Abstract
Mutations in the human SAMHD1 gene are known to correlate with the development of the Aicardi-Goutières Syndrome (AGS), which is an inflammatory encephalopathy that exhibits neurological dysfunction characterized by increased production of type I interferon (IFN); this evidence has lead to the concept that the SAMHD1 protein negatively regulates the type I IFN response. Additionally, the SAMHD1 protein has been shown to prevent efficient HIV-1 infection of macrophages, dendritic cells and resting CD4+ T cells. To gain insights on the SAMHD1 molecular determinants that are responsible for the deregulated production of type I IFN, we explored the biochemical, cellular and antiviral properties of human SAMHD1 mutants known to correlate with the development of Aicardi-Goutières Syndrome. Most of the studied SAMHD1 AGS mutants exhibit defects in the ability to oligomerize, decrease the levels of cellular dNTPs in human cells, localize exclusively to the nucleus, and restrict HIV-1 infection. At least half of the tested variants preserved the ability to be degraded by the lentiviral protein Vpx, and all of them interacted with RNA. Our investigations revealed that the SAMHD1 AGS variant p.G209S preserve all tested biochemical, cellular and antiviral properties, suggesting that this residue is a determinant for the ability of SAMHD1 to negatively regulate the type I interferon response in human patients with AGS. Overall, our work genetically separated the ability of SAMHD1 to negatively regulate the type I IFN response from its ability to restrict HIV-1.
Keywords: Aicardi-Goutières Syndrome, SAMHD1, interferon, HIV-1
INTRODUCTION
Mutations in the human SAMHD1 gene (MIM# 606754) have been linked to the development of Aicardi-Goutières Syndrome (AGS), a genetic disease characterized by increased expression of interferon alpha (IFN-α), which closely mimics congenital viral infection (Dale, et al., 2010; du Moulin, et al., 2011; Leshinsky-Silver, et al., 2011; Ramantani, et al., 2011; Rice, et al., 2009; Thiele, et al., 2010). AGS is considered an interferonopathie in which upregulation of type I interferon activity directly correlates with disease progression (Crow and Manel, 2015). AGS has also been shown to be caused by mutations in the 3′–5′ exonuclease TREX1 (Crow, et al., 2006), the components of the RNASEH2 endonuclease complex: RNASEH2A, RNASEH2B and RNASEH2C (Rice, et al., 2009), the cytosolic double-stranded RNA (dsRNA) sensor MDA5 (Miner and Diamond, 2014), and the dsRNA editing enzyme ADAR1 (Pestal, et al., 2015; Rice, et al., 2012). Since all known AGS mutations have been linked to proteins involved in nucleic acid metabolism, it has been hypothesized that AGS is caused by an increase in cellular nucleic acid debris which activates the cellular innate IFN-α-mediated immune response by mimicking viral infection (Crow, et al., 2006). The type I IFN response is tightly regulated and requires the cellular recognition of pathogen associated molecular patterns (PAMPs) via Pattern Recognition Receptors (PRR’s). PRR’s include Toll-Like Receptors (TLR’s) that sense nucleic acids, RIG-I-like receptors that sense RNA, and cyclic GMP-AMP synthase (cGAS), which senses DNA and activates the ER-associated stimulator of IFN genes protein (STING) pathway (Habjan and Pichlmair, 2015; Wu and Chen, 2014). Mutations in TREX1 linked to AGS have been shown to cause increased cellular nucleic acid levels and activate the innate immune response (Crow, et al., 2006; Fye, et al., 2011; Orebaugh, et al., 2011; Rice, et al., 2007; Rice, et al., 2013; Stetson, et al., 2008). The TREX1-deficient mice display chronic IFN production, as seen in AGS (Gray, et al., 2015). In addition, the TREX1-deficient mice exhibit a reduced survival and develop inflammatory myocarditis (Morita, et al., 2004). When the TREX1-deficient mice is crossed with interferon alpha-receptor 1 (IFNAR1)-deficient mice, progeny display no phenotypic differences compared to wild type mice demonstrating the need for IFN-α signaling in the development of the phenotype (Stetson, et al., 2008). In agreement with the hypothesis that SAMHD1 modulates the type I IFN response (Rice, et al., 2009), the SAMHD1-deficient mice also exhibit higher basal levels of type I IFN (Behrendt, et al., 2013; Rehwinkel, et al., 2013).
Recent work identified SAMHD1 as the protein that blocks infection of SIVΔVpx, HIV-2ΔVpx and HIV-1 before reverse transcription in macrophages, dendritic cells and resting CD4+ T cells (Baldauf, et al., 2012; Berger, et al., 2011; Descours, et al., 2012; Hrecka, et al., 2011; Laguette, et al., 2011). The SAMHD1 protein is comprised of the sterile alpha motif (SAM) and histidine-aspartic (HD) domains. The HD domain of SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase that decreases the cellular levels of dNTPs (Goldstone, et al., 2011; Kim, et al., 2012; Lahouassa, et al., 2012; Powell, et al., 2011). The sole HD domain is sufficient to potently restrict infection by different viruses (White, et al., 2013a). The HD domain is also necessary for the ability of SAMHD1 to oligomerize and to bind RNA (White, et al., 2013a). The antiviral activity of SAMHD1 is regulated by phosphorylation (Cribier, et al., 2013; Welbourn, et al., 2013; White, et al., 2013b). Interestingly, an antivirally active SAMHD1 is unphosphorylated on T592. By contrast, SAMHD1 phosphorylated on T592 is antivirally inactive. These findings showed that contrary to what happens in cycling cells, SAMHD1 is unphosphorylated in non-cycling cells. These results proposed an explanation for the reason that SAMHD1 is expressed in cycling and non-cycling cells, but it only exhibits antiviral activity in non-cycling cells.
The human population contains a large number of SAMHD1 variants that are associated with the development of AGS. We and others have previously defined the biochemical, cellular and antiviral properties of SAMHD1 by developing a series of robust assays (Brandariz-Nunez, et al., 2012b; Cribier, et al., 2013; Kim, et al., 2012; Lahouassa, et al., 2012; White, et al., 2013a; White, et al., 2013b). To gain insights on the molecular determinants of SAMHD1 that are involved in the development of AGS, we have characterized a large panel of SAMHD1 AGS mutants that are known to correlate with increase levels of type I IFN in human patients (Rice, et al., 2013). For this purpose, we tested the different human SAMHD1 AGS variants that increase levels of type I IFN in human patients for antiviral activity, oligomerization, RNA binding, subcellular localization, Vpx-mediated degradation, and their ability to decrease the cellular levels of dNTPs in human cells.
MATERIALS AND METHODS
Cell lines and Plasmids
Human U937 (ATCC# CRL-1593.2) cells were grown in RPMI suplemented with 10% (v/v) fetal bovine serum and1% (v/v) penicillin/streptomycin. Human HeLa (ATCC# CCL-2) cells were grown on DMEM suplemented with 10% (v/v) fetal bovine serum and 1% (v/v)penicillin/streptomycin. LPCX-SAMHD1-HA and LPCX-SAMHD1-FLAG plasmids expressing the codon optimized SAMHD1 (MIM# 606754, RefSeq NM_015474.3) fused to either HA or FLAG epitope were previously described (Brandariz-Nunez, et al., 2012b). The plasmids expressing SAMHD1 mutants were created using pLPCX-SAMHD1-FLAG as template and specific primers. PCR products were digested and cloned into the EcoRI and ClaI sites of pLPCX. Orientation of the inserts was confirmed by sequencing and restriction analysis.
Generation of U937 cells stably expressing SAMHD1 variants
Retroviral vectors encoding wild type or mutant SAMHD1 proteins fused to FLAG were created using the LPCX vector (Clontech). Recombinant viruses were produced in 293FT cells by co-transfecting the LPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells (Yee, et al., 1994). Transduced human monocytic U937 cells were selected in 0.4 μg /ml puromycin (Sigma).
Protein analysis
Cellular proteins were extracted with radioimmunoprecipitation assay (RIPA) as previously described (Lienlaf et al., 2011). Detection of proteins by Western blotting was performed using anti-FLAG (Sigma; Cat # F7425), anti-GAPDH (Ambion; Cat # 4300) or anti-HA (Sigma; Cat # 3663). Secondary antibodies against rabbit and mouse conjugated to Alexa Fluor 680 were obtained from Li-Cor. Bands were detected by scanning blots using the Li-Cor Odyssey Imaging System in the 700 nm channel.
Infection with retroviruses expressing the green fluorescent protein (GFP)
Recombinant retroviruses expressing GFP, pseudotyped with the VSV-G glycoprotein, were prepared as described (Diaz-Griffero et al., 2008). For infections, 6 × 104 cells seeded in 24-well plates and were either treated with either concentration 10 ng/mL phorbol-12-myristate-3-acetate (PMA) or DMSO for 16 hours. PMA stock solution was prepared in DMSO at 250 μg/mL. Subsequently, cells were incubated with the indicated retrovirus for 48 hours at 37°C. The percentage of GFP-positive cells was determined by flow cytometry (Becton Dickinson). Viral stocks were titrated by serial dilution on dog Cf2Th (ATCC# CRL-1430) cells.
SAMHD1 oligomerization assay
Approximately 1.0 × 107 human 293FT cells (Invitrogen) were cotransfected with plasmids encoding FLAG-tagged and HA-tagged mutant and wild type SAMHD1 proteins. Approximately 3ug of pLPCX containing FLAG-tagged wild type SAMHD1, p.[(Y146S; Y154S)], p.I201N, p.G209S, p.D311A, p.L369S, p.I448T or p.Q548A was cotransfected with 3ug of pLPCX containing wild type SAMHD1-HA and 12ug of empty pLPCX. For the less stable inserts p.H123P, p.R143C, p.R143H, p.R145Q, p.H167Y, p.F217C, p.R226G, p.M254V, p.M385V, p.R442X and p.Q548X, 15ug of pLPCX containing mutant SAMHD1-FLAG was cotransfected with pLPCX containing wild type SAMHD1-HA. After 24 h, cells were lysed in 0.5 ml of whole-cell extract (WCE) buffer (50 mM Tris [pH 8.0] 280 mM NaCl, 0.5% IGEPAL, 10% glycerol, 5mM MgCl2, 50 μg/ml ethidium bromide from a 10mg/ml solution [MP; Cat# 04802511], 50 U/ml benzonase [Novogen; Cat# 70746-3], EDTA-free protease inhibitor cocktail tail [Roche; Cat# 11836170001]). Lysates were incubated at 4°C with gentle rocking for 45min and centrifuged at 14,000 rpm for 1 h at 4°C. Post-spin lysates were then pre-cleared using protein A-agarose (bioWORLD; Cat# 20181028-1) for 1 h at 4°C with gentle rocking; a small aliquot of each of these lysates was stored as Input. Pre-cleared lysates containing the tagged proteins were incubated with anti-FLAG-agarose beads (Sigma; Cat# A2220) for 2 h at 4°C with gentle rocking. Anti-FLAG-agarose beads were washed three times in WCE buffer changing the tube between the second and third washes, and immune complexes were eluted using 100μl of 200 μg /ml FLAG tripeptide (Sigma; Cat# F4799) in WCE buffer for 1hr upright with gentle rocking at 4°C. The eluted samples were separated by SDS-PAGE and analyzed by Western blotting using either anti-HA or anti-FLAG antibodies.
Nucleic-acid binding assay
Nucleic-acid binding assay was performed as previously described (Goncalves et al., 2012). In brief, the synthetic DNA phosphorothioate-containing interferon-stimulatory DNA (ISD-PS), which is an RNA analog, was synthesized with a 5′-biotin tag using the following primers:
ISD sense 5′-tacagatctactagtgatctatgactgatctgtacatgatctaca-3′,
ISD antisense 5′-tgtagatcatgtacagatcagtcatagatcactagtagatctgta-3′
Sense and antisense primers were incubated at 65 °C for 20 minutes, and primers were allowed to anneal by cooling down to room temperature. Anealed primers were immobilized on a Ultralink Immobilized Streptavidin Plus Gel (Pierce). Cells were lysed using TAP lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 5% glycerol, 0.2% NP-40, 1.5 mM MgCl2, 25 mM NaF, 1 mM Na3VO4, protease inhibitors) and lysates were cleared by centrifugation. Cleared lysates (Input) were incubated with immobilized nucleic acids at 4°C on a rotary wheel for 2h in the presence of 10 μg/ml of Calf-thymus DNA (Sigma) as a competitor. Unbound proteins were removed by three consecutive washes in TAP lysis buffer. Bound proteins to nucleic acids (Immunoprecipitation) were eluted by boiling samples in SDS sample buffer (63 mM Tris HCl, 10% Glycerol 2% SDS, 0.0025% Bromophenol Blue) and analyzed by Western blotting using anti-FLAG antibodies (Sigma).
Transfections and Immunofluorescence microscopy
Transfections of cell monolayers were performed using Lipofectamine Plus reagent (Invitrogen), according to the manufacturer’s instructions. Transfections were incubated at 37 °C for 24 h. Indirect immunofluorescence microscopy was perfomed as previously described (Diaz-Griffero, 2002). Transfected monolayers grown on coverslips were washed twice with PBS and fixed for 15 min in 3.9 % paraformaldehyde in PBS. Fixed cells were washed twice in PBS, permeabilize for 4 min in permeabilizing buffer (0.5% Triton X-100 in PBS), and then blocked in PBS containing 2% bovine serum albumin(blocking buffer) for 1h at room temperature. Cells were then incubated for 1h at room temperature with primary antibodies diluted in blocking buffer. After three washes with PBS, cells were incubated for 30 min in secondary antibodies and 1μg of DAPI (49, 69-diamidino-2-phenylindole)/ml. Samples were mounted for fluorescence microscopy by using the ProLong Antifade Kit (Molecular Probes, Eugene, OR). Images were obtained with a Zeiss Observer.Z1 microscope using a 63X objective, and deconvolution was performed using the software AxioVision V4.8.1.0 (Carl Zeiss Imaging Solutions).
RESULTS
The ability of SAMHD1 AGS variants to restrict HIV-1
Although it is established that SAMHD1 deficient CD14+ cells derived from an AGS patient containing the SAMHD1 mutation p.R164X are susceptible to HIV-1 infection (Berger, et al., 2011), the ability of most SAMHD1 AGS mutants to restrict HIV-1 has not been studied. To this end, we generated a series of SAMHD1 AGS mutant constructs. For these studies we selected a comprehensive list of human SAMHD1 mutants expressed in patients that exhibit increased levels of type I IFN (Figure 1A and Table 1) (Rice, et al., 2009; Rice, et al., 2013). The indicated SAMHD1 variants were stably expressed in the human monocytic cell line U937 by using the LPCX vector system (Figure 1A and B), as previously described (Brandariz-Nunez, et al., 2012b). As shown in Figure 1B, the different SAMHD1 variants are well expressed in the human U937 cell line. U937 cells stably expressing SAMHD1 variants were induced to differentiate by PMA treatment to a noncycling state (Figure 1C) (Schwende, et al., 1996). Differentiated U937 cells were challenged with increasing amounts of HIV-1 viruses containing the green fluorescent protein as a reporter (HIV-1-GFP) (Figure 1C and Table 1). As a control, we performed similar infections in PMA-treated U937 cells containing the empty vector LPCX. As expected, wild type SAMHD1 blocked HIV-1 infection; however, all variants tested except for p.G209S lost the ability to restrict HIV-1 infection. p.G209S is among the first mutations found to correlate with the development of AGS (Rice, et al., 2009). Our results indicated that SAMHD1 variants p.H167Y, p.I201N, p.F217C, p.R226G, p.M254V, p.D311A, p.L369S, p.M385V, p.R442X, p.I448T, p.Q548A and p.Q548X, lost its ability to block HIV-1 infection when compared to wild type. In agreement with previous reports, the SAMHD1 variants p.H167Y, p.D311A and p.Q548A mutants do not restrict HIV-1 (Arnold, et al., 2015; Ryoo, et al., 2014). The HIV-1 restriction by the SAMHD1 AGS variants p.H123P, p.R143C, p.R143H, p.R145Q and p.R290H were not determined due to lack of expression.
Figure 1. Characterization of the ability of the different SAMHD1 AGS variants to restrict HIV-1 infection. (A).
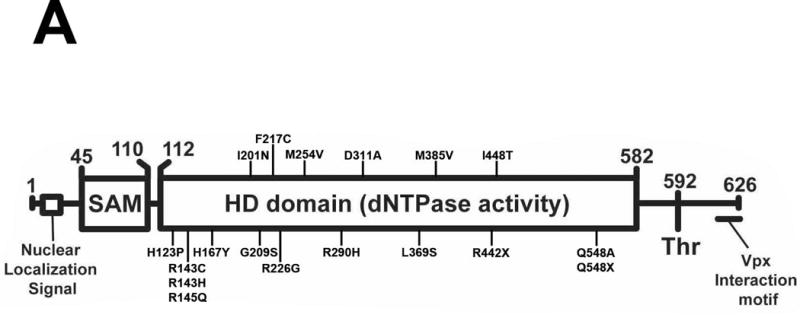
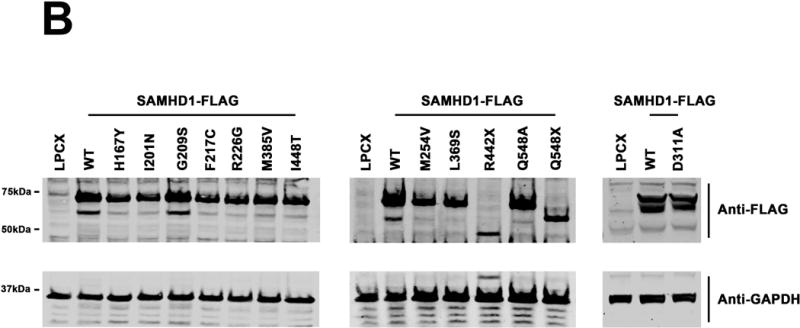
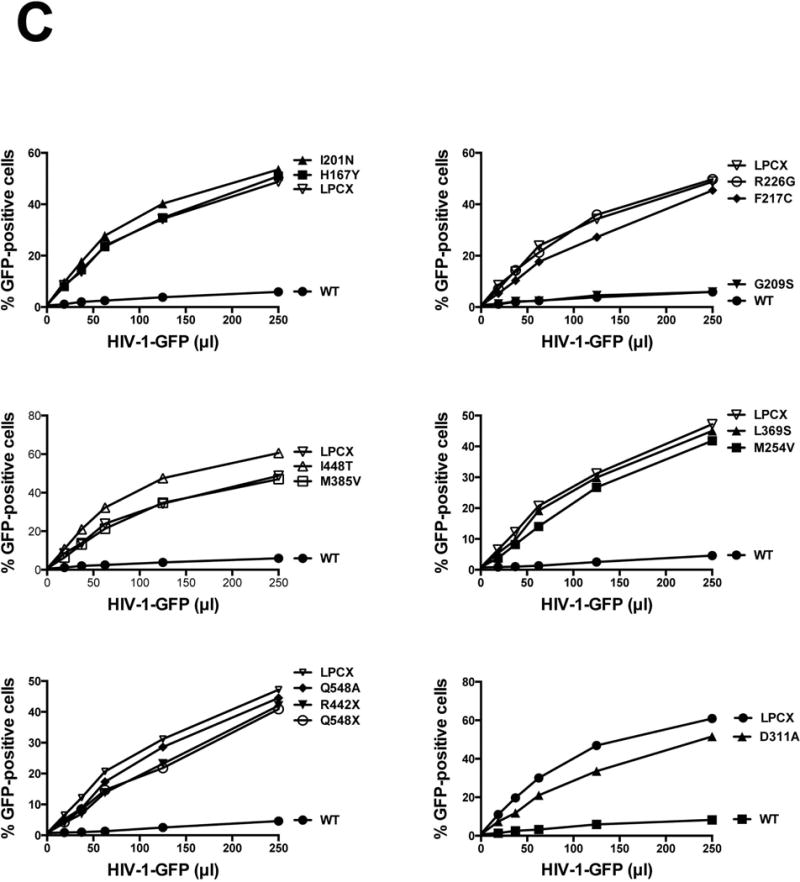
Human SAMHD1 protein is depicted showing the numbers of the amino acids residues at the boundaries of each domain along with the position and number of each AGS mutant studied here. AGS mutants studies in this work as indicates. The nuclear localization signal, the phosphorylation site on T592, and the Vpx interaction motif are depicted. Human monocytic U937 cells stably expressing the indicated mutant and wild type SAMHD1 proteins (B) were challenged with increasing amounts of HIV-1-GFP. Forthy hours post infection the percentage of GFP-positive cells was determined by flow cytometry (C). As control, U937 cells stably transduced with the empty vector LPCX were challenged with HIV-1-GFP. All SAMHD1 AGS mutant infection experiments were repeated at least 3 times and a representative figure is shown.
Table 1.
Phenotypes of SAMHD1 variants.
| SAMHD1 Variant | HIV-1 Restrictiona | Oligomerizationb | RNA Bindingc | Vpx Degradationd | dNTP Levele | Localizationf |
|---|---|---|---|---|---|---|
| WT | + | +++ | + | + | Low | N |
| p.H123P | ND | − | + | − | ND | N + C |
| p.R143C | ND | − | + | − | ND | N + C |
| p.R143H | ND | − | + | − | ND | N + C |
| p.R145Q | ND | − | + | + | ND | N + C |
| p.H167Y | − | − | + | + | High | N + C |
| p.1201 N | − | ++ | + | + | High | N + C |
| p.G209S | + | +++ | + | + | Low | N |
| p.F217C | − | − | + | + | High | N + C |
| p.R226G | − | + | + | + | High | N + C |
| p.M254V | − | +++ | + | − | High | N + C |
| p.R290H | ND | − | + | + | ND | N + C |
| p.D311A | − | +++ | + | + | High | N |
| p.L369S | − | ++ | + | + | High | N + C |
| p.M385V | − | − | + | − | High | N + C |
| p.R442X | − | + | − | High | N + C | |
| p.I448T | − | ++ | + | − | High | N + C |
| p.Q548A | − | +++ | + | + | Low | N + C |
| p.Q548X | − | − | + | − | High | N + C |
WT, wild type
ND, Not Determined
N, Nuclear; C, Cytoplasmic
Restriction was measured by infecting PMA-treated U937 cells stably expressing the indicated SAMHD1 (NM_015474.3) variants with HIV-1-GFP. After 48 h, the percentage of GFP-positive cells (infected cells) was determined by flow cytometry.
SAMHD1-FLAG variants were assayed for their ability to oligomerize, as described in Materials and Methods. “+++” indicates 100% oligomerization, which correspond to the amount of SAMHD1-HA that oligomerizes with wild type SAMHD1-FLAG; “++” indicates ~60% of binding; “+” indicates ~30% oligomerization; “−“ indicates that oligomerization was not detected.
SAMHD1-FLAG variants were assayed for their ability to interact with RNA as described in Materials and Methods. “+” is the binding achieved by wild type SAMHD1.
Vpx-dependent degradation of each human SAMHD1 polymorphism was determined as described in Materials and Methods. “+” indicates similar Vpx-mediated SAMHD1 degradation to the one observed for wild type SAMHD1; “−“indicates that degradation was not detected.
The cellular dNTP levels of U937 cells stably expressing the different SAMHD1 variants were determined as described in Materials and Methods. “High” indicates that SAMHD1 is unable to decrease the levels of dNTPs. ”Low” indicates similar dNTP levels to the ones observed for U937 cells stably expressing wild type SAMHD1.
Subcellular localization of SAMHD1 variants was determined as described in Materials and Methods and quantified (Supp. Table S1). “N” indicates nuclear localization; “C” indicates cytoplasmic localization; “N+C” indicates localization throughout the cell.
Oligomerization of SAMHD1 AGS variants
Previous reports have established the oligomerization nature of SAMHD1 (Brandariz-Nunez, et al., 2013; Ji, et al., 2014; White, et al., 2013a; Yan, et al., 2013). Next, we wanted to analyze the oligomerization ability of the SAMHD1 AGS mutations that correlate with the development of AGS. For this purpose, we compared the ability of the different SAMHD1-FLAG variants to interact with a wild type SAMHD1-HA (Figure 2), as we have previously described (White, et al., 2013a). As a negative control, we cotransfected the oligomerization deficient mutant p.[(Y146S; Y154S)] that we have previously reported (Brandariz-Nunez, et al., 2013). Our results indicated that SAMHD1 variants p.G209S, p.M254V, p.D311A and p.Q548A variants oligomerized with wild type SAMHD1-HA suggesting that these variants are intact for their ability to oligomerize in human cells. By contrast, SAMHD1 variants p.H123P, p.R143C, p.R143H, p.R145Q, p.H167Y, p.F217C, p.R290H, p.M385V, p.R442X, and p.Q548X lost their ability to oligomerize with wild type SAMHD1-HA. (Figure 2 and Table 1). In addition, we found that SAMHD1 variants p.I201N, p.L369S and p.I448T exhibit intermediate oligomerization ability when compared to wild type SAMHD1.
Figure 2. Oligomerization of mutant and wild type SAMHD1 proteins.
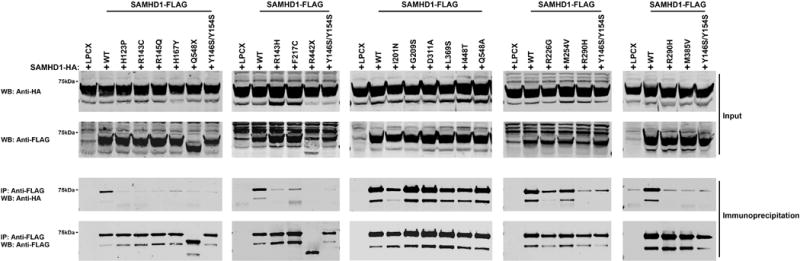
Human 293T cells were cotransfected with a plasmid expressing wild type SAMHD1-HA and a plasmid either expressing wild type or mutant SAMHD1-FLAG proteins. Cells were lysed 24 hours after transfection and analyzed by Western blotting using anti-HA and anti-FLAG antibodies (Input). Subsequently, lysates were immunoprecipitated by using anti-FLAG agarose beads, as described in Materials and Methods. Anti-FLAG agarose beads were eluted using FLAG peptide, and elutions were analyzed by Western blotting using anti-HA and anti-FLAG antibodies (Immunoprecipitation). Similar results were obtained in three independent experiments and representative data is shown. WB, Western blot; IP, Immunoprecipitation; WT, wild type.
SAMHD1 AGS variants binding to nucleic acids
Previous experiments demonstrated the ability of SAMHD1 to interact with nucleic acids (Goncalves, et al., 2012; Seamon, et al., 2015; Tungler, et al., 2013; White, et al., 2013a; White, et al., 2013b). Previous work has demonstrated that SAMHD1 preferentially binds RNA by testing the ability of SAMHD1 from total mammalian or bacterial extracts to bind the interferon-stimulatory DNA sequence containing a phosphorothioate backbone (ISD-PS), which is an RNA analog (Goncalves, et al., 2012; White, et al., 2013a; White, et al., 2013b). These experiments raised the possibility that SAMHD1 is directly interacting with RNA. Here we wanted to know whether the SAMHD1 variants described in this work bind RNA. To assay the ability of SAMHD1 variants to bind RNA, we tested the ability of SAMHD1 variants produced in human 293T cells to bind ISD-PS (Figure 3 and Table 1). Analysis of binding to RNA revealed that every SAMHD1 AGS variant tested binds RNA (Figure 3 and Table 1). In agreement with a previous report, we found that all SAMHD1 AGS variants containing an intact N-terminal region of the HD domain till residue R442 interact with RNA (Goncalves, et al., 2012) (Figure 3 and Table 1).
Figure 3. SAMHD1 AGS mutants binding to nucleic acids.
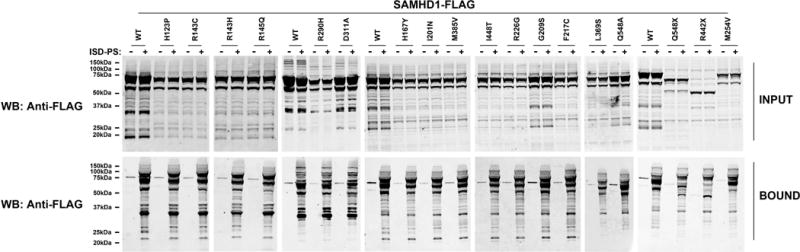
Human 293T cells were transfected with plasmid expressing the indicated wild type and mutant SAMHD1 protein. Cells were lysed 24 hours after transfection (Input), were incubated with ISD-PS immobilized in StrepTactin Superflow affinity resin. Similarly, eluted proteins from the resin were visualized by Western blotting using anti-FLAG antibodies (Bound). Similar results were obtained in three independent experiments and a representative experiment is shown. ISD-PS, interferon-stimulatory DNA sequence containing a phosphorothioate backbone; WB, Western blot; WT, wild type.
Subcellular Localization of SAMHD1 AGS variants
Previous work has demonstrated that SAMHD1 is a nuclearly localized protein (Brandariz-Nunez, et al., 2012b; Hofmann, et al., 2012; Rice, et al., 2009; Schaller, et al., 2014). Next we investigated the subcellular localization of the SAMHD1 variants studied on this work in the human HeLa cells (Figure 4, Supp. Figure S1, and Supp. Table S1). In agreement with previous findings that the NLS of SAMHD1 is located on the first 15 amino acids (11KRPR14) of the protein (Brandariz-Nunez, et al., 2012b; Hofmann, et al., 2012), all SAMHD1 variants containing the NLS localized to the nucleus of human HeLa cells. Most variants, with the exception of p.G209S and p.D311A, localized to the cytoplasm Figure 4, Supp. Figure S1, and Supp. Table S1). The nuclear and cytoplasmic localization for some of the SAMHD1 AGS variants is in agreement with a previous report (Goncalves, et al., 2012). Interestingly p.G209S and p.D311A showed nuclear localization similar to wild type SAMHD1.
Figure 4. Intracellular distribution of SAMHD1 AGS variant proteins in HeLa cells.
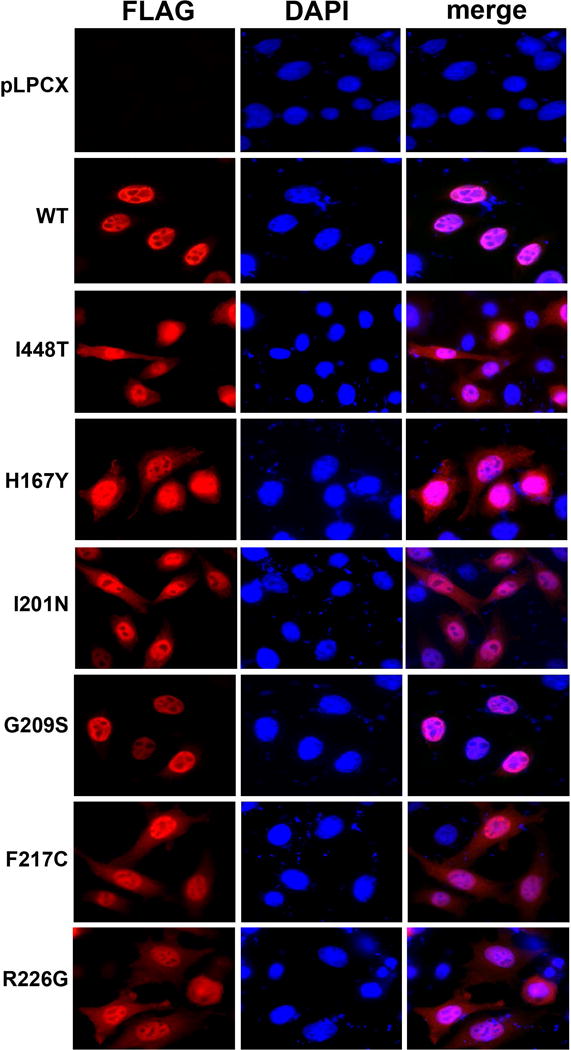
HeLa cells expressing the indicated SAMHD1-FLAG variants were fixed and immunostained using antibodies against FLAG (red). Cellular nuclei were stained by using DAPI (blue). Nuclear and cytosolic distribution was quantified(Supp. Table S1).
Ability of Vpx to degrade human SAMHD1 polymorphisms
Because the viral protein Vpx from different viruses induces the degradation of SAMHD1(Hrecka, et al., 2011; Laguette, et al., 2012; Laguette, et al., 2011), we tested the ability of Vpx from HIV-2ROD (VpxROD) to degrade the different SAMHD1 variants, as previously described (White, et al., 2013a). For this purpose, we cotransfected the different human SAMHD1 polymorphisms together with VpxROD and measured the expression level of SAMHD1 (Figure 5). As a control, we cotransfected the different human SAMHD1 variants with Vpx SIVrcm (Vpxrcm), which is unable to induce the degradation of human SAMHD1 (Brandariz-Nunez, et al., 2012a; White, et al., 2013a). As shown in Figure 5, we observed that the SAMHD1 variants p.R145Q, p.H167Y, p.I201N, p.G209S, p.F217C, p.R226G, p.R290H, p.D311A, p.L369S and p.Q548A were degraded by VpxROD (Table 1). By contrast, SAMHD1 variants p.H123P, p.R143C, p.R143H, p.M254V, p.M385V, and p.I448T were not degraded by VpxROD. Also, in agreement with previous reports, SAMHD1 p.R442X and p.Q548X were not degraded by VpxROD (Ahn, et al., 2012; Laguette, et al., 2012; Lim, et al., 2012; Zhang, et al., 2012).
Figure 5. Vpx-dependent degradation of SAMHD1 variants.
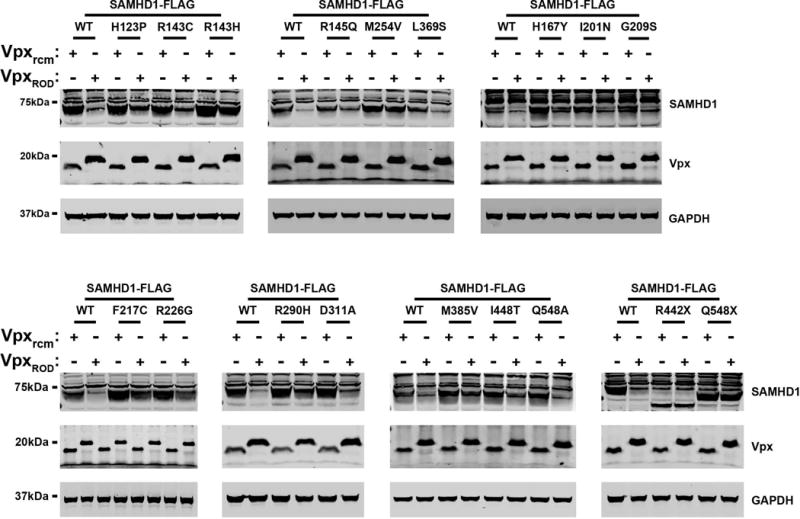
HeLa cells were cotransfected with plasmids expressing the indicated SAMHD1 variants and HA-tagged Vpx from HIV-2ROD (VpxROD) or SIVRCM (Vpxrcm). Thirty-six hours post-transfection cells were harvested, and the expression levels of SAMHD1 and Vpx were analyzed by Western blotting using anti-FLAG and anti-HA antibodies. As a loading control, cell extracts were Western blotted using antibodies against GAPDH. Similar results were obtained in three independent experiments and a representative experiment is shown.
Level of cellular dNTPs in U937 cells stably expressing the different SAMHD1 AGS mutants
Previous observations have suggested that SAMHD1 decreases the intracellular pool of deoxynucleotide triphosphates (dNTPs) (Goldstone, et al., 2011; Kim, et al., 2012; Lahouassa, et al., 2012; Powell, et al., 2011; White, et al., 2013a). To test whether the different human SAMHD1 proteins are affected in their ability to decrease the cellular levels of dNTPs, we measured the levels of dATP, dTTP and dGTP in U937 cells stably expressing the different SAMHD1 AGS variants. As shown in Figure 6, of all the tested SAMHD1 AGS polymorphisms, only p.G209S and p.Q548A were able to decrease the cellular levels of dNTPs. These findings are in agreement with a recent report showing that SAMHD1 p.Q548A retains the ability to decrease the cellular levels of dNTPs (Ryoo, et al., 2014). The ability of the SAMHD1 AGS variants p.H123P, p.R143C, p.R143H, p.R145Q and p.R290H to decrease cellular dNTP levels was not determined due to lack of expression.
Figure 6. Cellular dATP, dTTP and dGTP levels in U937 cells stably expressing the different SAMHD1 variants.
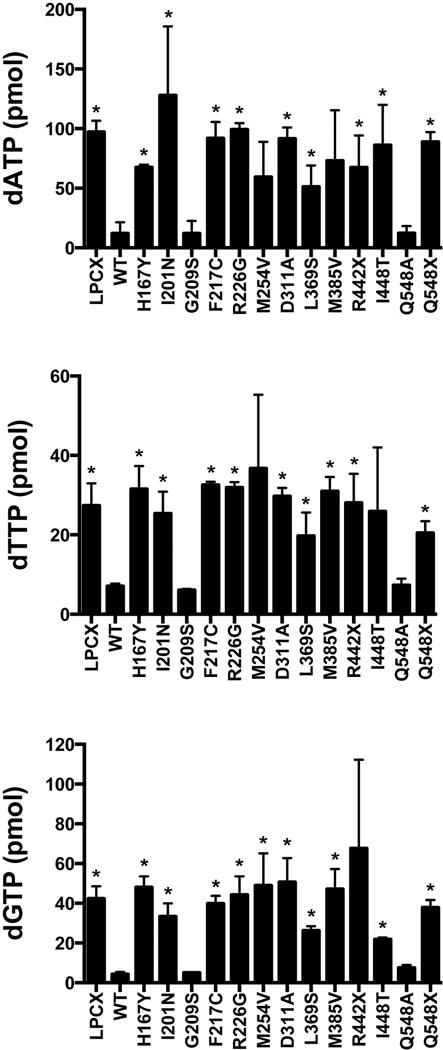
Quantification of dATP, dTTP and dGTP levels on PMA-treated U937 cells expressing the indicated SAMHD1 variants was performed by a primer extension assay as previously described. Similar results were obtained in three independent experiments and standard deviation is shown. “ * “ Indicates statistical significance compared to U937 cells expressing WT SAMHD1 as determined by an unpaired student t-test with P > 0.05. WT, wild type.
Discussion
This work explores the different properties of human SAMHD1 mutations associated with an increased production of type I IFN and the development of AGS in humans. We initially tested for the ability of these mutants to block HIV-1 infection. Of the tested SAMHD1 AGS mutants, only p.G209S retained the ability to block HIV-1 infection. Although most of the tested human SAMHD1 AGS mutations lose their ability to block HIV-1 infection, the discovery of an AGS mutant that retains the ability to restrict HIV-1 infection but loses the ability to protect individuals from AGS suggests that these two phenotypes are the result of distinguishable functions.
The SAMHD1 p.G209S variant along with a subset of the SAMHD1 AGS mutants characterized here have been shown to lose the ability to inhibit LINE-1 retrotransposition (Zhao, et al., 2013). Because human and mouse cells that are knockout for the expression of SAMHD1 exhibit elevated production of type I IFNs (Behrendt, et al., 2013; Kretschmer, et al., 2015; Rehwinkel, et al., 2013; Roesch and Schwartz, 2013; Zhang, et al., 2015), one hypothesis is that the inability of SAMHD1 p.G209S to interfere with LINE-1 replication allows innate immune sensors to recognize LINE-1 consequentially triggering an interferon response. This hypothesis is in agreement with recent findings suggesting that the spontaneous IFN response in SAMHD1-deficient mice requires the cGAS/STING cytosolic DNA-sensing pathway (Maelfait, et al., 2016). Altogether these experiments suggest that expression of the SAMHD1 p.G209S mutant or the absence of SAMHD1 expression in patients leads to the sensing of DNA, RNA or DNA/RNA species, by sensors such as cGAS/STING(DNA and DNA/RNA hybrids), RIG-I(RNA) or MDA5(RNA), which triggers a type I interferon response(Mankan, et al., 2014; Wu and Chen, 2014). This suggests that SAMHD1 might be preventing the production or the sensing of nucleic acids. Inhibition of LINE-1 retrotransposition is an example of preventing the formation of these nucleic acid species, and it is possible that SAMHD1 is preventing the formation of other yet unknown nucleic acid species that trigger inflammatory signals. In agreement with the observation that SAMHD1 interacts with nucleic acids (Goncalves, et al., 2012; Seamon, et al., 2015; White, et al., 2013a), it is also conceivable that SAMHD1 binds to nucleic acids sterically obstructing the detection of these nucleic acids by the sensors cGAS/STING, RIG-I or MDA5, preventing the induction of type I interferon. Future investigations will distinguish the mechanism that is used by SAMHD1 to negatively modulate the type I interferon response.
Previous reports have established the oligomerization nature of SAMHD1 (Brandariz-Nunez, et al., 2013; Ji, et al., 2014; White, et al., 2013a; Yan, et al., 2013). Next we thoroughly characterize the oligomerization properties of the different SAMHD1 mutants using our established assay to detect oligomerization of SAMHD1 proteins in mammalian cells (Brandariz-Nunez, et al., 2013; White, et al., 2013a; White, et al., 2013b). Our results indicated that most of the SAMHD1 AGS variants tested lost their oligomerization properties, at least to some extent, with the exception of p.G209S, p.M254V, p.D311A and p.Q548A, which were found to oligomerize similar to wild type SAMHD1.
Because SAMHD1 has the ability to bind RNA in vitro (Goncalves, et al., 2012; Seamon, et al., 2015; White, et al., 2013a), we tested the ability of SAMHD1 variants to bind RNA. All tested human SAMHD1 polymorphisms retained the ability to bind the RNA analogue ISD-PS in vitro. Overall, these results suggest that the affinity of SAMHD1 for RNA is difficult to abolish.
Because Vpx triggers the degradation of SAMHD1 (Hrecka, et al., 2011; Laguette, et al., 2011), we tested the ability of Vpx to induce the degradation of the different human SAMHD1 proteins. Consistent with the degradation requirement for VpxROD to bind to the C-terminus of SAMHD1 (Laguette, et al., 2012; Yan, et al., 2013), we found that the truncation mutants p.R442X and p.Q548X were not degraded by VpxROD. In addition, the full-length SAMHD1 variants p.H123P, p.R143C, p.R143H, p.M254V, p.M385V, and p.I448T were resistant to VpxROD-induced degradation when compared to wild type SAMHD1, which is in agreement with the notion that other regions of SAMHD1, besides the C-terminus, are required for Vpx induced degradation (Lim, et al., 2012). In a separate report, investigators have proposed a model in which Vpx interacts with the oligomeric interface of SAMHD1 suggesting that oligomeric SAMHD1 is required for its interaction with Vpx (Fregoso, et al., 2013). In agreement, we observed several SAMHD1 mutants that are defective for oligomerization and their ability to be degraded by Vpx (p.H123P, p.R143C, p.R143H and p.M385V). Interestingly, SAMHD1 variants p.R145Q, p.H167Y, p.I201N, p.G209S, p.F217C, p.R225G, p.R290H, p.D311A, p.L369S and p.Q548A were all degraded when co-expressed with VpxROD similar to wild type SAMHD1.
We also tested the nuclear localization of the different human SAMHD1 variants. In agreement with previous work, none of the studied human SAMHD1 variants are N-terminal truncations or contain mutations in the nuclear localization signal and therefore, none of the variants studied here lost the ability to localize to the nucleus. We did take note, however, that all SAMHD1 variants tested except for p.G209S and p.D311A also localized to the cytosol. In contrast to a previous report, we found that the p.G209S mutant localizes mostly to the nucleus (Goncalves, et al., 2012). These differences might be due to that we are using HeLa cells from different origin.
Next we tested the ability of the different SAMHD1 variants to decrease the cellular levels of dNTPs. All of the SAMHD1 variants tested here lost their ability to decrease cellular levels of dNTPs except for p.G209S and p.Q548A. Interestingly, the SAMHD1 variant p.Q548A decreases the levels of dNTPs in human cells, but do not restrict HIV-1, suggesting that the ability of SAMHD1 to decrease the cellular levels of dNTPs is necessary but not sufficient to restrict HIV-1 infection (Cribier, et al., 2013; Welbourn, et al., 2013; Welbourn and Strebel, 2016; White, et al., 2013b).
SAMHD1 has the ability to decrease the cellular levels of dNTPs in human cells (Goldstone, et al., 2011; Kim, et al., 2012; Lahouassa, et al., 2012; Powell, et al., 2011; White, et al., 2013a), which was initially correlated to the ability of SAMHD1 to block HIV-1 infection. This notion has been challenged by SAMHD1 mutations on T592 that decrease the cellular levels of dNTPs but no longer restrict HIV-1 infection (Cribier, et al., 2013; Welbourn, et al., 2013; White, et al., 2013b). Combined with previous observations, this suggests the existence of at least three SAMHD1 variants that decrease the cellular levels of dNTPs and do not restrict HIV-1: p.T592D, p.T592E, and p.Q548A. In agreement, the oligomerization defective mutant Y146S/Y154S displays a severe dNTPase defect in vitro, but is indistinguishable from wild type SAMHD1 in its ability to deplete cellular dNTP pools and to restrict HIV replication (Bhattacharya, et al., 2016; Brandariz-Nunez, et al., 2013), suggesting that the dNTPase activity of SAMHD1 might be necessary but is not sufficient for restriction.
Phosphorylation of SAMHD1 does not seem to regulate the enzymatic activity (dNTPase) of the protein in vitro. Several publications have demonstrated that recombinant SAMHD1-T592D/E or immunoprecipitated from human cells showed no effect on its enzymatic activity (dNTPase) when compared to the wild type protein (Arnold, et al., 2015; Bhattacharya, et al., 2016; Welbourn, et al., 2013; Welbourn and Strebel, 2016; White, et al., 2013b). Only one publication showed that purified SAMHD1-T592E exhibit a decreased dNTPase activity (Tang, et al., 2015). Collectively this evidence suggested that phosphorylation of T592 does not affect the dNTPase activity of the protein in vitro. Interestingly, in the case of human U937 cells, expression of phosphomimetic mutants pT592D or p.T592E in PMA-treated U937 cells deplete cellular dNTPs but do not restrict HIV-1, suggesting that depletion of dNTPs in cells is required but not sufficient for HIV-1 restriction (Bhattacharya, et al., 2016; White, et al., 2013b). Similarly, the oligomerization defective mutant p.[(Y146S; Y154S)] decreases the cellular levels of dNTPs and no longer block HIV-1 infection (Bhattacharya, et al., 2016; Brandariz-Nunez, et al., 2013). The work presented here adds to the growing number of SAMHD1 variants (p.Q548A) that decrease the levels of dNTPs and no longer block HIV-1 infection.
Finally, the SAMHD1 variant p.G209S blocks HIV-1 infection, but is unable to negatively regulate the type I IFN response in humans affected with AGS suggesting that p.G209S genetically separates the ability of SAMHD1 to restrict HIV-1 from the ability of SAMHD1 to negatively regulate the type I IFN response in humans. We believe that this mutant represents an instrumental genetic tool to understand the molecular role of SAMHD1 in the type I IFN response and in the development of AGS.
Supplementary Material
Acknowledgments
We are thankful to the NIH/AIDS repository program for providing valuable reagents such as antibodies and drugs. This work was funded by an NIH R01 GM123540 to F.D.-G. C.K., G.L. and B.K. were supported by R01 GM104198 and R01 AI049781-0 grants to B.K.
Grant Sponsor: National Institute of Health R01 GM123540 to F.D.-G.
References
- Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem. 2012;287(15):12550–8. doi: 10.1074/jbc.M112.340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold LH, Groom HC, Kunzelmann S, Schwefel D, Caswell SJ, Ordonez P, Mann MC, Rueschenbaum S, Goldstone DC, Pennell S, et al. Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction. PLoS Pathog. 2015;11(10):e1005194. doi: 10.1371/journal.ppat.1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012 doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt R, Schumann T, Gerbaulet A, Nguyen LA, Schubert N, Alexopoulou D, Berka U, Lienenklaus S, Peschke K, Gibbert K, et al. Mouse SAMHD1 Has Antiretroviral Activity and Suppresses a Spontaneous Cell-Intrinsic Antiviral Response. Cell Rep. 2013;4(4):689–96. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7(12):e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Wang Z, White T, Buffone C, Nguyen LA, Shepard CN, Kim B, Demeler B, Diaz-Griffero F, Ivanov DN. Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Sci Rep. 2016;6:31353. doi: 10.1038/srep31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology. 2012a;9(1):49. doi: 10.1186/1742-4690-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology. 2012b;9:49. doi: 10.1186/1742-4690-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Valle-Casuso JC, White TE, Nguyen L, Bhattacharya A, Wang Z, Demeler B, Amie S, Knowlton C, Kim B, et al. Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology. 2013;10:131. doi: 10.1186/1742-4690-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by Cyclin A2/CDK1 Regulates Its Restriction Activity toward HIV-1. Cell Rep. 2013;3(4):1036–43. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38(8):917–20. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N. Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15(7):429–40. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ. Familial Aicardi-Goutieres syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. Am J Med Genet A. 2010;152A(4):938–42. doi: 10.1002/ajmg.a.33359. [DOI] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Moulin M, Nurnberg P, Crow YJ, Rutsch F. Cerebral vasculopathy is a common feature in Aicardi-Goutieres syndrome associated with SAMHD1 mutations. Proc Natl Acad Sci U S A. 2011;108(26):E232. doi: 10.1073/pnas.1104699108. author reply E233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9(7):e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fye JM, Orebaugh CD, Coffin SR, Hollis T, Perrino FW. Dominant mutation of the TREX1 exonuclease gene in lupus and Aicardi-Goutieres syndrome. J Biol Chem. 2011;286(37):32373–82. doi: 10.1074/jbc.M111.276287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–82. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Hum Mutat. 2012;33(7):1116–22. doi: 10.1002/humu.22087. [DOI] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J Immunol. 2015;195(5):1939–43. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A. Cytoplasmic sensing of viral nucleic acids. Curr Opin Virol. 2015;11:31–7. doi: 10.1016/j.coviro.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Logue EC, Bloch N, Daddacha W, Polsky SB, Schultz ML, Kim B, Landau NR. The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J Virol. 2012 doi: 10.1128/JVI.01657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–61. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Tang C, Zhao Q, Wang W, Xiong Y. Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci U S A. 2014;111(41):E4305–14. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight Interplay among SAMHD1 Protein Level, Cellular dNTP Levels, and HIV-1 Proviral DNA Synthesis Kinetics in Human Primary Monocyte-derived Macrophages. J Biol Chem. 2012;287(26):21570–4. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S, Wolf C, Konig N, Staroske W, Guck J, Hausler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, et al. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis. 2015;74(3):e17. doi: 10.1136/annrheumdis-2013-204845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, et al. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe. 2012;11(2):205–17. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–7. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(6):621. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Malinger G, Ben-Sira L, Kidron D, Cohen S, Inbar S, Bezaleli T, Levine A, Vinkler C, Lev D, et al. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur J Hum Genet. 2011;19(3):287–92. doi: 10.1038/ejhg.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11(2):194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Bridgeman A, Benlahrech A, Cursi C, Rehwinkel J. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell Rep. 2016;16(6):1492–501. doi: 10.1016/j.celrep.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33(24):2937–46. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Diamond MS. MDA5 and autoimmune disease. Nat Genet. 2014;46(5):418–9. doi: 10.1038/ng.2959. [DOI] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24(15):6719–27. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orebaugh CD, Fye JM, Harvey S, Hollis T, Perrino FW. The TREX1 exonuclease R114H mutation in Aicardi-Goutieres syndrome and lupus reveals dimeric structure requirements for DNA degradation activity. J Biol Chem. 2011;286(46):40246–54. doi: 10.1074/jbc.M111.297903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity. 2015;43(5):933–44. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286(51):43596–600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramantani G, Hausler M, Niggemann P, Wessling B, Guttmann H, Mull M, Tenbrock K, Lee-Kirsch MA. Aicardi-Goutieres Syndrome and Systemic Lupus Erythematosus (SLE) in a 12-Year-Old Boy With SAMHD1 Mutations. J Child Neurol. 2011;26(11):1425–8. doi: 10.1177/0883073811408310. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, Bieniasz PD, Towers GJ, Moita LF, Crow YJ, et al. SAMHD1-dependent retroviral control and escape in mice. Embo J. 2013 doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O’Hara A, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;80(4):811–5. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–32. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U, et al. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 2013;12(12):1159–69. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet. 2012;44(11):1243–8. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch F, Schwartz O. The SAMHD1 knockout mouse model: in vivo veritas? Embo J. 2013 doi: 10.1038/emboj.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20(8):936–41. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Pollpeter D, Apolonia L, Goujon C, Malim MH. Nuclear import of SAMHD1 is mediated by a classical karyopherin alpha/beta1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation. Retrovirology. 2014;11:29. doi: 10.1186/1742-4690-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwende H, Fitzke E, Ambs P, Dieter P. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J Leukoc Biol. 1996;59(4):555–61. [PubMed] [Google Scholar]
- Seamon KJ, Sun Z, Shlyakhtenko LS, Lyubchenko YL, Stivers JT. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015;43(13):6486–99. doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Ji X, Wu L, Xiong Y. Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J Biol Chem. 2015;290(44):26352–9. doi: 10.1074/jbc.M115.677435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele H, du Moulin M, Barczyk K, George C, Schwindt W, Nurnberg G, Frosch M, Kurlemann G, Roth J, Nurnberg P, et al. Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Hum Mutat. 2010;31(11):E1836–50. doi: 10.1002/humu.21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungler V, Staroske W, Kind B, Dobrick M, Kretschmer S, Schmidt F, Krug C, Lorenz M, Chara O, Schwille P, et al. Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med (Berl) 2013 doi: 10.1007/s00109-013-0995-3. [DOI] [PubMed] [Google Scholar]
- Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of virus infection but not catalytic dNTPase activity are regulated by phosphorylation of SAMHD1. J Virol. 2013 doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Strebel K. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology. 2016;488:271–7. doi: 10.1016/j.virol.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology. 2013a;436(1):81–90. doi: 10.1016/j.virol.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The Retroviral Restriction Ability of SAMHD1, but Not Its Deoxynucleotide Triphosphohydrolase Activity, Is Regulated by Phosphorylation. Cell Host Microbe. 2013b;13(4):441–51. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–88. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- Yan J, Kaur S, DeLucia M, Hao C, Mehrens J, Wang C, Golczak M, Palczewski K, Gronenborn AM, Ahn J, et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem. 2013;288(15):10406–17. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Pt A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Zhang C, de Silva S, Wang JH, Wu L. Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS ONE. 2012;7(5):e37477. doi: 10.1371/journal.pone.0037477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature. 2015;517(7532):89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 2013;4(6):1108–15. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


