Abstract
Non-Hodgkin lymphoma (NHL) constitutes a diverse group of more than 40 subtypes, each characterized by distinct biologic and clinical features. Until recently, pinpointing genetic and epidemiologic risk factors for individual subtypes has been limited by the relative rarity of each. However, several large pooled case-control studies have provided sufficient statistical power for detecting etiologic differences and commonalities between subtypes and thus yield new insight into their unique epidemiologic backgrounds. Here, we review the subtype-specific medical, lifestyle, and biologic components identified in these studies, which suggest that a complex interplay between host genetics, autoimmune disorders, modifiable risk factors, and occupation contributes to lymphomagenesis.
Keywords: Non-Hodgkin lymphoma, Autoimmune disorders, Epidemiology, Biology, NHL subtypes
Introduction
Non-Hodgkin lymphoma represents the most common hematologic malignancy in the world and comprises a heterogeneous group of more than 40 different subtypes [1]. In addition to harboring distinct biologic and clinical features, these subtypes also vary in terms of incidence by age, sex, ethnicity, and geographic distribution. While population-based cancer registry data has been helpful in characterizing the epidemiology of NHL as a whole [2–4], identifying risk factors for individual NHL subtypes has been difficult given the relatively low number of patients affected by each one.
In the last decade, several large epidemiological studies have identified numerous environmental, medical, lifestyle, and genetic risk factors for developing NHL via geospatial analyses, surveys, and examination of single nucleotide variants in genome-wide association studies (GWAS) and other genetic analyses [5–20]. These approaches have been applied to identify clinical, environmental, occupational, and genetic risk factors for specific NHL subtypes. Herein, we review the biologic processes and epidemiologic factors thought to contribute to lymphomagenesis and examine how these factors may interact in the development of specific NHL subtypes.
Normal Lymphocyte Maturation: Setting the Stage for Lymphomagenesis
Since an understanding of physiologic B and T cell maturation lends insight into many of the mechanisms underlying lymphomagenesis, a brief review is provided here. During the process of differentiation, normal naïve lymphocytes undergo massive clonal expansion and mutagenesis. This requires major changes in gene expression, mediated in part by transcription factors, histonemodifying enzymes, and methylation changes in CpG dinucleotides. Some genetic instability is inherent to the physiologic processes responsible for producing antibody and T cell receptor diversity, as the involved enzymes introduce DNA double-strand breaks and point mutations to those genes. Lymphocytes are thus particularly predisposed to acquire chromosomal translocations and other oncogenic mutations when these processes are dysregulated, providing the groundwork for malignant transformation to lymphoma.
B cell development begins in the bone marrow, where progenitors progress through a series of steps resulting in the production of immunoglobulin (Ig) [21]. Most B cells undergo apoptosis in the bone marrow if they fail to produce surface IgM, or if their IgM binds self-antigen. Surviving mature B cells travel to secondary lymphoid tissues (e.g., lymph nodes and spleen) for exposure to antigen, migrate to the germinal center to proliferate, and undergo further Ig gene modification via somatic hypermutation and class switch recombination. Somatic hypermutation introduces point mutations, small deletions, and insertions into the variable region of the Ig gene to increase affinity for antigen. B cells with antibodies exhibiting high antigen affinity receive pro-survival signaling through their Ig receptors; however, most will die. Through class switch recombination, exons in the constant region of the Ig heavy chain are replaced by a recombination/deletion process, facilitating production of the secondary Ig isotypes IgG, IgA, and IgE [22]. Upon exiting the germinal center, B cells differentiate into either plasma cells or memory B cells.
Normal T cell development mirrors that of B cells: lymphocytes progress through a series of coordinated steps, undergoing positive and negative selection in the thymus until mature cells ultimately express a functional antigen receptor—the T cell receptor. While some T cell NHLs resemble normal lymphocytes at a distinct maturation stage (especially when gene expression profiling is considered), determining the cell of origin of many subtypes remains difficult. This is in sharp contrast to B cell lymphoma, in which each subtype corresponds with a normal cellular phenotype [23].
Genetic Lesions and Mechanisms of Lymphomagenesis
Balanced translocations leading to activated oncogenes are common events in many lymphoma subtypes. Frequently, the promoter of constitutively expressed Ig on chromosome 14 serves as a translocation partner in B cell lymphomas. For instance, in follicular lymphoma (FL), t(14,18) leads to overexpression of BCL2, an anti-apoptotic protein that promotes cell survival even in the harshly pro-death environment of the germinal center [24]. Similarly, constitutive expression of cyclin D1 in mantle cell lymphoma (MCL) results from t(11,14) and promotes proliferation [25].
The machinery involved in somatic hypermutation offers ample opportunity for gaining genetic lesions. In its ligation of DNA breaks induced by the enzyme activation-induced cytidine deaminase (AID), error-prone DNA polymerase generates point mutations. In fact, somatic hypermutation is known to target the coding regions of non-immunoglobulin genes such as BCL6, PAX5, and c-MYC, which may further explain why such genes are often involved in translocations.
AID-mediated DNA breaks also occur in class switch recombination, another process that may yield translocations if disrupted. Class switch recombination is implicated in the t(8,14) seen in sporadic Burkitt lymphoma, with translocation breakpoints involving the Ig heavy chain switch locus [26]. Additionally, defects in recombination are seen in activated B cell-like (ABC) diffuse large B cell lymphoma (DLBCL) but not in the germinal center B cell-like (GCB) subtype. This difference may thus contribute to the distinct mutations described in each subset (e.g., MYOM2, CD79 and MYD88 in ABC, and MLL2, BCL2, and CREBBP in GCB) [27].
The balanced translocations that often characterize B cell lymphoma subtypes are uncommon in T cell NHL. One notable exception is the anaplastic lymphoma kinase (ALK) translocation found in some anaplastic large cell lymphomas (ALCL). The ALK locus may be juxtaposed with a number of partners, each resulting in an oncogenic fusion protein. The most common of these, t(2,5), joins the promoter of nucleophosmin to the ALK catalytic domain, creating a constitutively active tyrosine kinase that contributes to malignant transformation and avoidance of apoptosis [28].
While balanced translocations may represent driving pathogenic events, unbalanced chromosomal gains and losses likely also cooperate to promote malignant transformation and cancer cell survival advantage. For instance, deletions and mutations of the p53 tumor suppressor gene have been reported in Burkitt lymphoma, FL, MCL, and chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) [29]. More recent studies also indicate that dysregulated epigenetic mechanisms contribute to lymphomagenesis [30].
Epidemiologic Studies by NHL Subtype
Identifying epidemiologic and genetic risk factors for developing specific NHL subtypes provides important correlates for the characterization of the particular molecular pathway abnormalities described in each. The International Lymphoma Epidemiology Consortium (InterLymph) was formed in 2001 to provide a comprehensive analysis of risk factors both particular to and shared by specific NHL subtypes. In a series of studies, InterLymph pooled cases and controls across studies and evaluated many exposures of interest, including medical and family history, lifestyle, and occupation factors, to provide well-powered comparisons of risk factors for specific NHL subtypes [31••, 32••, 33••, 34••, 35••, 36••, 37–40, 41••, 42]. InterLymph and others have also performed GWAS that have identified NHL subtype-specific single nucleotide variants (SNVs) associated with increased risk of developing lymphoma. We review recent key findings from these efforts by NHL subtype.
Diffuse Large B Cell Lymphoma
The most common NHL type, DLBCL comprises 25–30 % of NHL in Western countries [3, 4]. Although in many cases DLBCL is a curable disease, it is universally fatal if left untreated or treated improperly, and nearly 50 % of patients relapse [43, 44]. Gene expression profiling has provided some insight into this clinical heterogeneity, identifying two biologically distinct subtypes that appear to reflect unique cells of origin and harbor differing prognoses: the more favorable GCB and the more aggressive ABC DLBCL [45–47]. However, this subclassification represents only one step towards elucidating the complex underpinnings of lymphomagenesis and tumor behavior in DLBCL.
Genetic Risk Factors for DLBCL
In the past decade, gene expression profiling and cell line studies have yielded significant insight into a number of molecular pathways that are dysregulated in DLBCL, including B cell receptor and NF-κB signaling. GWAS have supplemented this work by identifying genetic variants associated with DLBCL. One study identified a novel susceptibility locus for DLBCL at 3q27 in the intergenic region between BCL6 and LPP known to contain strong regulatory elements [48]. Dysregulation of both genes has been implicated in malignant transformation, and BCL6 plays a particularly important role in germinal center formation during lymphocyte development [22]; thus, it is hypothesized that certain variants in the 3q27 locus may modulate the expression of one or both of these genes to contribute to lymphomagenesis.
Variants in several genes involved in augmenting the inflammatory milieu have been associated with increased DLBCL risk, including cytokines such as IL-10 [19, 49] and activators of the complement system, such as MASP2 [50]. SNVs in CTLA4, which codes for a negative regulator of Tcell activation, were found more commonly in DLBCL patients in an Egyptian study [51]. Importantly, the prevalence of hepatitis C virus (HCV) infection was higher in patients harboring these SNVs. As HCV is suspected to contribute to NHL development through antigenic stimulation of B cells, it is possible that some CTLA4 SNVs may cooperate with HCV infection to increase DLBCL risk above that conferred by either factor alone.
Interestingly, recent analyses suggest that the effect of certain SNVs may vary depending on ethnicity, highlighting possible interaction between genetic variants, environmental factors, and population genomics. For instance, while studies with predominantly Caucasian subjects identified the G308A SNV in TNF as a risk factor for DLBCL [12], the same variant was associated with decreased risk of DLBCL in Asian populations [52]. Another recent case-control study evaluated whether susceptibility to DLBCL increased in East Asian populations with the presence of SNVs previously identified as risk factors for the disease in European populations. Of the seven SNVs assessed (in EXOC2, PVT1, HLA-B, NCOA1, CD86, and ARAP3), only three were associated with increased DLBCL in those of East Asian ancestry [53•].
Several case-control studies indicate a possible link between SNVs in the methylenetetrahydrofolate reductase (MTHFR) gene and DLBCL, although these results have been controversial. A 2013 meta-analysis found that the C677T SNV in MTHFR was associated with increased DLBCL risk in East Asian populations, but not in populations of European heritage [54]. MTHFR functions as a key enzyme in folate metabolism and DNA synthesis, and reduced MTHFR activity may result in decreased methylation of deoxyuridine monophosphate. Thus, it is hypothesized that abnormal MTHFR function could lead to increased incorporation of uracil into DNA, thereby increasing the likelihood of DNA double-strand breaks and other aberrancies that could predispose to oncogenesis [55].
Epidemiological Risk Factors for DLBCL
Findings from the InterLymph NHL Subtypes Project confirmed several risk factors already reported for DLBCL and identified others never before described. The odds ratios and 95 % confidence intervals for epidemiological risk factors significantly associated with several NHL subtypes are shown in Table 1. Multivariate analyses of 4,667 cases and 22,639 controls revealed increased risk of DLBCL associated with B cell-activating autoimmune disease, HCV seropositivity, first-degree family history of NHL, and higher body mass index (BMI) as a young adult. Intriguingly, the study also suggested that DLBCL risk factors vary by gender and anatomical site of disease. For females, work as a field crop/vegetable farmer, seamstress/embroiderer, or hairdresser was associated with increased risk, while for males, risk increased with work as a driver or operator of material handling equipment. Smoking was found to be associated with central nervous system, testicular, and cutaneous DLBCL, inflammatory bowel disease with gastrointestinal DLBCL, and farming and hair dye use with mediastinal DLBCL. Other studies using geospatial analyses have identified environmental exposures associated with the risk of DLBCL [56•]. Emerging studies are beginning to integrate data from environmental exposures, epidemiological surveys, and genetic measures to examine the interactions between host genetics and environment in the development of NHL [57•], but additional comprehensive analyses are needed to identify target populations for prevention measures.
Table 1.
Odds ratios for all factors associated with significant increased risk of one or more B cell non-Hodgkin lymphoma subtypes
| Exposure category | Overall NHL OR (95 % CI) | DLBCL | BL | LPL/WM | FL | CLL/SLL | MZL | MCL |
|---|---|---|---|---|---|---|---|---|
| Autoimmune disease | ||||||||
| Any B cell-activating disease | 1.96 (1.60–2.40) | 2.45 (1.91–3.16) | 1.22 (0.29–5.04) | 2.61 (1.34–5.08) | 1.27 (0.90–1.79) | 1.07 (0.70–1.64) | 5.46 (3.81–7.83) | 1.03 (0.41–2.54) |
| Sjögren’s syndrome | 7.52 (3.68–15.4) | 8.77 (3.94–19.5) | – | 12.1 (3.16–46.6) | 3.32 (1.19–8.80) | 0.55 (0.07–4.43) | 38.1 (16.9–85.6) | |
| Systemic lupus erythematosus | 2.83 (1.82–4.41) | 2.49 (1.42–4.37) | – | 8.41 (2.81–25.2) | 1.81 (0.91–3.59) | 2.19 (0.92–5.25) | 6.54 (3.10–13.8) | 2.97 (0.68–13.1) |
| Any T cell-activating disease | 1.07 (0.95–1.21) | 1.08 (1.09–1.28) | 0.57 (0.25–1.32) | 1.00 (0.59–1.71) | 0.90 (0.72–1.11) | 1.04 (0.83–1.30) | 1.08 (0.75–1.56) | 1.01 (0.64–1.59) |
| Celiac disease | 1.77 (1.05–2.99) | 2.09 (1.04–4.18) | – | 1.28 (0.17–9.64) | 1.27 (0.53–3.01) | 0.60 (0.14–2.61) | – | 1.15 (0.15–8.71) |
| Systemic sclerosis/scleroderma | 1.03 (0.41–2.58) | 0.71 (0.16–3.24) | 20.2 (2.44–166) | – | 1.08 (0.23–5.00) | – | 2.69 (0.50–14.7) | – |
| HCV positive | 1.81 (1.369–2.37) | 2.33 (1.71–3.19) | 3.05 (0.90–10.3) | 2.70 (1.11–6.56) | 0.57 (0.30–1.10) | 2.08 (1.23–3.49) | 3.04 (1.65–5.60) | – |
| Cigarette smoking | ||||||||
| Duration≥20 years | 1.06 (0.99–1.14) | 1.02 (0.92–1.12) | 0.77 (0.51–1.17) | 1.50 (1.10–2.04) | 1.19 (1.06–1.33) | 0.84 (0.74–0.96) | 1.27 (1.03–1.57) | 1.24 (0.96–1.61) |
| Family history of hematologic malignancy | ||||||||
| Any | 1.72 (1.54–1.93) | 1.57 (1.34–1.83) | 0.51 (0.41–1.62) | 1.65 (1.03–2.65) | 1.48 (1.25–1.75) | 2.17 (1.77–2.65) | 1.73 (1.33–2.25) | 1.99 (1.39–2.84) |
| HL | 1.65 (1.18–2.29) | 2.08 (1.38–3.15) | – | 2.16 (0.50–9.36) | 1.48 (0.90–2.40) | 1.26 (0.60–2.64) | 2.74 (1.36–5.51) | 1.51 (0.46–5.03) |
| NHL | 1.79 (1.51–2.13) | 1.84 (1.46–2.33) | 0.75 (0.23–2.43) | 1.20 (0.52–2.76) | 1.99 (1.55–2.54) | 1.92 (1.42–2.61) | 1.65 (1.10–2.46) | 1.95 (1.14–3.34) |
| Leukemia | 1.51 (1.29–1.77) | 1.16 (0.92–1.47) | 0.88 (0.35–2.20) | 2.19 (1.21–3.96) | 0.97 (0.74–1.28) | 2.41 (1.85–3.14) | 1.66 (1.15–2.38) | 1.98 (1.21–3.24) |
| Multiple myeloma | 1.77 (1.15–2.72) | 1.35 (0.71–2.57) | – | – | 1.93 (1.06–3.51) | 1.99 (0.92–4.33) | 0.55 (0.13–2.37) | 3.10 (1.05–9.10) |
| Occupational history | ||||||||
| Painter | 1.22 (0.99–1.51) | 1.02 (0.76–1.37) | 2.28 (0.97–5.33) | 0.66 (0.16–2.72) | 1.31 (0.93–1.84) | 1.05 (0.66–1.67) | 1.68 (0.97–5.33) | 1.39 (0.67–2.91) |
| General farm worker | 1.28 (1.10–1.50) | 1.09 (0.87–1.37) | 1.49 (0.59–3.78) | 1.26 (0.64–2.47) | 1.18 (0.88–1.57) | 1.46 (1.15–1.85) | 1.21 (0.75–1.95) | 1.21 (0.67–2.19) |
Abbreviations: OR odds ratio, CI confidence interval, HL Hodgkin lymphoma, NHL non-Hodgkin lymphoma, DLBCL diffuse large B cell lymphoma, BL Burkitt lymphoma, LPL/WM lymphoplasmacytic lymphoma/Waldenström’s macroglobulinemia, FL follicular lymphoma, CLL/SLL chronic lymphocytic leukemia/small lymphocytic lymphoma, MZL marginal zone lymphoma, MCL mantle cell lymphoma
Genetic and Epidemiological Risk Factors for Follicular Lymphoma
Follicular lymphoma (FL) is an indolent lymphoma that accounts for about 15–20 % of NHL in the Western world [43, 58–60]. It is more common in Caucasians than in Black or Asian Americans but, unlike other NHL subtypes, has similar incidence rates in men and women, with the median age of diagnosis in the sixth decade of life [58, 61]. Interestingly, although FL is defined by t(14,18), which results in overexpression of the anti-apoptotic protein BCL2, this translocation has been identified in ostensibly normal B cells of healthy individuals, implying that the translocation alone is not sufficient to induce lymphomagenesis [62].
Multiple GWAS of FL have revealed susceptibility variants in human leukocyte antigen (HLA) class I and II regions, highlighting an important role of such variants in FL pathogenesis [63–67]. In 2014, InterLymph published a large-scale two-stage GWAS involving 4523 FL cases and 13,344 controls that identified susceptibility SNVs in the HLA region, and in 5 non-HLA loci [68]. These included regions near genes coding for BCL2, suggesting a role for SNVs in predisposing towards acquisition of the characteristic (14,18) translocation, and CXCR5, a receptor expressed on mature B cells, some T cells, and dendritic cells involved in B cell migration and activation. FL-associated SNVs near CXCR5 have been previously reported [69]. The authors proposed a dynamic function for CXCR5 in altering the immune milieu of the tumor microenvironment as a possible mechanism for influencing FL initiation and progression.
Epigenetic variants have also been identified by recent GWAS in FL. A 2014 study found that a specific G>T SNV in the stem-loop sequence of microRNA-618 was associated with increased risk of FL, with in vitro data suggesting that the variant results in decreased microRNA-618 expression [70]. Although the downstream effects of such a change remain incompletely characterized, targetome profiling of microRNA-618 lends hypothesis-generating insight into candidate genes for which post-translational dysregulation could contribute to follicular lymphomagenesis.
In its case-control study of 3530 FL cases and 22,639 controls, InterLymph identified family history of NHL, higher body mass index as a young adult, and work as a spray painter as risk factors. Interestingly, cigarette smoking and Sjögren’s syndrome were found to be associated with increased FL risk only in females.
Genetic and Epidemiological Risk Factors for Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
Chronic lymphocytic leukemia and small lymphocytic lymphoma (CLL/SLL) represent two manifestations of the same disease and harbor identical pathologic and immunophenotypic features. The distinction between the two relies on clinical presentation: in CLL, malignant cells appear primarily in the peripheral blood and bone marrow, while SLL presents predominantly in the lymph nodes [71]. Although CLL/SLL is relatively common in Western countries, it is quite rare in Asia [2–4]. However, recent studies show that the CLL/SLL incidence is increasing in Asian populations and that incidence is increased in people of Asian descent living in the USA compared to those who live in Asia, suggesting that components of the Western lifestyle may contribute to development of this disease [72, 73].
In the largest GWAS meta-analysis in CLL to date, nine novel risk loci for CLL were identified in addition to 13 previously described [74]. The proximity of some of these variants to genes involved in apoptosis, such as CASP10/CASP8, may provide a clue to their biological relevance, although more work is needed to elucidate this link. Variants in IRF8, which codes for a transcription factor integral to myeloid and B cell development, have also been recently associated with increased risk of CLL [75].
InterLymph confirmed previously identified risk factors for CLL/SLL such as HCV infection, family history of hematologic malignancy, work or residence on a farm, and increased height in a multivariate analysis of 2440 cases and 15,186 controls. In addition, occupation as a hairdresser was identified as a novel risk factor.
Genetic and Epidemiological Risk Factors for Marginal Zone Lymphoma
Marginal zone lymphoma (MZL) is a common indolent subtype that accounts for 5–10 % of NHL and comprises three distinct diseases: extranodal MZL of mucosa-associated lymphoid tissue (MALT), splenic MZL, and nodal MZL [58]. Several inflammatory diseases are known to be strongly associated with extranodal MZL of specific anatomic sites, including Helicobacter pylori infection in gastric MALT lymphoma, Chlamydia psittaci infection in ocular adnexal MALT lymphoma, Borrelia burgdorferi infection with cutaneous MALT lymphoma, Sjögren’s syndrome with salivary gland MALT lymphoma, and Hashimoto thyroiditis with thyroid MALT lymphoma. In fact, antibiotic eradication of H. pylori alone results in lymphoma regression in about half of gastric MALT lymphoma patients [76, 77].
A GWAS of 1281 MZL cases and 7127 controls of European ancestry identified independent loci in the HLA region significantly associated with MZL risk [78]. The authors proposed that these independent associations in the HLA region and their localization around known B cell genes associated with autoimmunity raise the possibility of shared genetic effects spanning B cell NHL and autoimmune diseases.
InterLymph represents the first large epidemiological study of MZL subtypes, evaluating 1052 MZL cases (extranodal 633, nodal 157, splenic 140, MZL-not otherwise specified 120) and 13,766 controls. It is worth noting that 60 % of cases examined were extranodal MZL, so results from analyses of nodal and splenic subtypes were based on a relatively small number of cases. B cell-activating autoimmune disorders were associated with all MZL subtypes, highlighting the contribution of chronic immune stimulation to this disease. Family history of hematologic malignancy was associated with increased risk for nodal and extranodal MZL, while novel associations were found between asthma and permanent hair dye use with splenic MZL and occupation as a metal worker with nodal MZL. Unsurprisingly, Sjögren’s syndrome was highly associated with salivary gland MALT lymphoma, and peptic ulcers with gastric MALT lymphoma.
Epidemiological Risk Factors for Burkitt Lymphoma
Burkitt lymphoma is a highly aggressive lymphoma characterized by dysregulation of MYC and constituting 1–5 % of all NHL in adults [4, 79]. It is categorized into three histologically indistinguishable subtypes: endemic, which is clearly associated with Epstein-Barr virus infection; immunodeficiency-associated, seen in immunosuppressed patients as a result of human immunodeficiency virus infection or solid organ transplant; and sporadic, which previously had no clear risk factors. InterLymph evaluated 295 cases and 21,818 controls to examine risk factors for sporadic Burkitt lymphoma, with age-stratified analyses to account for the two age peaks seen in disease incidence (during childhood and in the seventh decade of life). In patients younger than 50 years old, eczema, highest quartile of height, and work as a charworker or cleaner were associated with increased risk. In patients older than 50, only HCV was identified as a risk factor, emphasizing the age-specific nature of risk factors for this disease.
Epidemiological Risk Factors for Lymphoplasmacytic Lymphoma
Lymphoplasmacytic lymphoma (LPL) is a rare, indolent type of NHL with cells spanning the spectrum of plasmacytic differentiation from small lymphocytes to true plasma cells, which often produce paraproteins such as IgM or IgG. Waldenström’s macroglobulinemia represents a subset of LPL characterized by bone marrow involvement and monoclonal IgM gammopathy [58]. InterLymph analyzed 374 cases of LPL (of which only three were Waldenström’s macroglobulinemia) along with 23, 096 controls and confirmed a strong association with chronic immune stimulation (specifically, autoimmune diseases such as Sjögren’s syndrome and systemic lupus erythematosus), HCV infection, and family history of hematologic malignancy. The study also identified novel associations with cigarette smoking and occupation as a medical doctor.
Epidemiological Risk Factors for Mantle Cell Lymphoma
MCL is an aggressive disease with poor prognosis that remains essentially incurable with conventional chemotherapy. The vast majority of cases are characterized by t(11,14), which results in constitutive expression of cyclin D1. MCL is a rare disease constituting only 2–10 % of all NHL cases [58], which accounts for the paucity of known risk factors other than a 2:1 male predominance in this subtype. A recent study noted that MCLs had a mutational profile distinct from other B cell NHLs [80]. In its multivariate analyses of 557 MCL cases and 13,766 controls, InterLymph identified family history of hematologic malignancy and residence on a farm as risk factors for the disease [36••].
Peripheral T Cell Lymphoma
Peripheral T cell lymphoma (PTCL) accounts for 5–10 % of NHL and comprises a heterogeneous group of lymphomas derived from mature T and natural killer cells, with more than 20 distinct entities defined by characteristic morphologic, molecular, and clinical features, including site of disease [3, 58]. The most common subtypes are PTCL-not otherwise specified (PTCL-NOS), angioimmunoblastic T cell lymphoma (AITL), and anaplastic large cell lymphoma (ALCL). Unfortunately, most PTCL are characterized by poor response to treatment, with the notable exception of ALCL carrying an ALK translocation.
Genetic Risk Factors for PTCL
Given the difficulty in defining a genetic basis for most T cell lymphomas, recent efforts have employed gene expression profiling in an attempt to identify phenotypic differences among these tumors. For example, gene expression profiling studies demonstrate that hepatosplenic T cell lymphomas may be completely segregated from other T cell NHL, emphasizing the distinct clinicopathological characteristics of this subtype. Supervised analysis has also allowed for a distinction between αβ and γδ T cell lymphomas [81]. Interestingly, although TP53 mutations are uncommon in T cell NHL, whole-genome sequencing has revealed recurrent rearrangements of p53-related genes in PTCL such as a truncated p63 protein that inhibits p53 in a dominant negative fashion, indicating another mechanism for silencing p53 tumor suppression [82]. In AITL, gene expression profiling studies revealed mutations in a RHOA GTPase [83] as well as upregulation of genes related to vascular endothelial growth factor A [84], which may play roles in AITL pathogenesis. Gene expression profiling also helped characterize PTCL-NOS, a heterogeneous group of tumors. PTCL-NOS subgroups may be distinguished based on the level of expression of NF-κB pathway genes, and by correlation with the profiles of either CD4+ T cells or CD8+ T cells [85, 86].
Epidemiological Risk Factors for PTCL
To determine epidemiological risk factors for PTCL, InterLymph analyzed 584 cases (PTCL-NOS 234, AITL 81, ALCL 164, others 57) and 15,912 controls. Several factors were associated with higher risk of one or more PTCL subtypes, including family history of hematologic malignancies, history of psoriasis, smoking history, and occupation as an electrical fitter or in the textile industry (Table 2). As expected, celiac disease was strongly associated with enteropathy-associated T cell lymphoma, but also with PTCL-NOS and ALCL.
Table 2.
Odds ratios for all factors associated with significant increased risk of peripheral T cell lymphoma and/or mycosis fungoides/Sézary syndrome
| Exposure category | PTCL | MF/SS OR (95 % CI) |
|---|---|---|
| Autoimmune disease | ||
| Psoriasis | 2.41 (1.15–5.04) | – |
| Systemic lupus erythematosus | 3.90 (1.24–12.3) | 5.03 (1.17–21.6) |
| Any T cell-activating disease | 1.95 (1.37–2.77) | 1.66 (1.00–2.75) |
| Celiac disease | 14.8 (7.27–30.2) | – |
| Systemic sclerosis/scleroderma | – | 8.87 (1.11–71.3) |
| Eczema | ||
| Any diagnosis | – | 2.38 (1.73–3.29) |
| Diagnosis within 10 years of NHL | – | 4.87 (2.15–11.02) |
| Cigarette smoking | ||
| Duration≥20 years | 1.75 (1.33–2.30) | 1.22 (0.84–1.77) |
| Duration≥40 years | – | 1.55 (1.04–2.31) |
| BMI 35–50 kg/m2 | – | 1.57 (1.03–2.40) |
| Family history of hematologic malignancy | ||
| Any | 1.86 (1.26–2.74) | 1.16 (0.68–1.98) |
| Leukemia | 1.84 (1.09–3.13) | 1.06 (0.49–2.29) |
| Multiple myeloma | 2.86 (0.98–8.37) | 6.11 (2.36–15.8) |
| Occupational history | ||
| Painter | 1.45 (0.73–2.88) | 3.42 (1.81–6.47) |
| General farm worker | 0.79 (0.40–1.56) | 2.07 (1.06–4.07) |
| General carpenter | – | 4.07 (1.54–10.8) |
Abbreviations: OR odds ratio, CI confidence interval, PTCL peripheral T cell lymphoma, MF/SS mycosis fungoides/Sézary syndrome, BMI body mass index
Epidemiological Risk Factors for Mycosis Fungoides/Sézary Syndrome
Mycosis fungoides (MF) is the most common type of cutaneous T cell lymphoma (CTCL), representing nearly half of all CTCL cases [58]. It commonly follows an indolent clinical course but may progress to Sézary syndrome (SS), a rarer form of CTCL characterized by erythroderma, generalized lymphadenopathy, and peripheral blood involvement. The age-adjusted incidence of MF/SS is estimated at 0.5 per 100, 000 in the Western world, with the rate of MF about 1.5 times higher among African-Americans compared to Whites [87].
InterLymph evaluated 324 cases (MF 271, SS 13, MF/SS 40) and 17,217 controls to identify several risk factors, including smoking history ≥40 years, adult body mass index >30 kg/m [2], history of eczema, personal or family history of multiple myeloma, and occupation as a crop or vegetable farmer, painter, woodworker, or general carpenter. Notably, the association between eczema and MF/SS risk increased for those diagnosed with eczema within 10 years of their diagnosis of MF/SS, indicating that MF may have initially been misdiagnosed as eczema in some cases. It is also worth noting that since both multiple myeloma and MF carry higher incidence in African-Americans, their apparent association may be confounded by this shared racial predisposition.
Discussion
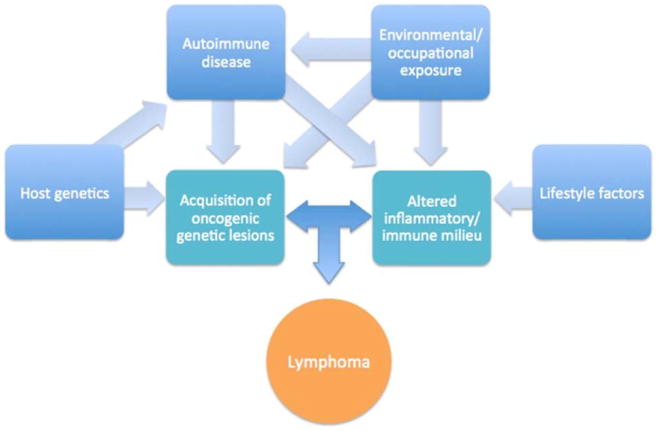
Recent pooled case-control studies examining genome-wide associations and epidemiological risk factors provide an enhanced understanding of subtype-specific risk factors that include genetics, medical history, family history, lifestyle factors, and occupations. These results suggest a complex interplay between host genetics, autoimmune disorders, modifiable risk factors, and occupation that contributes to lymphomagenesis in ways not fully understood (Fig. 1). It is important to note that despite their broad scope, several important factors were not assessed in these studies, including diet and infection other than HCV. Additionally, as in many studies, the populations analyzed were largely of Caucasian ancestry, and thus, it is unclear whether the risk factors identified are generalizable to non-Caucasian populations. In general, NHL risk factors among racial and ethnic minorities remain incompletely described [59, 87–93], and future analyses are needed to evaluate these factors and others not captured by the InterLymph study.
Fig. 1.
Proposed interactions between etiologic factors contributing to development of non-Hodgkin lymphoma subtypes
Identifying the interactions between epidemiologic components and genetic factors such as SNVs will help create a comprehensive model capable of predicting the probability of developing NHL subtypes, and could yield valuable in-sights regarding the biologic underpinnings of lymphomagenesis, which would provide targets for further molecular study and therapy. Recent work by Wang et al. utilized data from InterLymph to examine the relationship between history of autoimmune disease and five SNVs in common susceptibility immune-response gene loci found to confer increased risk of NHL (i.e., in IL10, TNFα, and HLA class genes), with the goal of determining whether these factors contributed to NHL risk independently or through a joint pathway [57•]. This group found that NHL risk across major subtypes increased in individuals with B cell-mediated autoimmune conditions who harbored TNF G308Avariants. This effect was especially pronounced in MZL, where risk increased from threefold with the GG genotype to eightfold with the AG or AA genotype. There was little evidence for interaction between autoimmune disease and four other SNVs. However, the apparent synergy between B cell-mediated autoimmunity and TNF G308A in augmenting NHL risk may imply a shared biologic pathway that promotes a chronic inflammatory state through increased expression of TNF-α and resultant NF-κB activation [94–96]. Indeed, a distinct pattern of immune response, termed T-helper 17 and characterized by inflammation, B cell activation, and production of certain inflammatory cytokines (including TNF-α), has been recognized as a central player in driving autoimmunity [97–99]. Given the proposed interaction between TNF G308A and autoimmune disease to increase NHL risk, it is possible that the T-helper 17 response may contribute to the pathogenesis of both autoimmune disease and NHL. Future studies investigating the link between autoimmune disorders and B cell NHL could complement such work by evaluating the B cell repertoire and specific mutations common between lymphoma subtypes and specific autoimmune diseases.
Conclusion
The unique glimpse into NHL subtype-specific epidemiology afforded by large case-control studies represents an important step towards understanding the complex network of factors underlying lymphomagenesis. Further work is needed to incorporate such epidemiologic studies with population genetics and lymphoma biology to build models for risk prediction and identify novel preventive and therapeutic targets.
Acknowledgments
This publication was supported in part by award number R21 CA158686 to Dr. Flowers. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with Ethics Guidelines
Conflict of interest The author(s) declare that they have no competing interests.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Ferlay J, SI, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet] 2013 doi: 10.1002/ijc.29210. < http://globocan.iarc.fr>. [DOI] [PubMed]
- 2.Chihara D, et al. Differences in incidence and trends of haematological malignancies in Japan and the United States. Br J Haematol. 2014;164:536–45. doi: 10.1111/bjh.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sant M, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood. 2010;116:3724–34. doi: 10.1182/blood-2010-05-282632. [DOI] [PubMed] [Google Scholar]
- 4.Morton LM, et al. Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood. 2006;107:265–76. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kane EV, et al. Postmenopausal hormone therapy and non-Hodgkin lymphoma: a pooled analysis of InterLymph case-control studies. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2013;24:433–41. doi: 10.1093/annonc/mds340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hjalgrim H, et al. Cigarette smoking and risk of Hodgkin lymphoma: a population-based case-control study. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored. Am Soc Prev Oncol. 2007;16:1561–6. doi: 10.1158/1055-9965.epi-07-0094. [DOI] [PubMed] [Google Scholar]
- 7.Gibson TM, et al. Smoking, variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2), and risk of non-Hodgkin lymphoma: a pooled analysis within the InterLymph consortium. Cancer Causes Control CCC. 2013;24:125–34. doi: 10.1007/s10552-012-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocco P, et al. Occupational exposure to trichloroethylene and risk of non-Hodgkin lymphoma and its major subtypes: a pooled InterLymph [correction of IinterLlymph] analysis. Occup Environ Med. 2013;70:795–802. doi: 10.1136/oemed-2013-101551. [DOI] [PubMed] [Google Scholar]
- 9.Nieters A, et al. PRRC2A and BCL2L11 gene variants influence risk of non-Hodgkin lymphoma: results from the InterLymph consortium. Blood. 2012;120:4645–8. doi: 10.1182/blood-2012-05-427989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kane EV, et al. Menstrual and reproductive factors, and hormonal contraception use: associations with non-Hodgkin lymphoma in a pooled analysis of InterLymph case-control studies. Ann Oncol Off J Eur Soc Med Oncol ESMO. 2012;23:2362–74. doi: 10.1093/annonc/mds171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner JJ, et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood. 2010;116:e90–8. doi: 10.1182/blood-2010-06-289561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skibola CF, et al. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph consortium. Am J Epidemiol. 2010;171:267–76. doi: 10.1093/aje/kwp383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vajdic CM, et al. Atopic disease and risk of non-Hodgkin lymphoma: an InterLymph pooled analysis. Cancer Res. 2009;69:6482–9. doi: 10.1158/0008-5472.can-08-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willett EV, et al. Non-Hodgkin lymphoma and obesity: a pooled analysis from the InterLymph consortium. Int J Cancer. 2008;122:2062–70. doi: 10.1002/ijc.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kricker A, et al. Personal sun exposure and risk of non Hodgkin lymphoma: a pooled analysis from the Interlymph consortium. Int J Cancer. 2008;122:144–54. doi: 10.1002/ijc.23003. [DOI] [PubMed] [Google Scholar]
- 16.Ekstrom Smedby K, et al. Autoimmune disorders and risk of non- Hodgkin lymphoma subtypes: a pooled analysis within the InterLymph consortium. Blood. 2008;111:4029–38. doi: 10.1182/blood-2007-10-119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SS, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10 211 cases and 11 905 controls from the international lymphoma epidemiology consortium (InterLymph) Blood. 2007;109:3479–88. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton LM, et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the pathology working group of the international lymphoma epidemiology consortium (InterLymph) Blood. 2007;110:695–708. doi: 10.1182/blood-2006-11-051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothman N, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/s1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 20.Morton LM, et al. Cigarette smoking and risk of non-Hodgkin lymphoma: a pooled analysis from the International Lymphoma Epidemiology Consortium (InterLymph) Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2005;14:925–33. doi: 10.1158/1055-9965.epi-04-0693. [DOI] [PubMed] [Google Scholar]
- 21.Taccioli GE, et al. Impairment of V(D)J recombination in double-strand break repair mutants. Science (New York, NY) 1993;260:207–10. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- 23.Costello R, et al. Peripheral T-cell lymphoma gene expression profiling and potential therapeutic exploitations. Br J Haematol. 2010;150:21–7. doi: 10.1111/j.1365-2141.2009.07977.x. [DOI] [PubMed] [Google Scholar]
- 24.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122:3424–31. doi: 10.1172/jci63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Galan P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117:26–38. doi: 10.1182/blood-2010-04-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920–32. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- 27.Lenz G, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204:633–43. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreri AJ, Govi S, Pileri SA, Savage KJ. Anaplastic large cell lymphoma, ALK-positive. Crit Rev Oncol Hematol. 2012;83:293–302. doi: 10.1016/j.critrevonc.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Xu-Monette ZY, et al. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood. 2012;119:3668–83. doi: 10.1182/blood-2011-11-366062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Vajdic CM, et al. Medical history, lifestyle, family history, and occupational risk factors for lymphoplasmacytic lymphoma/Waldenstrom’s macroglobulinemia: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:87–97. doi: 10.1093/jncimonographs/lgu002. The largest pooled analysis of epidemiological risk factors for patients with lymphoplasmacytic lymphoma/Waldenstrom’s macroglobulinemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Slager SL, et al. Medical history, lifestyle, family history, and occupational risk factors for chronic lymphocytic leukemia/small lymphocytic lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:41–51. doi: 10.1093/jncimonographs/lgu001. The largest pooled analysis of epidemiological risk factors for patients with chronic lymphocytic leukemia/small lymphocytic lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Mbulaiteye SM, et al. Medical history, lifestyle, family history, and occupational risk factors for sporadic Burkitt lymphoma/leukemia: the Interlymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:106–14. doi: 10.1093/jncimonographs/lgu003. The largest pooled analysis of epidemiological risk factors for patients with Burkitt lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Linet MS, et al. Medical history, lifestyle, family history, and occupational risk factors for follicular lymphoma: the InterLymph Non- Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:26–40. doi: 10.1093/jncimonographs/lgu006. The largest pooled analysis of epidemiological risk factors for patients with follicular lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Cerhan JR, et al. Medical history, lifestyle, family history, and occupational risk factors for diffuse large B-cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:15–25. doi: 10.1093/jncimonographs/lgu010. The largest pooled analysis of epidemiological risk factors for patients with diffuse large B- cell lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Smedby KE, et al. Medical history, lifestyle, family history, and occupational risk factors for mantle cell lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:76–86. doi: 10.1093/jncimonographs/lgu007. The largest pooled analysis of epidemiological risk factors for patients with mantle cell lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skibola CF, et al. Medical history, lifestyle, family history, and occupational risk factors for adult acute lymphocytic leukemia: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:125–9. doi: 10.1093/jncimonographs/lgu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton LM, et al. Rationale and design of the International Lymphoma Epidemiology Consortium (InterLymph) Non- Hodgkin lymphoma subtypes project. J Natl Cancer Inst Monogr. 2014;2014:1–14. doi: 10.1093/jncimonographs/lgu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morton LM, et al. Etiologic heterogeneity among non-Hodgkin lymphoma subtypes: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:130–44. doi: 10.1093/jncimonographs/lgu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monnereau A, et al. Medical history, lifestyle, and occupational risk factors for hairy cell leukemia: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:115–24. doi: 10.1093/jncimonographs/lgu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Bracci PM, et al. Medical history, lifestyle, family history, and occupational risk factors for marginal zone lymphoma: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:52–65. doi: 10.1093/jncimonographs/lgu011. The largest pooled analysis of epidemiological risk factors for patients with marginal zone lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aschebrook-Kilfoy B, et al. Medical history, lifestyle, family history, and occupational risk factors for mycosis fungoides and Sezary syndrome: the InterLymph Non-Hodgkin Lymphoma Subtypes Project. J Natl Cancer Inst Monogr. 2014;2014:98–105. doi: 10.1093/jncimonographs/lgu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flowers CR, Armitage JO. A decade of progress in lymphoma: advances and continuing challenges. Clin Lymphoma Myeloma Leuk. 2010;10:414–23. doi: 10.3816/CLML.2010.n.086. [DOI] [PubMed] [Google Scholar]
- 44.Sinha R, Nastoupil L, Flowers CR. Treatment strategies for patients with diffuse large B-cell lymphoma: past, present, and future. Blood Lymphat Cancer Targets Ther. 2012;2012:87–98. doi: 10.2147/blctt.s18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 46.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 47.Fu K, et al. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and non-germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:4587–94. doi: 10.1200/jco.2007.15.9277. [DOI] [PubMed] [Google Scholar]
- 48.Tan DE, et al. Genome-wide association study of B cell non- Hodgkin lymphoma identifies 3q27 as a susceptibility locus in the Chinese population. Nat Genet. 2013;45:804–7. doi: 10.1038/ng.2666. [DOI] [PubMed] [Google Scholar]
- 49.Cao HY, Zou P, Zhou H. Genetic association of interleukin-10 promoter polymorphisms and susceptibility to diffuse large B-cell lymphoma: a meta-analysis. Gene. 2013;519:288–94. doi: 10.1016/j.gene.2013.01.066. [DOI] [PubMed] [Google Scholar]
- 50.Hu W, et al. Polymorphisms in pattern-recognition genes in the innate immunity system and risk of non-Hodgkin lymphoma. Environ Mol Mutagen. 2013;54:72–7. doi: 10.1002/em.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khorshied MM, Gouda HM, Khorshid OM. Association of cytotoxic T-lymphocyte antigen 4 genetic polymorphism, hepatitis C viral infection and B-cell non-Hodgkin lymphoma: an Egyptian study. Leuk Lymphoma. 2014;55:1061–6. doi: 10.3109/10428194.2013.820294. [DOI] [PubMed] [Google Scholar]
- 52.Hosgood HD, 3rd, et al. IL10 and TNF variants and risk of non- Hodgkin lymphoma among three Asian populations. Int J Hematol. 2013;97:793–9. doi: 10.1007/s12185-013-1345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Bassig BA, et al. Genetic susceptibility to diffuse large B-cell lymphoma in a pooled study of three Eastern Asian populations. Eur J Haematol. 2015 doi: 10.1111/ejh.12513. A useful pooled analysis of genetic risk factors for diffuse large B-cell lymphoma. [DOI] [PubMed] [Google Scholar]
- 54.Sun YY, An L, Xie YL, Xu JY, Wang J. Methylenetetrahydrofolate reductase gene polymorphisms association with the risk of diffuse large B cell lymphoma: a meta-analysis. Tumour Biol J Int Soc Oncodev Biol Med. 2013;34:3587–91. doi: 10.1007/s13277-013-0938-1. [DOI] [PubMed] [Google Scholar]
- 55.Blount BC, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci. 1997;94:3290–5. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Bulka C, et al. Residence proximity to benzene release sites is associated with increased incidence of non-Hodgkin lymphoma. Cancer. 2013;119:3309–17. doi: 10.1002/cncr.28083. A geospatial analysis of the relationships between passive residential environmental exposures and risk of non-Hodgkin lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Wang SS, et al. Associations of non-Hodgkin lymphoma (NHL) risk with autoimmune conditions according to putative NHL loci. Am J Epidemiol. 2015 doi: 10.1093/aje/kwu290. An interesting exploration of the interactions between auto-immune diseases and genetic risk factors for non-Hodgkin lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swerdlow SH International Agency for Research on Cancer & World Health Organization. WHO classification of tumours of haematopoietic and lymphoid tissues. 4. International Agency for Research on Cancer; 2008. [Google Scholar]
- 59.Ambinder AJ, et al. Exploring risk factors for follicular lymphoma. Adv Hematol. 2012;13:2012. doi: 10.1155/2012/626035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedberg JW, et al. Follicular lymphoma in the United States: first report of the national LymphoCare study. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27:1202–8. doi: 10.1200/jco.2008.18.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solal-Celigny P, Cahu X, Cartron G. Follicular lymphoma prognostic factors in the modern era: what is clinically meaningful? Int J Hematol. 2010;92:246–54. doi: 10.1007/s12185-010-0674-x. [DOI] [PubMed] [Google Scholar]
- 62.Roulland S, et al. Follicular lymphoma-like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis. J Exp Med. 2006;203:2425–31. doi: 10.1084/jem.20061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smedby KE, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011;7:e1001378. doi: 10.1371/journal.pgen.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conde L, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42:661–4. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skibola CF, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–5. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vijai J, et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet. 2013;9:e1003220. doi: 10.1371/journal.pgen.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skibola CF, et al. A meta-analysis of genome-wide association studies of follicular lymphoma. BMC Genomics. 2012;13:516. doi: 10.1186/1471-2164-13-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Skibola CF, et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet. 2014;95:462–71. doi: 10.1016/j.ajhg.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charbonneau B, et al. CXCR5 polymorphisms in non-Hodgkin lymphoma risk and prognosis. Cancer Immunol Immunother CII. 2013;62:1475–84. doi: 10.1007/s00262-013-1452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu A, et al. Targetome profiling and functional genetics implicate miR-618 in lymphomagenesis. Epigenetics Off J DNA Methylation Soc. 2014;9:730–7. doi: 10.4161/epi.27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armitage JO, Weisenburger DD. New approach to classifying non- Hodgkin’s lymphomas: clinical features of the major histologic subtypes. Non-Hodgkin’s Lymphoma Classification Project. J Clin Oncol Off J Am Soc Clin Oncol. 1998;16:2780–95. doi: 10.1200/JCO.1998.16.8.2780. [DOI] [PubMed] [Google Scholar]
- 72.Wu SJ, et al. The incidence of chronic lymphocytic leukemia in Taiwan, 1986-2005: a distinct increasing trend with birth-cohort effect. Blood. 2010;116:4430–5. doi: 10.1182/blood-2010-05-285221. [DOI] [PubMed] [Google Scholar]
- 73.Clarke CA, et al. Lymphoid malignancies in U.S. Asians: incidence rate differences by birthplace and acculturation. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2011;20:1064–77. doi: 10.1158/1055-9965.epi-11-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berndt SI, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45:868–76. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slager SL, et al. Mapping of the IRF8 gene identifies a 3’UTR variant associated with risk of chronic lymphocytic leukemia but not other common non-Hodgkin lymphoma subtypes. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2013;22:461–6. doi: 10.1158/1055-9965.epi-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malfertheiner P, et al. Current concepts in the management of Helicobacter pylori infection: the Maastricht III consensus report. Gut. 2007;56:772–81. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chey WD, Wong BC. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808–25. doi: 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 78.Vijai J, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6:5751. doi: 10.1038/ncomms6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–96. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyazaki K, et al. Gene expression profiling of peripheral T-cell lymphoma including gammadelta T-cell lymphoma. Blood. 2009;113:1071–4. doi: 10.1182/blood-2008-07-166363. [DOI] [PubMed] [Google Scholar]
- 82.Vasmatzis G, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120:2280–9. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 84.de Leval L, et al. The gene expression profile of nodal peripheral T- cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–63. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 85.Martinez-Delgado B, et al. Expression profiling of T-cell lymphomas differentiates peripheral and lymphoblastic lymphomas and defines survival related genes. Clin Cancer Res Off J Am Assoc Cancer Res. 2004;10:4971–82. doi: 10.1158/1078-0432.ccr-04-0269. [DOI] [PubMed] [Google Scholar]
- 86.Piccaluga PP, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117:823–34. doi: 10.1172/jci26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imam MH, Shenoy PJ, Flowers CR, Phillips A, Lechowicz MJ. Incidence and survival patterns of cutaneous T-cell lymphomas in the United States. Leuk Lymphoma. 2013;54:752–9. doi: 10.3109/10428194.2012.729831. [DOI] [PubMed] [Google Scholar]
- 88.Abouyabis AN, Shenoy PJ, Lechowicz MJ, Flowers CR. Incidence and outcomes of the peripheral T-cell lymphoma subtypes in the United States. Leuk lymphoma. 2008;49:2099–107. doi: 10.1080/10428190802455867. [DOI] [PubMed] [Google Scholar]
- 89.Flowers CR, Glover R, Lonial S, Brawley OW. Racial differences in the incidence and outcomes for patients with hematological malignancies. Curr Probl Cancer. 2007;31:182–201. doi: 10.1016/j.currproblcancer.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 90.Flowers CR, Nastoupil LJ. Socioeconomic disparities in lymphoma. Blood. 2014;123:3530–1. doi: 10.1182/blood-2014-04-568766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flowers CR, Pro B. Racial differences in chronic lymphocytic leukemia. Digging deeper. Cancer. 2013;119:3593–5. doi: 10.1002/cncr.28233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shenoy PJ, et al. Racial differences in the presentation and outcomes of diffuse large B-cell lymphoma in the United States. Cancer. 2011;117:2530–40. doi: 10.1002/cncr.25765. [DOI] [PubMed] [Google Scholar]
- 93.Shenoy PJ, et al. Racial differences in the presentation and outcomes of chronic lymphocytic leukemia and variants in the United States. Clin Lymphoma Myeloma Leuk. 2011;11:498–506. doi: 10.1016/j.clml.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 94.Wang SS, et al. Immune mechanisms in non-Hodgkin lymphoma: joint effects of the TNF G308A and IL10 T3575A polymorphisms with non-Hodgkin lymphoma risk factors. Cancer Res. 2007;67:5042–54. doi: 10.1158/0008-5472.can-06-4752. [DOI] [PubMed] [Google Scholar]
- 95.Heesen M, Kunz D, Bachmann-Mennenga B, Merk HF, Bloemeke B. Linkage disequilibrium between tumor necrosis factor (TNF)- alpha-308 G/A promoter and TNF-beta NcoI polymorphisms: association with TNF-alpha response of granulocytes to endotoxin stimulation. Crit Care Med. 2003;31:211–4. doi: 10.1097/01.ccm.0000037167.19790.52. [DOI] [PubMed] [Google Scholar]
- 96.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proc Natl Acad Sci. 1997;94:3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Romagnani S. Human Th17 cells. Arthritis Res Ther. 2008;10:206. doi: 10.1186/ar2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oukka M. Interplay between pathogenic Th17 and regulatory T cells. Ann Rheum Dis. 2007;66(3):87–99. doi: 10.1136/ard.2007.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korn T, Oukka M, Kuchroo V, Bettelli E. Th17 cells: effector T cells with inflammatory properties. Semin Immunol. 2007;19:362–71. doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]