Abstract
Metabolic interactions between populations can influence patterns of spatial organization and diversity within microbial communities. Cross-feeding is one type of metabolic interaction that is pervasive within microbial communities, where one genotype consumes a resource into a metabolite while another genotype then consumes the metabolite. A typical feature of cross-feeding is that the metabolite may impose toxicity if it accumulates to sufficient concentrations. However, little is known about the effect of metabolite toxicity on spatial organization and local diversity within microbial communities. We addressed this knowledge gap by experimentally varying the toxicity of a single cross-fed metabolite and measuring the consequences on a synthetic microbial cross-feeding community. Our results demonstrate that metabolite toxicity slows demixing and thus slows local diversity loss of the metabolite-producing population. Using mathematical modeling, we show that this is because toxicity slows growth, which enables more cells to emigrate from the founding region and contribute towards population expansion. Our results show that metabolite toxicity is an important factor affecting local diversity within microbial communities and that spatial organization can be affected by non-intuitive mechanisms.
Introduction
The spatial organization of different microbial populations within communities not only affects the survival and functionality of populations, but also the diversity of microbial communities (Kim et al., 2008). An important factor that determines spatial organization is metabolic interactions between different microbial populations (Davey and O’Toole, 2000; Battin et al., 2007). For example, in a process called sequential cross-feeding (hereafter referred to as cross-feeding), one genotype consumes a resource into a metabolite while another genotype then consumes the metabolite (Schink, 2002; Pfeiffer and Bonhoeffer, 2004; Bull and Harcombe, 2009). Cross-feeding of metabolites is pervasive in nature and is a key component of important global biogeochemical processes. It affects carbon cycling (e.g., interspecies hydrogen transfer) (Schink, 1997; McInerney et al., 2009), nitrogen-cycling (Martienssen and Schöps, 1999) and pollutant degradation (de Souza et al., 1998; see also Morris et al. (2013) for a recent review).
Cross-feeding is thought to be an important driver of microbial diversity in nature because it permits several populations to co-exist by niche specialization, where each population specializes at consuming only part of the primary resource (Rainey et al., 2000). These cross-feeding populations can exhibit a range of interaction types. For example, if the consumption of the metabolite by the metabolite-consuming population provides a benefit to the metabolite-producing population, the cross-feeding interaction is mutualistic (i.e., each population benefits from the presence of the other). Such an interaction is sometimes referred to as reciprocal or cooperative cross-feeding (West et al., 2007; Estrela and Gudelj, 2010; Estrela et al., 2012). Such a mutualistic interaction may emerge if the cross-fed metabolite is toxic, where the metabolite-consuming population promotes the growth of the metabolite-producing population by consuming the toxic metabolite and preventing or reducing its negative effects (Pfeiffer and Bonhoeffer, 2004; Dolinšek et al., 2016).
While cross-feeding of toxic metabolites is pervasive, it is not clear how metabolite toxicity affects spatial organization and local diversity within cross-feeding microbial communities. In general, during colony growth, initially mixed populations of metabolically identical genotypes tend to demix into sectors, where only one population prevails within each sector. This results in a loss of local population diversity. Experimental and theoretical considerations suggest that this demixing is a consequence of genetic drift at the expansion front (Hallatschek et al., 2007; Excoffier et al., 2009). Random sampling at the expansion front, where population sizes are small, causes a repeated founder effect which reduces local population diversity (Excoffier et al., 2009). The strength of this demixing effect, however, depends on the size of the active population, which is influenced by the growth dynamics of the population (Müller et al., 2014). It was proposed theoretically and demonstrated experimentally that nutrient supply and expansion velocity can impact the size of the active population, and thus the strength of drift during expansion (Nadell et al., 2010; Mitri et al., 2015). Faster growing colonies with high nutrient supply tend to loose local diversity slower than colonies with low nutrient supply. In a cross-feeding ecosystem, resource availability and growth rates are coupled to the partner populations. The growth dynamics are therefore an emergent property of the entire system and are difficult to predict a priori. In general, toxicity is dose dependent and inhibits growth (Hodgson, 2004), which should slow the expansion velocity of the colonies. At first glance, this may suggest a stronger effect of drift, causing a faster loss of local diversity when cross-fed metabolites are toxic.
In this study we investigated how a conditionally toxic cross-fed metabolite affects local diversity. The main question was whether reduced growth rates caused by metabolite toxicity lead to faster demixing of neutral alleles within each population. Another question was whether a reduction of toxicity by the cross-feeding interaction itself could mitigate this effect. We previously constructed an experimental microbial cross-feeding system that allows us to control the toxicity of the cross-fed metabolite to test these hypotheses. The system consists of two strains of Pseudomonas stutzeri that differ in their ability to use nitrogen oxides. One strain consumes nitrate (NO3) to nitrite (NO2) while the other consumes the excreted nitrite (NO2) (Lilja and Johnson, 2016). We henceforth refer to these two cross-feeding strains as the producer and consumer, respectively, and to the ancestral strain that completely consumes nitrate (NO3) to nitrogen gas (N2) as the complete degrader.
Nitrite toxicity is pH dependent and can act via several mechanisms. As the pH is reduced, nitrite increasingly protonates to nitrous acid (HNO2) (Sijbesma et al., 1996). Nitrous acid can act as a protonophore, i.e. it increases proton permeability which inhibits ATP synthesis (Sijbesma et al., 1996; Zhou et al., 2011). In addition, as the pH is reduced, nitrite increasingly forms nitric oxide radicals (NO) that can cause enzyme damage (Zumft, 1993). The pH dependence of nitrite toxicity allowed us to experimentally manipulate the magnitude of nitrite toxicity, where toxicity is weak at pH 7.5 but strong at pH 6.5 (Lilja and Johnson, 2016). Importantly, in the absence of nitrite, pH itself has no statistically detectable effects on growth within this pH range, thus avoiding confounding effects that may be caused by differences in pH itself (Lilja and Johnson, 2016). We finally used an individual-based model (Nadell et al., 2010; Mitri et al., 2015) to simulate microbial growth dynamics under toxic conditions to assess the mechanism by which metabolite toxicity influences local diversity in expanding populations.
Materials and methods
Bacterial strains and growth conditions
We used the same strains and growth conditions as described elsewhere (Lilja and Johnson, 2016; Goldschmidt et al., 2017). In short, the nirS or narG and the comA genes were deleted from P. stutzeri A1501. The nirS gene, which encodes an enzyme for the reduction of nitrite to nitrous oxide (Zumft, 1997), was deleted from the producer, and the producer can therefore only convert nitrate to nitrite. The narG gene, which encodes an enzyme for the reduction of nitrate to nitrite (Zumft, 1997), was deleted from the consumer, and the consumer can therefore only convert nitrite to nitrogen gas. To prevent recombination during experimentation, the comA gene, which is a transporter required for competence, was deleted from all strains. To enable fluorescence microscopy, different fluorescent protein-encoding genes (gfp, cfp or mcherry (Minoia et al., 2008)) were introduced to each strain (Lilja and Johnson, 2016). To adjust the pH of the medium, we added 1 m HCl or 0.5 m NaOH to liquid lysogeny broth (LB) to achieve a final pH of 6.5 or 7.5. The LB medium was supplemented with sodium nitrate (NaNO3) to a final concentration of 1 mm and agar to a final concentration of 1.5% (w/v). A previous study demonstrated that, under these substrate supply conditions, nitrate is the growth limiting substrate (Lilja and Johnson, 2016). We then prepared LB plates in an anaerobic glove box (Coy Laboratory Products, Grass Lake, USA) as described elsewhere (Goldschmidt et al., 2017).
For the colony expansion experiments, we first inoculated liquid LB cultures from single colonies and grew the cultures aerobically at 30 °C overnight. We then adjusted the cell densities by measuring the optical density at 600 nm (OD600) and diluting the cultures in sterile 0.9% (w/v) NaCl solution accordingly. We mixed the different strains at a ratio of 2:1:1 (OD600:OD600:OD600) of consumer (carrying gfp):producer (carrying cfp):producer (carrying mcherry) and inoculated 2 μl of the mixture onto the middle of each anaerobic plate in an anaerobic glove box as described previously (Goldschmidt et al., 2017). We then stored the plates in closed plastic bags in order to minimize evaporation and incubated them for up to 4 weeks at ~21 °C.
Confocal microscopy
We imaged colonies using a Leica TCS SP5 II confocal microscope (Wetzlar, Germany) as described elsewhere (Goldschmidt et al., 2017). We exposed the colonies to ambient air for 1 h before imaging to allow maturation of the fluorescent proteins.
Sectors and dendrites
We measured the number of sectors and dendrites at the leading edge of the consumer expansion. To measure the sectors, we imported the images into ImageJ (Schneider et al., 2012) and drew a line at the leading edge of the consumer expansion. We then measured the profile of the blue and red channels along these lines with a line-width of 100 pixels. We next imported this data into R (R Core Team, 2015) to calculate the number of sectors. The profiles typically showed areas of high or low signal intensities with sharp transitions between them. We therefore defined a sector as an area where one channel had a stronger signal than the other and we calculated the number of these areas.
To calculate the decrease in sector number over expansion distance, we determined the number of transitions between two producers on concentric circles using the Sholl plugin (Ferreira et al., 2014) to ImageJ. We then imported this data into R (R Core Team, 2015) and fitted an exponential decrease model of the form a+b*e−c, where r is the radial distance from the inoculum, a corresponds to the final sector numbers, b to the initial sector numbers and c to the steepness of the decrease.
We measured the number of dendrites manually. The dendrites typically had a single stem at the edge of the inoculum and then divided into many branches during the expansion. We only counted a dendrite when it spanned the entire secondary consumer expansion, extending from the inoculation zone to the line that was drawn to measure the sectors. To reduce manual counting bias, we assigned all the images for both pH conditions a random identifier and two different people counted the dendrites in the first experiment. Because the results were similar between the two people, one person analyzed the remaining experiments manually. To account for the potential manual counting bias, we did not compare the total number of dendrites to, for example, producer sector numbers. Instead we used a rank-sum test to quantify differences between the toxicity levels.
Modeling
We simulated colony growth of the cross-feeding communities using a previously developed individual-based model. A detailed description of the model can be found in Nadell et al. (2010) and further adaptations of this model to simulate expanding colonies can be found in Mitri et al. (2015). The model simulates cells as round particles with a certain radius on a grid where nutrients can diffuse. If the radius of a cell becomes larger than a threshold, it divides into two and the cells are rearranged with a shoving algorithm. Initially the cells were inoculated randomly in a circle in the center of the grid containing only NO3. Then they were allowed to grow and expand. Producer cells were inoculated in red and blue but were otherwise identical while consumer cells were inoculated in green. The ratio of the cell numbers was 1:1:2 (producer:producer:consumer), which is identical to that used for experimentation. The producer used nitrate as a substrate and released nitrite, while the consumer used nitrite as a substrate. The growth rates of both populations were inhibited by local nitrite concentrations. A more detailed description of the model can be found in the Supplementary Information.
Results
Nitrite is toxic at low pH and reduces colony expansion velocity
To test the effect of pH on nitrite toxicity in spatially structured systems, which may differ from completely mixed systems studied previously (Lilja and Johnson, 2016), we grew co-cultures of nitrite cross-feeding P. stutzeri strains on agar plates. We previously demonstrated that at pH 7.5 nitrite is relatively non-toxic to P. stutzeri while at pH 6.5 it is severely toxic and reduces growth rates in liquid batch cultures (Lilja and Johnson, 2016). To quantify the effect of nitrite toxicity in spatially structured systems, we used expansion velocity as a measure of growth dynamics (Harcombe et al., 2014). We inoculated 30 replicated plates at the same time with a 1:1 (OD600:OD600) mixture of producer and consumer and then sacrificed three replicates per pH condition at different time-points. This allowed us to measure the extent of expansion as a function of time and nitrite toxicity.
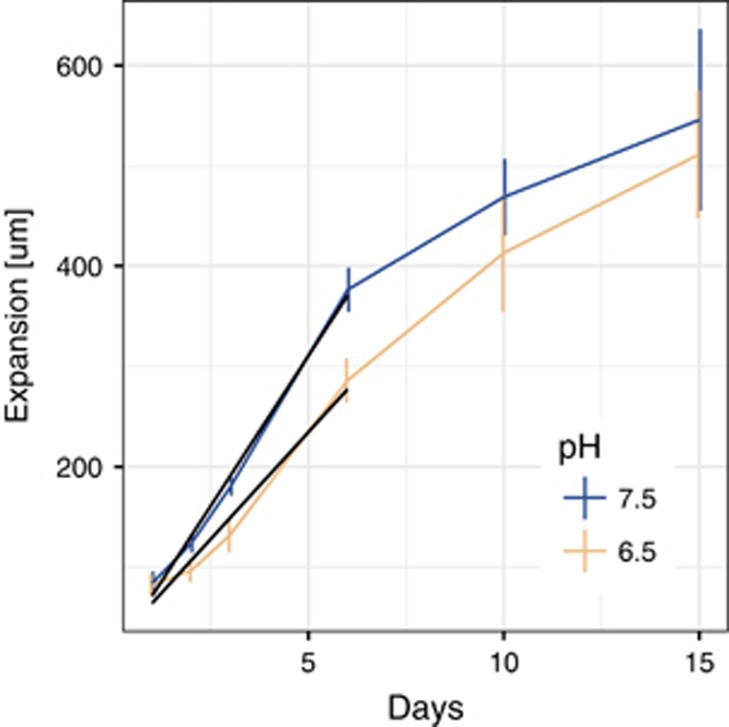
In general, the expansion velocity qualitatively slowed over time regardless of the pH, and thus regardless of the magnitude of nitrite toxicity (Figure 1). This was also true when we inoculated two complete degraders (Supplementary Figure 1). The decrease in expansion velocity could be caused by a combination of the depletion of nutrients and the hardening of the agar surface due to drying (although we did incubate the plates in sealed plastic bags). We thus estimated the initial expansion velocities by a linear regression of the velocity profiles over the first week. The estimated expansion velocities and the corresponding calculated apparent diffusion coefficients of the cells are given in Table 1. The initial expansion velocity was pH dependent, and thus coincided with the magnitude of nitrite toxicity. The cross-feeding colonies expanded slower at pH 6.5 (strong nitrite toxicity) than at pH 7.5 (weak nitrite toxicity) (Table 1; F-test, P=8 × 10−14). The pH dependence of nitrite toxicity is therefore qualitatively consistent with previous observations in liquid batch cultures (Lilja and Johnson, 2016).
Figure 1.
Expansion distances at different strengths of nitrite toxicity. Shown are the expansion distances over the first 15 days of growth at different pH values. pH 7.5 imposes weak nitrite toxicity while pH 6.5 imposes strong nitrite toxicity (n=3 per time point and condition, vertical bars are 95% confidence intervals). The black lines represent the linear regressions that were used to calculate the initial expansion velocities given in Table 1.
Table 1. Estimated initial expansion velocities.
| pH | Expansion velocity (m d−1) | s.e. | D (cm2 s−1) |
|---|---|---|---|
| 7.5 | 59.9 | 1.8 | 2.08E−10 |
| 6.5 | 42.7 | 2.2 | 1.06E−10 |
Abbreviation: D, apparent diffusion coefficient.
Nitrite toxicity slows local diversity loss of the nitrite producers
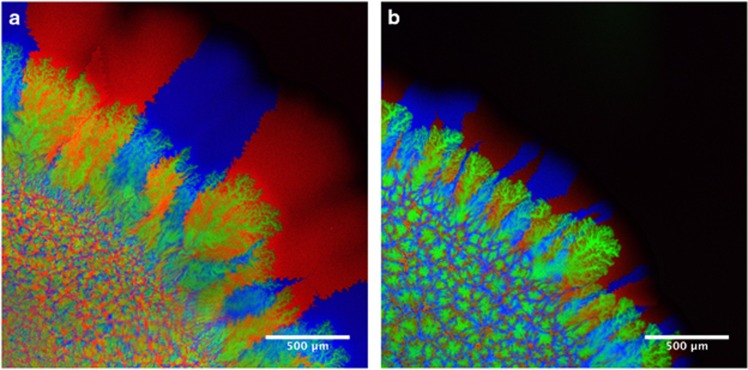
To assess the effect of nitrite toxicity on spatial organization and local diversity, we imaged the colonies after three weeks of growth. To measure local diversity of the producers, we inoculated a producer that expresses cyan fluorescent protein with a producer that expresses red fluorescent protein and a consumer that expresses green fluorescent protein. Figure 2 shows that the two sub-populations of the producer demixed into sectors when expanding from the inoculation zone, similar to previous reports for other organisms (Hallatschek et al., 2007; Mitri et al., 2015; Goldschmidt et al., 2017). The consumers succeeded the primary expansion of the producers and formed dendrites as described previously (Goldschmidt et al., 2017) under both conditions.
Figure 2.
Confocal microscopy images of the cross-feeding populations. (a) Co-cultures grown at pH 7.5 (weak nitrite toxicity). (b) Co-cultures grown at pH 6.5 (strong nitrite toxicity). The images show details of the edges of colonies. The inoculation zone is in the lower left and the colonies expand towards the upper right. Both expansions are shown at the same magnification. The producers expressed cyan fluorescent protein (cfp, blue) or mcherry fluorescent protein (mcherry, red) while the consumer expressed green fluorescent protein (gfp, green). The strains were inoculated at a ratio of 1:1:2 (cfp:mcherry:gfp) such that the total of the producers and the consumers had the same initial concentration.
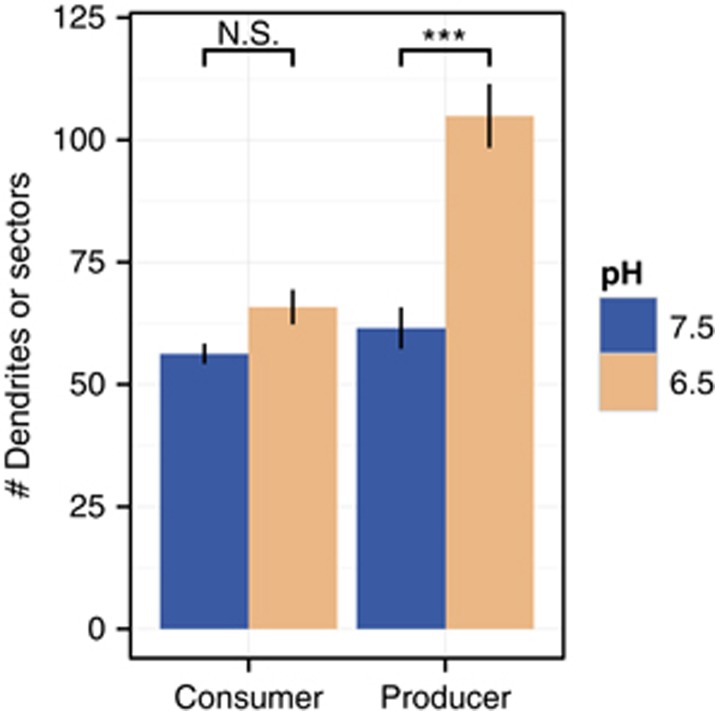
We used image analysis to measure the number of sectors for producers and the number of dendrites for consumers to investigate the effect of nitrite toxicity on intra-population diversity of the producers or consumers. While not being exactly the same, the number of dendrites, which typically also stem from a single point from the founding zone, behaves qualitatively similar to the number of producer sectors. However, the two cannot be directly compared. The number of dendrites of consumers was not affected by the pH (Mann–Whitney test, P=0.425), likely because the consumer reduced nitrite concentrations and thus reduced local nitrite toxicity when growing. In contrast, the number of producer sectors at pH 6.5 (strong nitrite toxicity) was greater than the number of producer sectors at pH 7.5 (weak nitrite toxicity) (Mann–Whitney test, P=0.000215; Figure 3). Local nitrite toxicity thus reduced the demixing effect of drift and slowed the loss of local diversity during expansion, but only for the producers.
Figure 3.
Mean numbers of sectors or dendrites. Consumer dendrites and producer sectors were measured radially at the position of the consumer front. The consumers and producers were measured on the same plates. N=13, error bars=standard error of the mean (s.e.m.), ***P of the Mann–Whitney test below 0.01.
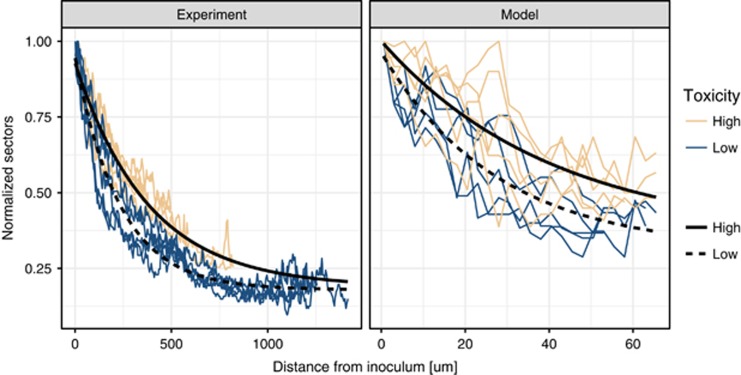
Recent experimental findings (Mitri et al., 2015) suggest that while the speed of local diversity loss due to drift might be affected by external factors (such as nutrient supply), the final sector numbers should remain the same. We thus measured the loss of sector numbers over expansion distance for the producer sectors. As can be seen in the left panel of Figure 4, sector numbers decreased exponentially and stabilized at values larger than 0. We fitted an exponential decay model to this data and found that, when nitrite toxicity was strong, the decrease in sector numbers was slower (−0.00250 at pH 6.5/−0.00442 at pH 7.5, F-test, P<2.2e−16). The final sector numbers, however, were not significantly affected by nitrite toxicity (final sector number=20, F-test, P-value=0.0709). In addition, there was a difference in initial sector numbers (115 at pH 6.5/107 at pH 7.5, F-test, P=0.00218). This indicates that nitrite toxicity slows the loss of sectors, but given enough time the final sector numbers will likely converge to similar values.
Figure 4.
Decrease of sector numbers with distance from inoculum (in pixels). (Left) Experimental data. (Right) Simulated data. Strong nitrite toxicity slows the loss of sector numbers during expansion, resulting in higher local diversity. Nitrite toxicity in the experimental data is manipulated via pH (7.5 for weak toxicity and 6.5 for strong toxicity), while in the model it represents the strength of the inhibition term (Supplementary Information). Note that here we normalized the sector numbers to the initial number of sectors in order to facilitate comparison between the experimental and modeled data.
To further assess whether the presence of the consumers influenced sector numbers of the producers, we performed control experiments where we grew the producers with and without the consumers and measured the number of sectors. Overall, we found that the presence of the consumers did not significantly affect the sector numbers of the producers (F-test, P=0.232). This result shows that the consumers did not significantly change the growth dynamics of the producers.
Negative correlation between expansion velocity and local diversity
The above results revealed two important effects of nitrite toxicity on the producers: first, nitrite toxicity slowed the expansion speed, and second, nitrite toxicity slowed the loss of sector numbers (and thus slowed the loss of local diversity of the producer). Combining these two results, we found a negative correlation (Spearman rank correlation: rho=−0.745, P=1.25 × 10−5) between sector numbers and the expansion velocity. At first glance, this may seem to contradict recent findings that showed a positive correlation between sector numbers and expansion velocity (Mitri et al., 2015). In that work, the authors found that colonies growing on high nutrient agar expanded faster and maintained more sectors over longer times than colonies growing on low nutrient agar. However, a more detailed analysis attributed this effect to the size of the actively growing population at the expansion edge, which corresponds to the effective population size in a growing colony (Korolev et al., 2011; Mitri et al., 2015). The smaller active population size, which occurs at lower nutrient supply, is more affected by drift, which results in fewer sectors in the expansion zone.
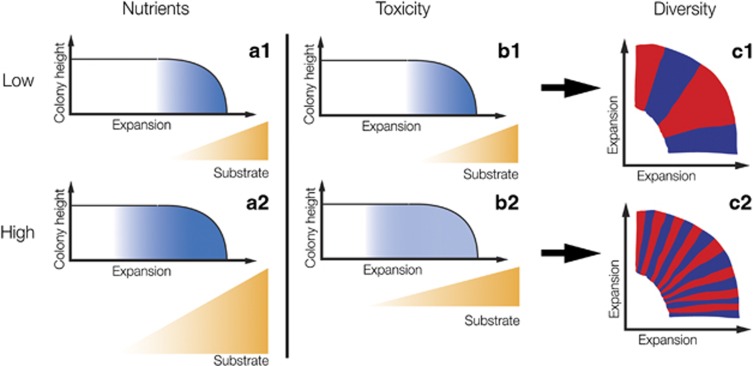
In our system, we did not manipulate the expansion speed by changing substrate supply, but we instead manipulated the expansion speed by changing the strength of nitrite toxicity. Because of spatial structure, the produced nitrite was locally inhibiting expansion. This could have non-intuitive effects on the size of the active population. We hypothesized that nitrite inhibition could slow the growth rate of the active population, which would enable the primary substrate nitrate to diffuse further into the colony before being consumed, and thus enable more cells to grow simultaneously and increase the active population size (see Figure 5 for a schematic representation). This implies that the active population size is, in our case, not just determined by nutrient availability, but also by nitrite toxicity, and that it should be larger for a slowly growing colony. The larger active population should be less affected by drift, which could explain the slower decrease of sectors numbers in the slower growing colonies when nitrite toxicity is strong.
Figure 5.
Schematic representation of the effect of substrate supply and metabolite toxicity on sector numbers and local diversity. The graphs show cross-sections of expanding colonies. The blue shading represents the activity of the cells at the expansion front. The size of the blue shaded area is thus equal to the effective population. Under low nutrient (a1) or weak toxicity (b1) conditions, the substrate is only available at a small edge of the colony. The effective population size is therefore small and drift is strong, which results in low local diversity (i.e., sector numbers, c1). At high nutrient supply (b2), more substrate is available which increases the effective population size. This reduces the effect of drift and increases local diversity (Mitri et al., 2015) (c2). Strong metabolite toxicity, in contrast, slows growth, which allows the substrate to diffuse deeper into the colony because it is consumed at a slower rate. The result is a larger effective population size that has a lower activity (b2) (indicated by a lighter shading). Nevertheless, the larger effective population size reduces drift, which leads to higher local diversity (c2). Note that in this study we only manipulated the strength of metabolite toxicity while Mitri et al. (2015) manipulated nutrient supply.
Nitrite toxicity increases the active population size, which in turn increases local diversity
To test the hypothesis described above (Figure 5), it is ideal to measure the size of the active population in the expanding colonies. While direct experimental measurements of the active population would provide further support for this conclusion, such measurements are not currently possible for our system. This is due to the anaerobic nature of our interaction, where we cannot track the fate of oxygen-dependent fluorescent proteins over time without creating potential confounding factors due to exposure to oxygen. We thus used a previously developed mathematical model (Nadell et al., 2010; Mitri et al., 2015) and incorporated toxicity of an intermediate metabolite to simulate expanding cross-feeding communities and predict the size of the active population. This allowed us to study the mechanism by which toxicity may influence sector numbers.
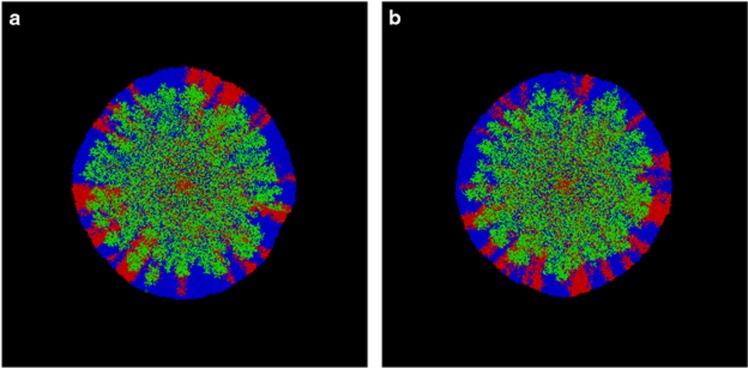
The model is an individual-based model and has successfully simulated sector dynamics in expanding colonies (Mitri et al., 2015). We extended this model by adding a toxicity term, which slows growth depending on the local concentration of nitrite. As can be seen in the simulations (Figure 6), the patterns with weak and strong nitrite toxicity were qualitatively similar. Under both conditions, the two sub-populations of the producers demixed into sectors and were succeeded by the consumers that formed structures that resemble dendrites. The dendrites were much less pronounced than in the experimental data, which is likely because of the much lower cell numbers and/or by the spherical shape of the cells used for simulations (instead of the natural rod-shape of P. stutzeri) (Goldschmidt et al., 2017). The model thus qualitatively reproduced the pattern and dynamics that we found in the experiments.
Figure 6.
Simulated colonies. (a) Weak nitrite toxicity. (b) Strong nitrite toxicity. The producer is shown in red and blue while the consumer is shown in green. The mixed area in the center is the inoculation zone. The two producer populations segregate into sectors and the consumer population forms dendrites during the expansion.
To assess the effect of nitrite toxicity on the active population size, we quantified the decrease in producer sector numbers during the simulated expansions. As can be seen in the right panel of Figure 4, the model qualitatively reproduced the experimentally observed dynamics of sector decrease. However, note that due to the much smaller population sizes, we did not expect the simulations to quantitatively represent the experiments. We fitted an exponential decrease model to the simulated data and found that, as in the experimental data, the steepness of the sector decrease was lower when nitrite toxicity was strong (−0.0232 strong nitrite toxicity/−0.0373 weak nitrite toxicity, F-test, P=2.132 × 10−8). The relative difference between the steepness was similar to that observed for the experiments (strong nitrite toxicity/weak nitrite toxicity=0.621 for model, 0.566 for experiments). As for the experimental data, the final sector numbers were the same (final sector number=17, F-test, P=0.862). However, in contrast to the experimental data, the initial sector numbers were not significantly affected by nitrite toxicity (initial sector number, 51 strong nitrite toxicity/52 weak nitrite toxicity, F-test, P=0.786). We thus concluded that the model simulated the experimentally observed dynamics with sufficient accuracy to infer the likely mechanism by which toxicity affects the loss of diversity in the producer.
In the next step, we quantified the size of the active population in the model. As stated above, the actively growing edge of an expanding colony corresponds to the effective population size (Korolev et al., 2011). We found that the number of active producer cells was larger in simulations when nitrite toxicity was strong (Mann–Whitney test, P=0.00794, see also Supplementary Figure 2). This result showed that, indeed, nitrite toxicity reduced growth rates and also increased active population sizes, which in turn reduced the effect of drift on the producer. This resulted in a slower loss of sectors and slower loss of local population diversity. The results therefore support our hypothesis that the purifying effects of drift may be countered not only by growth enhancing factors (e.g., high substrate supply), but also by growth-reducing factors (e.g., strong metabolite toxicity) (see Figure 5 for a schematic representation of the mechanism), where both mechanisms operate by affecting the active population size.
Discussion
We found that the toxicity of a single cross-fed metabolite led to a reduction in expansion velocity and slowed the loss of local diversity of a primary expansion. This resulted in a negative correlation between expansion velocity and local diversity. Mathematical modeling allowed us to investigate the mechanism that likely causes the loss of local diversity during expansion, which is genetic drift. Mitri et al. (2015) varied substrate supply to change the active population size. More nutrients enable the growth of more individuals at the expanding front. This is because the cells at the edge could not metabolize all the substrate sufficiently rapidly, such that substrate could diffuse further into the colony. This resulted in larger active population sizes and less drift.
The population dynamics in our system that contained metabolite toxicity caused less intuitive dynamics. By decreasing the pH, we made the cross-fed metabolite nitrite more toxic, which caused slower growth and expansion. The mathematical model showed that because the cells were growing slower, the cells at the front of the expansion could not consume all of the primary substrate nitrate sufficiently rapidly. This caused nitrate to diffuse further into the colony and increase the active population size. The larger active population size then slowed the demixing effect of drift, which slowed the loss of local population diversity (Figure 5). The speed at which local diversity was lost was therefore caused by the strength of genetic drift.
This mechanism is potentially not only limited to toxic metabolites, but might be a general ecological mechanism. We think that many growth-reducing factors could act via the same mechanism. Examples of important environmental factors that could have the same effect are temperature or salinity. Both can vary strongly over short distances and time scales, e.g., daily temperature changes due to sunlight or rainfall events, and both may reduce growth rates when they are not in the optimal range (Nichols et al., 2000). In fact, a recent study by Gralka et al. (2016) found larger sector numbers in colonies of Escherichia coli that were growing at lower than optimal temperatures. However, if these factors tend to extreme values, secondary effects (such as changes in cell morphology) might change the growth dynamics and spatial organization of the colonies. Nevertheless, the mechanism that we found here might broadly influence microbial colony growth and local diversity.
The presence of the consumer, which reduces toxicity by metabolizing nitrite, did not have a significant effect on local diversity of the producer. Likely, this is because the distance between the two expansions was too great to significantly reduce local nitrite concentrations within the primary producer expansion. Thus, in our case, the spatial separation of the two cross-feeding populations reduced the extent of interaction between them. There are many cases where spatial structure facilitates or stabilizes interactions (Dolinšek et al., 2016). A typical example being the spatial exclusion of cheaters from public good producers (Mitri et al., 2011; Momeni et al., 2013). However, it is important to keep in mind that spatial structure does not per se favor interactions, but can also hinder them by spatially segregating the interacting partners.
In conclusion, we investigated the effect of sequential cross-feeding of a conditionally toxic metabolite on spatial organization and local diversity in growing microbial colonies. We found that metabolite toxicity reduces expansion velocities and increases active population sizes, which slows the loss of local diversity. In addition, the spatial separation due to successive expansions reduced the strength of interaction between the two cross-feeding populations. These findings show that community dynamics can lead to non-trivial emergent properties of seemingly simple model microbial systems. The sum and balance of the substrates that enhance or reduce growth in such systems are often difficult to determine without deeper knowledge of the biogeochemistry at work. It is thus difficult to predict the emergent properties of complex microbial communities found in nature. A deeper knowledge of the fundamental ecological processes that govern simpler model microbial ecosystems could help us to understand and ultimately control microbial communities (Johnson et al., 2012; Dolinšek et al., 2016; Lindemann et al., 2016).
Acknowledgments
We acknowledge Martin Ackermann, Benedict Borer, Sara Mitri, Jonas Schulter and Simon Norrelykke for useful discussions and Simon van Vliet for helpful comments on a preliminary version of the manuscript. We also thank three anonymous reviewers for significantly improving the quality and clarity of this manuscript. This work was supported by grants from the Swiss National Science Foundation (31003A_132905, 31003A_149304) and SystemsX.ch, The Swiss Initiative in Systems Biology (MicroScapesX.ch).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Battin TJ, Sloan WT, Kjelleberg S, Daims H, Head IM, Curtis TP et al. (2007). Microbial landscapes: new paths to biofilm research. Nat Rev Microbiol 5: 76–81. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Harcombe WR. (2009). Population dynamics constrain the cooperative evolution of cross-feeding. PLoS One 4: e4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, O’Toole GA. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinšek J, Goldschmidt F, Johnson DR. (2016). Synthetic microbial ecology and the dynamic interplay between microbial genotypes. FEMS Microbiol Rev 40: 961–979. [DOI] [PubMed] [Google Scholar]
- Estrela S, Gudelj I. (2010). Evolution of cooperative cross-feeding could be less challenging than originally thought. PLoS One 5: e14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela S, Trisos CH, Brown SP. (2012). From metabolism to ecology: cross-feeding interactions shape the balance between polymicrobial conflict and mutualism. Am Nat 180: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Foll M, Petit RJ. (2009). Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40: 481–501. [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ et al. (2014). Neuronal morphometry directly from bitmap images. Nat Methods 11: 982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt F, Regoes RR, Johnson DR. (2017). Successive range expansion promotes diversity and accelerates evolution in spatially structured microbial populations. ISME J 11: 2112–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralka M, Stiewe F, Farrell F, Möbius W, Waclaw B, Hallatschek O. (2016). Allele surfing promotes microbial adaptation from standing variation. Ecol Lett 19: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. (2007). Genetic drift at expanding frontiers promotes gene segregation. PNAS 104: 19926–19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH et al. (2014). Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep 7: 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E. (2004) A Textbook of Modern Toxicology. John Wiley & Sons, Inc.: Hoboken, NJ, USA. [Google Scholar]
- Johnson DR, Goldschmidt F, Lilja EE, Ackermann M. (2012). Metabolic specialization and the assembly of microbial communities. ISME J 11: 1985–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. (2008). Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA 105: 18188–18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev KS, Xavier JB, Nelson DR, Foster KR. (2011). A quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am Nat 178: 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja EE, Johnson DR. (2016). Segregating metabolic processes into different microbial cells accelerates the consumption of inhibitory substrates. ISME J 10: 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann SR, Bernstein HC, Song HS, Frederickson JK, Fields MW, Shou W et al. (2016). Engineering microbial consortia for controllable outputs. ISME J 10: 2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen M, Schöps R. (1999). Population dynamics of denitrifying bacteria in a model biocommunity. Water Res 33: 639–646. [Google Scholar]
- McInerney MJ, Sieber JR, Gunsalus RP. (2009). Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol 20: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoia M, Gaillard M, Reinhard F, Stojanov M, Sentchilo V, van der Meer JR. (2008). Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc Natl Acad Sci U S A 105: 20792–20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S, Clarke E, Foster KR. (2015). Resource limitation drives spatial organization in microbial groups. ISME J, 1–12. [DOI] [PMC free article] [PubMed]
- Mitri S, Xavier JB, Foster KR. (2011). Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108: 10839–10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni B, Waite AJ, Shou W. (2013). Spatial self-organization favors heterotypic cooperation over cheating. Elife 2: e00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. (2013). Microbial syntrophy: Interaction for the common good. FEMS Microbiol Rev 37: 384–406. [DOI] [PubMed] [Google Scholar]
- Müller MJI, Neugeboren BI, Nelson DR, Murray AW. (2014). Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci USA 111: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Foster KR, Xavier JB. (2010). Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol 6: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DS, Olley J, Garda H, Brenner RR, McMeekin TA. (2000). Effect of temperature and salinity stress on growth and lipid composition of Shewanella gelidimarina. Appl Environ Microbiol 66: 2422–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Bonhoeffer S. (2004). Evolution of cross-feeding in microbial populations. Am Nat 163: E126–E135. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org/.
- Rainey PB, Buckling A, Kassen R, Travisano M. (2000). The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol Evol 15: 243–247. [DOI] [PubMed] [Google Scholar]
- Schink B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink B. (2002). Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81: 257–261. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbesma WFH, Almeida JS, Reis MAM, Santos H. (1996). Uncoupling effect of nitrite during denitrification by Pseudomonas fluorescens: an in vivo 31P-NMR study. Biotechnol Bioeng 52: 176–182. [DOI] [PubMed] [Google Scholar]
- de Souza ML, Newcombe D, Alvey S, Crowley DE, Hay A, Sadowsky MJ et al. (1998). Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl Environ Microbiol 64: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West Sa, Diggle SP, Buckling A, Gardner A, Griffin AS. (2007). The social lives of microbes. Annu Rev Ecol Evol Syst 38: 53–77. [Google Scholar]
- Zhou Y, Oehmen A, Lim M, Vadivelu V, Ng WJ. (2011). The role of nitrite and free nitrous acid (FNA) in wastewater treatment plants. Water Res 45: 4672–4682. [DOI] [PubMed] [Google Scholar]
- Zumft WG. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. (1993). The biological role of nitric oxide in bacteria. Arch Microbiol 160: 253–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.