Abstract
Background
Tongue squamous cell carcinoma (TSCC) is a major type of oral cancers and has remained an intractable cancer over the past decades. The aim of this study was to identify differentially expressed genes (DEGs) during TSCC and reveal their potential mechanisms.
Material/Methods
The gene expression profiles of GSE13601 were downloaded from the GEO database. The GSE13601 dataset contains 57 samples, including 31 tongue SCC samples and 26 matched normal mucosa samples. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analyses were performed; Cytoscape software was used for the protein-protein interaction (PPI) network and module analysis of the DEGs.
Results
We identified a total of 1,050 upregulated DEGs (uDEGs) and 702 downregulated DEGs (dDEGs) of TSCC. The GO analysis results showed that uDEGs were significantly enriched in the following biological processes (BP): signal transduction, positive or negative regulation of cell proliferation, and negative regulation of cell proliferation. The dDEGs were significantly enriched in the following biological processes: signal transduction, cell adhesion, and apoptotic process. The KEGG pathway analysis showed that uDEGs were enriched in metabolic pathways, pathways in cancer, and PI3K-Akt signaling pathway, while the dDEGs were enriched in focal adhesion and ECM-receptor interaction. The top centrality hub genes RAC1, APP, EGFR, KNG1, AGT, and HRAS were identified from the PPI network. Module analysis revealed that TSCC was associated with significant pathways, including neuroactive ligand-receptor interaction, calcium signaling pathway, and chemokine signaling pathway.
Conclusions
The present study identified key genes and signal pathways, which deepen our understanding of the molecular mechanisms of carcinogenesis and development of the disease, and might be used as diagnostic and therapeutic molecular biomarkers for TSCC.
MeSH Keywords: Computational Biology, Signal Transduction, Tongue Neoplasms
Background
Oral cancer is one of the most prevalent malignancies around the world, with an estimated 300,000 new cases and 130,000 deaths every year worldwide [1]. Tongue squamous cell carcinoma (TSCC) is a major type of oral cancer, which is characterized by remarkably aggressive biological behavior with a high incidence of lymph node and distant metastasis [2]. Although, the 5-year survival rate is reported to be up to 50% with early detection, most patients are diagnosed at a late stage, leading to poorer prognosis [3] and resulting in complications such as the malfunction of mastication, speech, and deglutition, or death. Despite advances in surgical procedures and chemo-radiotherapy, as well as the advent of targeted therapy, clinical outcomes have remained unchanged for decades [4]. Therefore, it is of primary importance to identify the etiological factors, molecular mechanisms, and pathways of carcinogenesis to discover novel diagnostic and treatment strategies for TSCC.
The molecular pathogenesis of carcinogenesis may be a combination of somatic mutations [5], and epigenetic and transcriptional alterations. Aberrant genetic alterations in gene expression may lead to the malignant transformation of TSCC. With advances of sequencing and high-throughput DNA microarray analyses, numerous gene alterations manifesting differentially expressed genes (DEGs) have been demonstrated to be correlated with the genesis and progression of tumors [6]. For example, Nadia et al. [7] found a significant association between methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and p16 and O6-methylguanine-DNA methyltransferase (MGMT) gene promoter methylation in oral squamous cell cancer (OSCC) patients, which suggests that hypermethylation of cancer-related genes may be affected by MTHFR polymorphisms. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long non-coding RNA (lncRNA), may play an oncogenic role by increasing proliferation and metastasis of tongue cancer via miR-124-dependent jagged1 (JAG1) regulation [8]. Obvious genetic genes changes, including loss of TRAF3 and amplification of E2F1 (a cell cycle gene), have been found in head and neck squamous carcinoma [5]. Chaisaingmongkol et al. [9] found that NEIL1 (Nei endo-nuclease VIII-like 1 gene) promoter hypermethylation might have a function in mediating the response to treatment of head and neck squamous cell carcinoma (HNSCC). Also, various signaling pathways have been shown to be important, such as loss-of-function alterations of the WNT pathway [5]; in addition, the COX-2 (cyclooxygenase-2) signaling pathway has been shown to be closely related to tumor angiogenesis [10] in TSCC. Moreover, the importance of inflammation in carcinogenesis of TSCC has been proven [11]; and Giovanni et al. [12] found that transglutaminase 2 (TG2) played a key role in periodontal inflammatory disease through the nuclear factor-kappa B (NF-κB) pathway. Therefore, identifying DEGs and elucidating the interactions network among them, and the signal pathways, is essential for TSCC. Novel therapeutic biomarkers are needed for TSCC for predictive and curative purposes.
In the present study, the DEGs of TSCC and normal tissue samples were analyzed to achieve a better understanding of TSCC. GO and KEGG enrichment analyses of DEGs were applied, and the protein-protein interaction (PPI) network and module of these DEGs was also constructed. The aim of this study was to identify key genes and pathways in TSCC using bioinformatics analysis, and then to explore the intrinsic mechanisms of TSCC and distinguish novel potential diagnostic therapeutic biomarkers of TSCC. We anticipated that these studies will provide further insight of TSCC pathogenesis and development at the molecular level.
Material and Methods
Datasets
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) is a public functional genomics data repository including array- and sequence-based data, and is freely available for users. The gene expression profiles of GSE13601 were obtained from the GEO database. GSE13601, which was based on the GPL8300 platform [HG_U95Av2] Affymetrix Human Genome U95 Version 2 Array, was submitted by Estilo et al. [13]. The GSE13601 dataset has 57 samples, including 31 TSCC samples and 26 matched normal mucosa samples.
Data processing
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) is an online tool where different groups of samples from the GEO series can be compared so as to identify genes that are differentially expressed across experimental conditions [14]. The analysis of screening DEGs between TSCC and normal mucosa samples was carried out by GEO2R. The adjusted p values (adj. p) were applied to correct the false positive results by default Benjamini-Hochberg false discovery rate method. The adj. p<0.01 and |log2FC|>1 were considered as the cutoff values.
Gene ontology (GO) and pathway enrichment analysis of DEGs
The GO (http://www.geneontology.org) [15] database can provide functional classification for genomic data, including categories of biological processes (BP), cellular component (CC), and molecular function (MF). GO analysis is a common genes and gene products annotating method. The Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.ad.jp/kegg/) [16] database is a knowledge base for systematic analysis, annotation, and visualization of gene functions. The Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) [17] is an online tool for gene functional classification, which is an essential foundation for high-throughput gene analysis to understand the biological significance of genes. In the present study, in order to analyze the functions of DEGs, GO enrichment and KEGG pathway analysis were conducted using the DAVID online tool; p<0.05 was set as the cutoff point.
Integration of protein-protein interaction (PPI) network and module analysis
The Search Tool for the Retrieval of Interacting Genes (STRING, http://string.embl.de/) [18] is a biological database designed to predict protein-protein interaction (PPI) information. The DEGs were mapped to STRING to evaluate the interactive relationships, with a confidence score >0.9 defined as significant. Then, we use Cytoscape [19], a biological graph visualization tool for integrated models of biologic molecular interaction networks software, to construct PPI networks. The Molecular Complex Detection (MCODE) [20], a plugin for Cytoscape, was used to screen the modules of the PPI network. The criteria were set as follows: degree cutoff=2, node score cutoff=0.2, k-core=2 and maximum depth=100. Moreover, the function and pathway enrichment analysis were performed for DEGs in the modules.
Analysis of key nodes in the PPI network
The key genes in the PPI network were investigated topologically. Three centrality methods including degree, closeness, and subgraph centrality [21] were used to explore key genes in the PPI network. The three centrality methods were calculated using the Cytoscape plugin: CytoNCA [22].
Results
Identification of DEGs
A total of 31 TSCC samples and 26 matched normal mucosa samples were analyzed, the mean age was 57 years (36–97 years). Based on the GEO2R analysis, using the adj. p<0.01 and |log2FC|>1 criteria, a total of 1,752 DEGs were identified, consisting of 1,050 upregulated DEGs (uDEGs) and 702 downregulated DEGs (dDEGs) in TSCC tissues compared with normal tissues.
GO term enrichment analysis
To acquire further understanding of the functions of identified DEGs, all DEGs were uploaded to DAVID to identify significant GO categories and KEGG pathways. GO analysis results showed that uDEGs were markedly enriched in BP, including signal transduction, positive or negative regulation of cell proliferation, and negative regulation of cell proliferation (Table 1); the dDEGs were enriched in signal transduction, cell adhesion, and apoptotic process (Table 1). For MF, the uDEGs were enriched in protein, ATP, and calcium ion binding; and the dDEGs were enriched in protein, ATP, and poly (A) RNA binding (Table 1). In addition, GO CC analysis showed that uDEGs were significantly enriched in the plasma membrane, cytosol, and extracellular exosome; and dDEGs were enriched in cytoplasm, nucleus, and cytosol (Table 1).
Table 1.
Gene ontology analysis of differentially expressed genes associated with TSCC.
| Expression | Category | Term/gene function | Gene count | % | P value |
|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_DIRECT | GO: 0007165~signal transduction | 95 | 9.59 | 8.67E-05 |
| GOTERM_BP_DIRECT | GO: 0045944~positive regulation of transcription from RNA polymerase II promoter | 82 | 8.27 | 1.72E-04 | |
| GOTERM_BP_DIRECT | GO: 0000122~negative regulation of transcription from RNA polymerase II promoter | 60 | 6.05 | 0.002 | |
| GOTERM_BP_DIRECT | GO: 0008285~negative regulation of cell proliferation | 45 | 4.54 | 1.50E-05 | |
| GOTERM_BP_DIRECT | GO: 0007155~cell adhesion | 45 | 4.54 | 2.35E-04 | |
| GOTERM_CC_DIRECT | GO: 0005886~plasma membrane | 263 | 26.54 | 6.71E-05 | |
| GOTERM_CC_DIRECT | GO: 0005829~cytosol | 223 | 22.50 | 8.89E-05 | |
| GOTERM_CC_DIRECT | GO: 0070062~extracellular exosome | 215 | 21.70 | 1.05E-09 | |
| GOTERM_CC_DIRECT | GO: 0005576~extracellular region | 141 | 14.23 | 5.09E-10 | |
| GOTERM_CC_DIRECT | GO: 0005615~extracellular space | 134 | 13.52 | 5.39E-13 | |
| GOTERM_MF_DIRECT | GO: 0005515~protein binding | 372 | 37.53 | 0.004 | |
| GOTERM_MF_DIRECT | GO: 0005524~ATP binding | 106 | 10.70 | 0.003 | |
| GOTERM_MF_DIRECT | GO: 0005509~calcium ion binding | 77 | 7.77 | 1.41E-08 | |
| GOTERM_MF_DIRECT | GO: 0042803~protein homodimerization activity | 66 | 6.66 | 5.29E-05 | |
| GOTERM_MF_DIRECT | GO: 0003700~transcription factor activity, sequence-specific DNA binding | 64 | 6.46 | 0.003 | |
| Down-regulated | GOTERM_BP_DIRECT | GO: 0007165~signal transduction | 83 | 12.56 | 6.42E-08 |
| GOTERM_BP_DIRECT | GO: 0007155~cell adhesion | 57 | 8.62 | 1.90E-14 | |
| GOTERM_BP_DIRECT | GO: 0006915~apoptotic process | 47 | 7.11 | 1.69E-06 | |
| GOTERM_BP_DIRECT | GO: 0008284~positive regulation of cell proliferation | 44 | 6.66 | 2.66E-07 | |
| GOTERM_BP_DIRECT | GO: 0008285~negative regulation of cell proliferation | 42 | 6.35 | 2.26E-08 | |
| GOTERM_CC_DIRECT | GO: 0005737~cytoplasm | 273 | 41.30 | 3.74E-13 | |
| GOTERM_CC_DIRECT | GO: 0005634~nucleus | 242 | 36.61 | 3.76E-05 | |
| GOTERM_CC_DIRECT | GO: 0005829~cytosol | 235 | 35.55 | 1.36E-26 | |
| GOTERM_CC_DIRECT | GO: 0070062~extracellular exosome | 227 | 34.34 | 9.35E-36 | |
| GOTERM_CC_DIRECT | GO: 0005886~plasma membrane | 202 | 30.56 | 2.95E-07 | |
| GOTERM_MF_DIRECT | GO: 0005515~protein binding | 350 | 52.95 | 1.77E-20 | |
| GOTERM_MF_DIRECT | GO: 0005524~ATP binding | 89 | 13.46 | 3.73E-05 | |
| GOTERM_MF_DIRECT | GO: 0044822~poly(A) RNA binding | 65 | 9.83 | 0.001 | |
| GOTERM_MF_DIRECT | GO: 0005509~calcium ion binding | 52 | 7.867 | 2.02E-05 | |
| GOTERM_MF_DIRECT | GO: 0042802~identical protein binding | 46 | 6.96 | 2.95E-05 |
BP – biological process; CC – cellular component; MF – molecular function; Count – numbers of DEGs; GO – gene ontology.
KEGG pathway analysis
The most significantly enriched pathways of uDEGs and dDEGs analyzed by KEGG analysis are shown in Table 2. The uDEGs were enriched in metabolic pathways, pathways in cancer, PI3K-Akt signaling pathway, calcium signaling pathway, and MAPK signaling pathway, while the dDEGs were enriched in pathways in cancer, PI3K-Akt signaling pathway, focal adhesion, and ECM-receptor interaction.
Table 2.
KEGG pathway analysis of differentially expressed genes associated with TSCC.
| Expression | Pathway ID | Name | Gene count | % | P value |
|---|---|---|---|---|---|
| Up-regulated | hsa01100 | Metabolic pathways | 110 | 11.10 | 0.029 |
| hsa05200 | Pathways in cancer | 47 | 4.74 | 0.001 | |
| hsa04151 | PI3K-Akt signaling pathway | 39 | 3.94 | 0.011 | |
| hsa04020 | Calcium signaling pathway | 37 | 3.73 | 3.28E-08 | |
| hsa04010 | MAPK signaling pathway | 36 | 3.63 | 3.35E-04 | |
| Down-regulated | hsa05200 | Pathways in cancer | 47 | 7.11 | 4.92E-07 |
| hsa04151 | PI3K-Akt signaling pathway | 42 | 6.35 | 1.47E-06 | |
| hsa04510 | Focal adhesion | 38 | 5.75 | 6.87E-11 | |
| hsa04512 | ECM-receptor interaction | 29 | 4.39 | 4.14E-15 | |
| hsa05205 | Proteoglycans in cancer | 29 | 4.39 | 3.33E-06 |
hsa – Homo sapiens; KEGG – Kyoto Encyclopedia of Genes and Genomes.
PPI network construction and modules selection
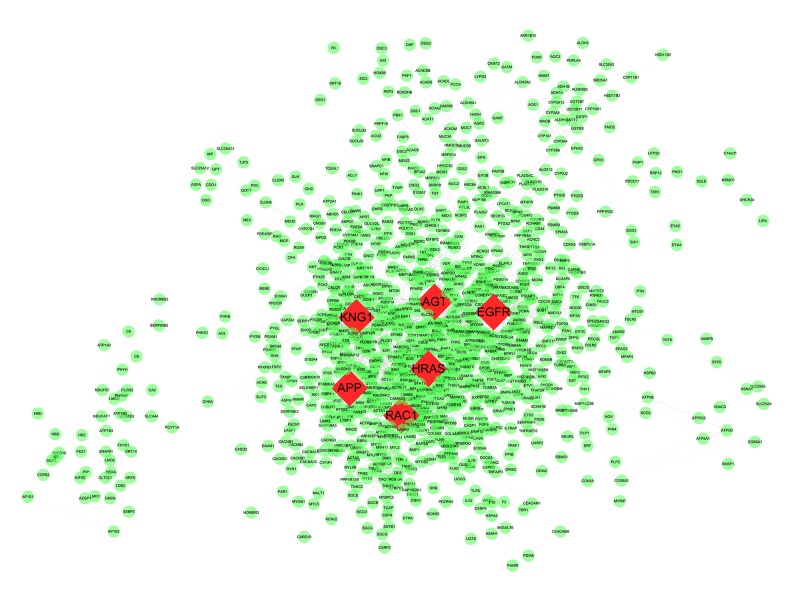
The PPI network of DEGs consisted of 1,616 nodes and 5,866 edges constructed in the STRING database (version 10.5) and visualized using Cytoscape software (Figure 1). Degree >10 was set as the cutoff criterion. Based on the STRING database, the DEGs with the highest PPI scores identified by the three centrality methods are shown in Table 3. After repeated genes were removed, the hub genes (shown in Figure 1, highlighted in red and shaped in diamond) were obtained using the three centrality methods, including RAC1 (ras-related C3 botulinum toxin substrate 1), APP (amyloid beta precursor protein), EGFR (epidermal growth factor receptor), KNG1 (kininogen 1), AGT (angiotensinogen), and HRAS (HRas proto-oncogene, GTPase). Among these genes, RAC1 showed the highest node degree, which was 78. A significant module was constructed from the PPI network of the DEGs using MCODE, including 43 nodes and 462 edges (Figure 2). Biological functional enrichment analysis showed that genes in this module were markedly enriched in signal transduction, single organism signaling, and cell communication (Table 4). Neuroactive ligand-receptor interaction, calcium signaling pathway, and chemokine signaling pathway were enriched in the KEGG pathway analysis.
Figure 1.
Protein-protein interaction network for products of DEGs. A total of 1616 nodes and 5866 interaction associations were identified. The nodes with highest PPI scores were shaped as diamond in red.
Table 3.
The top 10 differentially expressed genes with higher scores, respectively, identified by the three centrality methods.
| Subgraph | Degree | Closeness | |||
|---|---|---|---|---|---|
| APP | 7.93E10 | RAC1 | 78.0 | RAC1 | 0.01866 |
| KNG1 | 7.47E10 | APP | 74.0 | EGFR | 0.01864 |
| AGT | 7.20E10 | EGFR | 59.0 | HRAS | 0.01863 |
| RGS19 | 4.86E10 | KNG1 | 58.0 | BCL2 | 0.01861 |
| GNAI1 | 3.64E10 | CDK1 | 56.0 | CDC42 | 0.01860 |
| GCG | 3.51E10 | AGT | 55.0 | HIF1A | 0.01860 |
| ADCY8 | 3.27E10 | HRAS | 55.0 | PRKCA | 0.01860 |
| ADCY1 | 3.03E10 | CDC42 | 55.0 | CDKN1A | 0.01859 |
| CXCL12 | 2.97E10 | ADCY8 | 54.0 | MMP9 | 0.01859 |
| GNA15 | 2.74E10 | GCG | 50.0 | MAX | 0.01858 |
Figure 2.
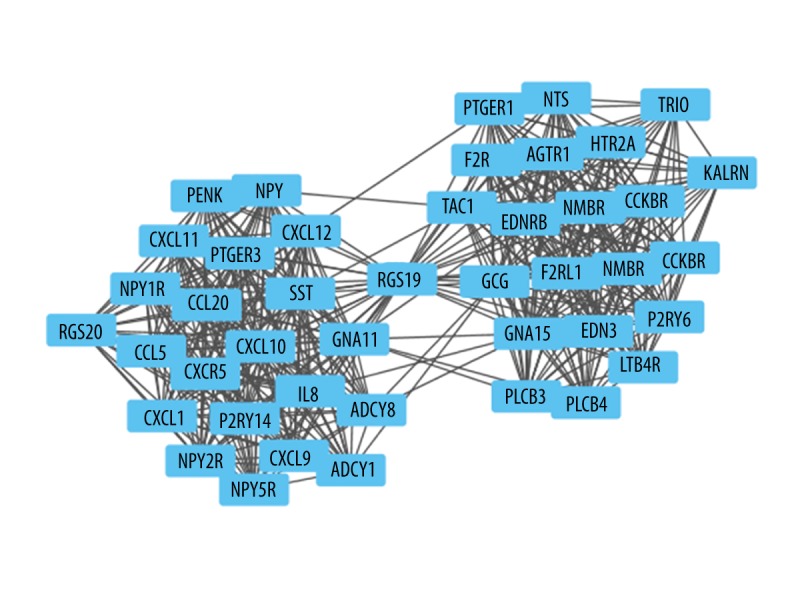
Sub network screened from protein protein interaction network. Nodes re-fer to the products of the differentially expressed genes.
Table 4.
GO and pathway analysis of genes in selected module.
| Category | Pathway ID | Term/gene and function | Count | P-value |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa4080 | Neuroactive ligand-receptor interaction | 16 | 2.83E-17 |
| hsa4020 | Calcium signaling pathway | 12 | 1.50E-13 | |
| hsa4062 | Chemokine signaling pathway | 12 | 1.50E-13 | |
| GOTERM_BP_DIRECT | GO.0007165 | Signal transduction | 31 | 1.74E-10 |
| GO.0044700 | Single organism signaling | 29 | 3.40E-08 | |
| GO.0007154 | Cell communication | 29 | 5.69E-08 | |
| GOTERM_CC_DIRECT | GO.0005886 | Plasma membrane | 28 | 1.20E-07 |
| GO.0071944 | Cell periphery | 28 | 1.38E-07 | |
| GO.0044459 | Plasma membrane part | 21 | 1.20E-07 | |
| GOTERM_MF_DIRECT | GO.0005515 | Protein binding | 20 | 0.0176 |
| GO.0005102 | Receptor binding | 19 | 7.39E-11 | |
| GO.0004871 | Signal transducer activity | 16 | 5.68E-06 |
Discussion
Despite advances in current therapeutics, TSCC has remained an intractable cancer over the past decades. Uncovering the etiological and molecular mechanisms of TSCC is of vital importance for therapy and prevention. Nowadays, with the rapid developing of DNA microarrays and high-throughput sequencing techniques, it is possible to research diseases, including cancers, at the gene level. DNA microarray gene expression profiling has been widely used to explore differentially expressed genes involved in tumor genesis, diagnosis, and therapeutic approaches [23,24].
In this study, we extracted the data from GSE13601 and identified 1,050 uDEGs and 702 dDEGs between TSCC and normal tissue samples using bioinformatics analysis. These uDEGs were obviously enriched in metabolic pathways, pathways in cancer, and the PI3K-Akt signaling pathway which are intimately related to cancer. The dDEGs were predominantly enriched in pathways in cancer, the PI3K-Akt signaling pathway, and focal adhesion.
The uDEGs were shown to be mostly involved in signal transduction, positive or negative regulation of cell proliferation, and negative regulation of cell proliferation, while dDEGs were shown to be concerned with signal transduction, cell adhesion, and the apoptotic process in the GO term analysis. This conforms to the knowledge that signal transduction, regulating of cell proliferation, cell adhesion, and apoptotic process are all important mechanisms of tumor genesis, development, and progression [25–30]. Moreover, the enriched KEGG pathways of uDEGs included metabolic pathways, pathways in cancer, and the PI3K-Akt signaling pathway. Numbers of studies have shown that metabolic pathways and the PI3K-Akt signaling pathway play an important role in genesis and growing of squamous cell carcinoma of the oral tongue [31–35]. Zhang et al. [36] reported that by rewiring alternative metabolic pathways, oral cancer cells may still survive when metabolic enzymes were silenced by siRNAs. Downregulated DEGs were also found to be involved in focal adhesion and ECM-receptor interaction. Exceedingly abnormal expression of focal adhesion kinase affected cellular proliferation and apoptosis [37], served as a marker of cervical lymph node metastasis, and a potential therapeutic target of TSCC [38]. Therefore, studying these signaling pathways could assist in the prediction of cancer progression.
The PPI network was constructed with DEGs and the top centrality hub genes were obtained: RAC1, APP, EGFR, KNG1, AGT, and HRAS. The genesis of tumor is an extremely complicated process during which lots of genetic and epigenetic modifications of driving genes occur. RAC1 was identified as one of the hub genes with the highest degree of connectivity. The protein encoded by RAC1 is a GTPase belonging to the RAS superfamily, members of which appear to be regulated widely in cellular events, such as controlling the cell growth and the activation of protein kinases. As an oncogene, RAC1 was associated with various cancers, such as melanoma, colorectal cancer, breast cancer, and glioma [39]. Increased expression and subcellular localization of RAC1 could lead to lower early response rate and higher recurrences in head and neck squamous cell carcinomas (HNSCC), suggesting that it seems to be a potential therapeutic target for HNSCC patients of chemo-radiotherapy resistant [40]. Patel et al. [41] found that most HNSCC cells showed an outstandingly high level of RAC1, and the EGFR/Vav2/Rac1 axis was a critical pathway for the ability of invasion and metastasis of most HNSCC cells. APP has been well studied in the pathogenesis of Alzheimer disease. However, little is known concerning the role of APP in carcinogenesis. Gain-of-function studies have shown that APP overexpression leads to increased cellular proliferation. Loss-of-function studies, either by APP knockdown or blockage of APP function by antibody application, have demonstrated regression of carcinoma growth in vitro and in vivo [42]. Recently, it was shown that APP was upregulated in several cancer species, including pancreatic [43], colon [44], melanoma [45], and prostate [46] cancer and had growth-promoting features. APP expression was found to be involved in the carcinogenesis and proliferation of oral SCC cells, and could serve as a marker indicating oral cancer genesis [47]. EGFR is a protein located on the cell surface binding to epidermal growth factor [48]. When a ligand binds to EGFR, the receptor will dimerize and tyrosine will autophosphorylate, leading to cell proliferation. EGFR was overexpressed in about 30% of human epithelial tumors [49], including HNSCCs [48]. Ansell et al. found that the amount of EGF had a determinant function in cell proliferation and the response to treatment of cetuximab in tongue cancer, so EGF was a potential predictive biomarker of poor cetuximab response and a possible target of treatment [50]. EGFR copy number alteration, rather than overexpression, was a better prognostic indicator in TSCC [51]. Thus, EGFR was particularly important in the pathogenesis of TSCC. There is more and more evidence demonstrating a role for KNG1 in carcinogenesis [52]. Liu et al. [53] showed that KNG1 seemed to have a function of anti-angiogenesis and blocked the proliferation of endothelial cells. In addition, lower expression of KNG1 was detected in the serum of cancer patients, which was associated with cancer cells survival [54]. Furthermore, KNG1 was shown to be a potential serum predictor of advanced colorectal adenoma and cancer [55]. AGT, encoding angiotensinogen, has been shown to be a suppressor of tumor progression and metastasis. Overexpression of human AGT decreased angiogenesis and prohibited remodeling and neovascularization of tumor cells, thus delayed tumor advancement in vivo [24,56]. Bouquet et al. [57] demonstrated that AGT had a very powerful antiangiogenic function in vivo, independent of angiotensin II generation, representing a promising novel strategy to inhibit growth and metastasis of primary tumors. HRAS belongs to the Ras oncogene family; obvious mutations of the HRAS gene were found in oral cancer, suggesting that RAS may affect the tumorigenesis process [58]. There was another interesting finding: activated HRAS mutations could overcome the resistance to erlotinib in an HNSCC cell line with HRAS mutation [59]. In summary, the top centrality hub genes (RAC1, APP, EGFR, KNG1, AGT, and HRAS) obtained from the PPI network are all deeply involved in cancer genesis or progression process, which suggesting that these hub genes may serve as prognostic biomarkers or therapeutic targets for this disease.
Module analysis of the PPI network showed that TSCC was associated with neuroactive ligand-receptor interaction, calcium signaling pathway, and chemokine signaling pathway. Neuroactive ligand-receptor interaction has been shown to be involved in various kinds of cancers such as renal cell carcinoma [60], breast cancer [61], bladder cancer [62], and lung adenocarcinoma [63] when using pathways and gene interaction networks analysis. Moreover, the intracellular calcium overload could initiate mitochondrial-dependent apoptosis [64], which is the most frequent strategy for inhibiting cancer cell proliferation. Accumulating evidence has shown that chemokines are involved in tumor growth and metastasis. Abnormal function of chemokines in cancer promotes cell survival, facilitated proliferation, angiogenesis, and metastasis in multiple types of tumors. Furthermore, it is believed that chronic inflammatory conditions facilitate oral carcinogenesis, and functions of cytokine-dependent and chemokine-dependent immunoregulatory pathways are apparent in oral carcinoma [3]. Thus, neuroactive ligand-receptor interaction and calcium and chemokine signaling pathways represent promising candidates for therapeutic intervention in TSCC patients.
Conclusions
The present study provided an extensive bioinformatics analysis of DEGs and revealed a series of targets and pathways, which may affect the carcinogenesis and progression of TSCC, for future investigation. These findings add to significant insights into the diagnosis and treatment of this disease. However, the absence of experimental validation was a limitation to our study conclusions. Therefore, further experimental studies, with larger sample sizes, are required to validate these findings.
Acknowledgements
We thank Dr. Gangjun Yuan and Dr. Xiaopin Zhong for their technical assistance.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Yu X, Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J Cell Mol Med. 2016;20(1):10–16. doi: 10.1111/jcmm.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie N, Wang C, Liu X, et al. Tumor budding correlates with occult cervical lymph node metastasis and poor prognosis in clinical early-stage tongue squamous cell carcinoma. J Oral Pathol Med. 2015;44(4):266–72. doi: 10.1111/jop.12242. [DOI] [PubMed] [Google Scholar]
- 3.Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol. 2015;6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu H, Wang Y, Li Z, et al. Overexpression of suppressor of zest 12 is associated with cervical node metastasis and unfavorable prognosis in tongue squamous cell carcinoma. Cancer Cell Int. 2017;17:26. doi: 10.1186/s12935-017-0395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B, Bao X. Identification of genes associated with laryngeal squamous cell carcinoma samples based on bioinformatic analysis. Mol Med Rep. 2015;12:3386–92. doi: 10.3892/mmr.2015.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferlazzo N, Curro M, Zinellu A, et al. Influence of MTHFR genetic background on p16 and MGMT methylation in oral squamous cell cancer. Int J Mol Sci. 2017;18(4) doi: 10.3390/ijms18040724. pii: E724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang TH, Liang LZ, Liu XL, et al. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol Rep. 2017;37:2087–94. doi: 10.3892/or.2017.5445. [DOI] [PubMed] [Google Scholar]
- 9.Chaisaingmongkol J, Popanda O, Warta R, et al. Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma. Oncogene. 2012;31(49):5108–16. doi: 10.1038/onc.2011.660. [DOI] [PubMed] [Google Scholar]
- 10.Gallo O, Masini E, Bianchi B, et al. Prognostic significance of cyclooxygenase-2 pathway and angiogenesis in head and neck squamous cell carcinoma. Hum Pathol. 2002;33(7):708–14. doi: 10.1053/hupa.2002.125376. [DOI] [PubMed] [Google Scholar]
- 11.Lao XM, Liang YJ, Su YX, et al. Distribution and significance of interstitial fibrosis and stroma-infiltrating B cells in tongue squamous cell carcinoma. Oncol Lett. 2016;11:2027–34. doi: 10.3892/ol.2016.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matarese G, Curro M, Isola G, et al. Transglutaminase 2 up-regulation is associated with RANKL/OPG pathway in cultured HPDL cells and THP-1-differentiated macrophages. Amino Acids. 2015;47:2447–55. doi: 10.1007/s00726-015-2039-5. [DOI] [PubMed] [Google Scholar]
- 13.Estilo CL, O-charoenrat P, Talbot S, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: Archive for functional genomics data sets – update. Nucleic Acids Res. 2013;41(Database issue):D991–95. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von MC, Huynen M, Jaeggi D, et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–61. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Li M, Wang J, et al. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Mohr S, Leikauf GD, Keith G, Rihn BH. Microarrays as cancer keys: An array of possibilities. J Clin Oncol. 2002;20(14):3165–75. doi: 10.1200/JCO.2002.12.073. [DOI] [PubMed] [Google Scholar]
- 24.Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33(10):111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Goto K, Iino M. Diverse functions and signal transduction of the exocyst complex in tumor cells. J Cell Physiol. 2017;232(5):939–57. doi: 10.1002/jcp.25619. [DOI] [PubMed] [Google Scholar]
- 26.Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nat Rev Cancer. 2015;15(9):515–27. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 27.Chung W, Kim M, de la Monte S, et al. Activation of signal transduction pathways during hepatic oncogenesis. Cancer Lett. 2016;370(1):1–9. doi: 10.1016/j.canlet.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croce CM, Reed JC. Finally, an apoptosis-targeting therapeutic for cancer. Cancer Res. 2016;76(20):5914–20. doi: 10.1158/0008-5472.CAN-16-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonastre E, Brambilla E, Sanchez-Cespedes M. Cell adhesion and polarity in squamous cell carcinoma of the lung. J Pathol. 2016;238(5):606–16. doi: 10.1002/path.4686. [DOI] [PubMed] [Google Scholar]
- 30.Xin M, Dong XW, Guo XL. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomed Pharmacother. 2015;69:179–85. doi: 10.1016/j.biopha.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Kahlert UD, Mooney SM, Natsumeda M, et al. Targeting cancer stem-like cells in glioblastoma and colorectal cancer through metabolic pathways. Int J Cancer. 2017;140(1):10–22. doi: 10.1002/ijc.30259. [DOI] [PubMed] [Google Scholar]
- 32.Benatti P, Chiaramonte ML, Lorenzo M, et al. NF-Y activates genes of metabolic pathways altered in cancer cells. Oncotarget. 2016;7(2):1633–50. doi: 10.18632/oncotarget.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J, Hu X, Shao W, et al. Metabolomic analysis reveals altered metabolic pathways in a rat model of gastric carcinogenesis. Oncotarget. 2016;7(37):60053–73. doi: 10.18632/oncotarget.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Lin C, Wang C, et al. Silencing Kif2a induces apoptosis in squamous cell carcinoma of the oral tongue through inhibition of the PI3K/Akt signaling pathway. Mol Med Rep. 2014;9(1):273–78. doi: 10.3892/mmr.2013.1804. [DOI] [PubMed] [Google Scholar]
- 35.Kozaki K, Imoto I, Pimkhaokham A, et al. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;97(12):1351–58. doi: 10.1111/j.1349-7006.2006.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Chai YD, Brumbaugh J, et al. Oral cancer cells may rewire alternative metabolic pathways to survive from siRNA silencing of metabolic enzymes. BMC Cancer. 2014;14:223. doi: 10.1186/1471-2407-14-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J, Lv N, Hong Y, et al. Increased expression of focal adhesion kinase correlates with cellular proliferation and apoptosis during 4-nitroquinoline-1-oxide-induced rat tongue carcinogenesis. J Oral Pathol Med. 2009;38(6):524–29. doi: 10.1111/j.1600-0714.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, Liu L, Ye J, et al. Focal adhesion kinase serves as a marker of cervical lymph node metastasis and is a potential therapeutic target in tongue cancer. J Cancer Res Clin Oncol. 2010;136(9):1295–302. doi: 10.1007/s00432-010-0780-4. [DOI] [PubMed] [Google Scholar]
- 39.Zou T, Mao X, Yin J, et al. Emerging roles of RAC1 in treating lung cancer patients. Clin Genet. 2017;91(4):520–28. doi: 10.1111/cge.12908. [DOI] [PubMed] [Google Scholar]
- 40.Skvortsov S, Dudas J, Eichberger P, et al. Rac1 as a potential therapeutic target for chemo-radioresistant head and neck squamous cell carcinomas (HNSCC) Br J Cancer. 2014;110(11):2677–87. doi: 10.1038/bjc.2014.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel V, Rosenfeldt HM, Lyons R, et al. Persistent activation of Rac1 in squamous carcinomas of the head and neck: Evidence for an EGFR/Vav2 signaling axis involved in cell invasion. Carcinogenesis. 2007;28(6):1145–52. doi: 10.1093/carcin/bgm008. [DOI] [PubMed] [Google Scholar]
- 42.Venkataramani V, Rossner C, Iffland L, et al. Histone deacetylase inhibitor valproic acid inhibits cancer cell proliferation via down-regulation of the alzheimer amyloid precursor protein. J Biol Chem. 2010;285(14):10678–89. doi: 10.1074/jbc.M109.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansel DE, Rahman A, Wehner S, et al. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032–37. [PubMed] [Google Scholar]
- 44.Meng JY, Kataoka H, Itoh H, Koono M. Amyloid beta protein precursor is involved in the growth of human colon carcinoma cell in vitro and in vivo. Int J Cancer. 2001;92(1):31–39. [PubMed] [Google Scholar]
- 45.Botelho MG, Wang X, Arndt-Jovin DJ, et al. Induction of terminal differentiation in melanoma cells on downregulation of beta-amyloid precursor protein. J Invest Dermatol. 2010;130(5):1400–10. doi: 10.1038/jid.2009.296. [DOI] [PubMed] [Google Scholar]
- 46.Takayama K, Tsutsumi S, Suzuki T, et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Res. 2009;69(1):137–42. doi: 10.1158/0008-5472.CAN-08-3633. [DOI] [PubMed] [Google Scholar]
- 47.Ko SY, Lin SC, Chang KW, et al. Increased expression of amyloid precursor protein in oral squamous cell carcinoma. Int J Cancer. 2004;111(5):727–32. doi: 10.1002/ijc.20328. [DOI] [PubMed] [Google Scholar]
- 48.Campbell NP, Hensing TA, Bhayani MK, et al. Targeting pathways mediating resistance to anti-EGFR therapy in squamous cell carcinoma of the head and neck. Expert Rev Anticancer Ther. 2016;16:847–58. doi: 10.1080/14737140.2016.1202116. [DOI] [PubMed] [Google Scholar]
- 49.Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr Relat Cancer. 2001;8(2):83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 50.Ansell A, Jedlinski A, Johansson AC, Roberg K. Epidermal growth factor is a potential biomarker for poor cetuximab response in tongue cancer cells. J Oral Pathol Med. 2016;45(1):9–16. doi: 10.1111/jop.12310. [DOI] [PubMed] [Google Scholar]
- 51.Nakata Y, Uzawa N, Takahashi K, et al. EGFR gene copy number alteration is a better prognostic indicator than protein overexpression in oral tongue squamous cell carcinomas. Eur J Cancer. 2011;47(15):2364–72. doi: 10.1016/j.ejca.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Kawasaki M, Maeda T, Hanasawa K, et al. Effect of His-Gly-Lys motif derived from domain 5 of high molecular weight kininogen on suppression of cancer metastasis both in vitro and in vivo. J Biol Chem. 2003;278(49):49301–7. doi: 10.1074/jbc.M308790200. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Cao DJ, Sainz IM, et al. The inhibitory effect of HKa in endothelial cell tube formation is mediated by disrupting the uPA-uPAR complex and inhibiting its signaling and internalization. Am J Physiol Cell Physiol. 2008;295(1):C257–67. doi: 10.1152/ajpcell.00569.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdul-Rahman PS, Lim BK, Hashim OH. Expression of high-abundance proteins in sera of patients with endometrial and cervical cancers: Analysis using 2-DE with silver staining and lectin detection methods. Electrophoresis. 2007;28(12):1989–96. doi: 10.1002/elps.200600629. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Wang X, Lin S, et al. Identification of kininogen-1 as a serum biomarker for the early detection of advanced colorectal adenoma and colorectal cancer. PLoS One. 2013;8(7):e70519. doi: 10.1371/journal.pone.0070519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent F, Bonnin P, Clemessy M, et al. Angiotensinogen delays angiogenesis and tumor growth of hepatocarcinoma in transgenic mice. Cancer Res. 2009;69(7):2853–60. doi: 10.1158/0008-5472.CAN-08-2484. [DOI] [PubMed] [Google Scholar]
- 57.Bouquet C, Lamandé N, Brand M, et al. Suppression of angiogenesis, tumor growth, and metastasis by adenovirus-mediated gene transfer of human angiotensinogen. Mol Ther. 2006;14(2):175–82. doi: 10.1016/j.ymthe.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Koumaki D, Kostakis G, Koumaki V, et al. Novel mutations of the HRAS gene and absence of hotspot mutations of the BRAF genes in oral squamous cell carcinoma in a Greek population. Oncol Rep. 2012;27(5):1555–60. doi: 10.3892/or.2012.1653. [DOI] [PubMed] [Google Scholar]
- 59.Hah JH, Zhao M, Pickering CR, et al. HRAS mutations and resistance to the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in head and neck squamous cell carcinoma cells. Head Neck. 2014;36(11):1547–54. doi: 10.1002/hed.23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, Wang J, Sun G. Identification of key genes and pathways in renal cell carcinoma through expression profiling data. Kidney Blood Press Res. 2015;40(3):288–97. doi: 10.1159/000368504. [DOI] [PubMed] [Google Scholar]
- 61.Huan J, Wang L, Xing L, et al. Insights into significant pathways and gene interaction networks underlying breast cancer cell line MCF-7 treated with 17beta-estradiol (E2) Gene. 2014;533(1):346–55. doi: 10.1016/j.gene.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 62.Fang ZQ, Zang WD, Chen R, et al. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet Mol Res. 2013;12(2):1479–89. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- 63.Wu X, Zang W, Cui S, Wang M. Bioinformatics analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. Eur Rev Med Pharmacol Sci. 2012;16(11):1582–87. [PubMed] [Google Scholar]
- 64.Pinton P, Giorgi C, Siviero R, et al. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27(50):6407–18. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]