss DS, Ghigo JM, Beckwith J. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol 181:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harry EJ, Stewart BJ, Wake RG. 1993. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol Microbiol 7:611–621. doi: 10.1111/j.1365-2958.1993.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Y-H, Tang N, Aspiras MB, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaspar J, Ahn S-J, Palmer SR, Choi SC, Stanhope MJ, Burne RA. 2015. A unique ORF within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol Microbiol 96:463–482. doi: 10.1111/mmi.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung V, Lévesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol 194:2265–2274. doi: 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, Hamilton IR. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J Bacteriol 182:6055–6065. doi: 10.1128/JB.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spatafora GA, Sheets M, June R, Luyimbazi D, Howard K, Hulbert R, Barnard D, el Janne M, Hudson MC. 1999. Regulated expression of the Streptococcus mutans dlt genes correlates with intracellular polysaccharide accumulation. J Bacteriol 181:2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapot-Chartier M-P, Kulakauskas S, Neef J, Audouy S, Roosmalen M van, Steen A, Buist G, Kok J, Kuipers O, Robillard G, Leenhouts K. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13:S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardt P, Engels I, Rausch M, Gajdiss M, Ulm H, Sass P, Ohlsen K, Sahl H-G, Bierbaum G, Schneider T, Grein F. 2017. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int J Med Microbiol 307:1–10. doi: 10.1016/j.ijmm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Mir M, Asong J, Li X, Cardot J, Boons G-J, Husson RN. 2011. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog 7:e1002182. doi: 10.1371/journal.ppat.1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Libby EA, Goss LA, Dworkin J, Lowy F, Uhlemann A. 2015. The eukaryotic-like Ser/Thr kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis. PLoS Genet 11:e1005275. doi: 10.1371/journal.pgen.1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stipp RN, Boisvert H, Smith DJ, Höfling JF, Duncan MJ, Mattos-Graner RO. 2013. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One 8:e58271. doi: 10.1371/journal.pone.0058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho KH. 2017. The structure and function of the Gram-positive bacterial RNA degradosome. Front Microbiol 8:154. doi: 10.3389/fmicb.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman JA, Hewitt L, Rodrigues C, Solovyova AS, Harwood CR, Lewis RJ. 2012. Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J Mol Biol 416:121–136. doi: 10.1016/j.jmb.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 47.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci U S A 96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lampe DJ, Churchill ME, Robertson HM. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 49.Shields RC, Mokhtar N, Ford M, Hall MJ, Burgess JG, Elbadawey MR, Jakubovics NS. 2013. Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS One 8:e55339. doi: 10.1371/journal.pone.0055339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.