Abstract
Background
Macrocephaly, which is defined as a head circumference greater than or equal to + 2 standard deviations, is a feature commonly observed in children with developmental delay and/or autism spectrum disorder. Although PTEN is a well-known gene identified in patients with this syndromic presentation, other genes in the PI3K-AKT-mTOR signalling pathway have also recently been suggested to have important roles. The aim of this study is to characterise the mutation spectrum of this group of patients.
Methods
We performed whole-exome sequencing of 21 patients with macrocephaly and developmental delay/autism spectrum disorder. Sources of genomic DNA included blood, buccal mucosa and saliva. Germline mutations were validated by Sanger sequencing, whereas somatic mutations were validated by droplet digital PCR.
Results
We identified ten pathogenic/likely pathogenic mutations in PTEN (n = 4), PIK3CA (n = 3), MTOR (n = 1) and PPP2R5D (n = 2) in ten patients. An additional PTEN mutation, which was classified as variant of unknown significance, was identified in a patient with a pathogenic PTEN mutation, making him harbour bi-allelic germline PTEN mutations. Two patients harboured somatic PIK3CA mutations, and the level of somatic mosaicism in blood DNA was low. Patients who tested positive for mutations in the PI3K-AKT-mTOR pathway had a lower developmental quotient than the rest of the cohort (DQ = 62.8 vs. 76.1, p = 0.021). Their dysmorphic features were non-specific, except for macrocephaly. Among the ten patients with identified mutations, brain magnetic resonance imaging was performed in nine, all of whom showed megalencephaly.
Conclusion
We identified mutations in the PI3K-AKT-mTOR signalling pathway in nearly half of our patients with macrocephaly and developmental delay/autism spectrum disorder. These patients have subtle dysmorphic features and mild developmental issues. Clinically, patients with germline mutations are difficult to distinguish from patients with somatic mutations, and therefore, sequencing of buccal or saliva DNA is important to identify somatic mosaicism. Given the high diagnostic yield and the management implications, we suggest implementing comprehensive genetic testing in the PI3K-AKT-mTOR pathway in the clinical evaluation of patients with macrocephaly and developmental delay and/or autism spectrum disorder.
Electronic supplementary material
The online version of this article (10.1186/s13229-017-0182-4) contains supplementary material, which is available to authorized users.
Keywords: Somatic mosaicism, Macrocephaly, Megalencephaly, Developmental delay, Autism spectrum disorder, PTEN, MTOR, PIK3CA, PPP2R5D
Background
Macrocephaly is defined as a disproportionally enlarged head size with an occipitofrontal circumference greater than or equal to +2 standard deviations (SDs). On the other hand, megalencephaly is defined as hyperplasia of the brain parenchyma observed in a radiological examination together with clinical features of macrocephaly. Both conditions are associated with developmental delay (DD) and/or autism spectrum disorder (ASD). Moreover, a recent neuroimaging study has shown that brain volume overgrowth is linked to the emergence and severity of autistic social deficits [1].
ASD is a complex, behaviourally defined disorder characterised by impairments in communication and reciprocal social interaction, restrictive interests and repetitive stereotypic behaviours [2]. It is known that ASD has a strong genetic basis [3, 4], and environmental factors might also influence the development of ASD [5–7]. Previous studies have reported a genetic diagnosis in 10% to 40% of patients with ASD [8–11]. According to the American Academy of Pediatrics guidelines for ASD published in 2000, genetic testing is a standard diagnostic test for children with ASD and dysmorphic features or intellectual disability (ID).
As shown in previous studies, 14–34% of children with ASD also have macrocephaly [12–17], and a meta-analysis revealed that 15.7% have macrocephaly and 9.1% have brain overgrowth [18]. PTEN is a well-known gene associated with ASD and macrocephaly [19–21]. Hence, genetic testing for PTEN mutations is recommended as part of the clinical evaluation in this group of patients [22–24]. Recently, mutations in other genes in the PI3K-AKT-mTOR signalling pathway, including PIK3CA, PIK3R2, MTOR, CCND2 and PPP2R5D, were also reported in patients with ASD/DD and macrocephaly [25–30]. Although most PTEN mutations reported in this group of patients were germline mutations [20, 31], mutations in other genes in the PI3K-AKT-mTOR signalling pathway were frequently detected with a low level of mosaicism, which are undetectable using conventional Sanger sequencing. The use of next-generation sequencing, such as whole-exome sequencing (WES) or target panel sequencing, enables the detection of the low level of mosaicism in these patients. In this study, we aim to define the mutation spectrum in a cohort of patients with ASD/DD and macrocephaly using WES.
Methods
Patient recruitment
We recruited patients from January 2013 to December 2016 at the Duchess of Kent Children’s Hospital Child Assessment Center (DKCAC). Patients were initially assessed by a developmental paediatrician and relevant allied health professionals, including clinical psychologists, physiotherapists, occupational therapists and speech therapists. The developmental profile of patients less than 72 months of age was assessed using the Griffiths Mental Developmental Scales-Extended Revised (GMDS-ER). The developmental quotient (DQ) score was calculated based on the neurodevelopmental assessment to compare the developmental profiles between mutation-positive and mutation-negative patients. Because many patients were examined using more than one developmental assessment, the DQ scores from the first assessment were used. DQ scores were estimated for four patients based on their allied health assessment records and a clinical assessment by a developmental paediatrician because the patient was either too sick for the formal assessment with GMDS-ER (patient 3) or data were missing (patients 5, 15 and 19). Intellectual functioning of children ≥72 months of age was assessed using the Hong Kong Wechsler Intelligence Scale. ASD was diagnosed based on the Diagnostic and Statistical Manual of Mental Disorders—Fourth Edition (DSM-IV) criteria for ASD. Suspected cases were further assessed using the Autism Diagnostic Observation schedule (ADOS).
Developmental paediatricians at DKCAC referred patients with negative findings in the chromosomal microarray to the clinical geneticist when the patients also presented with macrocephaly (head circumference ≥ + 2 SD). These patients were recruited by the clinical geneticist, with the exception of patients presenting with obvious syndromic diseases who were assessed using targeted genetic tests and excluded from this study. Twenty-one unrelated patients were recruited in this study. Anthropometric data used for the measurements are specific to Hong Kong. Buccal swab or saliva sample was obtained in addition to the blood sample from five patients. This study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 12–211), and written consent was obtained from the patients’ parents.
WES
WES was first performed at Macrogen Inc. and then at our university at a later stage of the project using either a TruSeq Exome Enrichment Kit (n = 9), SeqCap EZ Exome + UTR Exome Kit (n = 9) or TruSeq Rapid Exome Library Prep Kit (n = 3). The choice of the enrichment kit depended on the library preparation kit available at the time of sequencing. We aimed to obtain a minimum of 50X depth for WES performed with genomic DNA obtained from the blood and a minimum of 100X depth for WES performed with DNA derived from buccal mucosa or saliva samples. The increased sequencing depth in buccal and saliva samples served to increase the chance of detecting somatic mutations. Details of the library preparation method, sequencer used and average depth after sequence alignment of each individual is presented in Additional file 1.
Variant calling and data analysis
Raw reads were aligned to the UCSC hg19 reference human genome using BWA 0.5.7, and variant calling was performed according to the best practices of GATK 3.4. The output VCF files were than annotated using ANNOVAR. The following criteria were applied to filter the variants and generate a list of high-quality variants for subsequent analyses: variants located outside the exonic regions and splice sites, synonymous variants, population frequency ≥ 1% or total depth < 10X. Genes involved in the PI3K-AKT-mTOR signalling pathway (PTEN, AKT1, AKT3, PIK3CA, MTOR, PIK3R2, CCND2, PPP2R5D and PPP2R1A) were prioritised for analysis because of their known pathogenicity in patients with macrocephaly and DD/ASD, and other genes in the PI3K-AKT signalling pathway obtained from Kyoto Encylopedia of Genes and Genomes (KEGG; reference: hsa04151) were also analysed. Mutations identified in this pathway that are associated with diseases are expected to be rare, and activating mutations are likely to be found in the Catalogue of Somatic Mutations in Cancer (COSMIC) database [27, 32]. In addition, genes involved in epigenetic regulation have recently been reported to be an important cause of macrocephaly/overgrowth and DD/ASD, and thus, genes related to epigenetic regulation (CHD8, DNMT3A, EED, EZH2, HIST1H1E and NSD1) were also prioritised for analysis [33].
Confirmation of variants
For germline mutations, variants were confirmed by Sanger sequencing, and parental sequencing was also performed to determine whether the mutation was de novo or inherited. For somatic mosaic mutations, droplet digital PCR was used for validation, as described previously [32].
Statistical analysis
Unpaired t tests were performed to assess the significance between the DQ scores of patients with and without mutations. A p-value < 0.05 was considered statistically significant. Analyses were performed using SPSS Statistics version 19 (IBM).
Results
Twenty-one patients (17 males and 4 females, 4 to 108 months of age at the time of clinical assessment/recruitment) with macrocephaly and DD/ID/ASD were recruited. All patients had DD at the time of recruitment, and three patients were diagnosed with ID in subsequent assessments. Among the 21 patients, ten were also diagnosed with ASD and two with suspected ASD (i.e., patients with autistic features who had not yet satisfied all the DSM-IV criteria for a diagnosis of ASD). A summary of the patients’ clinical presentations is presented in Table 1. Prior to WES, the chromosomal microarray was performed on these patients, as described previously [34], and no pathogenic/likely pathogenic copy number variations were identified in these patients. WES identified ten pathogenic/likely pathogenic mutations in ten patients (Fig. 1, Table 2), corresponding to a diagnostic yield of 47.6%. All pathogenic mutations were located in genes involved in the PI3K-AKT-mTOR signalling pathway, including PTEN (n = 4), PIK3CA (n = 3), MTOR (n = 1) and PPP2R5D (n = 2). Although most variants were germline mutations, two somatic PIK3CA mutations were identified. No pathogenic mutations were identified in genes related to epigenetic regulation, such as CHD8, DNMT3A, EED and NSD1, as reported by Tatton-Brown K et al. [33], and the analysis of the remainder of the exome did not reveal other variants of interest.
Table 1.
Clinical presentations of all patients recruited in this study
| Patient ID | Sex | First assessment (years) | Most recent assessment (years) | HC (z-score) | DQ | ASD | Developmental problem(s) | Other clinical features | MRI findings | Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 3.0 | 5.2 | 3.6 | 75 | Mild GDD with severe language delay | 2/3/4 syndactyly | MEG, PG, VM, PWMSA | PIK3CA | |
| 2 | M | 1.5 | 4.1 | 3.2 | 55 | Moderate GDD | Mild 2/3 syndactyly | MEG, PG, PWMSA | PIK3CA | |
| 3 | M | 0.8 | 1.6 | 5.9 | 50* | Moderate GDD | Recurrent respiratory infections, hypoglycaemia | MEG, PG, VM, PWMSA | PTEN | |
| 4 | M | 2.3 | 6.5 | 4.0 | 73 | Suspected ASD | Mild GDD | NA | MEG, PG, PWMSA | PTEN |
| 5 | M | 0.4 | 0.7 | 6.7 | 62* | Mild GDD | Liver haemangioma | MEG, PG, VM, PWMSA | MTOR | |
| 6 | M | 2.7 | 8.2 | 4.5 | 80 | ASD | Mild GDD | 2/3 syndactyly | NA | PTEN |
| 7 | M | 2.0 | 11.6 | 2.6 | 49 | ASD | Moderate GDD, mild ID | Hypertelorism, epilepsya | MEG | PPP2R5D |
| 8 | M | 1.8 | 5.3 | 2.2 | 54 | Suspected ASD | Moderate GDD, mild ID | Hypertelorism, epilepsyb | MEG | PPP2R5D |
| 9 | M | 2.0 | 3.5 | 3.5 | 62 | ASD | Mild GDD with moderate language delay | Left lower limb hypertrophy with 2/3 syndactyly | MEG | PIK3CA |
| 10 | M | 2.6 | 6.0 | 3.4 | 68 | Mild GDD | Frontal bossing | MEG | PTEN | |
| 11 | M | 4.4 | 10.1 | 3.4 | 95 | ASD | Language delay | z-scores for height: 3.0; weight: 5.6 | Normal | Negative |
| 12 | M | 2.3 | 6.3 | 3.3 | 79 | ASD | Borderline delay with language delay | Left ptosis, scoliosis | Dilated cisterna magna, relative atrophy of the right cerebellar hemisphere | Negative |
| 13 | F | 9.8 | 10.3 | 3.1 | NA | ASD | NA | Normal | Negative | |
| 14 | F | 1.6 | 7.3 | 3.3 | 75 | Borderline delay with language delay | Intra-cranial haemorrhage in the perinatal period | Aqueduct stenosis with hydrocephalus | Negative | |
| 15 | F | 4.0 | 6.0 | 4.2 | 90* | ASD | Language delay | NA | NA | Negative |
| 16 | M | 2.2 | 23.4 | 3.2 | 71 | ASD | Borderline delay | NA | NA | Negative |
| 17 | M | 1.8 | 14.4 | 4.8 | 84 | ASD | Borderline delay | Epilepsyc | Normal | Negative |
| 18 | F | 2.4 | 6.2 | 2.2 | 78 | Borderline delay | NA | NA | Negative | |
| 19 | M | 2.0 | 19.8 | 2.1 | 50* | ASD | Moderate GDD, moderate ID | Epilepsyd | NA | Negative |
| 20 | M | 2.0 | 2.3 | 2.8 | 67 | Mild GDD | NA | Dilated ventricles with external hydrocephalus | Negative | |
| 21 | M | 2.4 | 10.8 | 3.0 | 72 | Mild GDD | Cleft lip and palate, nasal deformity | Normal | Negative |
*Estimated DQ scores
aEpilepsy: onset at 4 years of age, generalised tonic-clonic seizure
bEpilepsy: onset at 4 days of age, generalised epileptic spasm
cEpilepsy: onset at 11 months, focal seizure with impaired awareness
dEpilepsy: onset 11 years of age, generalised tonic-clonic seizure
MEG megalencephaly, PG polymicrogyria, VM ventriculomegaly, PWMSA periventricular white matter signal abnormalities, NA not available
Fig. 1.
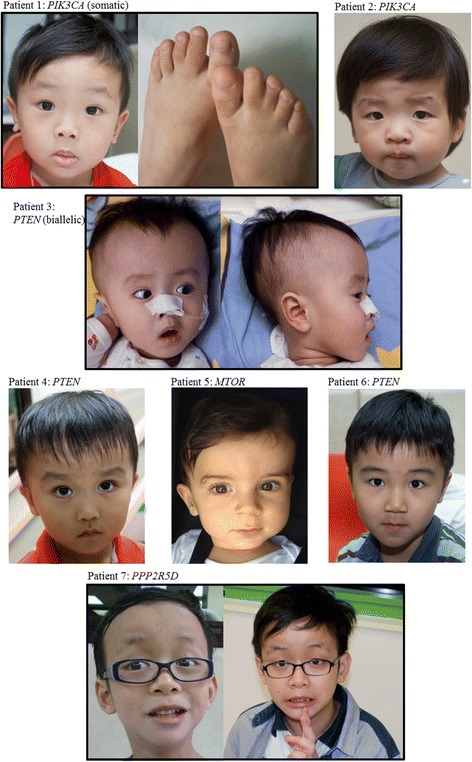
Clinical photographs of patients with mutations in the PI3K-AKT-mTOR pathway. Clinical photographs of patients with mutations identified in the PI3K-AKT-mTOR pathway are shown. For patient 1, syndactyly is shown. Patient 7 presented with hypertelorism and frontal bossing
Table 2.
Mutations in genes involved in the PI3K-AKT-mTOR pathway identified in the ten patients with macrocephaly and DD/ASD
| Patient ID | Gene | Nucleotide change | Amino acid change | Inheritance | Mutation type | Allelic count in ExAC | No. of cases in COSMIC | Reported mutation |
|---|---|---|---|---|---|---|---|---|
| 1 | PIK3CA | c.G263A | p.(Arg88Gln) | De novo | Mosaic | 0 | 137 | 43 |
| 2 | PIK3CA | c.G1030A | p.(Val344Met) | Maternal | Germline | 0 | 15 | 27 |
| 3 | PTEN | c.G314T | p.(Cys105Phe) | De novo | Germline | 0 | 6 | 39b |
| c.G492T | p.(Lys164Asn) | Maternal | Germline | 1 in 120,466 | 0b | |||
| 4 | PTEN | c.G509C | p.(Ser170Thr) | De novo | Germline | 0 | 0b | 35.36b |
| 5a | MTOR | c.G5395A | p.(Glu1799Lys) | De novo | Germline | 0 | 10 | 28.45 |
| 6 | PTEN | c.546delA | p.(Lys183ArgfsTer16) | De novo | Germline | 0 | 0b | |
| 7 | PPP2R5D | c.G592A | p.(Glu198Lys) | De novo | Germline | 0 | 1 | 29.46 |
| 8 | PPP2R5D | c.G592A | p.(Glu198Lys) | De novo | Germline | 0 | 1 | 29.46 |
| 9 | PIK3CA | c.G2740A | p.(Gly914Arg) | De novo | Mosaic | 0 | 2 | 25.27 |
| 10 | PTEN | c.A203G | p.(Tyr68Cys) | De novo | Germline | 0 | 4 | 37.38 |
The COSMIC database (June 2017) was accessed to retrieve the number of somatic mutations identified in cancer samples
aThis patient was reported in a previous study [28] with the patient ID LR15-065
bOther mutations in the same codon have been reported
Mutations in the PTEN gene were the most frequently identified mutations in our patients, with four pathogenic variants found in four patients (19% among 21 patients). Both missense and frameshift mutations were identified. Although the PTEN p.(Ser170Thr) mutation detected in patient 4 has not previously been reported, a mutation in the same codon, resulting in PTEN p.(Ser170Arg), has been reported in multiple patients with PTEN cancer syndrome [35, 36], suggesting the pathogenicity of mutations in this amino acid. The mutation in patient 6 was a frameshift mutation and therefore a pathogenic mutation, because loss of function mutations is known to cause disease. The PTEN p.(Tyr68Cys) mutation identified in patient 10 has already been reported in multiple patients with Cowden Syndrome [37, 38].
In addition, we report here a second patient with bi-allelic germline PTEN mutations. Two PTEN mutations were identified in patient 3, where p.(Cys105Phe) was a de novo mutation and p.(Lys164Asn) was maternally inherited. Based on the sequencing data, the two mutations did not occur in the same allele (Additional file 2, Fig. S1a). Exon 5 was cloned to confirm that the mutations were located on different alleles, and clonal sequencing showed that the two mutations occurred on different alleles (Additional file 2, Fig. S1b). The mutation p.(Cys105Phe) has not been reported, but a mutation in the same codon resulting in p.(Cys105Tyr) has been reported in patients with Bannayan-Riley-Ruvalcaba Syndrome [39], suggesting the pathogenicity of mutations in this amino acid. The maternally inherited p.(Lys164Asn) mutation has not been reported in a disease-specific database and has only been reported in the Exome Aggregation Consortium (ExAC) database with an allelic frequency of 1 in 120,466. Familial testing showed that this mutation was also detected in the patient’s mother and elder sister, and both the mother and elder sister had macrocephaly (z-scores for the head circumference were 2.6 and 3.7, respectively). Both had unremarkable developmental issues. Active cancer surveillance was recommended, and at the age of 38, the patient’s mother was diagnosed with multifocal papillary carcinoma. Based on the above evidence, although the variant p.(Lys164Asn) being pathogenic was compelling, it was still classified as a variant of unknown significance. It was because the mother did not meet the diagnostic criteria for PTEN hamartoma tumour syndrome, who only fulfilled one major criterion (macrocephaly) and one minor criterion (papillary carcinoma) [40]. Patient 3 who had bi-allelic mutations, however, displayed a severe clinical presentation despite one of the mutation being classified as a variant of unknown significance. In addition to megalencephaly, polymicrogyria and developmental delay, he suffered from recurrent sinopulmonary infections and colitis, resulting in persistent fever and septic shock that required care in the intensive care unit. The immune workup showed hypogammaglobulinaemia, specifically, a low level of IgG subclass 3. During a salmonella gastrointestinal infection at 19 months of age, a dihydrorhodamine test showed a suppressed oxidative burst with only half of the function compared to the control. However, a specific primary immunodeficiency syndrome was not identified. Second, this patient had suffered from recurrent hypoglycaemia since 19 months of age that required high glucose infusions; however, his insulin level was normal and an extensive endocrine workup was unremarkable. The patient died at 25 months of age due to sepsis. This case showed that patients with bi-allelic PTEN mutations can present with other PI3K-AKT-mTOR pathway-related features, including the recurrent respiratory infections observed in patients with PIK3CD mutations [41, 42] and hypoglycaemia observed in patients with AKT2 or AKT3 mutations [43, 44].
The PIK3CA mutation was the second most common mutation identified in our patients (patients 1, 2 and 9). The germline mutation in patient 2 was inherited from his mother, who had macrocephaly (z-score of head circumference was 4.6) but no history of developmental issues. In addition to the germline mutation, two somatic mutations were identified, and all mutations have been reported previously [25, 27]. WES detected a p.(Arg88Gln) mutation in patient 1, with a percentage of 4.5% (4 of 89 reads) in the blood and 27.1% (29 of 107 reads) in the buccal mucosa, whereas confirmation using droplet digital PCR showed that the percentages of p.(Arg88Gln) mutations in the blood and buccal mucosa samples were 8.6 and 22.8%, respectively. For patient 9, WES detected a p.(Gly914Arg) mutation with a percentage of 2.8% (3 of 109 reads) in the blood and 11.9% (13 of 109 reads) in the saliva. Again, droplet digital PCR confirmed the WES results, showing that the percentages of mutations were 2.6, 9.3 and 22.8% in the blood, saliva and buccal mucosa samples from patient 9, respectively. Our results confirmed previous findings that the mutation load in saliva or buccal mucosa is higher than the mutation load in the blood [25–27].
Finally, known pathogenic variants in the MTOR [28, 45] and PPP2R5D [29, 46] genes were also identified in our patients. Patient 5, who had a MTOR mutation, has already been reported in another publication (referred to as LR15-065 in the publication) describing a wide spectrum of patients with germline/somatic MTOR mutations [28]. In addition, PPP2R5D p.(Glu198Lys) was identified twice in two unrelated patients as de novo mutation. Both patients had a clinical presentation compatible with other patients with PPP2R5D mutations, including hypertelorism, frontal bossing, and a history of epilepsy.
At the time when genetic counselling was provided to patients with mutations, patients were re-examined to determine whether they had features of PTEN hamartoma tumour syndrome [40] such as macular pigmentation of the glans penis, mucocutaneous lesions and lipomas. For megalencephaly-capillary malformation syndrome (MCAP)/megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome (MPPH) [47], features of syndactyly, signs of overgrowth and vascular anomalies were assessed. Most of these features were absent in our patients. Only a small minority of patients presented with additional clinical features, such as syndactyly, hypertelorism and epilepsy. Our findings demonstrated the diversity of the clinical spectrum in this group of patients (see Table 1). Overall, patients with identified mutations had only subtle dysmorphic features (Fig 1). The mean DQ scores for mutation-positive and mutation-negative patients were 62.8 and 76.1, respectively, and the difference was statistically significant (p = 0.021). The prevalence of ASD/autistic features was similar between the two groups. It was noted that except for the two patients with PPP2R5D mutations, the head circumference of other mutation-positive patients were > + 3 SD. However, patients who tested positive for mutations in the PI3K-AKT-mTOR pathway did not had a significant difference in head circumference than mutation-negative patients. We also reviewed the brain magnetic resonance imaging (MRI) findings. Of the ten patients with an identified mutation, nine underwent MRI (Fig. 2). Megalencephaly was present in all nine of these patients; in addition, polymicrogyria was also identified in five patients, periventricular white matter signal abnormalities were identified in five patients, and ventriculomegaly was identified in three patients. With the exception of brain overgrowth, mutation-positive patients had no structural brain abnormalities. In contrast, three of the seven mutation-negative patients also had brain abnormalities, such as Dandy-Walker variant (n = 1) or aqueduct stenosis with hydrocephalus (n = 2). The brain MRI findings of the remaining four patients were normal.
Fig. 2.
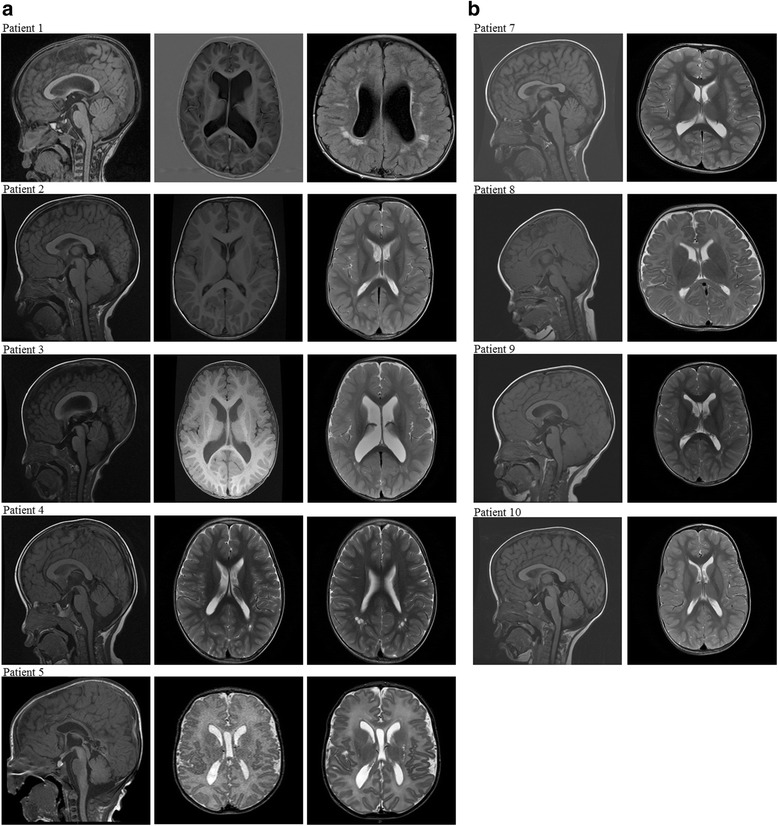
MRI of patients with mutations in the PI3K-AKT-mTOR pathway. a MRI in patients 1–5 showing megalencephaly, polymicrogyria and periventricular white matter signal abnormalities. Ventriculomegaly was observed in patients 1, 3 and 5. b MRI in patients 7–10 showing megalencephaly, without other abnormalities. MRI was not available for patient 6 because the family declined the MRI
Discussion
In this study, we aimed to characterise the mutation spectrum of patients with macrocephaly and DD/ASD. Among the 21 patients, ten had mutations in the PI3K-AKT-mTOR signalling pathway, indicating the importance of this pathway in macrocephaly with DD/ASD (Table 2). Our overall diagnostic yield was 47.6%, and PTEN mutations were detected in 19% of patients (n = 4), similar to previous studies detecting PTEN mutations/deletions in patients with DD/ASD [20, 21, 48, 49]. The higher diagnostic yield in this study is because multiple genes in the PI3K-AKT-mTOR pathway in addition to PTEN were considered, and in selected patients, WES of reasonably high depth was performed using additional sources of DNA, including saliva or buccal mucosa samples, rather than blood samples alone. Findings from our study suggest the need to refine the recommendation of current guidelines for the genetic evaluation of patients with macrocephaly and DD/ID/ASD. The American Academy of Pediatrics guidelines do not specifically mention an evaluation of children with macrocephaly and DD/ID [50], but the ASD evaluation proposed by the Autism Consortium Clinical Genetics/DNA Diagnostics Collaboration [22], the American College of Medical Genetics and Genomics [23] and other experts [24] only suggests genetic testing for PTEN mutations. From a practical perspective, children with DD/ASD will be referred for genetic consultation when their developmental problem is moderate to severe or when they present with dysmorphic features. However, based on our findings, most patients with mutations present with mild to moderate developmental problems, and dysmorphism is mild and non-specific. The absence of typical features of PTEN hamartoma tumour syndrome (such as glans penis pigmentation, mucocutaneous lesions and lipomas) may be due to the relatively young age of the patients or to the variable presentation of these features. Thus, genetic tests should be considered for patients with DD/ASD and macrocephaly, regardless of the degree of DD/ASD and the presence/absence of dysmorphic features. A panel of genes in the PI3K-AKT-mTOR pathway, including but not limited to PTEN, should be tested, and a low level of mosaicism for variants should be considered when collecting samples from patients for DNA extraction and determining the methodology to use to detect mutations [25–28]. Additional sources of DNA obtained from the buccal mucosa, saliva or brain (if available) should also be used for sequencing, and next-generation sequencing with reasonably high depth and coverage of genes in the PI3K-AKT-mTOR pathway should be performed. Although our use of WES successfully identified somatic mutations in two patients, a targeted gene panel has the advantage of higher depth than WES and therefore is a better choice for testing.
The genetic diagnosis of mutations in genes involved in the PI3K-AKT-mTOR pathway is clinically important. First, monogenic mutations in the PI3K-AKT-mTOR pathway are important in the pathogenesis of a subset of patients with DD/ASD. The genetic information may facilitate genetic counselling and estimate the risk of occurrence. Second, the genetic diagnosis facilitates a determination of prognosis. For example, patients with PPP2R5D mutations are expected to have poor language and locomotor performance, moderate to severe ID/DD and epilepsy [29, 46]. Third, Riviere et al. recommended that brain MRI should be performed on these children, with special attention to abnormal patterns of headache, changes in gait or other neurological problems [25]. Fourth, long-term cancer surveillance should be provided for these patients, because the PI3K-AKT-mTOR pathway is an important cancer-related pathway and is frequently mutated in tumours [51]. Patients with PTEN mutations have an increased risk of breast cancer, thyroid cancer, melanoma and endometrial cancer [52, 53], and recently, Peterman et al. found that patients with somatic PIK3CA mutations had an increased risk of Wilms tumour [54]. Finally, genetic counselling and family cascade testing should be provided to patients with germline mutations, because mutations may have been inherited from parents with macrocephaly yet without a remarkable history of DD/ID. One of the PTEN mutations in patient 3 was maternally inherited, but his mother had a clinically unremarkable presentation, except for macrocephaly. She was counselled, and after a year of cancer surveillance, she was diagnosed with early-stage thyroid cancer. This finding illustrates the importance of family cascade testing and cancer surveillance. Nevertheless, because of the complexity of genetic tests (such as the choice of tissue and depth of sequencing) and diverse clinical presentations in patients, we emphasise that genetic testing should only be offered by clinical geneticists who provide comprehensive pre- and post-test counselling to ensure the quality of the test, data interpretation and standard of care.
Here, we reported a second patient with bi-allelic germline PTEN mutations. Although one of the mutations was classified as variant of unknown significance, his clinical presentation was more severe than typical patients with heterozygous PTEN mutations and siblings with homozygous mutations, as reported by Schwerd et al. [55]. According to these authors, the homozygous p.Leu182Ser mutation is functionally hypomorphic, and thus, the patients have a recessive form of macrocephaly syndrome with a milder clinical course and lower risk of malignancy. Our patient (patient 3) serves as a contrasting example, showing that patients with bi-allelic PTEN mutations can present with a more severe clinical course involving multiple systems and display early lethality.
Historically, different nomenclatures have been used in this group of patients, including but not limited to macrocephaly-capillary malformation [56], MCAP [25], MPPH [25, 30], hemimegalencephaly [26], focal cortical dysplasia [28], megalencephaly [28, 57], and PIK3CA-related overgrowth spectrum [58]. The overlapping phenotypic presentation makes a differential diagnosis difficult, and the use of different nomenclatures is confusing to clinicians and patients. For example, MCAP and MPPH are usually associated with PIK3CA and PIK3R2 mutations, respectively. However, patients 1 and 9 in our study, who had somatic PIK3CA mutations, did not present with somatic features observed in MCAP other than syndactyly [47], whereas patients 3, 4 and 5 who did not have PIK3R2 mutations, presented with megalencephaly, polymicrogyria or ventriculomegaly, consistent with MPPH. Thus, a differential diagnosis is difficult, and the clinical presentation is a spectrum. A consensus on the nomenclature for this group of patients should be reached among international clinicians and scientists to facilitate communication, management, determination of prognoses, and further research and clinical trials [58]. Although the umbrella term “PIK3CA-related overgrowth spectrum” has been proposed to encompass patients with PIK3CA mutations [58], it is not sufficiently comprehensive to describe patients with macrocephaly who are complicated with DD/ID/ASD, because mutations in genes other than PIK3CA have also been identified in this group of patients. Since these patients share overlapping phenotypes and mutations in the same pathway, we propose the umbrella term “mTOR pathway-related macrocephaly spectrum” to encompass patients with macrocephaly and DD/ID/ASD associated with germline or somatic mutations in the PI3K-AKT-mTOR signalling pathway.
The limitation of the present study is that we only included a small number of patients, and long-term follow up was not available for all patients. In addition, the sequencing strategy was not uniform throughout the study because we made changes to improve the sequencing depth and to include DNA obtained from saliva and buccal mucosa samples, in addition to blood samples. We believe that brain MRI findings may be an indicator for genetic testing because all patients with mutations in the PI3K-AKT-mTOR pathway had features of megalencephaly and/or brain overgrowth, rather than macrocephaly alone. Nevertheless, our findings should be confirmed in larger studies, given our small sample size. Because the association between the PI3K-AKT-mTOR pathway and macrocephaly and DD/ID/ASD is relatively new, we have limited knowledge of this disease spectrum. We hope that the identification of more patients will enable the better characterisation of the clinical presentation of this group of diseases, and hence, clinicians will be able to provide better clinical management for these patients.
Conclusions
In summary, nearly 50% of children with macrocephaly and developmental delay/ASD had mutations in the PI3K-AKT-mTOR pathway, suggesting the importance of this pathway in this patient group. The presence of somatic mosaicism increases the difficulty in providing a molecular diagnosis, and therefore, DNA samples from different tissues should be sequenced. Finally, we propose the use of the umbrella term “mTOR pathway-related macrocephaly spectrum” to emphasise the overlapping clinical phenotypes and genotypes associated with this spectrum of patients.
Additional files
Summary of sequencing data for all patients with macrocephaly and DD/ASD. The data consist of the sequencing information for all patients recruited in this study, including the kit used to prepare the library, sequencers, and the depth of coverage. (XLSX 18 kb)
Bi-allelic PTEN mutations in patient 3. The file consists of WES reads and clonal sequencing data to show that patient 3 has two mutations in different PTEN alleles. (DOCX 936 kb)
Acknowledgements
We thank all the participating patients and their families.
Funding
This study was supported by grants from the SK Yee Medical Research Fund, The Society for the Relief of Disabled Children and HKU Seed Funding for Basic Research. The funding agencies were not involved in the design of the study; in the collection, analysis and interpretation of the data; or in the writing of the manuscript.
Availability of data and materials
The datasets used and/or analysed in the current study are available from the corresponding author upon reasonable request.
Abbreviations
- ASD
Autism spectrum disorder
- COSMIC
Catalogue of Somatic Mutations in Cancer
- DD
Developmental delay
- DKCAC
Duchess of Kent Children’s Hospital Child Assessment Center
- DQ
Developmental quotient
- ExAC
Exome Aggregation Consortium
- GDD
Global developmental delay
- ID
Intellectual disability
- MCAP
Megalencephaly-capillary malformation syndrome
- MPPH
Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome
- MRI
Magnetic resonance imaging
- SD
Standard deviation
- WES
Whole-exome sequencing
Authors’ contributions
KSY and WWYT performed the data analysis and wrote the manuscript. KSY performed the Sanger validations. JJKI performed the MRI analysis. KSY, CCYM, GKCL, MHYT, DY and SLCP assisted with the study design, data management and WES analysis. WWYT, SLL and BHYC recruited and managed the patients and provided facilities and resources for the study. BHYC designed and supervised the study. All authors read the manuscript, revised it critically for important intellectual content and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 12–211), and written consent was obtained from the patients’ parents.
Consent for publication
Written informed consent was obtained from the patients’ parents for publication of their children’s details and accompanying images in this manuscript. The consent form is held by the authors and is available for review by the Editor-in-Chief.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13229-017-0182-4) contains supplementary material, which is available to authorized users.
Contributor Information
Kit San Yeung, Email: jsks.kit@gmail.com.
Winnie Wan Yee Tso, Email: wytso@hku.hk.
Janice Jing Kun Ip, Email: janicejkip@gmail.com.
Christopher Chun Yu Mak, Email: ccymak@hku.hk.
Gordon Ka Chun Leung, Email: gordon.leungkc@gmail.com.
Mandy Ho Yin Tsang, Email: mandyhytsang@gmail.com.
Dingge Ying, Email: jonsonfox@gmail.com.
Steven Lim Cho Pei, Email: peisteven@yahoo.com.hk.
So Lun Lee, Email: slleem@hku.hk.
Wanling Yang, Email: yangwl@hkucc.hku.hk.
Brian Hon-Yin Chung, Phone: (852) 22554482, Email: bhychung@hku.hk.
References
- 1.Hazlett HC, Gu H, Munsell BC, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature. 2017;542(7641):348–351. doi: 10.1038/nature21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Constantino JN, Charman T. Diagnosis of autism spectrum disorder: reconciling the syndrome, its diverse origins, and variation in expression. Lancet Neurol. 2016;15(3):279–291. doi: 10.1016/S1474-4422(15)00151-9. [DOI] [PubMed] [Google Scholar]
- 3.Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113(5):e472–e486. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- 4.Vorstman JAS, Parr JR, Moreno-De-Luca D, RJL A, Nurnberger JI, Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18(6):362–376. doi: 10.1038/nrg.2017.4. [DOI] [PubMed] [Google Scholar]
- 5.Williams G, King J, Cunningham M, Stephan M, Kerr B, Hersh JH. Fetal valproate syndrome and autism: additional evidence of an association. Dev Med Child Neurol. 2001;43(3):202–206. doi: 10.1111/j.1469-8749.2001.tb00188.x. [DOI] [PubMed] [Google Scholar]
- 6.Isaksson J, Pettersson E, Kostrzewa E, Diaz Heijtz R, Bolte S. Brief report: association between autism spectrum disorder, gastrointestinal problems and perinatal risk factors within sibling pairs. J Autism Dev Disord. 2017;47(8):2621–2627. doi: 10.1007/s10803-017-3169-2. [DOI] [PubMed] [Google Scholar]
- 7.Juul-Dam N, Townsend J, Courchesne E. Prenatal, perinatal, and neonatal factors in autism, pervasive developmental disorder-not otherwise specified, and the general population. Pediatrics. 2001;107(4):E63. doi: 10.1542/peds.107.4.e63. [DOI] [PubMed] [Google Scholar]
- 8.Herman GE, Henninger N, Ratliff-Schaub K, Pastore M, Fitzgerald S, McBride KL. Genetic testing in autism: how much is enough? Genet Med. 2007;9(5):268–274. doi: 10.1097/GIM.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer GB, Lutz RE. Diagnostic yield in the clinical genetic evaluation of autism spectrum disorders. Genet Med. 2006;8(9):549–556. doi: 10.1097/01.gim.0000237789.98842.f1. [DOI] [PubMed] [Google Scholar]
- 10.Tammimies K, Marshall CR, Walker S, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314(9):895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 11.CY RK, Merico D, Bookman M, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20(4):602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courchesne E, Karns CM, Davis HR, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57(2):245–254. doi: 10.1212/WNL.57.2.245. [DOI] [PubMed] [Google Scholar]
- 13.Aylward EH, Minshew NJ, Field K, Sparks BF, Singh N. Effects of age on brain volume and head circumference in autism. Neurology. 2002;59(2):175–183. doi: 10.1212/WNL.59.2.175. [DOI] [PubMed] [Google Scholar]
- 14.Dementieva YA, Vance DD, Donnelly SL, et al. Accelerated head growth in early development of individuals with autism. Pediatr Neurol. 2005;32(2):102–108. doi: 10.1016/j.pediatrneurol.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Dissanayake C, Bui QM, Huggins R, Loesch DZ. Growth in stature and head circumference in high-functioning autism and Asperger disorder during the first 3 years of life. Dev Psychopathol. 2006;18(2):381–393. doi: 10.1017/S0954579406060202. [DOI] [PubMed] [Google Scholar]
- 16.Sacco R, Curatolo P, Manzi B, et al. Principal pathogenetic components and biological endophenotypes in autism spectrum disorders. Autism Res. 2010;3(5):237–252. doi: 10.1002/aur.151. [DOI] [PubMed] [Google Scholar]
- 17.Sacco R, Militerni R, Frolli A, et al. Clinical, morphological, and biochemical correlates of head circumference in autism. Biol Psychiatry. 2007;62(9):1038–1047. doi: 10.1016/j.biopsych.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 18.Sacco R, Gabriele S, Persico AM. Head circumference and brain size in autism spectrum disorder: a systematic review and meta-analysis. Psychiatry Res. 2015;234(2):239–251. doi: 10.1016/j.pscychresns.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Parada LFPTEN. Signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22(5):873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Varga EA, Pastore M, Prior T, Herman GE, KL MB. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11(2):111–117. doi: 10.1097/GIM.0b013e31818fd762. [DOI] [PubMed] [Google Scholar]
- 21.Butler MG, Dasouki MJ, Zhou XP, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42(4):318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Dies KA, Holm IA, et al. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics. 2010;125(4):e727–e735. doi: 10.1542/peds.2009-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer GB, Mendelsohn NJ, Professional P, Guidelines C. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15(5):399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 24.Anagnostou E, Zwaigenbaum L, Szatmari P, et al. Autism spectrum disorder: advances in evidence-based practice. CMAJ. 2014;186(7):509–519. doi: 10.1503/cmaj.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riviere JB, Mirzaa GM, O’Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44(8):941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirzaa G, Timms AE, Conti V, et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI Insight. 2016;1(9):e87623. [DOI] [PMC free article] [PubMed]
- 28.Mirzaa GM, Campbell CD, Solovieff N, et al. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA neurology. 2016;73(7):836–845. doi: 10.1001/jamaneurol.2016.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houge G, Haesen D, Vissers LE, et al. B56delta-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. J Clin Invest. 2015;125(8):3051–3062. doi: 10.1172/JCI79860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirzaa GM, Parry DA, Fry AE, et al. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet. 2014;46(5):510–515. doi: 10.1038/ng.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frazier TW, Embacher R, Tilot AK, Koenig K, Mester J, Eng C. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. 2015;20(9):1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeung KS, Ip JJ, Chow CP, et al. Somatic PIK3CA mutations in seven patients with PIK3CA-related overgrowth spectrum. Am J Med Genet A. 2017;173(4):978–984. doi: 10.1002/ajmg.a.38105. [DOI] [PubMed] [Google Scholar]
- 33.Tatton-Brown K, Loveday C, Yost S, et al. Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am J Hum Genet. 2017;100(5):725–736. doi: 10.1016/j.ajhg.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao VQ, Chan KY, Chu YW, et al. The clinical impact of chromosomal microarray on paediatric care in Hong Kong. PLoS One. 2014;9(10):e109629. doi: 10.1371/journal.pone.0109629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh DJ, Dahia PL, Zheng Z, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16(4):333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 36.de Leon MP, Di Gregorio C, Giunti L, et al. Duodenal carcinoma in a 37-year-old man with Cowden/Bannayan syndrome. Dig Liver Dis. 2013;45(1):75–78. doi: 10.1016/j.dld.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Lobo GP, Waite KA, Planchon SM, Romigh T, Nassif NT, Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum Mol Genet. 2009;18(15):2851–2862. doi: 10.1093/hmg/ddp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browning MJ, Chandra A, Carbonaro V, Okkenhaug K, Barwell J. Cowden’s syndrome with immunodeficiency. J Med Genet. 2015;52(12):856–859. doi: 10.1136/jmedgenet-2015-103266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsh DJ, Kum JB, Lunetta KL, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8(8):1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 40.Pilarski R, Burt R, Kohlman W, Pho L, Shannon KM, Swisher E. Cowden syndrome and the PTEN hamartoma tumor syndrome: systematic review and revised diagnostic criteria. J Natl Cancer Inst. 2013;105(21):1607–1616. doi: 10.1093/jnci/djt277. [DOI] [PubMed] [Google Scholar]
- 41.Angulo I, Vadas O, Garcon F, et al. Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas CL, Kuehn HS, Zhao F, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hussain K, Challis B, Rocha N, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334(6055):474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nellist M, Schot R, Hoogeveen-Westerveld M, et al. Germline activating AKT3 mutation associated with megalencephaly, polymicrogyria, epilepsy and hypoglycemia. Mol Genet Metab. 2015;114(3):467–473. doi: 10.1016/j.ymgme.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Baynam G, Overkov A, Davis M, et al. A germline MTOR mutation in aboriginal Australian siblings with intellectual disability, dysmorphism, macrocephaly, and small thoraces. Am J Med Genet A. 2015;167(7):1659–1667. doi: 10.1002/ajmg.a.37070. [DOI] [PubMed] [Google Scholar]
- 46.Shang L, Henderson LB, Cho MT, et al. De novo missense variants in PPP2R5D are associated with intellectual disability, macrocephaly, hypotonia, and autism. Neurogenetics. 2016;17(1):43–49. doi: 10.1007/s10048-015-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirzaa GM, Riviere JB, Dobyns WB. Megalencephaly syndromes and activating mutations in the PI3K-AKT pathway: MPPH and MCAP. Am J Med Genet C Semin Med Genet. 2013;163C(2):122–130. doi: 10.1002/ajmg.c.31361. [DOI] [PubMed] [Google Scholar]
- 48.Buxbaum JD, Cai G, Chaste P, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(4):484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McBride KL, Varga EA, Pastore MT, et al. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010;3(3):137–141. doi: 10.1002/aur.132. [DOI] [PubMed] [Google Scholar]
- 50.Moeschler JB, Shevell M. Committee on G. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134(3):e903–e918. doi: 10.1542/peds.2014-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18(2):400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bubien V, Bonnet F, Brouste V, et al. High cumulative risks of cancer in patients with PTEN hamartoma tumour syndrome. J Med Genet. 2013;50(4):255–263. doi: 10.1136/jmedgenet-2012-101339. [DOI] [PubMed] [Google Scholar]
- 54.Peterman CM, Fevurly RD, Alomari AI, et al. Sonographic screening for Wilms tumor in children with CLOVES syndrome. Pediatric blood & cancer. 2017;64(12):e26684. [DOI] [PubMed]
- 55.Schwerd T, Khaled AV, Schurmann M, et al. A recessive form of extreme macrocephaly and mild intellectual disability complements the spectrum of PTEN hamartoma tumour syndrome. Eur J Hum Genet. 2016;24(6):889–894. doi: 10.1038/ejhg.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toriello HV, Mulliken JB. Accurately renaming macrocephaly-cutis marmorata telangiectatica congenita (M-CMTC) as macrocephaly-capillary malformation (M-CM) Am J Med Genet A. 2007;143A(24):3009. doi: 10.1002/ajmg.a.31971. [DOI] [PubMed] [Google Scholar]
- 57.Tapper WJ, Foulds N, Cross NC, et al. Megalencephaly syndromes: exome pipeline strategies for detecting low-level mosaic mutations. PLoS One. 2014;9(1):e86940. doi: 10.1371/journal.pone.0086940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keppler-Noreuil KM, Rios JJ, Parker VE, et al. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet A. 2015;167A(2):287–295. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of sequencing data for all patients with macrocephaly and DD/ASD. The data consist of the sequencing information for all patients recruited in this study, including the kit used to prepare the library, sequencers, and the depth of coverage. (XLSX 18 kb)
Bi-allelic PTEN mutations in patient 3. The file consists of WES reads and clonal sequencing data to show that patient 3 has two mutations in different PTEN alleles. (DOCX 936 kb)
Data Availability Statement
The datasets used and/or analysed in the current study are available from the corresponding author upon reasonable request.


