Abstract
Massive biofilms have been discovered in the cave of an iodine-rich former medicinal spring in southern Germany. The biofilms completely cover the walls and ceilings of the cave, giving rise to speculations about their metabolism. Here we report on first insights into the structure and function of the biofilm microbiota, combining geochemical, imaging and molecular analytics. Stable isotope analysis indicated that thermogenic methane emerging into the cave served as an important driver of biofilm formation. The undisturbed cavern atmosphere contained up to 3000 p.p.m. methane and was microoxic. A high abundance and diversity of aerobic methanotrophs primarily within the Methylococcales (Gammaproteobacteria) and methylotrophic Methylophilaceae (Betaproteobacteria) were found in the biofilms, along with a surprising diversity of associated heterotrophic bacteria. The highest methane oxidation potentials were measured for submerged biofilms on the cavern wall. Highly organized globular structures of the biofilm matrix were revealed by fluorescent lectin staining. We propose that the extracellular matrix served not only as an electron sink for nutrient-limited biofilm methylotrophs but potentially also as a diffusive barrier against volatilized iodine species. Possible links between carbon and iodine cycling in this peculiar habitat are discussed.
Introduction
Natural microbiota often organize as biofilms, where structural features and microbial interactions give rise to an enhanced ability of biofilm microbiota to be active and persist under challenging environmental conditions. Extensive biofilm production has been previously reported mostly for energy-rich surface water systems dominated by phototrophic primary production (Battin et al., 2016) or in engineered water systems (Boltz et al., 2017). In subsurface and groundwater systems, biofilms are largely considered oligotrophic (Griebler and Lueders, 2009; Ortiz et al., 2014). Nonetheless, a number of caves and karstic systems have been reported to host biofilms rich in microbial diversity and with elevated, mostly lithotrophic biogeochemical activities (Holmes et al., 2001; Engel et al., 2010; Jones et al., 2010; Rusznyák et al., 2012; Barton et al., 2014; Riquelme et al., 2015).
In this study, we report on an exceptionally extensive and massive biofilm formation that has recently been discovered in a semiartificial cave of a historic medicinal spring in Sulzbrunn (Schott, 1858), situated in prealpine southern Germany. Subaerial and submersed microbial biofilms completely cover the walls and ceiling of this seminatural cave (Figure 1), giving rise to extensive pendulous, mucous structures of up to 15 cm in length also known as snottites (Hose and Pisarowicz, 1999). To date, microbial snottites have mostly been described to harbor low-diversity communities of lithotrophs in acidophilic, thermophilic or sulfidic habitats (Bond et al., 2000; Holmes et al., 2001; Northup et al., 2003; Jones et al., 2010; Ziegler et al., 2013). Such extreme conditions do not seem to prevail in Sulzbrunn. Thus our objective was to understand the primary biogeochemical drivers of this peculiar microbial habitat.
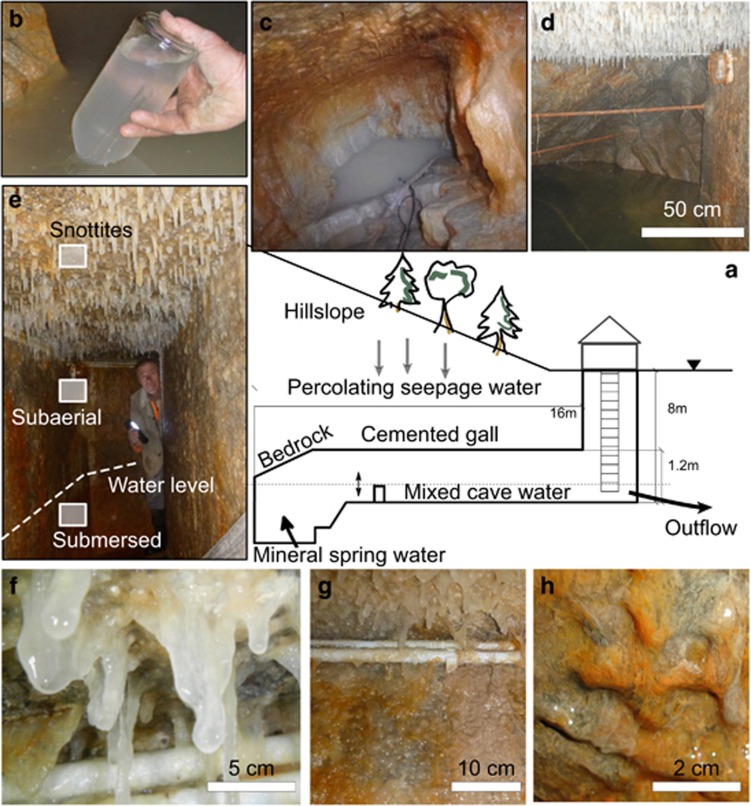
Figure 1.
(a) Conceptual cross-section of the Sulzbrunn spring cavern and its water flows (not to scale). Inset images: (b) Collecting natural gas seeping from the spring pool in an inverted glass bottle using a funnel. (c) Natural bedrock and the emptied spring pool during biofilm and mineral spring water sampling. (d) The filled spring pool with surrounding natural bedrock and man-made gallery covered with biofilms. (e) The three distinct biofilm compartments sampled in this study: submersed and subaerial wall biofilms, snottites at the ceiling. (f) Close-up of thick, slimy snottites. (g) Close-up of subaerial biofilms at upper wall and ceiling. (h) Close-up of submersed wall biofilms.
The Sulzbrunn cave is located in the Allgäu Alps (Bavaria, Germany) at an altitude of 875 m above sea level. The cave lies in a well-jointed sandstone of the Weissach-Schichten of the subalpine Lower Freshwater Molasse. Within a radius of 18 km from Sulzbrunn, natural gas has been repeatedly observed to emerge from deep drill holes that reach Tertiary formations of the subalpine Molasse. The porous sandstone of Bausteinschichten from the Lower Marine Molasse is identified as a reservoir for migrating hydrocarbons. The subjacent organic-rich clays are a potential source of brines with high iodine concentrations (Hesse and Schmidt-Thomé, 1975).
Water enters the cave in the form of upwelling mineral spring water, as well as percolating seepage water. Under normal hydraulic regimes, this semiartificial cave is approximately half-filled with water, with the water level controlled by a simple overflow system. Historic (Schott, 1858), as well as recent water analyses (LfU—Bavarian Environment Agency, personal communication 2014), report high iodine loads of up to 23 mg l−1 emerging with the mineral spring water, which mixes with recent meteoric groundwater in the spring cavern. These high iodine levels, which exceed regular freshwater concentrations by a thousand-fold (Whitehead, 1984), as well as elevated salinity in the mineral spring water, are an indicator of upwelling formation water, which has been in contact with oil- and gas-laden sediment deposits (Lu et al., 2015). The sources of hydrocarbons in the subalpine Molasse basin are autochthonic, originating from mesozoic sediments (Hiltmann et al., 1999) overthrusted by Molasse formations during the alpine orogeny. Fossilized algal biomass is typically highly enriched in iodine, which has been found at concentrations of up to 150 mg l−1 in a drilled artesian well in the area (LfU, personal communication 2014). In Sulzbrunn, the sediments of the Lower Marine Molasse are situated >1000 m below the surface. It can be assumed that the upwelling of iodine-rich waters occurs together with natural gases seeping from deeper hydrocarbon formations along deeply penetrating fault systems.
Although the microbiota of marine gas seeps have been intensively investigated (Ruff et al., 2015; Paul et al., 2017), comparably little information is available about such systems in the terrestrial subsurface. Aerobic methanotrophs and biofilms have been previously found in groundwater and drinking water systems, where they can be involved in the oxidation of methane or methylated compounds (Newby et al., 2004; Stoecker et al., 2006). The Movile Cave in Romania, receiving deep thermal waters rich in hydrogen sulfide (Sarbu et al., 1996), also hosts microbial mats of active methanotrophs (Hutchens et al., 2004; Chen et al., 2009). Recently, the role of microbial methane oxidation within cave and karst ecosystems has been addressed globally (Fernandez-Cortes et al., 2015; McDonough et al., 2016; Lennon et al., 2017). Methane-driven communities can comprise a multitude of interactions between methanotrophs, methylotrophs and heterotrophs (Beck et al., 2013; Kalyuzhnaya et al., 2013; Oshkin et al., 2015; Paul et al., 2017). Energetic constraints imposed by the various pathways of carbon assimilation under low concentrations of oxygen and methane have been shown to trigger substantial exopolysaccharide production in methanotrophs (Linton et al., 1986; Strong et al., 2015), which could potentially explain such massive biofilm production.
The appearance and uniform distribution of the snottites (Figure 1) pointed toward the use of a gaseous substrate for growth. Thus we posit that deep gaseous energy inputs emerging with the upwelling water, possibly light alkanes or methane, could be a major driver of biofilm formation in the Sulzbrunn cavern. We hypothesize that the snottites, as well as subaerial and submersed biofilms, on the wall should be dominated by a low diversity of autotrophs capitalizing on the available energy inputs. The compartmentalization of the cave and possible distinctions in substrate supply should be reflected in distinct biofilm subtypes, substrate turnover rates and isotopic signatures. Finally, we ask whether possible links between methane and iodine cycling can be inferred for this peculiar microbial habitat.
Materials and methods
Site and sampling
The semiartificial Sulzbrunn cave (47.67° N; 10.39° E) is accessible by descending 8 m via a metal ladder (Figure 1). Entering the cave without breathing equipment is only possible during cold seasons, as limited air exchange through the chuted entrance causes microoxic conditions, especially during warmer outside temperatures. Biofilms from both the man-made cemented gallery and the distal bedrock walls were sampled in 2 consecutive years (November 2012 and December 2013) for molecular analyses. In October 2015, additional biofilm samples were taken for microscopy. In order to sample biofilms and mineral spring water from rock fissures at the end of the cave, water filling the gallery was pumped out (Figure 1b). Biofilm samples were collected from three different compartments: submersed biofilms (~30 cm from the bottom of the wall), subaerial biofilms (~20 cm below ceiling), and snottites taken from the ceiling (Figures 1e and f). At each sampling time point, the three compartments were sampled in replicates along a horizontal transect spanning 16 m of the cave gallery. Biofilm samples were directly transferred by scraping into sterile polypropylene tubes (Falcon, Becton Dickinson, Franklin Lakes, NJ, USA), and all samples were frozen (−20 °C) within 6 h after sampling for molecular analyses.
Water samples of percolating seepage water, mineral spring water and the mixed cave water were collected in sterile, 1-liter glass bottles for microbiological and physicochemical analyses. Mineral spring water was repeatedly taken (November 2012, December 2013) from a stainless-steel sampling flume installed at a fissure at the back of the spring cave, whereas seepage water was collected (December 2013) with a sterile glass funnel from the ceiling. Mixed cave water (Figure 1d) was collected during several occasions (October 2012, November 2013, December 2014, October 2015) using a Ruttner sampler (KC Denmark A/S, Silkeborg, Denmark). Water samples were filtered using sterile filter tops (0.2 μm; Corning, Corning, NY, USA). Microbial cells within the water samples were counted using SybrGreen for DNA staining on a flow cytometer (Beckmann Coulter FC 500, Beckmann Coulter, München, Germany) as previously described (Bayer et al., 2016).
Physicochemical analyses
Temperature, pH, dissolved oxygen and specific conductivity of water samples were measured with calibrated field sensors (Hach, Düsseldorf, Germany). Water samples were analyzed for dissolved organic carbon (DOC) using a TOC-V (Shimadzu, Neufahrn, Germany). Prior to injection, DOC samples (0.45 μm filtered) were automatically acidified (pH <2), sparged with oxygen to remove inorganic carbon and analyzed by high-temperature combustion (Mathis et al., 2007). Major cations (calcium, magnesium, potassium, ammonium, sodium) and anions (nitrite, nitrate, chloride, bromide, sulfate) were measured on a DX-100 (Dionex, Germering, Germany) ion chromatograph as described (Stoewer et al., 2015). Total iodine concentrations in water samples were analyzed by ion chromatography–inductively coupled plasma mass spectrometry (Michalke and Witte, 2015). Analysis of water stable isotopes was carried out by cavity ring-down spectrometry (Picarro L2120-I, Picarro, Santa Clara, CA, USA) for 2H and 16O and with liquid scintillation counting for 3H and was used to estimate water mixing ratios (Stoewer et al., 2015).
Gas samples were either taken directly by collecting gas bubbles emerging from the distal spring pool into water-filled, inverted bottles or by pumping from the undisturbed cave atmosphere to the outside of the cavern via tubing installed at the ceiling. Tubing volume was flushed 5 × before sample collection. Methane and CO2 were quantified within 24 h after gas sampling, while gas for isotope analysis was kept in the dark at 4 °C in appropriate glass containers until measurement. CH4 and CO2 were quantified by injecting 250 μl of gas via a HayeSep D column (80–100 mesh, 6 m × 1/8') to a gas chromatograph (GC) equipped with helium ionization and thermal conductivity detectors (SRI Instruments, Bad Honnef, Germany). Methane dissolved in water was quantified using the headspace equilibration method (Kampbell and Vandegrift, 1998). 13C and 2H abundance of gases was measured by separation on a RT –QPLOT (30 m × 0.32, Restek, Bad Homburg, Germany) column and measured using a TRACE GC Ultra (Thermo Fisher Scientific, München, Germany), coupled to a Finnigan MAT 253 IRMS (Thermo Fisher Scientific) via a Finnigan GC Combustion III Interface as previously described (Bergmann et al., 2011).
For isotopic and elemental analysis, biofilms were lyophilized and ground to powder. 13C and 15N abundance was measured on an elemental analyzer (EA; Euro Vector SPA, Redavalle, Italy) with a combustion unit (Hekatech, Wegberg, Germany) connected to a Mat 253 IRMS as previously described (Bernstein et al., 2010). Trace element analysis of lyophilized biofilms was carried out for total iodine, phosphorous, iron and sulfur by inductively coupled plasma–optical emission spectrometry (Hou et al., 2006). Further details on the physicochemical and isotopic analyses are available in Supplementary Information.
Activity measurements
Methane oxidation rates were measured for triplicates of fresh biofilm samples taken from the three different compartments. For this, ~6 g of biofilm samples were dispersed in 20 ml of nitrate mineral salt methanotroph medium (Whittenbury et al., 1970) filled into 250 ml glass bottles and sealed with butyl rubber stoppers. In all, 16 000 p.p.m. of CH4 (Linde, München, Germany) was added to the air-filled bottles and methane oxidation was followed over time using GC measurements described above. The incubations were continuously shaken (150 r.p.m.) and kept in the dark at 12 °C. Gas analyses were carried out after 0, 24, 48 and 72 h of incubation. Methane uptake rates were normalized to biofilm fresh weight (gFW).
Biofilm visualization
Structural features of the biofilms were examined by confocal laser scanning microscopy of extracellular polymeric substances and glycoconjugates with fluorescently labeled lectins as described (Zhang et al., 2015). Biofilms were used fresh, fixed in paraformaldehyde or embedded in O.C.T. compound (Tissue-Tek, Sakura GmbH, Staufen, Germany). Samples were cut into thin sections using a razor blade or a CM 3050 S cryotome (Leica, Wetzlar, Germany). Various nucleic acid-specific stains, for example, SybrGreen, Syto 9 and Syto 60 (Molecular Probes, Leiden, The Netherlands) were used for staining of bacterial cells. Glycoconjugates were contrasted with fucose-specific AAL lectin (Vector Laboratories, Burlingame, CA, USA) labeled with Alexa-488, Alexa-568 or Alexa-633 fluorochromes (Molecular Probes). A confocal laser scanning microscope (TCS SP5X, Leica) equipped with a super continuum light source and controlled by the LAS AF software (ver. 2.6.1., Leica) was used for imaging. Images were collected at 1 μm sectioning intervals using the 25 × NA 0.95 wi and 63 × NA 1.2 wi objective lenses. Signal-to-noise-ratios were optimized using the glow-over-under lookup table. Multichannel image data sets were projected by using the Imaris software (ver. 8.2.0, Bitplane, Zürich, Switzerland).
DNA extraction and molecular analyses
DNA was extracted from biologically replicated samples (n=2–4). Frozen biofilms were re-suspended in 1 × phosphate-buffered saline buffer and disrupted by sonication (Cury and Koo, 2007). Repetitive (3 ×) sonication (35 kHz, Sonorex RK102; Bandelin Electronic GmbH & Co., Berlin, Germany), shaking and spinning (5500 g for 10 min at 4 °C) was used to interrupt the extracellular matrix. DNA from cell pellets and filters from water sampling was extracted following the previously published protocols (Pilloni et al., 2012).
Quantitative PCR of bacterial rRNA genes in DNA extracted from biofilm and water samples was performed as published (Pilloni et al., 2011). In addition, a preliminary screening of the diversity of methanotroph marker genes in biofilms was carried out by terminal restriction fragment length polymorphism fingerprinting as described in Supplementary Methods and Supplementary Table S1. Barcoded amplicons of bacterial 16S rRNA genes, covering the V1–3 region, were generated and sequenced on a FLX+ Genome Sequencer (454 Life Sciences, Roche, Indianapolis, IN, USA) as previously described (Pilloni et al., 2012; Karwautz and Lueders, 2014) but analyzed and classified using the SILVAngs data analysis platform (Pruesse et al., 2012; Quast et al., 2013). Default settings were used for quality control, de-replication, operational taxonomic unit (OTU) clustering and classification on a 97% sequence identity level. Taxonomic assignments were based on the SILVA database release 123 (24 July 2014). The sequencing data set was further processed using the phyloseq package (McMurdie and Holmes, 2013) within the R environment version 3.1.2 (R Development CoreTeam, 2013). Mean abundance of the most prevalent taxa (>3% relative abundance) from replicate samples were plotted as Krona plots (Ondov et al., 2011). Further details on molecular analyses are available in Supplementary Information. All sequencing data have been deposited with the EBI sequence read archive under the BioProject ID PRJEB14605.
Results
Geochemical characterization of cave water, atmosphere and biofilms
The mineral spring water was saline, microoxic and carried only low amounts of DOC, nutrients, phosphate and alternative electron acceptors, such as nitrate or sulfate (Table 1). Total iodine concentration was ~20 mg l−1 in the spring water and diluted to ~3 mg l−1 in the mixed cave water. The temperature of the mixed cave water was between 8 and 11 °C depending on the time points of sampling, while the mineral spring seemed constant at ~7 °C. The pH of the mixed cave water was circumneutral (pH ~7.5). Analysis of the water stable isotopes (δ18O and δ2H) allowed estimation of ratios of mixing between deep spring water and recent meteoric water in the cavern, based on end member mixing calculations. On two of the water sampling dates, the contribution of meteoric seepage water from the surface was estimated to be at 58±17% or 54±18% of the mixed cave water, respectively.
Table 1. Water physicochemistry and microbial cell counts for different water bodies in the cave.
|
Mixed cave water |
Mineral spring water |
Meteoric seepage water |
||||
|---|---|---|---|---|---|---|
| Median | Min.–max. | Median | Min.–max. | Median | Min.–max. | |
| Physicochemistry | ||||||
| Temperature | 7.9 | 7.2 to 11.7 | 7.2 | 8.6a | ||
| EC (μS cm−1) | 2100 | 2020 to 2200 | 6200b | 5890 to 6900b | 526a | |
| pH | 7.5 | 7.2 to 7.6 | 7.6b | 7.2 to 8.3b | 8.3a | |
| O2 (mg l−1) | 4.3 | 2.3 to 5.5 | 3.2 | 2.3 to 3.4 | 8.5a | |
| Nutrients, electron acceptors | ||||||
| DOC (mg l−1) | 1.2 | 0.8 to 1.4 | 0.7b | 0.5 to 0.9b | 0.6a | |
| NO3− (mg l−1) | 1.9 | 0.8 to 3.4 | bd | bd to 0.2 | 5.6 | 4 to 5.6 |
| PO43− (mg l−1) | bd | bd | bd | bd to 0.02 | 0.01 | 0.01 to 0.02 |
| SO42− (mg l−1) | 2.1 | 1.1 to 3 | bd | bd to 1.7 | 4.2 | 3.4 to 4.2 |
| FeTotal (mg l−1) | NA | 0.87c | 0.46 to 1c | NA | ||
| Water mineralization | ||||||
| Na+ (mg l−1) | 93.9 | 51.9 to 328.6 | 649.8 | 581.3 to 1131.1 | 5 | 5 to 18.8 |
| K+ (mg l−1) | 1.6 | 1.2 to 3 | 4.6 | 4.2 to 8.6 | 1 | 1 to 1.4 |
| Mg2+ (mg l−1) | 22.8 | 20.7 to 140.7 | 24.4 | 23.1 to 55.3 | 23 | 22.2 to 23.3 |
| Ca2+ (mg l−1) | 97.2 | 62.1 to 205.1 | 51.5 | 50.2 to 111.9 | 82.2 | 76.9 to 82.5 |
| Cl− (mg l−1) | 154 | 77.5 to 606.6 | 1301.7 | 1237.9 to 2248.3 | 1.1 | 1.1 to 4.6 |
| Br− (mg l−1) | 1.3 | 0.7 to 4.6 | 19.2 | 16.1 to 29.4 | 0.01 | 0.01 to 0.03 |
| I (mg l−1)c | 3.2 | 0.9 to 5 | 20.5 | 20 to 30.7 | NA | |
| Water isotopes | ||||||
| δ18Od | −10.5 | −10.8 to −10.2 | −8 | −9.6 to −7.7 | −11 | −11.8 to −10 |
| δ2Hd | −73.9 | −74.7 to −73.2 | −67.2 | −69.1 to −66.5 | −75.4 | −82.3 to −68 |
| TU | 7.1 | 4.8 | 8.3a | |||
| Cell counts | ||||||
| 1.6 × 106 | 3.7 × 105 to 1.9 × 106 | 3.1 × 103 | 2.7 × 103 to 1.9 × 106 | 6.7 × 103 | 6.6 × 103 to 2.4 × 105 | |
Abbreviations: bd, below detection limit; DOC, dissolved organic carbon; EC, electric conductivity; NA, not analyzed; TU, tritium units defined as ratio of 1 3H atom to 1018 H atoms.
Single measurement in December 2012.
This study (n=4) and routine monitoring data from LfU (n=4).
LfU monitoring data (n=4) from October 2011 to April 2012.
This study (n=3) and LfU monitoring data (n=4).
Natural gas directly bubbling from rock fissures into the distal spring pool was captured and characterized by GC-isotope ratio mass spectrometry. Isotopic signatures identified it as thermogenic methane, indicated by its δ13C and δ2H values of −43.6±0.2‰ (Figure 2) and -164.9±2.2‰, respectively. High methane concentrations of up to 50% were measured in the emerging gas. Average methane concentrations in the mixed cave water and in the cave atmosphere were 6000 and 3000 p.p.m., respectively. The carbon isotope signature of CH4 in the undisturbed cave atmosphere was −36.8±0.1‰ δ13C. Mean carbon dioxide concentrations of 8000 p.p.m. in the cave air were also clearly elevated compared with ambient backgrounds. The δ13C of CO2 in the seeping gas and in the cave atmosphere was −33.2±0.1‰ and −25.2±0.1‰, respectively (Figure 2). Dissolved oxygen concentrations in the spring water and mixed cave water were 3.2% and 4.3% in average, respectively (Table 1). Sampling of the undisturbed cave atmosphere via gas tubes indicated microoxic conditions (O2: 15.1±4%, n=5) under steady state.
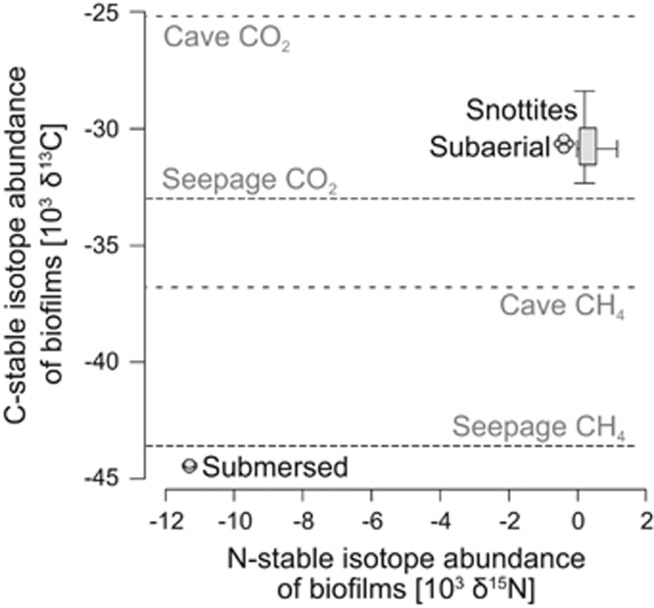
Figure 2.
Carbon and nitrogen stable isotope ratios measured for distinct biofilm compartments. Stable isotope signatures of methane and carbon dioxide either in gas bubbles seeping from the spring pool or in the cave atmosphere are indicated in gray shading. The length of the boxplots depicts the quartiles and the crosshair the range (min., max.) of isotope measurements for snottites.
The pH of biofilm samples was similar to that of the water (pH ~7.5). Elemental analysis of freeze-dried biofilms recovered a light carbon isotope signature (−44.4‰), directly corresponding to the seeping methane itself, especially for the submersed biofilms (Figure 2). Snottites and subaerial biofilms showed a significantly heavier δ13C (−30.7±1.1‰). This was in between the signatures of CH4 and CO2 in the cave atmosphere, rather than directly reflecting seeping CH4. A similar pattern of isotope signatures, albeit less pronounced, was recovered also for nitrogen stable isotopes (Figure 2). Nitrate was the most prevalent nitrogen species in the water (Table 1), whereas ammonium and nitrite were below detection limits. Total nitrogen content within the biofilms decreased toward the ceiling (Table 2). However, distinct nitrogen species in water and biofilms were not independently analyzed.
Table 2. Elemental composition of dried biofilms and potential methane oxidation rates of fresh biofilms at 12 °C in the laboratory.
| Biofilm | C (mg g−1) | N (mg g−1) | P (mg g−1) | Fe (mg g−1) | S (mg g−1) | I (μg g−1) | CH4 ox.a (μM g−1 day−1) |
|---|---|---|---|---|---|---|---|
| Snottitesb | 372.3±80 | 6.1±1 | 0.1±0.1 | 1.7±1.1 | 1.9±0.6 | 0.5±0.2 | 3 |
| Subaerialc | 283.8 | 8.7 | 0.2 | 18.1 | 4.4 | 0.2 | 3.1 |
| Submersedc | 183 | 11.9 | 0.5 | 80 | 5.6 | 0.4 | 25.7 |
n=3.
n=6.
n=2.
A high total carbon content of ~372 mg gDW−1 on average was measured for the snottites (Table 2). Nitrogen and phosphorus concentrations were low, especially for snottites on the ceiling, resulting in an elemental composition of 37% C, 0.6% N and 0.01% P and high C:N (61), C:P (4727) and N:P (82) ratios. These nutrients, plus iron and sulfur, were comparably more abundant in submersed biofilms. In contrast, total iodine concentrations of biofilms were highest at the ceiling but varied substantially throughout sampling locations and were 482.1±197.6 μg gDW−1 on average (Table 2).
Methane oxidation potentials in laboratory biofilm incubations
Potential methane oxidation rates were quantified for biofilms of the three compartments to substantiate our hypothesis of methane as an important driver of biofilm formation. Fresh biofilm samples dispersed in nitrate mineral salt medium were amended with 16000 p.p.m. CH4 in headspace. For all biofilm samples, substantial methane oxidation rates were recorded under laboratory conditions. They were ~3 μmol gbiofilm FW−1 day−1 for the subaerial biofilms and snottites and almost an order of magnitude higher (~26 μmol gbiofilm FW−1 day−1) for the submersed biofilms (Table 2).
Visualization of biofilm structures
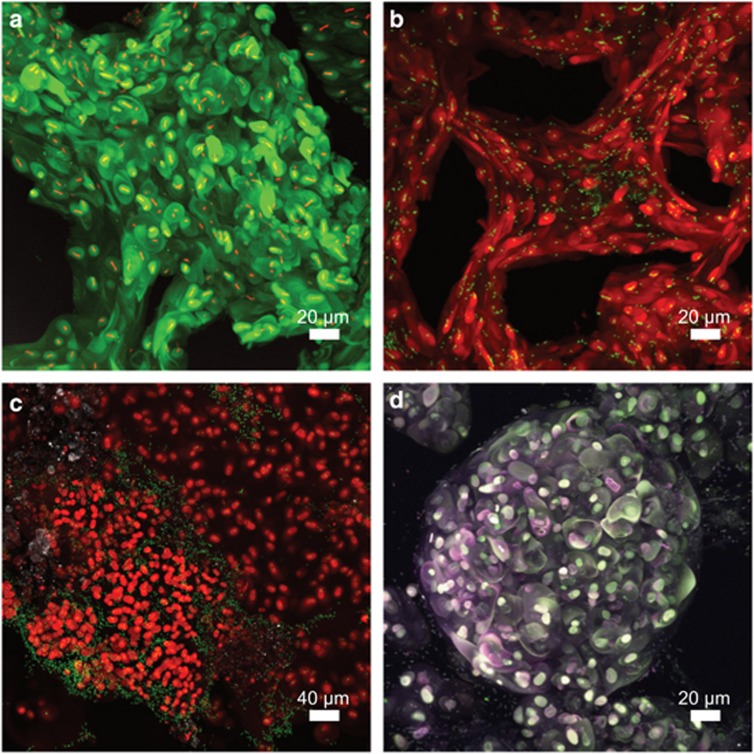
First structural insights into the biofilm matrices were generated by confocal laser microscopy in combination with lectin staining of glycoconjugates (Figure 3). Image data revealed unusually large capsule-like structures, which seemed to embed small numbers of cells and which were partly connected (Figure 3a). Often these glycoconjugates formed larger multilayer strands with voids (non-lectin-stained zones) in between (Figure 3b). Glycoconjugate structures appeared evenly distributed with single embedded bacterial cells in some images (Figures 3a and b), whereas only part of the cells formed AAL-specific glycoconjugates in others (Figure 3c). Multilayer globular features with clusters of capsule-like structures, as well as globules with higher levels of organization, were also observed (Figure 3d).
Figure 3.
Visualization of biofilm structures in snottites by laser microscopy in combination with lectin staining. (a) Globular glycoconjugate signals (green) surrounding individual bacterial cells (red) (67 sections). (b) Strands of glycoconjugate signals (red) with bacterial cells (green) and interspersed voids (58 sections). (c) Biofilm section where only part of the bacterial cells (green) are shown to be associated with glycoconjugate signals (red) (62 sections). (d) Multilayer glycoconjugates (triple-stained AAL) indicating three orders of organization in snottites: glycoconjugate capsules surrounding individual bacterial cells, embedded clusters of capsules, and higher-order spheres of glycoconjugate clusters (59 sections). The selected 3D image series are shown as 2D maximum intensity projections. Samples were stained with AAL lectin (a–d), as well as nucleic acid-specific stains (a–c).
Molecular analysis of water and biofilm microbiota
Microbial abundances as determined by flow cytometry were ~3.1 × 103 cells ml−1 in the mineral spring water and a much higher ~1.6 × 106 in the mixed cave water (Table 1). Mixed cave water cell counts were largely consistent with 16S rRNA gene quantification (Supplementary Figure S1), but quantitative PCR counts for the spring water were higher than cell counts. Gene quantification revealed a high abundance of up to ~3.6 × 109 bacterial 16S rRNA genes gbiofilm FW−1 for the submersed biofilms.
PCR screening for functional marker genes indicative of methanotrophs or methylotrophs was conducted as a first qualitative test for the presence of such microbes in the system. All tested water and biofilm samples were PCR positive (data not shown) for genes encoding the methanol dehydrogenase (mxaF) and particulate methane monooxygenase (pmoA) but not for soluble methane monooxygenase. Terminal restriction fragment length polymorphism fingerprinting of the pmoA gene pool revealed a considerable diversity of pmoA T-RFs in all biofilms. In total, 13 of the 30 detected pmoA fragments overlapped between samples, while 14 appeared unique for the mixed cave water (not shown).
Bacterial community composition in water and biofilms
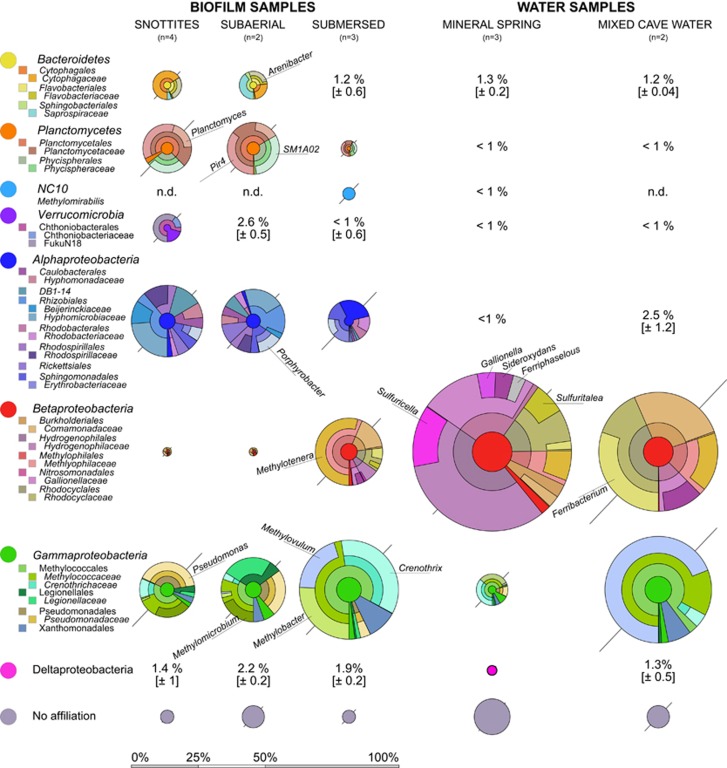
Replicate amplicon sequencing libraries were generated for the three biofilm compartment and water samples (Figure 4). Data processing resulted in 482 OTUs affiliated with the bacteria (Supplementary Table S2). Out of these, 363 OTUs were found in biofilm samples and 297 OTUs in water, with an overlap of 178 OTUs. Besides the Proteobacteria (especially Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria), Planctomycetes, Bacteroidetes and Verrucmicrobia also contributed substantially to the communities. A total of 8 phyla were present at >1% abundance in at least 1 of the samples. Betaproteobacteria were especially abundant (59%) directly in the spring water, detectable at notable abundance in mixed cave water and submersed biofilms (44% and 25%, respectively), but almost absent from subaerial biofilms and snottites. In contrast, members of the Alphaproteobacteria, Planctomycetes, Bacteroidetes and Verrucomicrobia were generally more abundant in subaerial biofilms compared with submersed samples (Figure 4).
Figure 4.
Bacterial community composition in biofilm and water samples as shown by 16S rRNA gene amplicon sequencing. All abundant phyla or proteobacterial classes (relative abundance >3%) are shown as composite Krona plots (Ondov et al., 2011) resolved down to the family level. Selected taxa mentioned in the text are shown at the genus level. The diameter of Krona circles is scaled to the mean abundance of phyla or classes. Variations of taxon abundances between replicate samples, either given as s.d. or the range from the mean (if n=2), is shown as diagonal bars.
In-depth taxonomic analysis revealed many known methylotrophic and methanotrophic populations (Knief, 2015) within the biofilms. Potential methylotrophic Alphaproteobacteria were apparent as Beijerinckiaceae, Hyphomicrobiaceae, Rhodobacteraceae, Erythrobacteraceae and Sphingomonadaceae, detected especially in subaerial biofilms (Figure 4). Several facultative methanotrophs within the Alphaproteobacteria, such as Methylocella, Methylorosula and Methylobacterium spp. were also identified but only at low abundance (all <1%).
Within the Betaproteobacteria, members of the Methylophilaceae (many of them obligate methylotrophs) were abundant in submersed biofilms (14%) but were also found in mineral spring water (4%) and mixed cave water (8%) samples. Dominant taxa within this group were affiliated to Methylotenera spp. Most notably, a dominance of Hydrogenophilaceae (27%) and Gallionellaceae (14%) was observed in the spring water, while they were almost absent from all other samples. The Rhodocyclaceae also contributed to the bacterial community of the spring water and mixed cave water and submersed biofilms.
The Methylococcales (Gammaproteobacteria) represented the most abundant known methanotrophic group (Knief, 2015) across all samples (Figure 4). In total, 15 divergent lineages were found, with the Crenotrichaceae and Methylococcaceae being most abundant. Dominant taxa within the mixed cave water were affiliated to Methylovulum sp. (28%). Crenothrix and Methylobacter spp. were abundant in submersed biofilms (13% and 10%, respectively) but apparently replaced by Methylomicrobium spp. in the subaerial biofilms (4%). Members of the Pseudomonadaceae were also abundant in the snottites and subaerial biofilms (8% and 6%, respectively). Unexpectedly, the aerial communities appeared more diverse (Supplementary Table S1) and less clearly dominated by known methanotrophic or methylotrophic taxa than the submersed biofilms and mixed cave water.
The Planctomycetes in aerial biofilms were mostly affiliated to the Pir4 lineage and Planctomyces spp. (Planctomycetaceae), as well as the Phycisphaeraceae. The recently discovered NC10 candidate phylum, including the denitrifying, methanotrophic Methylomirabilis spp. (Ettwig et al., 2010), was detected only in submersed biofilms. The Bacteroidetes, mostly present in subaerial biofilms and snottites, included a notable population of Arenibacter spp.
Summing up the abundance of the most prominent known methanotrophic and methylotrophic taxa (Figure 5), subaerial biofilms and snottites contained significantly fewer respective lineages than the submersed biofilms and mixed cave water (t=3.9, df=2.7, P<0.05). Besides known C1-oxidizers, other taxa of clear functional connotation were also detected, such as Sulfuritalea (Betaproteobacteria), Sulfurimonas (Epsilonproteobacteria) and Sulfuricella spp. (Betaproteobacteria) in the mineral spring water, all well known to be capable of sulfur oxidation. Moreover, several putatively iron-oxidizing taxa such as Siderooxydans, Ferriphaselus and Gallionella spp. (all Gallionellaceae), as well as close relatives of the the iron-reducing Ferribacterium spp. (Rhodocyclaceae), were found with distinct distribution between spring and mixed cave water (Figure 4).
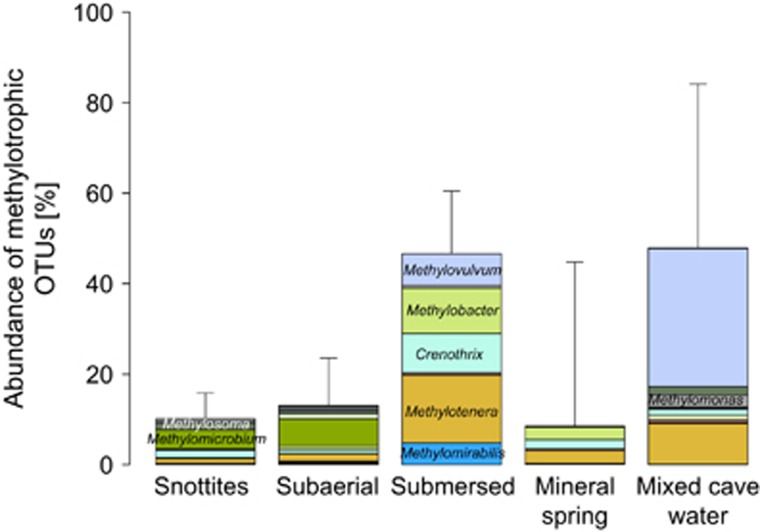
Figure 5.
Summed relative abundance of well-known methanotrophic and methylotrophic taxa recovered from biofilm and water samples. Shown are members of the Beta- and Gammaproteobacteria and the NC10 phylum. Color coding is identical to Figure 4.
Discussion
Methane as the driver of biofilm formation
Here we provide first insights into the biogeochemistry and microbiology of a peculiar biofilm system discovered in prealpine southern Germany. We show that deep formation water enters the spring cave together with appreciable amounts of methane, as indicated by the elevated mixing ratios of methane in the water and atmosphere of the cave. Carbon isotope signatures clearly identified the methane as thermogenic in origin (Aelion et al., 2009), consistent with the well-established presence of fossil deposits and gas reservoirs in this region of the subalpine Molasse (Hiltmann et al., 1999; Etiope, 2009). In Sulzbrunn, the seeping gas can be speculated to actually lift up the deep briny mineral water that ascends into the cave. Although seeping gas bubbles have been reported to induce porewater flow velocities of up to several meters per day in coastal seeps (O'Hara et al., 1995), further investigations will be necessary to delineate the hydrogeological setting in Sulzbrunn.
End member mixing calculations based on water isotopes showed that the influx of deep formation water contributed roughly 40–50% to the spring water in Sulzbrunn. It is likely that this mixing of distinct water inputs, at least in part, contributes to the definition of the unique biogeochemical system in the cave. Despite the detection of low amounts of oxygen directly in the mineral spring water (Table 1), we assume the upwelling formation water to be anoxic, and oxygen exposure or mixing with more aerated surface water to take place only in the last meters before entering the cave. Were the mineral spring water aerated, we would have expected to detect a high abundance of aerobic methanotrophs in these samples, which was not the case. Apart from methane, inputs of DOC into the cave via the different water fluxes seemed negligible. Still, comparably low amounts of DOC in seeping surface water have been shown to support appreciable populations of heterotrophic bacteria in caves (Ortiz et al., 2014). Also, we cannot exclude potential seasonality in DOC inputs from the surface, which might have been missed during our time points of sampling.
The unambiguous δ13C signature identified thermogenic methane to be the main driver of biofilm formation, especially for the submersed biofilms. As shown above, this had the highest methane oxidation rates and methanotroph/methylotroph abundance. The CH4 consumption rates of up to 25 μmol gbiofilm FW−1 day−1 were >4 orders of magnitude higher than rates recently reported for the water column above methane seeps in Lake Constance (Bornemann et al., 2016) and in a similar high range as reported for other methane-venting geothermal sites (Gagliano et al., 2016; Lennon et al., 2017). Upscaling this for biofilm mass estimates in the cave, a potential methane turnover of ~1.6 mol day−1 (~35.8 l CH4 day−1) can be extrapolated for the submersed biofilms alone. Methane oxidation rates were unfortunately not determined for the mixed cave water in this study. The high abundance of methanotrophs in the water (Figure 5) and the reduced oxygen concentrations compared with surface seepage water (Table 1) suggested that at least part of the methane oxidation could also have been allocated to the water body itself. However, the much lower total bacterial abundances in the water (Supplementary Figure S1) combined with a steady discharge and thus purging of lotic populations could still result in biofilms being the most relevant for total methane turnover.
In contrast to submersed biofilms, the δ13C signature of subaerial biofilms and snottites less directly pointed toward thermogenic methane as being their main carbon input. However, the isotopic signatures of CH4 and CO2 in the cave atmosphere were both substantially heavier than directly in seeping gas (Figure 2), suggesting strong stable isotope fractionation (Preuss et al., 2013) to occur during oxidation between the compartments. The placement of the aerial biofilms at approximately −31‰ δ13C in between the signature of both gaseous end members in the cave atmosphere indicated an equal importance of both methanotrophy and autotrophic or heterotrophic CO2 fixation for biofilm buildup, as previously inferred for other biofilms in caves (Sarbu et al., 1996; Chen et al., 2009). Nevertheless, the lower methane oxidation rates and lower abundance of potentially C1-oxidizing microbes both seem to point toward a possible role of electron donors other than methane in aerial biofilms. This will be discussed further down.
The interpretation of observed nitrogen isotope signatures in biofilms was not possible due to the lack of defined input signals. The depleted δ15N isotope values of the submersed biofilms (−11‰) were comparable to values found in Movile Cave biofilms (Sarbu et al., 1996). The much higher values of aerial biofilms suggested distinct inputs, possibly connected to the known capacity of many methanotrophs to fix atmospheric dinitrogen (Knief, 2015). Nevertheless, the major sources and routes of nitrogen cycling in the cave system remain to be specifically elucidated.
Biofilm community composition
The formation of pendulous snottites and other macroscopic biofilm structures in caves has been observed before but mostly under acidic or otherwise extreme conditions. In comparison to the massive biofilm structures (pH ~7.5) now reported for the Sulzbrunn cave, previously discovered snottites appeared much thinner in shape, were less densely distributed and formed lower amounts of extracellular polymeric substances (Bond et al., 2000; Holmes et al., 2001; Northup et al., 2003; Jones et al., 2010; Ziegler et al., 2013). Together with the rich diversity of methylotrophs and other bacterial lineages now discovered in the Sulzbrunn biofilms, this points toward distinct biogeochemical and ecophysiological drivers of biofilm formation in the different systems.
We suggest the identified biofilm compartments to be a function of methane influx, water submersion and oxygen and nutrient supply within the cave. The high abundance of methanotrophs and methylotrophs of up to ~45% in submersed biofilms and between 10 and ~20% in the other biofilm compartments was consistent with the importance of methane as the driver of biofilm formation. An abundance of aerobic methanotrophs of up to 40% has been reported previously for a terrestrial methane seep (Gagliano et al., 2016). However, central questions remain how carbon and energy flows are shared between the methanotrophs, other methylotrophs and the diverse non-methylotrophic lineages discovered in the Sulzbrunn biofilms.
Distinguishing between obligate and facultative methanotrophs requires genomic and proteomic information, which is not yet available for the investigated system. However, the ecophysiology of some of the microbes detected can be cautiously extrapolated from the literature. For example, Methylobacter spp. are generally considered as obligate methanotrophs (Knief, 2015), while Methylotenera spp. and other Methylophilaceae are mostly known as obligate non-methane-utilizing methylotrophs (Kalyuzhnaya et al., 2012). The co-occurrence of these methanotrophs and methylotrophs, especially in the submersed biofilms, suggest methane-fueled cooperation (Chen et al., 2009). Members of the Methylococcaceae and Methylophilaceae have been previously reported to trophically interact in methane-fueled systems under oxic and microoxic conditions (Beck et al., 2013; Oshkin et al., 2015). Members of the Methanococcaceae have been shown to shunt carbon to diverse non-methylotrophic community members in microbial mats situated at marine hydrocarbon seeps (Paul et al., 2017). Thus, in the Sulzbrunn biofilms, complex communities and interaction networks can be considered to drive methane oxidation rather than single microbial species.
Methanotrophs are also well known as producers of abundant extracellular polysaccharides (Linton et al., 1986; Strong et al., 2015). Many of them possess the ribulose monophosphate pathway to fix methyl-group-derived carbon. The production of extracellular polysaccharides from methanol is balanced in terms of adenosine triphosphate and reducing equivalents (Linton et al., 1986). The observed exopolysaccharide production is conceivable as an energy-spilling reaction, preventing the buildup of toxic formaldehyde under excess methane supply, and providing methane-derived reduced carbon to other heterotrophic members of the biofilm community. Some methanotrophs have been shown to ferment methane and release large amounts of reduced carbon under oxygen-limited conditions (Kalyuzhnaya et al., 2013). Growth limitation by limited nitrogen or phosphorous supply, as suggested especially for the aerial biofilms by high C:N:P ratios, would support this scenario. Although many methanotrophs are capable of fixing dinitrogen (Knief, 2015), P limitation will not be readily complemented in aerial biofilms. The C:P (~4700) and N:P (~80) ratios observed in biofilms are clearly much higher than canonical Redfield ratios or than C:N:P ratios suggested to indicate bacterial P limitation (Vrede et al., 2002). Therefore, we propose that the massive extracellular matrix formed in the Sulzbrunn biofilms serves, at least in part, as an electron sink for nutrient-limited methylotrophs.
Besides well-known proteobacterial methanotrophs and methylotrophs, members of the candidate genus Methylomirabilis were also detected but appeared restricted to submersed biofilms in Sulzbrunn. These are known as nitrate-dependent anaerobic methane oxidizers proposed to intra-aerobically oxidize methane under NO dismutation (Ettwig et al., 2010). Their detection points toward the possible occurrence of anaerobic methane oxidation in specific microniches of the Sulzbrunn biofilms. Furthermore, we are currently investigating whether Archaea could also possibly be involved in methane cycling in the system. Preliminary data suggest that a low abundance of largely uncultured archaeal lineages can be found in the submersed biofilms and water samples but not in aerial biofilms. Furthermore, the abundant detection of putatively sulfur-oxidizing (Watanabe et al., 2014) and iron-oxidizing (Emerson et al., 2013), as well as iron-reducing (Cummings et al., 1999), members of the Betaproteobacteria, especially in spring and mixed cave waters, points toward active sulfur and iron cycling in the cave. However, these processes, as well as their possible link to carbon cycling, could not be further traced in the present study but will be the subject of future work.
Possible role of iodine
The visualization of the biofilm matrix revealed unique structural features of the biofilms. Although large globular structures have been previously reported for biofilms in technical systems (Okabe et al., 1999; Weissbrodt et al., 2013), a comparably massive embedding of single or small numbers of cells in capsules and networks of glycoconjugates, to the best of our knowledge, has not been observed. It is tempting to speculate that besides a possible role as an electron sink, the biofilm matrix could also serve as protective barrier against harmful agents possibly present in the Sulzbrunn system. The concept of biofilms as a diffusive barrier against antimicrobials is well established (Flemming et al., 2016). In the iodine-rich waters and biofilms of the Sulzbrunn cave, the possibility of bactericidal activity of iodine species should be discussed.
Iodine is well known as a disinfectant, but interestingly, the mechanisms of its toxicity are still not fully elucidated, probably owing to its complex chemistry (Küpper et al., 2011). Elemental iodine (I2) is not stable in aqueous solution, where it readily hydrolyses to iodide (I−), hypoiodous acid (HOI) and several other iodine species (Gottardi, 1999). Under elevated pH, the formation of iodate (IO3−) by chemical disproportionation is also possible. Iodide and iodate are considered as non-toxic, while elemental iodine, hypoiodous acid and triiodide (I3−) are suggested as bactericidal oxidizing agents (Gottardi, 1999). Owing to its complex behavior as a solute, comprehensive iodine speciation is challenging and has not yet been accomplished for different Sulzbrunn samples.
Although we assume that most of the total iodine emerging with the reduced mineral water was iodide, this could undergo a number of microbially driven oxidation and volatilization reactions in the cave. The oxidation of iodide to elemental iodine in the presence of polysaccharides has been shown for Pseudomonas iodooxidans (Gozlan and Margalith, 1974) and distinct Alphaproteobacteria (Amachi et al., 2005), which were even stimulated under high iodide concentrations (Arakawa et al., 2012). An Arenibacter sp. (Bacteroidetes) has been reported to accumulate iodine during that process (Ito et al., 2016). Moreover, various isolates from iodine-rich habitats, including Erythrobacter, Pseudomonas and Rhizobium spp., have been shown to methylate iodide, thus volatilizing it as highly reactive iodomethane (CH3I) (Amachi et al., 2005; Fujimori et al., 2012). And finally, Pseudomonas sp. SCT has been shown capable of anaerobic growth with iodate as sole electron acceptor or while simultaneously reducing nitrate (Amachi et al., 2007).
The abundant detection of all of the above genera in the Sulzbrunn biofilms, as well as the higher iodine concentrations found in the snottites (Table 2), imply that volatilization processes may actually have been ongoing in the cave. The volatilization as iodomethane and subsequent oxidation by methyl halide oxidizers in aerial biofilms (McDonald et al., 2002) seems plausible and could establish a link between the cycling of methane and iodine in the system. It can be cautiously speculated that iodide released upon iodomethane oxidation by methylotrophs in snottites could then be oxidized to iodine by other community members, thus possibly contributing to iodine stress and glycoconjugate production in biofilms. As a first step to follow-up on this, we have tried to quantify iodomethane in the cave atmosphere by gas chromatography. Although we were able to detect it upon several occasions (data not shown), a consistent and reproducible quantification was not accomplished so far, possibly due to the highly reactive nature of this methylating agent.
Conclusions
Here the Sulzbrunn spring cave is described as a unique habitat for microbial biofilm growth. Although the cave is situated just several meters below the surface, microbial communities largely independent from surface carbon and energy inputs were discovered. In contrast to our initial expectation, biofilm microbiota were surprisingly diverse, with a host of populations closely related to well-known methanotrophs, methylotrophs and also potentially iodine-cycling bacteria. These findings provide further evidence for the relevance of subterranean methane sinks by microbes (McDonough et al., 2016; Lennon et al., 2017). We propose that the massive extracellular polymeric substance production observed could serve as an electron sink for the nutrient-limited and therefore growth-limited methylotrophs. Although these first insights into an apparently unique subsurface biofilm system are very intriguing, many open research questions remain. The application of 13C-labeled methane and methylotrophic substrates in combination with nucleic acid-based stable isotope probing is currently ongoing and will help to further unravel the complex patterns of carbon and energy sharing to be expected within the biofilms. Future research should also address the spatial organization and metagenomic repertoire of biofilm microbiota, as well as the possible role of Archaea, Protozoa, phage and fauna, in the food web of this ecosystem apparently dominated by prokaryotes.
Acknowledgments
We thank Franz Hösle, Sulzbrunn for site access and sampling support. Roland Eichhorn of the LfU in Hof is acknowledged for his continued support of our site investigation and for scientific discussion. We thank Christine Stumpp, Petra Seibel, Harald Lowag and Armin Meyer (Helmholtz München) for the analysis of stable isotopes and Bernhard Michalke (Helmholtz München) for inductively coupled plasma analytics. Technical support of Kathrin Hörmann in sequencing, Nina Weber in flow cytometry (both at Helmholtz München) and of Ute Kuhlicke in confocal laser scanning microscopy (UFZ Magdeburg) is also greatly acknowledged. Lauren Bradford (Helmholtz München) is thanked for expert language editing. This research was supported by funds of the Helmholtz-Zentrum München and the Helmholtz Center for Environmental Research – UFZ.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aelion CM, Höhener P, Hunkeler D, Aravena R. (2009) Environmental Isotopes in Biodegradation and Bioremediation. CRC Press: Boca Raton, FL, USA. [Google Scholar]
- Amachi S, Muramatsu Y, Akiyama Y, Miyazaki K, Yoshiki S, Hanada S et al. (2005). Isolation of iodide-oxidizing bacteria from iodide-rich natural gas brines and seawaters. Microbial Ecol 49: 547–557. [DOI] [PubMed] [Google Scholar]
- Amachi S, Kawaguchi N, Muramatsu Y, Tsuchiya S, Watanabe Y, Shinoyama H et al. (2007). Dissimilatory iodate reduction by marine Pseudomonas sp. strain SCT. Appl Environ Microbiol 73: 5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y, Akiyama Y, Furukawa H, Suda W, Amachi S. (2012). Growth stimulation of iodide-oxidizing α-Proteobacteria in iodide-rich environments. Microbial Ecol 63: 522–531. [DOI] [PubMed] [Google Scholar]
- Barton HA, Giarrizzo JG, Suarez P, Robertson CE, Broering MJ, Banks ED et al. (2014). Microbial diversity in a Venezuelan orthoquartzite cave is dominated by the Chloroflexi (Class Ktedonobacterales and Thaumarchaeota Group I.1c. Front Microbiol 5: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI. (2016). The ecology and biogeochemistry of stream biofilms. Nat Rev Microbiol 14: 251–263. [DOI] [PubMed] [Google Scholar]
- Bayer A, Drexel R, Weber N, Griebler C. (2016). Quantification of aquatic sediment prokaryotes—a multiple-steps optimization testing sands from pristine and contaminated aquifers. Limnologica 56: 6–13. [Google Scholar]
- Beck DA, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina Del Rio T, Ivanova N et al. (2013). A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 1: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann FD, Abu Laban NMFH, Meyer AH, Elsner M, Meckenstock RU. (2011). Dual (C, H) isotope fractionation in anaerobic low molecular weight (poly)aromatic hydrocarbon (PAH) degradation: potential for field studies and mechanistic implications. Environ Sci Technol 45: 6947–6953. [DOI] [PubMed] [Google Scholar]
- Bernstein A, Adar E, Ronen Z, Lowag H, Stichler W, Meckenstock RU. (2010). Quantifying RDX biodegradation in groundwater using [delta]15N isotope analysis. J Contam Hydrol 111: 25–35. [DOI] [PubMed] [Google Scholar]
- Boltz JP, Smets BF, Rittmann BE, van Loosdrecht MCM, Morgenroth E, Daigger GT. (2017). From biofilm ecology to reactors: a focused review. Water Sci Technol 75: 1753–1760. [DOI] [PubMed] [Google Scholar]
- Bond PL, Smriga SP, Banfield JF. (2000). Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66: 3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornemann M, Bussmann I, Tichy L, Deutzmann J, Schink B, Pester M. (2016). Methane release from sediment seeps to the atmosphere is counteracted by highly active Methylococcaceae in the water column of deep oligotrophic Lake Constance. FEMS Microbiol Ecol 92: fiw123–fiw123. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H et al. (2009). Life without light: microbial diversity and evidence of sulfur-and ammonium-based chemolithotrophy in Movile Cave. ISME J 3: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Cummings DE, Caccavo Jr F, Spring S, Rosenzweig RF. (1999). Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch Microbiol 171: 183–188. [Google Scholar]
- Cury JA, Koo H. (2007). Extraction and purification of total RNA from Sreptococcus mutans biofilms. Anal Biochem 365: 208–214. [DOI] [PubMed] [Google Scholar]
- Emerson D, Field E, Chertkov O, Davenport K, Goodwin L, Munk C et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AS, Meisinger DB, Porter ML, Payn RA, Schmid M, Stern LA et al. (2010). Linking phylogenetic and functional diversity to nutrient spiraling in microbial mats from Lower Kane Cave (USA). ISME J 4: 98–110. [DOI] [PubMed] [Google Scholar]
- Etiope G. (2009). Natural emissions of methane from geological seepage in Europe. Atmos Environ 43: 1430–1443. [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cortes A, Cuezva S, Alvarez-Gallego M, Garcia-Anton E, Pla C, Benavente D et al. (2015). Subterranean atmospheres may act as daily methane sinks. Nat Commun 6: 7003. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat Rev Micro 14: 563–575. [DOI] [PubMed] [Google Scholar]
- Fujimori T, Yoneyama Y, Taniai G, Kurihara M, Tamegai H, Hashimoto S. (2012). Methyl halide production by cultures of marine proteobacteria Erythrobacter and Pseudomonas and isolated bacteria from brackish water. Limnol Oceanogr 57: 154–162. [Google Scholar]
- Gagliano AL, Tagliavia M, D'Alessandro W, Franzetti A, Parello F, Quatrini P. (2016). So close, so different: geothermal flux shapes divergent soil microbial communities at neighbouring sites. Geobiol 14: 150–162. [DOI] [PubMed] [Google Scholar]
- Gottardi W. (1999). Iodine and disinfection: theoretical study on mode of action, efficiency, stability, and analytical aspects in the aqueous system. Arch Pharm (Weinheim) 332: 151–157. [DOI] [PubMed] [Google Scholar]
- Gozlan RS, Margalith P. (1974). Iodide oxidation by Pseudomonas iodooxidans. J Appl Bacteriol 37: 493–499. [DOI] [PubMed] [Google Scholar]
- Griebler C, Lueders T. (2009). Microbial biodiversity in groundwater ecosystems. Freshw Biol 54: 649–677. [Google Scholar]
- Hesse R, Schmidt-Thomé P. (1975). Neue Jodwasser-Vorkommen im Bereich der bayerischen Alpenrand-strukturen bei Bad Tölz (aufgrund von Tiefbohrungen 1957 - 1967). Geol Jahrb C11: 31–66. [Google Scholar]
- Hiltmann W, Kuckelkorn K, Wehner H. (1999). Das Inkohlungsprofil der Bohrung Grambach 1 - erster Hinweis auf eine 'Ölküche' im Molassebecken. Erdöl Erdgas Kohle 115: 294–297. [Google Scholar]
- Holmes AJ, Tujula NA, Holley M, Contos A, James JM, Rogers P et al. (2001). Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ Microbiol 3: 256–264. [DOI] [PubMed] [Google Scholar]
- Hose LD, Pisarowicz JA. (1999). Cueva de Villa Luz, Tabasco, Mexico: reconnaissance study of an active sulfur spring cave and ecosystem. J Cave Karst Stud 61: 13–21. [Google Scholar]
- Hou X, Amais RS, Jones BT, Donati GL. (2006) Inductively coupled plasma optical emission spectrometry. In: Meyers RA (ed), Encyclopedia of Analytical Chemistry. John Wiley & Sons, Ltd: Hoboken, NJ, USA. [Google Scholar]
- Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. (2004). Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol 6: 111–120. [DOI] [PubMed] [Google Scholar]
- Ito K, Nakajima N, Yamamura S, Tomita M, Suzuki H, Amachi S. (2016). Draft genome sequence of Arenibacter sp. strain C-21, an iodine-accumulating bacterium isolated from surface marine sediment. Genome Announc 4: e01155–e01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DS, Tobler DJ, Schaperdoth I, Mainiero M, Macalady JL. (2010). Community structure of subsurface biofilms in the thermal sulfidic caves of Acquasanta Terme, Italy. Appl Environ Microbiol 76: 5902–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Beck DAC, Vorobev A, Smalley N, Kunkel DD, Lidstrom ME et al. (2012). Novel methylotrophic isolates from lake sediment, description of Methylotenera versatilis sp. nov. and emended description of the genus Methylotenera. Int J Syst Evol Microbiol 62: 106–111. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A et al. (2013). Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4: 2785. [DOI] [PubMed] [Google Scholar]
- Kampbell DH, Vandegrift SA. (1998). Analysis of dissolved methane, ethane, and ethylene in ground water by a standard gas chromatographic technique. J Chromatogr Sci 36: 253–256. [DOI] [PubMed] [Google Scholar]
- Karwautz C, Lueders T. (2014). Impact of hydraulic well restoration on native bacterial communities in drinking water wells. Microbes Environ 29: 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper FC, Feiters MC, Olofsson B, Kaiho T, Yanagida S, Zimmermann MB et al. (2011). Commemorating two centuries of iodine research: an interdisciplinary overview of current research. Angew Chem Int Ed 50: 11598–11620. [DOI] [PubMed] [Google Scholar]
- Lennon JT, Nguyê~n-Thùy D, Pham TM, Drobniak A, Ta PH, Phạm NÐ et al. (2017). Microbial contributions to subterranean methane sinks. Geobiol 15: 254–258. [DOI] [PubMed] [Google Scholar]
- Linton J, Watts P, Austin R, Haugh D, Niekus H. (1986). The energetics and kinetics of extracellular polysaccharide production from methanol by micro-organisms possessing different pathways of C1 assimilation. Microbiology 132: 779–788. [Google Scholar]
- Lu Z, Hummel ST, Lautz LK, Hoke GD, Zhou X, Leone J et al. (2015). Iodine as a sensitive tracer for detecting influence of organic-rich shale in shallow groundwater. Appl Geochem 60: 29–36. [Google Scholar]
- Mathis JT, Hansell DA, Kadko D, Bates NR, Cooper LW. (2007). Determining net dissolved organic carbon production in the hydrographically complex western Arctic Ocean. Limnol Oceanogr 52: 1789–1799. [Google Scholar]
- McDonald IR, Warner KL, McAnulla C, Woodall CA, Oremland RS, Murrell JC. (2002). A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ Microbiol 4: 193–203. [DOI] [PubMed] [Google Scholar]
- McDonough LK, Iverach CP, Beckmann S, Manefield M, Rau GC, Baker A et al. (2016). Spatial variability of cave-air carbon dioxide and methane concentrations and isotopic compositions in a semi-arid karst environment. Environ Earth Sci 75: 700. [Google Scholar]
- McMurdie PJ, Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalke B, Witte H. (2015). Characterization of a rapid and reliable method for iodide biomonitoring in serum and urine based on ion chromatography–ICP-mass spectrometry. J Trace Elem Med Biol 29: 63–68. [DOI] [PubMed] [Google Scholar]
- Newby DT, Reed DW, Petzke LM, Igoe AL, Delwiche ME, Roberto FF et al. (2004). Diversity of methanotroph communities in a basalt aquifer. FEMS Microbiol Ecol 48: 333–344. [DOI] [PubMed] [Google Scholar]
- Northup DE, Barns SM, Yu LE, Spilde MN, Schelble RT, Dano KE et al. (2003). Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ Microbiol 5: 1071–1086. [DOI] [PubMed] [Google Scholar]
- O'Hara SCM, Dando PR, Schuster U, Bennis A, Boyle JD, Chui FTW et al. (1995). Gas seep induced interstitial water circulation: observations and environmental implications. Cont Shelf Res 15: 931–948. [Google Scholar]
- Okabe S, Satoh H, Watanabe Y. (1999). In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol 65: 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Bergman NH, Phillippy AM. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 12: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M, Legatzki A, Neilson JW, Fryslie B, Nelson WM, Wing RA et al. (2014). Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave. ISME J 8: 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshkin IY, Beck DAC, Lamb AE, Tchesnokova V, Benuska G, McTaggart TL et al. (2015). Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 9: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BG, Ding H, Bagby SC, Kellermann MY, Redmond MC, Andersen GL et al. (2017). Methane-oxidizing bacteria shunt carbon to microbial mats at a marine hydrocarbon seep. Front Microbiol 8: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloni G, von Netzer F, Engel M, Lueders T. (2011). Electron acceptor-dependent identification of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol Ecol 78: 165–175. [DOI] [PubMed] [Google Scholar]
- Pilloni G, Granitsiotis MS, Engel M, Lueders T. (2012). Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS One 7: e40467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss I, Knoblauch C, Gebert J, Pfeiffer EM. (2013). Improved quantification of microbial CH4 oxidation efficiency in arctic wetland soils using carbon isotope fractionation. Biogeosci 10: 2539–2552. [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development CoreTeam. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, http://www.R-project.org. [Google Scholar]
- Riquelme C, Rigal F, Hathaway JJM, Northup DE, Spilde MN, Borges PAV et al. (2015). Cave microbial community composition in oceanic islands: disentangling the effect of different colored mats in diversity patterns of Azorean lava caves. FEMS Microbiol Ecol 91: fiv141. [DOI] [PubMed] [Google Scholar]
- Ruff SE, Biddle JF, Teske AP, Knittel K, Boetius A, Ramette A. (2015). Global dispersion and local diversification of the methane seep microbiome. PNAS 112: 4015–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusznyák A, Akob DM, Nietzsche S, Eusterhues K, Totsche KU, Neu TR et al. (2012). Calcite biomineralization by bacterial isolates from the recently discovered pristine karstic Herrenberg Cave. Appl Environ Microbiol 78: 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbu SM, Kane TC, Kinkle BK. (1996). A chemoautotrophically based cave ecosystem. Science 272: 1953. [DOI] [PubMed] [Google Scholar]
- Schott AC. (1858) Wildbad Sulzbrunn bei Kempten in Bayern nebst der neuesten vom Freiherrn Dr Von Liebig vollzogenen authentischen Analyse seiner Jodquellen, sowie der dadurch auf organischem Wege erzeugten Jodmilch und Jodmolke. Vömel: Frankfurt, Germany. [Google Scholar]
- Stoecker K, Bendinger B, Schöning B, Nielsen PH, Nielsen JL, Baranyi C et al. (2006). Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. PNAS 103: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoewer MM, Knöller K, Stumpp C. (2015). Tracing freshwater nitrate sources in pre-alpine groundwater catchments using environmental tracers. J Hydrol 524: 753–767. [Google Scholar]
- Strong PJ, Xie S, Clarke WP. (2015). Methane as a resource: can the methanotrophs add value? Environ Sci Technol 49: 4001–4018. [DOI] [PubMed] [Google Scholar]
- Vrede K, Heldal M, Norland S, Bratbak G. (2002). Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl Environ Microbiol 68: 2965–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kojima H, Fukui M. (2014). Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of Betaproteobacteria. Syst Appl Microbiol 37: 387–395. [DOI] [PubMed] [Google Scholar]
- Weissbrodt D, Neu T, Kuhlicke U, Rappaz Y, Holliger C. (2013). Assessment of bacterial and structural dynamics in aerobic granular biofilms. Front Microbiol 4: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead D. (1984). The distribution and transformations of iodine in the environment. Environ Int 10: 321–339. [Google Scholar]
- Whittenbury R, Phillips K, Wilkinson J. (1970). Enrichment, isolation and some properties of methane-utilizing bacteria. Microbiology 61: 205–218. [DOI] [PubMed] [Google Scholar]
- Zhang R, Neu TR, Zhang Y, Bellenberg S, Kuhlicke U, Li Q et al. (2015). Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl Microbiol Biotechnol 99: 7343–7356. [DOI] [PubMed] [Google Scholar]
- Ziegler S, Dolch K, Geiger K, Krause S, Asskamp M, Eusterhues K et al. (2013). Oxygen-dependent niche formation of a pyrite-dependent acidophilic consortium built by archaea and bacteria. ISME J 7: 1725–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.