Abstract
Methylation at cytosine (5mC) is a fundamental epigenetic DNA modification recently associated with iAs-mediated carcinogenesis. In contrast, the role of 5-hydroxymethylcytosine (5hmC), the oxidation product of 5mC in iAs-mediated carcinogenesis is unknown. Here we assess the hydroxymethylome in iAs-transformed cells, showing that dynamic modulation of hydroxymethylated DNA is associated with specific transcriptional networks. Moreover, this pathologic iAs-mediated carcinogenesis is characterized by a shift toward a higher hydroxymethylation pattern genome-wide. At specific promoters, hydroxymethylation correlated with increased gene expression. Furthermore, this increase in hydroxymethylation occurs concurrently with an upregulation of ten-eleven translocation (TET) enzymes that oxidize 5-methylcytosine (5mC) in DNA. To gain an understanding into how iAs might impact TET expression, we found that iAs inhibits the binding of CTCF at the proximal, weak CTCF binding sites of the TET1 and TET2 gene promoters and enhances CTCF binding at the stronger distal binding site. Further analyses suggest that this distal site acts as an enhancer, thus high CTCF occupancy at the enhancer region of TET1 and TET2 possibly drives their high expression in iAs-transformed cells. These results have major implications in understanding the impact of differential CTCF binding, genome architecture and its consequences in iAs-mediated pathogenesis.
Keywords: Inorganic arsenic, 5hmC, Epigenetics, EMT, TET, RRHP
1. Introduction
Each year millions of people worldwide are exposed to the ubiquitous carcinogen, inorganic arsenic (iAs) (IARC, 2004; Salnikow and Zhitkovich, 2008; Cheng et al., 2012; IARC, 2012). This exposure occurs mainly through drinking water and food, and can result in acute toxicity and malignant transformation, among other consequences. The severity of these consequences is related to the dose and duration of exposure to this toxicant (Huang et al., 2004; Ren et al., 2011). Many mechanisms have been implicated in the altered gene expression in iAs-induced toxicity and pathogenesis, including oxidative stress. An equally important mechanism - changes to the epigenome - is also known to alter gene expression in iAs-induced pathogenesis (Bird, 2002; Feng et al., 2010).
Epigenetic mechanisms include changes in DNA methylation, posttranslational modification of histones, histone variants and microRNAs. Disruption of these epigenetic processes can lead to altered gene function and malignant transformation. Several recent studies showed that the iAs exposure can alter these epigenetic marks (Rea et al., 2016; Eckstein et al., 2017a; Rea et al., 2017). These changes to the epigenome drive specific gene expression patterns relevant to disease pathogenesis (Arita and Costa, 2009; Treas et al., 2012; Miao et al., 2015; Chen et al., 2016). Among these epigenetic marks, DNA methylation, the addition of a methyl group to the 5′ carbon of cytosine residues, is considered the most stable (Okano et al., 1999). DNA methylation within a promoter region is repressive for gene expression, while DNA methylation within a gene body is generally thought to increase gene expression, though it may also play a role alternative splicing (Herman and Baylin, 2003; Ball et al., 2009; Jjingo et al., 2012; Jones, 2012; Nestor et al., 2012; Rea et al., 2017). With respect to tumorigenesis, changes in methylation levels have been linked to differential expression of genes, that lead cells through epithelial-to mesenchymal transition (EMT) and into a cancerous state (Jones and Baylin, 2002; Berman et al., 2012; Hon et al., 2012; Jones, 2012; Ashktorab et al., 2014).
Until recently, DNA methylation was thought to be permanent and removed only if there was a mistake in the replication process, or a malfunction of the DNA methyltransferases - enzymes that write or maintain this mark - or through passive demethylation (Mayer et al., 2000; Guo et al., 2014). However, recent studies found that this process is dynamic with demethylases excising the modified cytosine residue (Cortellino et al., 2011; Song and He, 2012). In plants, multiple families of DNA glycosylases (DME and ROS) are responsible for the removal of 5-methylCytosine (5mC) followed by the base excision repair pathway replacing the cytosine (Kinoshita et al., 2004; Xiao et al., 2006; Feng et al., 2010; Rea et al., 2012). In mammals, active demethylation is a complex process involving several modifications of the 5mC with its eventual removal by thymine DNA glycosylase (TDG) (He et al., 2011; Kohli and Zhang, 2013). First, a TET (ten-eleven translocation methylcytosine dioxygenase) family member oxidizes 5mC converting it to 5-hydroxymethylcytosine (5hmC) (He et al., 2011; Zhang et al., 2013; Chen and Wu, 2016). This newly modified cytosine, now considered the sixth base, is a stable intermediate, playing a role in gene regulation (Bachman et al., 2014). For DNA demethylation, 5hmC is further modified to 5-formylcytosine and 5-carboxylcytosine, which then allows for its removal by the mammalian TDGs through the base excision repair pathway. And lastly, an unmodified cytosine is added to the DNA sequence (He et al., 2011; Ito et al., 2011; Zhang et al., 2013; Hu et al., 2014).
We previously reported that chronic low-dose iAs exposure leads to iAs-induced cellular transformation through genome-wide loci specific reprogramming of DNA methylation (Rea et al., 2017), that drive specific gene expression patterns. Here we report that during iAs-induced cellular transformation, 5hmC levels increase globally, as well as at specific loci. Many of the genes and pathways affected by the differential DNA methylation as well as differential hydroxymethylation between NT and iAs-T cells, are involved in the cellular adhesion, cell communication and neurogenesis. We attribute this increase in the global 5hmC levels to the increased expression of the TETs, which appears to be driven through increased binding of CTCF to the distal promoter. Our data therefore presents a platform to begin to tease the differential binding of ‘master regulators’ such as CTCF to drive specific epigenetic patterns critical in disease pathogenesis.
2. Experimental procedures
2.1. Cell growth conditions and transformation with sodium arsenite
BEAS-2B were grown in Dulbecco’s modified Eagle medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (Sigma-Aldrich), 1% MEM non-essential amino acids (Sigma-Aldrich) and 1% penicillin-streptomycin (Sigma-Aldrich). Cells were grown to ~80% confluency in a humidified chamber at 37 °C, 5% CO2 prior to use. Cells were transformed with low-dose Sodium Arsenite (0.5 µM-Sigma-Aldrich) for 8 weeks and phenotypic changes were observed indicating epithelial-to-mesenchymal transformation. This transformation was confirmed at both transcript and protein levels of known EMT markers (Riedmann et al., 2015; Rea et al., 2016).
2.2. qRT-PCR
RNA was isolated from 1 × 107 BEAS-2B cell with RNeasy MiniKit (Qiagen). 1 µg of RNA was used in a reverse transcriptase reaction with iScript Reverse Transcriptase (Bio-Rad #1708891) to prepare cDNA. 25 ng of the cDNA was used in each of the qRT-PCR reactions. The reaction protocol was as follows: 1) 94 °C 5 min; 2) 94 °C 30 s; 3) 55–63 °C (dependent on primer pair) 30 s; 4) 72 °C 45 s; 5) repeat steps 2–4 for 40 cycles; 6) 72 °C 10 min. Primers for the housekeeping gene (GAPDH) and primers used for genes with differential 5hmC were created for this study using PrimerBank (Supplemental Table 5) (Spandidos et al., 2008, 2010).
2.3. Gene ontology
Genes with 20 or more reads of differential 5hmC reads in the promoter regions were subjected to a gene ontology annotation using the Consensus PathDatabase (CPDB-Max Plank Institute; http://consensuspathdb.org) using terms or pathways that had at least 10 genes and p-values < 0.05 (Kamburov et al., 2011, 2013). Resulting -log10 (p-value) are graphed.
2.4. Reduced Representation Hydroxymethylcytosine Profiling (RRHP) library preparation
Genomic DNA was fragmented overnight at 37 °C with a hydroxymethyl-insensitive enzyme, MspI, and purified using the DNA Clean and Concentrator kit (Zymo Research). Modified Illumina TruSeq P5 and P7 adapters containing 5′-CG overhangs were ligated onto the digested DNA using T4 DNA ligase (2 h at 16 °C). Libraries were then strand extended at 72 °C with Taq DNA Polymerase. The adapters were designed to regenerate the 5′-CCGG site at the P5 junction while the P7 adapter generates a 5′-TCGG junction, making it insensitive to MspI digestion. Adapterized libraries were treated with β-glucosyltransferase to label 5-hmC modifications and purified using the DNA Clean and Concentrator kit. The glucosylated libraries were then subjected to an overnight MspI digestion at 37 °C, cutting any fragments not containing a glucosyl-5hmC site at the P5 CCGG junction. After incubation, the libraries were size-selected from 100 bp to 500 bp and purified using the ZymoClean Gel DNA Recovery Kit (Zymo Research). The fragments were amplified using OneTaq 2× Master Mix (NEB), and the PCR conditions include an initial denaturation of 94 °C for 30 s followed by 12 cycles of 94 °C for 30 s, 58 °C for 30 s, and 68 °C for 1 min. Fragments containing 5-hmC were positively selected during PCR amplification with adapter-specific indexing primers whereas fragments lacking glucosylated-5-hmC at the P5 junction were cleaved and, therefore, not amplified by PCR. Amplified libraries were purified using the DNA Clean and Concentrator kit, and multiplexed using equal volume of the libraries. Integrated DNA Technologies synthesized all adapters and primers used in these studies. RRHP analysis was performed with 2 biological replicates. To confidently identify 5hmC-enriched regions and sites, we used algorithm based on local likelihood smoothing (BSmooth) to identify consensus 5hmC regions and sites for each NT and iAs-T cells. We have previously used this method successfully to identify differentially methylated regions especially between groups with low sequencing coverage (Rea et al., 2016).
2.5. Bioinformatics processing and statistical analyses
Sequence reads from RRHP libraries were first processed to trim off the low-quality bases and the P7CG adapter at the 3′ end of the reads. Reads were then aligned to the reference genome (GRCh37/hg19) using the bowtie1 default parameters and -best. Aligned reads with the MspI tag (CCGG) were counted. The correlation analysis between different RRHP libraries was performed by comparing the presence of the tagged reads at each profiled MspI site, and Pearson’s coefficient was calculated accordingly.
2.6. Chromatin immunoprecipitation
ChIP analyses were performed as previously described (Rea et al., 2017) using 25 µg of crosslinked chromatin and 2 µg of specific antibodies for the immunoprecipitations, which are listed in Supplemental Table 4. 2 µL of eluate was used for qPCR (described above) to determine the CTCF occupancy differences and the histone modifications in the TET family promoter regions. Primers used in ChIP analyses are listed in Supplemental Table 5.
2.7. 5hmC DNA immunoprecipitation (5hme-DIP)
Genomic DNA was purified from 5 × 106 cells (BEAS-2B NT and iAs-T) using Qiagen DNeasy Blood and Tissue Kit (Qiagen 69506) and 5hme-DIP was performed using the EpiQuik hMeDIP Kit (Epigentek P-1038) following the manufacturer’s protocol. 2 µL of eluate was used in qRT-PCR reactions to determine changes in 5hmC occupancy at loci showing differential 5hmC levels from RRHP. Primers used are listed in Supplemental Table 5.
2.8. Dot blots
Purified genomic DNA from non-treated (NT) and iAs-T cells were diluted to 50 ng/µl. 25 ng - 125 ng of genomic DNA were spotted on Positively charged Nylon Membrane (Invitrogen AM10104). These were allowed to air dry, then DNA was UV-crosslinked to the membrane by placing on a UV transilluminator (Biorad) for 3 min. Membranes were blocked with 5% Milk + PBST, then incubated with the 5hmC antibody (Abcam AB106918-1:500 dilution) overnight at 4 °C. Alkalinephosphate conjugated anti-rat secondary antibody was added and incubated for 1 h at room temperature. After washing three times with PBST, the blots were developed with ECF (Vistra ECF Western blotting system, GE Health Care) following the manufacturer’s protocol. The Western bands were scanned with the GE-Typhoon FLA9500, and intensities of each dot were quantified using ImageQuant (GE Health Care).
2.9. TET hydroxylase activity assay
Total nuclear protein fraction was extracted from NT and iAs-T cells. In details, cells were gently resuspended in hypotonic solution (20 mM Tris-Cl, pH 7.4; 10 mM NaCl; 3 mM MgCl2) and allow to sit for 5 min on ice. Cells were then lysed by adding NP-40 to a final concentration of 1% and vortexed. Thereafter, nuclei were pelleted by centrifugation at 4 °C for 10 min. Nuclei were then lysed with 1× RIPA buffer (Thermofisher Scientific; Cat # 89900) for 30 min on ice and centrifuged for 10 min at full speed. Supernatant was used to estimate the protein concentration using the BCA assay. For each experiment, 20 µg of nuclear protein extract was used to determine TET activity using the Epigentek-Epigenase 5mC Hydroxylase TET Activity/Inhibition Assay Kit (Colorimetric) (Epigentek #P-3086). TET activity was calculated in two ways: 1) Specific Activity through a standard curve (expressed in ng/min/mg - Fig. 6B) and 2) Simple Calculation (expressed in OD/min/mg - Supplemental Fig. 5) as suggested by manufacturer.
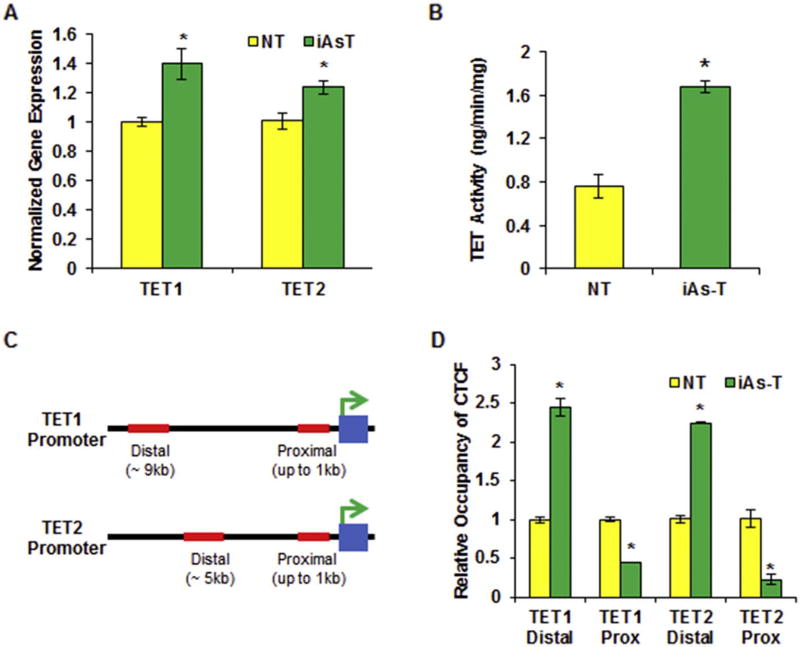
Fig. 6.
CTCF binding is preferentially increased at the distal binding site of the TET promoter in iAs-T cells directing increased expression. (A) Graphic representation of qRT-PCR results showing an increase in gene expression for TET1 and TET2 in iAs-T cells. (B) Graphic representation of increase in TET hydroxylase activity in iAs-T cells compared o NT cells. Shown here is Specific Activity as measured using hydroxylated product produced (ng/min/mg). Experiments were done in triplicates (C) Cartoon showing the distal and proximal CTCF binding sites of TET1 (top) and TET2 (bottom) promoters (D) Graphic representation of ChIP-qRT-PCR results for CTCF occupancy at TET promoters reveals increased relative occupancy binding at distal promoter sites and decreased binding at the proximal CTCF binding sites. The * denotes p < 0.05, error bars represent the SEM.
3. Results
3.1. Increase in 5hmC in cells transformed with chronic, low-dose iAs
We previously showed using ELISA (enzyme-linked immunosorbent assay), that there was little or no change in global DNA methylation between NT and iAs-T BEAS-2B cells. However, in the same studies using high-resolution single nucleotide MiniMethyl-Seq analysis, we showed many loci-specific hypermethylation and hypomethylation changes. These studies suggested that the iAs-mediated DNA methylation reprogramming directed differential gene expression in iAs-transformed cells, both at the transcription initiation and splicing levels (Rea et al., 2017). DNA methylation although a more stable modification, can be removed in mammals, through the conversion of 5mC to 5hmC by the TET family of proteins. The 5hmC is then further modified to 5-formylcytosine, and eventually to 5carboxycytosine, before being recognized by the Base Excision Repair Pathway, removed, and replaced with a non-modified cytosine.
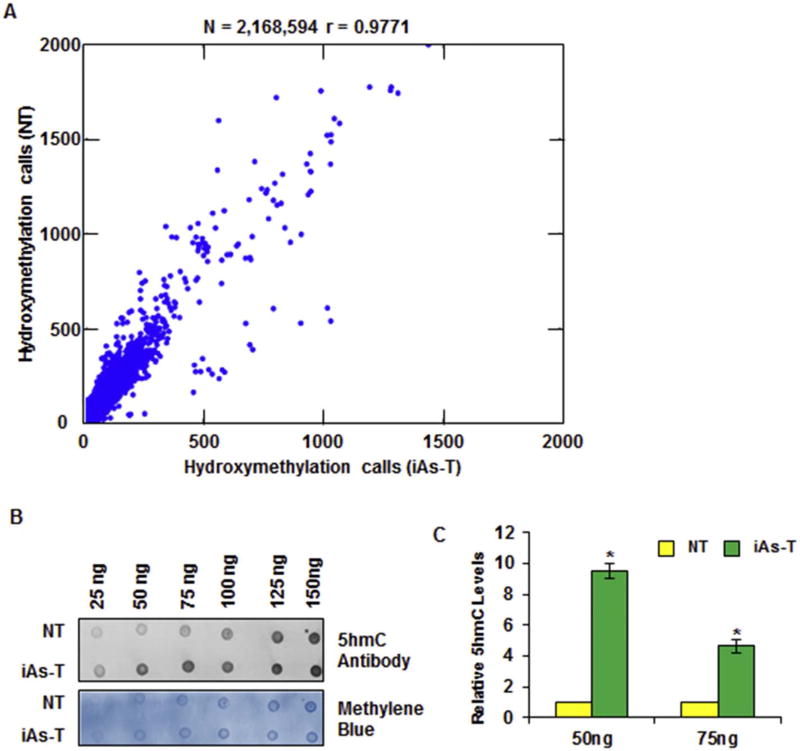
Our previous studies showing both hypermethylation and hypo-methylation at many sites, prompted us to ask whether some of the hypomethylation sites are due to removal of the 5mC by TETs to 5-hydroxymethylation. We therefore carried out a high-resolution RRHP protocol followed by sequencing, to determine the regions with differential 5hmC in time-matched NT and iAs-BEAS-2B transformed cells (iAs-T). RRHP makes use of the unique glucosylation of 5hmC, and a second MspI digestion; the glucosylated 5hmC can be distinguished from 5mC at CCGG sequences (Petterson et al., 2014). After sequencing, we obtained a library of 38.27 × 106 reads covering 1.93 × 106 unique CpG sites for NT and 58.81 × 106 reads covering 2.04 × 106 unique CpG sites for iAsT with average read counts of 9 and 13 per CpG, respectively (Table 1). These mapped to the human genome (GRCh37/hg19) at 63% and 68% for NT and iAsT, respectively (Table 1). A total of 2,165,849 unique loci were found with 5hmC, which is about 0.07% of the total genome. This value is in line with previous observations that 5hmC is relatively low in non-neural tissue humans (Globisch et al., 2010; Ye and Li, 2014). Comparison of NT and iAs-T results showed a high correlation between these samples (Pearson correlation r = 0.9771), with some strong outliers, in the iAs-T sample (Fig. 1A).
Table 1.
Read efficiency of RRHP.
| Total number of reads | Mapping efficiency | Detected CpGs | Average read count (coverage) | Loci with> 20 reads | Loci with> 50 reads | |
|---|---|---|---|---|---|---|
| NT | 38.27 × 106 | 63% | 1.93 × 106 | 9 | 10,542 | 340 |
| iAsT | 58.81 × 106 | 68% | 2.04 × 106 | 13 | 96,538 | 30,665 |
Fig. 1.
Profiling analysis through RRHP reveals a global increase in 5hmC. (A) Scatter Plot from RRHP analysis showing an increase in global 5hmC levels in iAs-T cells. (B) Representative Dot Blot shows global 5hmC levels in NT BEAS-2B (NT) cells compared with cells exposed for 8 weeks with chronic, low dose iAs (iAs-T) shows higher overall 5hmC levels in iAs-T cells (top), and methylene blue staining for genomic DNA (bottom) for loading control. (C) Quantification of 3 independent Dot Blots showing an increase in 5hmC levels with the increasing concentrations of genomic DNA. Shown is one biological replicate with 3 technical replicates, a second biological replicate with technical replicates showed similar results. The * denotes p < 0.05 and error bars represent the SEM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
RRHP profiling is not an absolute quantification of 5hmC, but it is a positive read assay, so that increased 5hmC levels at a specific locus returns higher read numbers. Thus based on this assertion, the total global number of reads in iAs-T cells was ~20 million more than in NT cells, indicating an increase in hydroxymethylation in these cells. This increase in 5hmC reads is interesting as the distribution is not limited to any specific chromosome or region, but is well distributed across the genome. Further analysis showed an increase of 5-hydroxymethylation at 65.77% of loci, while 24.93% of loci showed decreased 5hmC levels in the iAs-T compared to NT cells. The remaining 9% of CCGG sites showed no difference in 5hmC levels (Supplemental Table 1). We confirmed this global increase in 5hmC through Dot Blot analysis using an antibody directed toward 5hmC (Fig. 1B). Genomic DNA isolated from both NT and iAs-T cells was titrated linearly by blotting 25 ng to 125 ng and probed with antibody specific for 5hmC. The staining was quantified and results show that at 50 ng DNA concentration, a 10-fold in 5hmC was seen in iAs-T compared to NT, while at 75 ng a 4-fold increase was seen (Fig. 1C). These results validate the global increase in 5hmC levels measured through with RRHP.
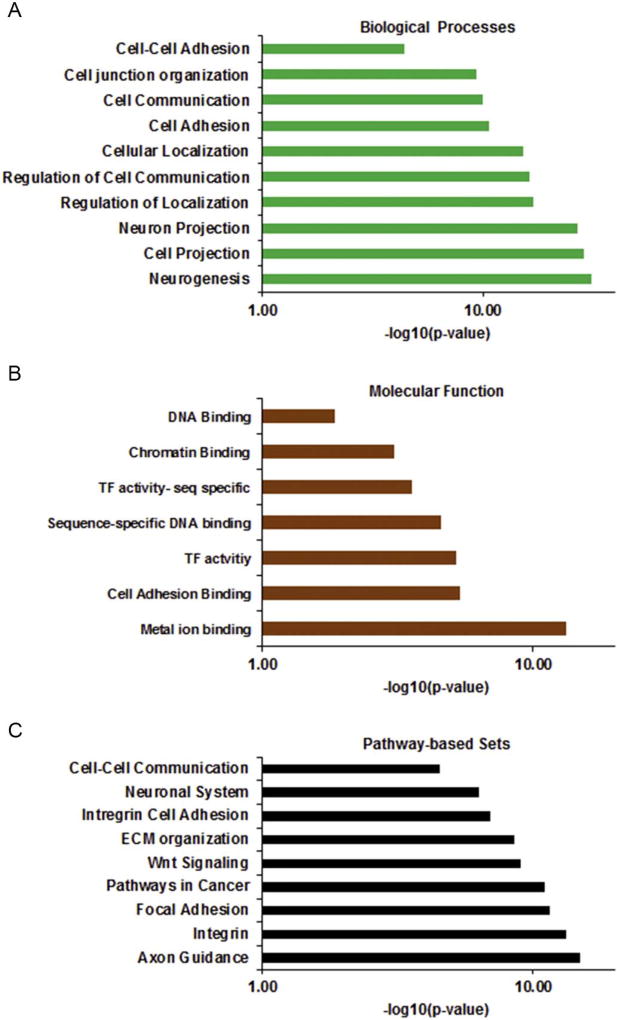
To begin to understand the functional impact of these differential hydroxymethylation patterns, we analyzed the genes with changes in 5hmC pattern. We performed a gene ontology (GO) analysis using the Consensus PathDatabase (CPDB-Max Plank Institute) (Kamburov et al., 2011, 2013), and the most enriched GO Biological Process and Molecular function for these genes are cell communication, cell adhesion, neurogenesis, transcription factor activity and metal ion binding (Fig. 2A and B). These same GO terms were associated with genes with altered DNA methylation patterns from our previous studies (Rea et al., 2017). We next carried out the Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis to identify the enriched metabolic or signaling pathways in which these genes are involved. This analysis revealed cell communication pathways, integrin cell adhesion, focal adhesion and multiple cellular adhesion pathways as the most enriched pathways (Fig. 2C). Interestingly, many of these adhesion genes had lower 5-hydroxymethylation and were down-regulated in gene expression (Supplemental Table 2).
Fig. 2.
Enrichment analysis of genes with differential 5hmC. Gene Ontology Analysis of global 5hmC changes show alterations to genes involved in cellular adhesion, cellular communication, transcription factor activity, and neural cell growth for (A) Biological Processes (B) Molecular Function (C) Pathway-based Sets.
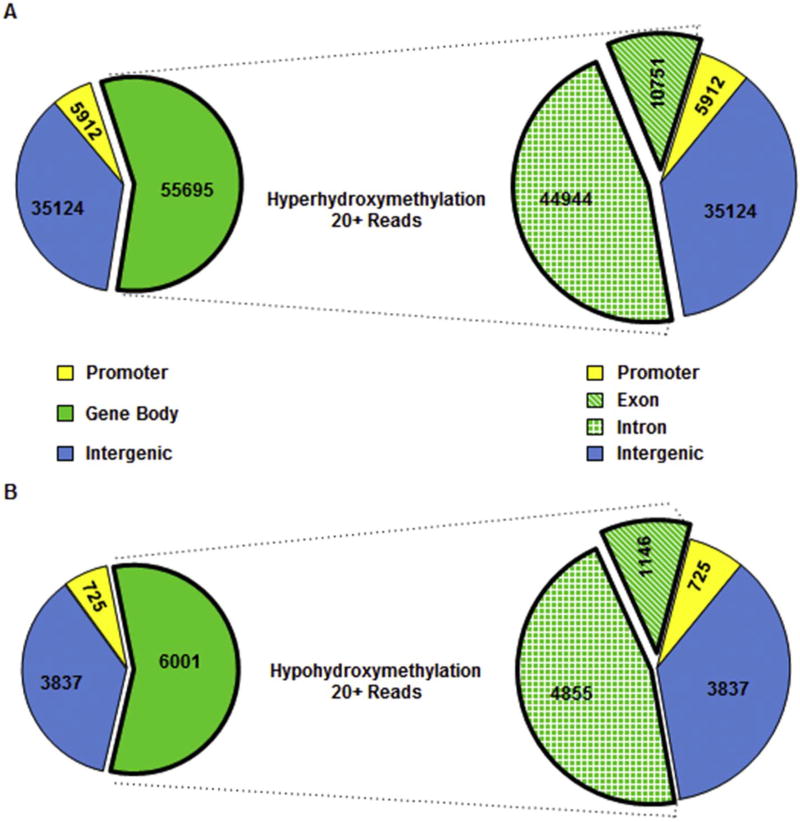
We and others have shown that DNA methylation occurs not only at promoter regions but also at other regulatory regions (Khalid et al., 2014; Kubo et al., 2015; Rea et al., 2017). We therefore asked if these changes in 5hmC occur at other functional genomic regions as well. For these analyses, we used a stringent read number of > 20 (Table 1) allowing for a deeper assessment of the 5hmC distribution in the genome (Petterson et al., 2014; Grosser et al., 2015; Tekpli et al., 2016). We observed a similar differential of 5hmC distribution at functional genomic regions - 46% differential hypo- and hyper-5hydroxymethylation within introns, 11% within exons, 36% in intergenic regions, and about 7% at the promoters (Fig. 3A and B). While this distribution of hypo- and hyper-hydroxymethylation is similar, there were more loci (9.5× more reads) for hyper-hydroxymethylation compared to hypo-hydroxymethylation in iAs-T compared to NT cells (96,538 vs. 10,542 - Table 1), again confirming increased hydroxymethylation in iAs-T cells. If we increase the stringency to > 50 reads we again see a similar distribution in functional genomic regions (Supplemental Fig. 1), however 30,665 loci with more reads indicating increased 5hmC and 340 loci with less reads or decreased 5hmC (Table 1).
Fig. 3.
Distribution of differential 5hmC in functional genomic regions with stringent read counts of > 20. Number of loci with 20+ reads, are low within promoter regions and high in the Introns. (A) Number of loci that show increased 5hmC levels in iAs-T. (B) Number of loci that show decreased 5hmC levels in iAs-T. Increased 5hmC loci total 96,538, while only 10,542 loci show decreased 5hmC levels. Even though there is a difference in loci, the relative distribution between promoter, intron, exon, and intergenic regions is very similar.
3.2. Differential promoter 5hmC levels correlate with gene expression changes
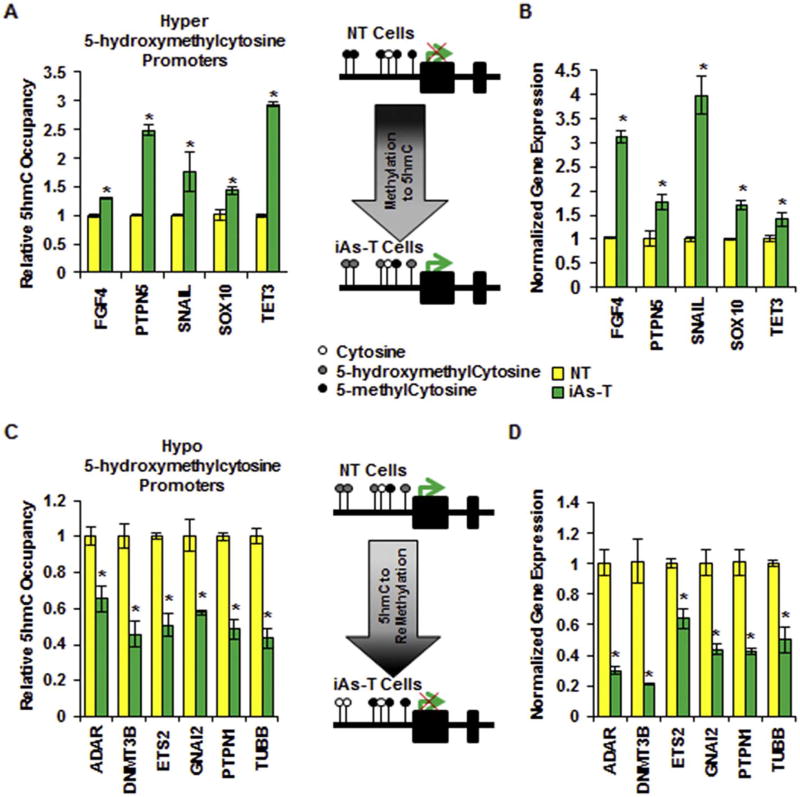
Changes in promoter 5mC levels correlate with changes in gene expression (Eckstein et al., 2017b; Rea et al., 2017). This is an inverse correlation relationship - increased promoter 5mC correlates with decrease gene expression while decreased promoter 5mC levels correlates with increase gene expression. Since hydroxylation of 5mC to 5hmC is the first step in the active DNA demethylation process, we next asked if the observed changes in promoter 5hmC levels correlate to changes in gene expression - if increased 5hmC correlates to increased gene expression and vice versa. First, we analyzed and validated the change in occupancy of 5hmC at 5 different promoters with RRPH identified increased 5hmC in iAs-T cells compared to NT cells (Fig. 4A). We used hydroxymethylated DNA immunoprecipitation (5hme-DIP) followed by quantitative real-time PCR (qRT-PCR) for these analyses. In detail, genomic DNA was sonicated, and then immunoprecipitated with 5hmC-specific antibody. Resulting DNA was purified and subjected qRT-PCR to determine the relative occupancy of 5hmC at specific gene loci. Hyper-5hmC was confirmed at the promoters of FGF4, PTPN5, SNAIL, SOX10, and TET3 (Fig. 4A). We then asked whether this change in 5hmC correlated with increased gene expression. Using quantitative RT-PCR, we observed an increase in gene expression in iAs-T compared to NT for all of the 5 hyper-hydroxymethylated promoters, cells (Fig. 4B).
Fig. 4.
Differential 5hmC levels in the promoter correlate with changes in gene expression. Validation of promoter regions that showed (A) Hyper and (C) Hypo 5-hydroxymethylation changes in RRHP dataset. Using 5hme-DIP followed by qRT-PCR five genes were validated with an increase in 5hmC levels (A) in their promoters in iAs-T cells and six were validated as having a decrease in 5hmC levels (C) in their promoters in iAs-T cells. Hydroxymethylation levels in the promoter correlate with gene expression levels; genes with an increase in promoter 5hmC levels show increased expression (B) and genes with a decrease in promoter 5hmC levels show decreased expression (D). The * denotes p < 0.05, error bars represent the SEM.
Next, we also validated the observed decreased 5hmC at 6 different promoter loci - ADAR, DNMT3B, ETS2, GNAI2, TUBB, and PTPN1 (Fig. 4C). Many of these with decreased 5hmC levels in the promoter are involved with cellular adhesion. Here too, the 6 promoters with reduced hydroxymethylation in iAs-T compared to NT cells, showed a decrease in gene expression levels (Fig. 4D). These results are in line with other studies showing a positive correlation of hydroxymethylation levels with levels of gene expression (Colquitt et al., 2013; Tsai et al., 2014; Skvortsova et al., 2017). Indeed, other genes that had differential promoter hydroxymethylation from RRHP (though we were not able to amplify products from 5hme-DIP) also showed correlating changes in gene expression levels. MMP2, FOXC2, BAG2 and UBB all show increased gene expression correlating with the hyper-hydroxymethylation levels observed from RRHP, while PRKACA, and OLFM1 show decreased expression levels correlating with decreased promoter 5hmC levels observed in the RRHP data (Supplemental Fig. 2). These genes used for validation were chosen based on the fact that they showed both high differential 5hmC and gene expression as seen in our microarray dataset (GSE90211).
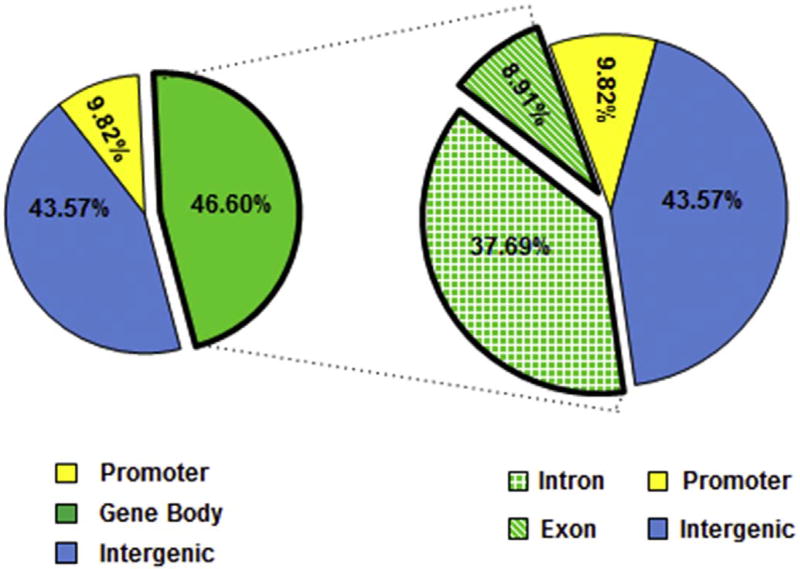
3.3. Overlap of 5mC (MethylMIni-Seq) and 5hmC (RRHP)
Bisulfite sequencing is a widely used technique to map 5mC and 5hmC sites. However, neither bisulfite-treated 5mC nor 5hmC can undergo the C → T transitions, rendering this method unable to distinguish between 5mC and 5hmC sites (Huang et al., 2010; Yu et al., 2012; Petterson et al., 2014). The additional step of glucosylation of 5hmC in the RRPH technique, gives it the ability to distinguish between 5mC and 5hmCs. Since we had shown previously using MethylMini-Seq that iAs-mediated transformation resulted in extensive DNA methylation reprogramming (Rea et al., 2017), we asked if some of those previously identified 5mC sites were indeed 5hmC sites (Booth et al., 2012; Chen et al., 2013; Petterson et al., 2014). We carried out an overlap of our RRPH data with the methyl-seq data and found that 7532 loci showed both significant 5mC changes as well as differential 5hmC levels (Supplemental Table 3). The distribution of these changes was as follows: 46.6% in gene bodies (37.7% in intron, and 8.91% exon), 43.6% in intergenic regions and 9.8% at promoters (Fig. 5). This distribution of changed sites is similar to the overall changes seen in 5hmC levels (Fig. 3). We categorize the overlapping sites into six categories: 1) Hypermethylation and increased 5hmC - could indicate sites of 5hmC, 2) Hypermethylation and decreased 5hmC - could indicate sites of 5mC, 3) Hypermethylation and no change in 5hmC - could indicate sites of 5mC, 4)Hypomethylation and increased 5hmC - could indicate 5hmC sites, 5) Hypomethylation and decreased 5hmC - could indicate the possible removal of the 5mC completely in the treated cells or 6) Hypomethylation and no change in 5hmC - could indicate loci where 5mC is removed (Supplemental Table 3 Browser tracks for representative four genes (GDF2, CLN5, CHST13, and DISP2) in the UCSC genome browser provide examples of categories 1, 2, 4, and 5 (Supplemental Fig. 3). Gene ontology analysis showed similar over-represented functional groups - cell adhesion, cell communication, cell signaling, neural development, chromatin binding, epithelium development, EMT, KRAS signaling and extracellular Matrix (Supplemental Fig. 4).
Fig. 5.
Functional Genomic Region analysis of 5mC and 5hmC overlapping loci. Pie charts show the distribution of loci (7532) at functional genomic regions reveal that both differential 5mC (measured via MethyMini-Seq) and differential 5hmC (measured via RRHP) occur. 46.60% of changes were within the gene body; 43.57% were in the inter-genic region and 9.82% were in the promoter region.
3.4. Differential CTCF binding regulates TET expression in iAs-T cells
Active DNA demethylation occurs mainly through the conversion of 5mC to 5hmC by TET enzyme-mediated oxidation (Hill et al., 2014; Wu and Zhang, 2017). Since we observed increased global and site-specific hydroxymethylation in iAs-T cells, we hypothesized there would be an increase in the expression of TETs. Using qRT-PCR we determined the relative gene expression of TET1, TET2 and TET3. We found that the levels increased by about 1.2 to 1.4-fold in iAs-T cells compared to NT cells (Figs. 6A and 4B). We next tested whether this increase in TET gene expression correlated with increased TET activity. To answer this question, we used the 5mC hydroxylase TET activity Assay Kit (Epigentek) to measure the activity of TETs in nuclear extracts obtained from NT and iAs-T cells. Our results show increase in TET activity in iAs-T cells compared to NT cells as measured using either the level of hydroxymethylated product (1.67 vs 0.75 ng/min/mg) (Fig. 6B) or the amount of hydroxylase activity present in the cells (Supplemental Fig. 5).
To a large degree, the expression of genes is regulated at the level of transcription initiation mediated by the specific binding of protein transcription factors (TFs) to short DNA sequence motifs located in gene promoter regions, the DNA-sequence region upstream of genes. We asked if iAs could regulate the binding of some key TF to alter gene expression at the TET promoters. We had previously shown that iAs inhibition of CTCF binding at the promoters of DNA methyltransferases (DNMTs) reduced their expression. Interestingly, using the ModEncode consortium database (UCSC Genome browser), we identified two potential binding sites for CTCF at the promoters of the TET1 and TET2 genes. One of the sites, which is henceforth called the proximal site, is near the transcription start site (TSS) while the second site is 5 kb and 9 kb upstream of the TSS (henceforth distal site) for TET2 and TET1 respectively (Fig. 6C). We measured the relative occupancy of CTCF in NT and iAs-T cells at both loci using chromatin immunoprecipitation, with anti-CTCF antibodies, followed by qRT-PCR (ChIP-qRT-PCR). We found that the relative occupancy of CTCF at the distal sites of TET1 and TET2 was increased by at least 2-fold while the occupancy at the proximal CTCF sites decreased by 2.5- and 5-fold respectively (Fig. 6D). iAs is known to specifically target CTCF binding to weak CTCF binding sites (Zhou et al., 2011; Kasowski et al., 2013; Rojas et al., 2015; Rea et al., 2017). Using in silico analysis, we calculated the strength of these CTCF binding sites using the CTCFBSDB database: (http://insulatordb.uthsc.edu/home_new.php) (Bao et al., 2008; Ziebarth et al., 2013). This database uses motifs from multiple sources to generate Position Weight Matrices (PWM) and produces log-odds for best fit for sequence against the background. Using this in silico tool, we found that the distal binding site of TET1 promoter returned a score of 14.125, while the proximal binding site returned a score of 6.419. Likewise, the proximal CTCF binding site at the TET2 promoter the distal region returned a score of 12.32, while the proximal site returns a score of 8.0. These data indiate that the distal CTCF binding sites of both TET1 and TET2 are stronger binding sites than the proximal site.
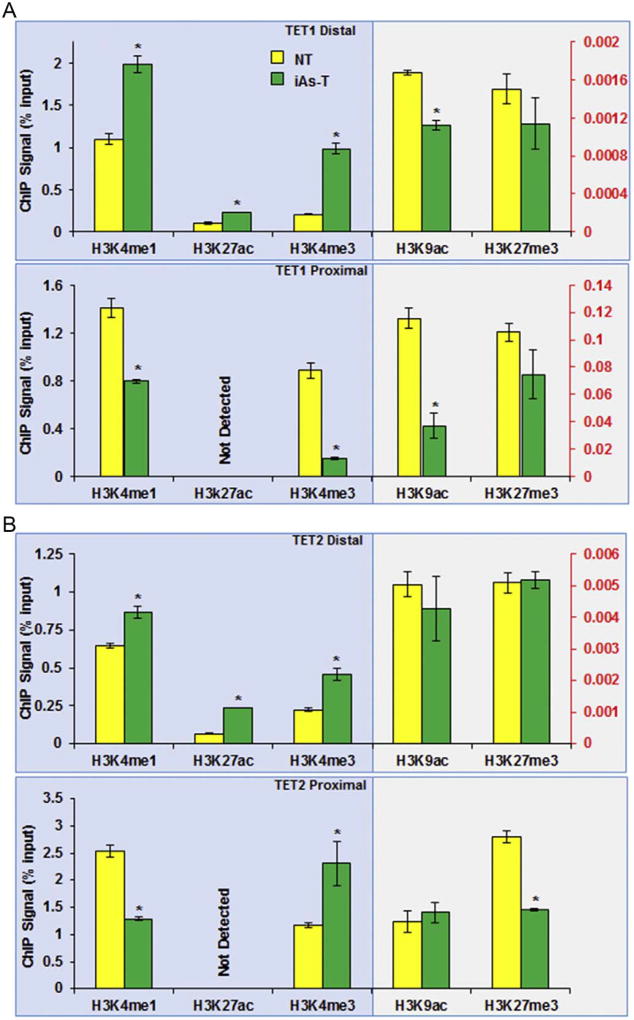
The results outlined above suggest that these distal CTCF binding sites may act as enhancer regions for the TET1 and TET2 genes. To test this hypothesis, we performed ChIP-qRTPCR assays for histone posttranslational modifications (PTMs) known to be associated with promoter (H3K4me3, H3K9ac, H3K27me3) and enhancer regions (H3K4me1, H3K27ac). First we confirmed the presence of enhancer associated PTMs at the distal sites while the promoter-associated PTMs such as H3K9ac and H3K27me3 are low (Fig. 7A and B top panel). H3K4me3 though typically associated with promoters, can also be found at enhancers. Nonetheless, compared to promoters, H3K4me3 levels at enhancers are low, and a high ratio of H3K4me1 to H3K4me3 broadly distinguishes enhancers from promoters (Heintzman et al., 2007; Koch and Andrau, 2011; Djebali et al., 2012). This differential level of H3K4me3 further confirmed that this site is most likely an enhancer region (Fig. 7A and B bottom panel). A further comparison between NT and iAsT cells, showed an increase in the relative occupancy of the enhancer-associated histone PTMs (Supplemental Fig. 6, left panels) at the TET distal promoter region. For instance, H3K4me1, H3K4me3 and H3K27ac levels are significantly increased at the distal region, while the promoter associated-histone PTMs (H3K9ac, H3K27me3), was decreased slightly or not all (Fig. 7 top panels and Supplemental Fig. 6, left panels).
Fig. 7.
Changes in enhancer-associated histone PTMs at the TET promoter suggest enhancer activity of the distal CTCF binding site. Distal Region (top panel) of the TET1 (A) and TET2 (B) promoters, have elevated enhancer-associated marks (H3k4me1 and H3k27ac). The proximal promoter regions (A, B bottom panel) show decreased levels of promoter-associated modifications, however a decrease in the repressive H3K27me3 is observed. The * denotes p < 0.05, error bars represent the SEM.
At the TET1 proximal promoter region, a decrease in H3Kme1, H3K4me3, H3K9ac and H3K27me3 was observed while H3K27ac was not detected when iAs-T was compared to NT cells, (Fig. 7A bottom panel, Supplemental Fig. 6B, right panel). For the proximal site of TET2, a similar decrease in H3K4me1, H3K9ac, and H3K27me3 was observed and again H3k27ac was not detected. Contrary though to TET1 promoter, we observed an increase in H3K4me3 was observed (Fig. 7B bottom, Supplemental Fig. 6A, right panel). These results suggest that the increased binding of CTCF to the distal sites is regulatory for TET1 and TET2 expression, and support the hypothesis of these distal CTCF binding sites maybe potential enhancer regions.
4. Discussion
This study represents the first comprehensive genome-wide analysis of the differential distribution of 5hmC in response to iAs-mediated EMT. Most studies investigating changes to epigenome in response to iAs focused on DNA methylation and histone PTMs (Salnikow and Zhitkovich, 2008; Rea et al., 2016; Eckstein et al., 2017b; Rea et al., 2017). In this study, we report a global increase in 5-hydroxymethylcytosine during iAs-mediated transformation. These results are in line with several studies showing increased 5hmC levels and increased TET levels during the process of carcinogenesis (Tsai et al., 2014; Kao et al., 2016; Tian et al., 2017). These 5hmC genome-wide changes were not random but found at specific targeted loci. We identified these 5hmC-targeted loci with changes between NT and iAs-T cells (Fig. 1, Table 1). Pathway enrichment analyses of genes with differential 5hmC sites revealed pathways such as cellular communication, cellular adhesion, and neurogenesis (Fig. 2). Interestingly, these same pathways were targeted by iAs through DNA reprogramming (Rea et al., 2017). Our findings that the same pathways are targeted by both 5mC and 5hmC are consistent with the idea 5hmC methylation is an important constituent of the DNA demethylation pathways, contributing to epigenetic plasticity and altered gene expression in iAs-mediated carcinogenesis.
We next examined the genome-wide distribution of 5mC and 5hmC (Fig. 3) for two main reasons: 1) demethylation of 5mCs leads to other products including 5hmCs; and 2) the widely used bisulfite sequencing is unable to distinguish between methylation and hydroxymethylation, since both 5mC and 5hmC do not undergo deamination after reacting with sodium bisulfite. Thus in the RRHP technique, a glucosylation and a second restriction enzyme digest helps in distinguishing the 5hmC from 5mCs (Huang et al., 2010). We therefore asked whether the results from our previous MethylMini-Seq did contain some 5hmC sites. An overlay of both data showed that there were about 7600 sites that were identified previously as 5mCs (Fig. 5) that are most likely 5hmCs. Further analysis also helped to delineate regions previously methylated that potentially change to a demethylated state with iAs- transformation (Supplemental Table 3). Furthermore, we illustrate a positive correlation between 5hmC promoter enrichment and RNA abundance for several genes (Fig. 4). Not surprisingly, genes with low promoter 5hmC enrichment are least expressed, while genes with high 5hmC at the promoter are highly expressed. This observation is in agreement with earlier reports of positive correlation of 5hmC levels with promoter gene expression (Colquitt et al., 2013; Tsai et al., 2014; Skvortsova et al., 2017). We also observed low 5hmC levels within high CpG regions (only 16.25% of our dataset), which is in line with previous studies (Booth et al., 2012; Putiri et al., 2014). One limitation, however of the RRHP analysis, is that it relies on MspI digest, thus only 5hmC sites within a CCGG context will be read (Petterson et al., 2014)
This study also reveals a dynamic interplay between TET expression, CTCF and different chromatin marks (Fig. 6). The simplest explanation for this observation is that CTCF binding inhibition will inhibit gene expression of TET genes. After investigating the CTCF occupancy at the proximal promoter region of the TETs we observed a decrease in the occupancy at TET1 and TET2 (Fig. 6D). Based on our previous studies, this decrease in CTCF occupancy should lead to a decrease in gene expression as we had seen with at the promoters of DNMTs (Rea et al., 2017). However, the expression of TET genes was increased in iAs-T cells as well as an increase in the hydroxylase activity (Fig. 6A and B). Further analyses of the promoter of the TET genes, showed a second CTCF binding site ~6 to 10 kb upstream of the TSSs (distal site). Analysis of the relative occupancy of CTCF at these distal sites, showed an increase in CTCF binding in iAs-T cells. CTCF binds DNA sequence motifs with specific sequences that influence the strength of CTCF binding (Nakahashi et al., 2013). Due to this differential binding, we analyzed the strength of these CTCF binding sites. Thus, using CTCFBSDB, we determined that the proximal binding site had a motif weak for CTCF-binding while the motif found at the distal site is a strong binder. Taken together, these results suggest that iAs blocks CTCF binding to the weaker target sites, and thus to promote the observed increase in gene expression, the block at the proximal weaker site must be compensated by increased binding at a stronger binding site.
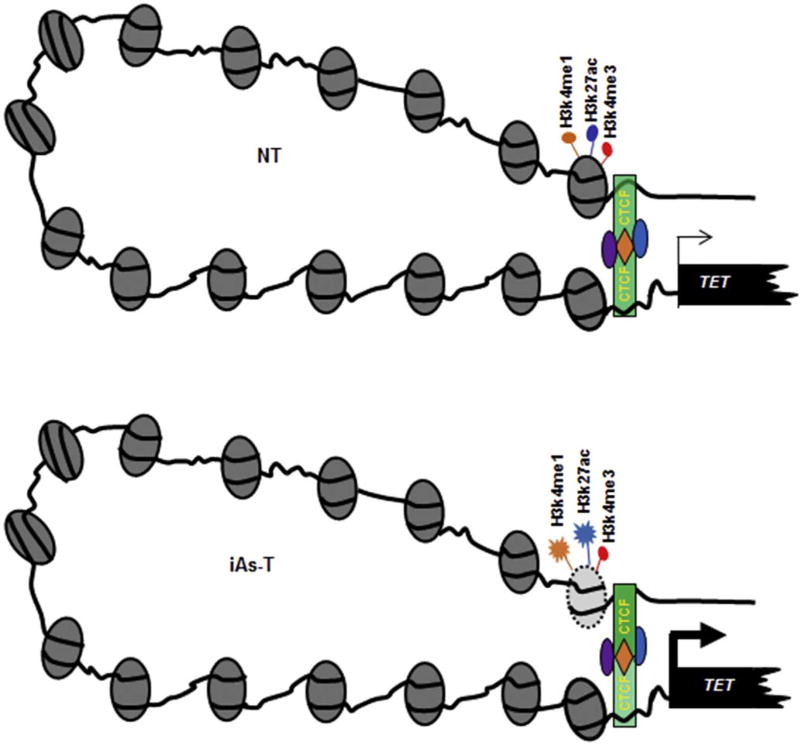
CTCF is known to act in several ways to regulate gene expression. It can act as a transcriptional activator, repressor and insulator, or it can pause transcription. CTCF is also an architectural protein creating short-range and long-range loops. In forming short-range loops, CTCF links promoters to transcription start sites, while in its formation of long-range loops, CTCF links enhancers to promoters; also acting as an insulator, creating boundaries between topologically associated domains (TADs). Our data suggest that the binding of CTCF to the stronger site might loop the regulatory region to the promoter region, to drive TET expression. To test this idea that the stronger binding site is an enhancer, we analyzed enhancer-associated histone PTMs. At both the TET1 and TET2 distal promoter regions we detected H3K4me1 and H3K27ac, as well as H3K4me3. Interestingly in iAs-T cells there was an increase in occupancy of these marks (Fig. 7 top). This finding seems to suggest that in NT cells, the occupancy of enhancer-associated histone PTMs, and CTCF binding at the proximal site, drive a ‘normal’ TET expression. On the otherhand, in iAs-T, because of the reduction of positive marks (specifically, CTCF binding at the proximal sites), a compensatory mechanism is occurring, promoting increased CTCF binding to the distal site to enhance TET expression (Fig. 8).
Fig. 8.
Model for CTCF binding at TET promoters during iAs exposure. In NT cells (Top), CTCF has occupancy at both distal and proximal binding sites, and TET gene expression occurs normally. In iAs-T cells (Bottom), depletion of CTCF at the weaker proximal binding site is associated with an increased binding of CTCF at the stronger CTCF distal binding site, thus driving increased gene expression at the TET genes. CTCF is a chromatin architectural protein that forms both short-distance and long-distance loops, bringing regulatory elements that are often separated by considerable distances. In this figure, we depict CTCF control of transcription at the TET gene through interactions between enhancer region and promoter regions in a three-dimensional nuclear space (Right). To illustrate this, the figure depicts regulatory elements at the TET locus and the CTCF, histone modifications within these regions.
An understanding of the differential binding of CTCF to target sites is probably more complicated than the simple model we depict in Fig. 8. CTCF is known to bind with associated complexes such as condensin and BORIS (Pugacheva et al., 2010; Nakahashi et al., 2013; Ali et al., 2016). It is possible that some of these complexes make CTCF an insulator, activator or repressor and that some of these complexes aid in stronger CTCF binding to the distal site. However, a clear mechanistic insight into selective inhibition of CTCF by iAs requires a detailed genome-wide functional analysis of CTCF binding in iAs-T cells. Future studies will help delineate such selectivity. Interestingly, other heavy metals have also been shown to inhibit CTCF binding at weaker binding sites (Jose et al., 2014). Whether this is a mechanism for all heavy metals remains still to be determined.
In conclusion, this study together with our previous methylation studies (Rea et al., 2017), demonstrates that selective inhibition of CTCF binding, regulates epigenomic reprogramming to drive specific gene expression profiles. And since CTCF insulates TADs and controls proper genomic architecture and chromosome folding, our studies may provide a mechanistic insight into the rules governing mammalian genome organization. Disruptions of these rules may have profound implications in understanding disease pathogenesis.
Supplementary Material
Acknowledgments
We would like to thank the Markey Cancer Center’s Research Communications Office for manuscript editing and assistance with graphic design (P30 CA177558). GEO accession numbers for Methyl-MiniSeq data is GSE85012 and RRHP data is GSE103626 this work was supported by NSF grant MCB 1517986 to YFN-M, NIEHS grant R01-ES024478, TG was supported by NIEHSR25ES027864 to the SURES program at UK, and NIH T32 grant 165990 to MR, through Markey Cancer Center at University of Kentucky.
Abbreviations
- 5mC
5-methylCytosine
- 5hmC
5-hydroxymethylCytosine
- EMT
epithelial-to-mesenchymal transition
- iAs
inorganic arsenic
- iAs-T
inorganic arsenic transformed cells
- NT
non-treated cell
- qRT-PCR
quantitative reverse transcription-PCR
- ChIP-qRT-PCR
chromatin immunoprecipitation followed by quantitative real time PCR
- Histone PTMs
histone posttranslational modifications
- TSS
transcription start site
- GO
gene ontology
- H3K4me1
histone H3 lysine 4 monomethylation
- H3K4me3
histone H3 lysine 4 trimethylation
- H3K9ac
histone H3 lysine 9 acetylation
- H3K27ac
histone H3 lysine 27 acetylation
- H3K27me3
histone H3 lysine 27 trimethylation
Footnotes
Conflict of interest statement
The authors state no conflicts of interest pertaining to this work.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.taap.2017.11.015.
References
- Ali T, Renkawitz R, Bartkuhn M. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016;37:17–26. doi: 10.1016/j.gde.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, Sun X, Brim H. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9:503–512. doi: 10.4161/epi.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 2014;6 doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methy-lation signatures in human cells. Nat. Biotechnol. 2009;27 doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–D87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336 doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Chen H-F, Wu K-J. Epigenetics, TET proteins, and hypoxia in epithelial-mesenchymal transition and tumorigenesis. Biomedicine. 2016;6:1–8. doi: 10.7603/s40681-016-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Shen F, Huang W. Quantification of 5-methylcytosine and 5-hy-droxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clin. Chem. 2013;59:824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Zhang J, Guo Z, Ma Q, Xu Z, Zhou Y, Xu Z, Li Z, Liu Y, Ye X, Li X, Yuan B, Ke Y, He C, Zhou L, Liu J, Ci W. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26:103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T-F, Choudhuri S, Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. JAT. 2012;32:643–653. doi: 10.1002/jat.2717. [DOI] [PubMed] [Google Scholar]
- Colquitt BM, Allen WE, Barnea G, Lomvardas S. Alteration of genic 5-hy-droxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc. Natl. Acad. Sci. 2013;110:14682–14687. doi: 10.1073/pnas.1302759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146 doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See L-H, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein M, Eleazer R, Rea M, Fondufe-Mittendorf Y. Epigenomic reprogramming in inorganic arsenic-mediated gene expression patterns during carcinogenesis. Rev. Environ. Health. 2017a;32:93–103. doi: 10.1515/reveh-2016-0025. [DOI] [PubMed] [Google Scholar]
- Eckstein M, Rea M, Fondufe-Mittendorf YN. Transient, permanent changes in DNA methylation patterns in inorganic arsenic-mediated epithelial-to-mesenchymal transition. Toxicol. Appl. Pharmacol. 2017b doi: 10.1016/j.taap.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser C, Wagner N, Grothaus K, Horsthemke B. Altering TET dioxygenase levels within physiological range affects DNA methylation dynamics of HEK293 cells. Epigenetics. 2015;10:819–833. doi: 10.1080/15592294.2015.1073879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15 doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333 doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39 doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hill PWS, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics. 2014;104:324–333. doi: 10.1016/j.ygeno.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22 doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14 doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Huang C, Ke Q, Costa M, Shi X. Molecular mechanisms of arsenic carcinogenesis. Mol. Cell. Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. Some drinking-water disinfectants and contaminants, including arsenic. IARC Monogr. Eval. Carcinog. Risks Hum. 2004;84:39–267. [PMC free article] [PubMed] [Google Scholar]
- IARC. Cadmium and Cadmium Compounds. IARC Monogr. Eval. Carcinog. Risks Hum. 2012:121–146. [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333 doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan KI. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jose CC, Xu B, Jagannathan L, Trac C, Mallela RK, Hattori T, Lai D, Koide S, Schones DE, Cuddapah S. Epigenetic dysregulation by nickel through repressive chromatin domain disruption. Proc. Natl. Acad. Sci. 2014;111:14631–14636. doi: 10.1073/pnas.1406923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S-H, Wu K-J, Lee W-H. Hypoxia, Epithelial-Mmesenchymal Transition, and TET-Mediated Epigenetic Changes. J. Clin. Med. 2016;5:24. doi: 10.3390/jcm5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, Boyle AP, Zhang Q, Zakharia F, Spacek DV, Li J, Xie D, Olarerin-George A, Steinmetz LM, Hogenesch JB, Kellis M, Batzoglou S, Snyder M. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid O, Kim JJ, Kim H-S, Hoang M, Tu TG, Elie O, Lee C, Vu C, Horvath S, Spigelman I, Kim Y. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in ArabidopsisEndosperm by DNA methylation. Science. 2004;303:521. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Koch F, Andrau J-C. Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Toh H, Shirane K, Shirakawa T, Kobayashi H, Sato T, Sone H, Sato Y, Tomizawa S-i, Tsurusaki Y, Shibata H, Saitsu H, Suzuki Y, Matsumoto N, Suyama M, Kono T, Ohbo K, Sasaki H. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16:624. doi: 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403 doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Miao Z, Wu L, Lu M, Meng X, Gao B, Qiao X, Zhang W, Xue D. Analysis of the transcriptional regulation of cancer-related genes by aberrant DNA methylation of the cis-regulation sites in the promoter region during hepatocyte carcinogenesis caused by arsenic. Oncotarget. 2015 doi: 10.18632/oncotarget.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi H, Kieffer Kwon K-R, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, Qian J, Dubois W, Welsh S, Phair RD, Pugh BF, Lobanenkov V, Hager GL, Casellas R. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Petterson A, Chung TH, Tan D, Sun X, Jia X-Y. RRHP: a tag-based approach for 5-hydroxymethylcytosine mapping at single-site resolution. Genome Biol. 2014;15:1–13. doi: 10.1186/s13059-014-0456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Suzuki T, Pack SD, Kosaka-Suzuki N, Yoon J, Vostrov AA. The structural complexity of the human BORIS gene in gametogenesis and cancer. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi JH. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15 doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M, Zheng W, Chen M, Braud C, Bhangu D, Rognan TN, Xiao W. Histone H1 affects gene imprinting and DNA methylation in Arabidopsis. Plant J. 2012;71:776–786. doi: 10.1111/j.1365-313X.2012.05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M, Jiang T, Eleazer R, Eckstein M, Marshall AG, Fondufe-Mittendorf YN. Quantitative mass spectrometry reveals changes in histone H2B variants as cells undergo inorganic arsenic-mediated cellular transformation. Mol. Cell. Proteomics. 2016;15:2411–2422. doi: 10.1074/mcp.M116.058412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea M, Eckstein M, Eleazer R, Smith C, Fondufe-Mittendorf YN. Genome-wide DNA methylation reprogramming in response to inorganic arsenic links inhibition of CTCF binding, DNMT expression and cellular transformation. Sci. Rep. 2017;7:41474. doi: 10.1038/srep41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ. Health Perspect. 2011;119:11–19. doi: 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedmann C, Ma Y, Melikishvili M, Godfrey S, Zhang Z, Chen K, Rouchka EC, Fondufe-Mittendorf YN. Inorganic arsenic-induced cellular transformation is coupled with genome wide changes in chromatin structure, transcriptome and splicing patterns. BMC Genomics. 2015;16:212. doi: 10.1186/s12864-015-1295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobna Z, Rubio-Andrade M, Styblo M, Garcia-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci. 2015;143:97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem. Res. Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsova K, Zotenko E, Luu P-L, Gould CM, Nair SS, Clark SJ, Stirzaker C. Comprehensive evaluation of genome-wide 5-hydroxymethylcytosine profiling approaches in human DNA. Epigenetics Chromatin. 2017;10:16. doi: 10.1186/s13072-017-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-X, He C. Balance of DNA methylation and demethylation in cancer development. Genome Biol. 2012;13:1–3. doi: 10.1186/gb-2012-13-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekpli X, Urbanucci A, Hashim A, Vågbø CB, Lyle R, Kringen MK, Staff AC, Dybedal I, Mills IG, Klungland A, Staerk J. Changes of 5-hydroxymethylcytosine distribution during myeloid and lymphoid differentiation of CD34+ cells. Epigenetics Chromatin. 2016;9:1–13. doi: 10.1186/s13072-016-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Pan F, Sun X, Gan M, Lin A, Zhang D, Zhu Y, Lai M. Association of TET1 expression with colorectal cancer progression. Scand. J. Gastroenterol. 2017;52:312–320. doi: 10.1080/00365521.2016.1253767. [DOI] [PubMed] [Google Scholar]
- Treas JN, Tyagi T, Singh KP. Effects of chronic exposure to arsenic and estrogen on epigenetic regulatory genes expression and epigenetic code in human prostate epithelial cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y-P, Chen H-F, Chen S-Y, Cheng W-C, Wang H-W, Shen Z-J, Song C, Teng S-C, He C, Wu K-J. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017;18(9):517–534. doi: 10.1038/nrg.2017.33. (advance online publication) [DOI] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell. 2006;18:805–814. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Li L. 5-hydroxymethylcytosine. Cancer Biol. Ther. 2014;15:10–15. doi: 10.4161/cbt.27144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149 doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Huang B, Xu X, Sessa WC. Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a. Biochem. Biophys. Res. Commun. 2013;437 doi: 10.1016/j.bbrc.2013.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun X, Cooper KL, Wang F, Liu KJ, Hudson LG. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J. Biol. Chem. 2011;286:22855–22863. doi: 10.1074/jbc.M111.232926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth JD, Bhattacharya A, Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41:D188–D194. doi: 10.1093/nar/gks1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.