Abstract
The association between corals and photosynthetic dinoflagellates (Symbiodinium spp.) is the key to the success of reef ecosystems in highly oligotrophic environments, but it is also their Achilles‘ heel due to its vulnerability to local stressors and the effects of climate change. Research during the last two decades has shaped a view that coral host–Symbiodinium pairings are diverse, but largely exclusive. Deep sequencing has now revealed the existence of a rare diversity of cryptic Symbiodinium assemblages within the coral holobiont, in addition to one or a few abundant algal members. While the contribution of the most abundant resident Symbiodinium species to coral physiology is widely recognized, the significance of the rare and low abundant background Symbiodinium remains a matter of debate. In this study, we assessed how coral–Symbiodinium communities assemble and how rare and abundant components together constitute the Symbiodinium community by analyzing 892 coral samples comprising >110 000 unique Symbiodinium ITS2 marker gene sequences. Using network modeling, we show that host–Symbiodinium communities assemble in non-random ‘clusters‘ of abundant and rare symbionts. Symbiodinium community structure follows the same principles as bacterial communities, for which the functional significance of rare members (the ‘rare bacterial biosphere’) has long been recognized. Importantly, the inclusion of rare Symbiodinium taxa in robustness analyses revealed a significant contribution to the stability of the host–symbiont community overall. As such, it highlights the potential functions rare symbionts may provide to environmental resilience of the coral holobiont.
Introduction
Corals and coral reef ecosystems are immediately threatened by global climate change and local anthropogenic impacts (Hoegh-Guldberg et al., 2007). This is because the obligate association of corals with dinoflagellate endosymbionts of the genus Symbiodinium represents both the success and the Achilles’ heel of these ecosystems. Fueled by their endosymbiont’s provision of energy in the form of photosynthates, corals provide the structural foundation for the ecologically and economically important reef ecosystems (Roberts et al., 2002). Although reef ecosystems cover only a small percentage of the world’s oceans, almost a third of global marine biodiversity is dependent on their functional integrity (Reaka-Kudla, 1997). But ocean warming and other impacts lead to the breakdown of the coral–Symbiodinium symbioses (coral bleaching) and the ongoing decline of coral reef habitats on a global scale (Hoegh-Guldberg et al., 2014; Hughes et al., 2017). Based on this premise, fundamental questions to help forecast the future of coral reefs emerge. For instance, how are Symbiodinium communities structured in their respective coral hosts and across reef ecosystems, how will this change under future ocean conditions, and does this affect coral host and ecosystem resilience?
Research on the physiology of Symbiodinium is typically focused on the most abundant symbiont(s) within the host (Cooper et al., 2011; Ziegler et al., 2015) or on cultured clonal isolates (Schoenberg and Trench, 1980; Suggett et al., 2015; Parkinson et al., 2016). For instance, the bleaching response and stress tolerance of the coral host are affected by the associated Symbiodinium type (Berkelmans and van Oppen, 2006; Abrego et al., 2008), where a type is defined based on ribosomal ITS2 DNA sequences, which, in combination with other genetic markers, is used to distinguish Symbiodinium species (LaJeunesse et al., 2012, 2014). However, these host–symbiont associations are not static and may change as a mechanism to increase the size of the ecological niche (Ziegler et al., 2015) or in response to environmental perturbation (Silverstein et al., 2015; Boulotte et al., 2016). The replacement of stress-susceptible with stress-resistant Symbiodinium according to the adaptive bleaching hypothesis (Buddemeier and Fautin, 1993) is accomplished by the substitution of dominant symbionts with rare types already present within hosts and within the broader environment (Berkelmans and van Oppen, 2006; Boulotte et al., 2016).
Although most coral colonies are primarily associated with one Symbiodinium type at a given time, a large cryptic diversity of low abundant Symbiodinium is becoming increasingly apparent through the combined use of next-generation sequencing (NGS) and large-scale sampling of coral–symbiont assemblages (Quigley et al., 2014; Thomas et al., 2014; Hume et al., 2016; Ziegler et al., 2017). These NGS studies have in common that they describe the existence of a rare and diverse Symbiodinium community, in addition to an abundant community composed of one or a few symbiont taxa. This observation of few abundant and many rare algal symbionts associated with a coral host parallels the structure of bacterial communities where the so-called rare bacterial biosphere (sensu Sogin et al., 2006; Pedrós-Alió, 2012) supposedly represents the predominant sector of bacterial diversity (Skopina et al., 2016). The rare bacterial biosphere has been shown to fulfill essential functions associated with nutrient cycling, the degradation of pollutants, host health, and rare microbes may further also enhance functionality of abundant microbes (reviewed in Jousset et al., 2017). In ocean and freshwater environments, rare members of bacterial and protist communities tend to be more active (as measured by RNA to DNA ratios) and thus contribute overproportionally to ecosystem function (Campbell et al., 2011; Debroas et al., 2015). In the case of corals and their algal endosymbionts, the putative significance of a rare background Symbiodinium community remains largely unresolved (Boulotte et al., 2016; Lee et al., 2016). A key question is whether the rare biosphere of microbial symbionts in general, and that of algal endosymbionts in particular, has a functional role in the coral holobiont.
Building onto approaches and insights from other study systems, we further investigated this research question. For instance, the diversity of species and the complexity of their interactions are linked to the stability of ecosystems in a dynamic relationship with environmental drivers (Kondoh, 2003; Ives and Carpenter, 2007). This has been particularly well-studied for plant communities in which phylogenetic and functional species diversity promote ecosystem stability (for example, Cadotte et al., 2012). Mathematical approaches to describe and understand relationships introduce the concepts of (1) ‘networked buffering‘, which relies on community components to perform multiple functional roles and a partial overlap between their functional capabilities (Whitacre and Bender, 2010) and (2) ‘buffered qualitative stability', which predicts robustness of a network (of genes or other biological entities) by its responses to arbitrary (that is, modeled) perturbations (Albergante et al., 2014). Following this, in this study we used network modeling to understand the composition and structure of coral host-associated algal endosymbiont communities. Our aim was to explore the potential role of rare Symbiodinium community members in regard to Symbiodinium community composition as a whole and in regard to the robustness (that is, stress tolerance) of the coral holobiont community composition under disturbance.
Materials and methods
Sample collection and ITS2 sequencing
For the Symbiodinium community modeling in this study, we used an NGS data set of the ITS2 rRNA marker gene from 892 coral samples that were previously collected and sequenced from three regions and 22 reef sites around the Arabian Peninsula in the Red Sea, the Sea of Oman, and the Persian/Arabian Gulf (for sample information refer to Supplementary Table S1 in Ziegler et al., 2017). Two sites each were sampled at Subri reef (Persian/Arabian Gulf) and Fahal Island (Sea of Oman), which were previously grouped together, but treated as distinct sampling locations in this study. The Symbiodinium ITS2 marker was sequenced using standard methods and quality control was conducted as detailed in Ziegler et al. (2017). Briefly, sequence reads were denoised (Quince et al., 2011) and filtered for low-quality scores in mothur v 1.34.1 (Schloss et al., 2009), and then screened for chimeric sequences (Edgar et al., 2011). Because a large proportion of Symbiodinium ITS2 sequencing data are comprised of intragenomic ITS2 variants (Arif et al., 2014; Thomas et al., 2014), sequence data were subsampled to 1000 sequences per sample, and clustered into Operational Taxonomic Units (OTUs) at 97% similarity cutoff. This cutoff was shown to capture the taxonomic level typically associated with ‘species’ (or above) (Arif et al., 2014). The final data thus contained information on the abundance of each of 92 Symbiodinium OTUs, which ranged in abundance from 1 to 418 068 sequences.
Evenness of the Symbiodinium community: abundance and richness scaling
OTU abundance data were used to create a rank-abundance distribution, where OTUs were ranked by decreasing abundance (that is, the OTU with the highest number of reads received rank 1). Next, we tested for a correlation of the abundance of each OTU with its degree, that is, with the number of samples the respective OTU was found in (the more samples the OTU was found in, the higher the degree). The empirical distribution of OTU abundance indicated that the number of reads per OTU was described by a power-law distribution. These broad distributions require a large range of samples for a proper characterization. To further support this observation, here we developed a theoretical framework that allowed us to relate the richness and abundance of the Symbiodinium community with the assumption of a power-law distribution. The scaling of the Symbiodinium OTU abundances was best captured by the Yule model (Yule, 1925). In terms of the abundance of OTUs, this model considered two processes: specific mutations, producing new sequences of the same OTU, occurring at a rate p, and generic mutations, leading to new OTUs, at a rate q. Thus, the governing equations in the continuous limit were:
where Ai was the population (abundance) of OTU i and NOTU was the number of different OTUs. Starting with a single OTU of population 1 at time t=0, the solution of these equations led to the following distribution of abundances:
and the population of the most abundant OTU, the initial OTU, was given by 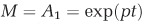 , and the total number of different OTUs
, and the total number of different OTUs 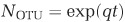 . Combining these results we found
. Combining these results we found 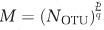 .
.
The total number of sequences N was given by (Simkin and Roychowdhury, 2011)
Thus in the limit of large t, we obtained for p>q, M~N (and thus NOTU~Nq/p), while for p<q, NOTU~N (and thus M~Np/q).
Applying these operations yielded a rank-abundance that scaled as A~r-s. From this rank-abundance scaling, the distribution of abundances was given by P(A)~A−1−1/s. Comparing with Equation (3), s=p/q>1 led to the scaling for the most abundant OTU M~N and the richness S(=NOTU)~N1/s (and S~A1/s).
Accounting for ITS2 gene copy number differences
Differences in intragenomic ITS2 gene copy numbers between Symbiodinium OTUs may cause a potential bias in our model, as they may lead to over- or underestimation of relative OTU abundance. We addressed this issue by implementing a simulation to account for potential ITS2 gene copy number differences between OTUs. In this approach, OTU counts were first randomly assigned an integer value between 1 and 10 and then the OTU count was randomly multiplied or divided by the assigned value. This corresponds to the reported two orders of magnitude difference in single cell-based intragenomic variability of the ITS2 region in Symbiodinium within types (Mieog et al., 2009; Wilkinson et al., 2015) and represents a conservative approach in light of estimated ITS2 copy number ratios of 1:3 to 1:5 between clades (Mieog et al., 2007; unpublished reference in Thornhill et al., 2007). Based on the resulting values for each OTU, we ranked the new relative abundances and re-calculated the OTU community distribution. The simulation was repeated with 1000 iterations to create confidence intervals for an OTU community distribution plot (Supplementary Figure S1). Our approach suggested that the rank of individual low abundant OTUs may change, but that the Symbiodinium community structure was not heavily affected by differences in ITS2 copy numbers, which was attributed to the high unevenness of the Symbiodinium community.
Host–Symbiodinium bipartite networks and Symbiodinium co-occurrence networks
Network modeling was applied to assess composition and structure of coral host-associated algal symbiont communities. To do this, the abundance of each OTU per sample was represented in a bipartite network where each link connected an OTU and a sample with a weight corresponding to the number of sequences. This bipartite network was then transformed into a co-occurrence network where two OTUs were connected if they co-occurred in the same sample and the weight was the number of samples in which they co-occured. The co-occurrence matrix contained 419 links.
Similarly, a bipartite network connecting OTUs and host taxa was constructed by connecting OTUs and the hosts in which they were observed with the total number of sequences. The data contained 46 host taxa. The host co-occurrence network was then constructed by linking pairs of OTUs observed in the same host taxon; the weight reported the number of hosts in which they co-occurred. In this case the host co-occurrence matrix contained 1492 links.
Identification of co-occurrence clusters
To further elucidate Symbiodinium community structure, we tested for statistically significant associations of Symbiodinium OTUs that co-occurred with each other in so-called ‘clusters’. We applied Order Statistics Local Optimization Method (OSLOM) (Lancichinetti et al., 2011) as the algorithm to detect topological communities (clusters) from the co-occurrence matrix. OSLOM reported statistical significant clusters for a predefined P-value that we set at P<0.01. It also reported overlapping OTUs, that is, OTUs could be assigned to more than one community. As the weight informed about the number of co-occurrences we used the ansatz of a weighted network where considering weights as multiple links, which translated into a null model that randomly rewired all these links together with all the links of the network (option –uw).
Host–Symbiodinium community robustness to perturbations
Next, we explored the potential role of rare Symbiodinium community members in the robustness (that is, stress tolerance) of the coral host–symbiont community composition under disturbance. We conducted a robustness analysis of the host–Symbiodinium network by sequential extinction (that is, removal) of OTUs according to three scenarios: (a) random, (b) generalists, and (c) specialists. In the random scenario, one of the existing OTUs was selected at random for extinction; in the generalist scenario, the OTU with the largest degree (that is, the highest presence over all samples) was selected (in case of >1 OTU complying to this condition, one of them was selected at random); in the specialist scenario, the OTU with the smallest degree was selected (in case of >1 OTU complying to this condition, one of them was selected at random). After selection of an OTU, it was removed from the system together with its links. In cases where samples became devoid of OTUs, the samples also became extinct. After a number of sequential OTU extinctions, the system could split into several connected components. We evaluated the robustness of the host–Symbiodinium community against removal of Symbiodinium OTUs as a measure of ecosystem resilience against perturbations by quantifying the number of remaining samples connected through the presence of OTUs and the size of largest connected component. All simulations were written in FORTRAN by the authors.
Adaptive robustness analysis
Recent studies suggest that rare Symbiodinium members may replace dominant ones upon environmental change, attesting to a buffering capacity of the Symbiodinium community that provides resilience to the host–symbiont system (Boulotte et al., 2016; Hume et al., 2016). To address possible scenarios of emerging rare endosymbionts, we introduced a probability (p) that an alternative OTU can take over the prevalence and abundance of a removed OTU. We reduced the set of possible replacement OTUs to those OTUs that originally co-occurred with the removed OTU (in at least one sample). The rationale of this approach was that if the two OTUs co-occurred in at least one sample, there is an increased likelihood that they have close niche spaces allowing the less frequent co-occurring OTU to replace the lost OTU. Proceeding as for the static robustness analysis (above), we then allowed the possibility that each link, indicating the presence of an OTU in a sample, was replaced by another OTU in the adaptive robustness analysis. The replacement was done with probability p.
Results
Symbiodinium community distribution reveals high number of rare species
We explored community patterns of host–Symbiodinium assemblages using ITS2 NGS data of Symbiodinium from 892 hard corals and other symbiotic anthozoans covering 46 genera across the Arabian Seas (that is, Red Sea, Sea of Oman, Persian/Arabian Gulf) (Ziegler et al., 2017). The resulting data set comprised 92 OTUs, which were clustered from >7 million sequence reads representing >110 000 distinct Symbiodinium ITS2 sequences, illustrating the large intragenomic diversity of the ITS2 marker (Ziegler et al., 2017).
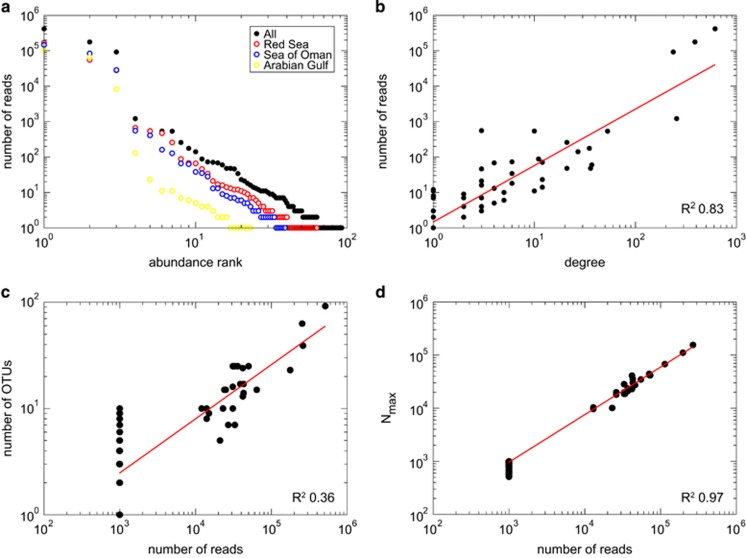
The rank-abundance distribution of Symbiodinium OTUs in the data set was highly skewed with a few abundant OTUs and a long tail composed of many rare OTUs (OTU abundance range 1-418 068), characterized by a scaling exponent s=2.52 (Figure 1a). Symbiodinium community composition followed a rank-abundance distribution, best described by a Zipf-like distribution, where the abundance, A, scaled with the rank order of the OTUs, r, as A~r–s, where s was the scaling exponent characterizing the distribution (Clauset et al., 2009; Saichev et al., 2009). In other words, 90% of the less abundant OTUs accounted for less than 1% of the population; in the other extreme, the two most abundant OTUs captured more than 99% of the population (Figure 1a). Thus, the community was composed of few abundant and many rare members (Figure 1a) confirming the Symbiodinium diversity to be dominated by a rare symbiont assemblage comparable to the rare biosphere reported for bacteria in microbial communities in the ocean (Pedrós-Alió, 2012) and the human gut (Arumugam et al., 2011). A complementary result on the rarity of the Symbiodinium community was obtained by analyzing the scaling of the richness, S, and the abundance of the most abundant OTU, M, with the total number of sequences (Figure 1c). The agreement of the scaling exponents of the richness and the most abundant OTU with the total number of reads N with the rank-abundance distribution based on a power-law distribution supported the hypothesis of a rare coral–symbiont assemblage. Correlation between prevalence in corals, P (measured by degree), and abundance (measured by the number of reads) for a symbiont OTU also followed a power-law relationship, P~Ab with a scaling exponent, b, of 1.59 (±0.08) (s.d.), further providing evidence for a low evenness of OTU-based community structure (Figure 1d). Hence, not only were the majority of the OTUs rare in terms of relative abundance, but they were each also detected in only a small fraction of all coral hosts analyzed, although most host taxa were harboring rare symbiont OTUs (Supplementary Table S1). This resulted in a strong correlation between abundance and prevalence for Symbiodinium OTUs (Figure 1b).
Figure 1.
Rank-abundance distribution of Symbiodinium taxa (that is, OTUs) in hard corals and other symbiotic anthozoans from the Arabian Seas (a) and correlation between presence in coral samples (measured by degree) and the number of reads per OTU (b). The power-law relationship indicates that the Symbiodinium community is comprised of few abundant and many rare members with a prevalence that scales sub-linearly with abundance, that is, Symbiodinium OTUs are not randomly distributed, but concentrated across hosts. (c) Richness, S, measured as the number of distinct OTUs and (d) number of reads of the most abundant OTUs as a function of the total number of reads. The fit shows S~N0.5 and Nmax~N0.9.
Symbiodinium communities form discrete clusters of co-occurrence
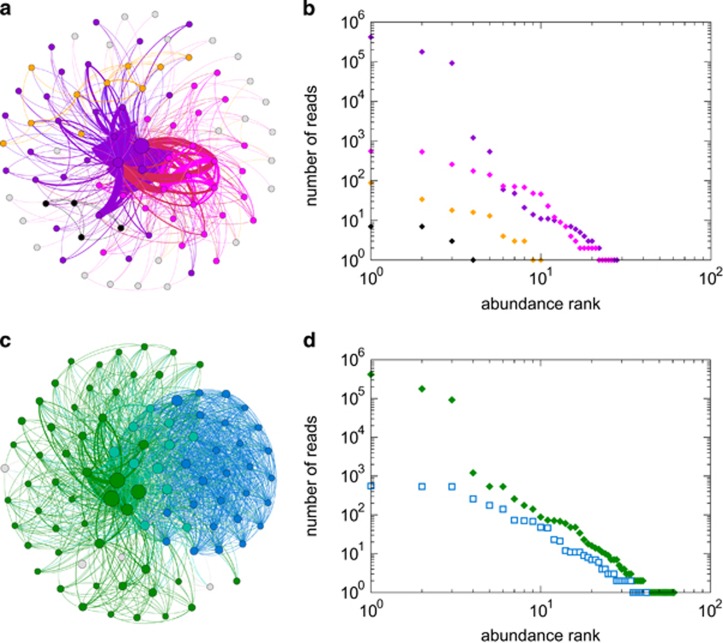
We next examined the patterns of co-occurrence of Symbiodinium OTUs at the individual sample (that is, colony) and at the host genus level. The co-occurrence network represented the weighted projection in the OTU set, that is, a link connected two OTUs if they co-occurred in at least one sample (Figure 2a) or host taxon (Figure 2c), and the weight of the link was determined by the number of samples or host genera, respectively, in which they co-occurred. From these co-occurrence networks, we statistically analyzed the Symbiodinium community.
Figure 2.
(a) Co-occurrence of Symbiodinium OTUs in hard corals and other symbiotic anthozoans from the Arabian Seas form four significant clusters (P<0.01). Each symbol represents an OTU with respective sizes denoting relative abundance and colors (purple, pink, orange, black) denoting cluster membership; red connections between OTUs represent overlaps between the purple and pink clusters. Lines indicate co-occurrence of two OTUs in a sample, and increasing line thickness indicates a higher number of samples in which the OTUs co-occur. (b) The rank-abundance distribution of Symbiodinium OTUs in each co-occurrence cluster displays a structured assembly that is comprised of rare and abundant members. (c) Two co-occurrence clusters (blue and green) of Symbiodinium OTUs were detected when samples were grouped by host taxon, with an overlap of 11 OTUs (turquoise) shared between both clusters. (d) The two host taxon-based co-occurrence clusters were of comparable size and followed the power-law scaling behavior of rank abundance. Colors denote separate clusters, colors in (b) and (d) correspond to clusters in (a) and (c), respectively, gray=OTUs not part of a cluster (P >0.01).
Overall, we observed highly structured co-occurrence patterns using the sample-based data set, which resulted in four clusters of OTUs comprising 68 of the 92 OTUs (Figure 2a). The two larger clusters consisted of 28 and 26 OTUs, respectively, and the two smaller clusters consisted of 10 and 4 OTUs. Despite differences in the number of Symbiodinium OTUs in the co-occurrence clusters, each of the clusters was equally comprised of rare and abundant OTUs (Figures 2b and d) and conserved the scaling behavior of rank abundance observed for the entire Symbiodinium community (Figure 1a). To exclude the possibility that the co-occurrence of abundant and rare OTUs was an artifact caused by the 97% similarity cutoff that may sort intragenomic ITS2 variants into discrete OTUs, we considered the OTUs’ identity based on their most abundant sequence (Supplementary Table S2). The largest co-occurrence cluster consisted of OTUs from five clades (A–D, F), making it unlikely that intragenomic variants were represented in distinct OTUs. Nevertheless, several OTUs had best hits to the same Symbiodinium type (for example, C15), indicating that potentially some OTUs may represent intragenomic variants of the same Symbiodinium type. The opposite effect may happen if diversity is underestimated with an OTU-based approach, due to the clustering of similar sequences that may de facto be representative of distinct Symbiodinium types. This might have affected the number and size of the co-occurrence clusters we observed, but potentially has a minor effect on our analysis of Symbiodinium community composition because of the large unevenness and robust scaling behavior of the community. When samples were grouped by host taxon, two co-occurrence clusters of Symbiodinium OTUs of comparable size were detected (Figure 2c).
Bipartite host–symbiont networks distinguish specialist and generalist community members
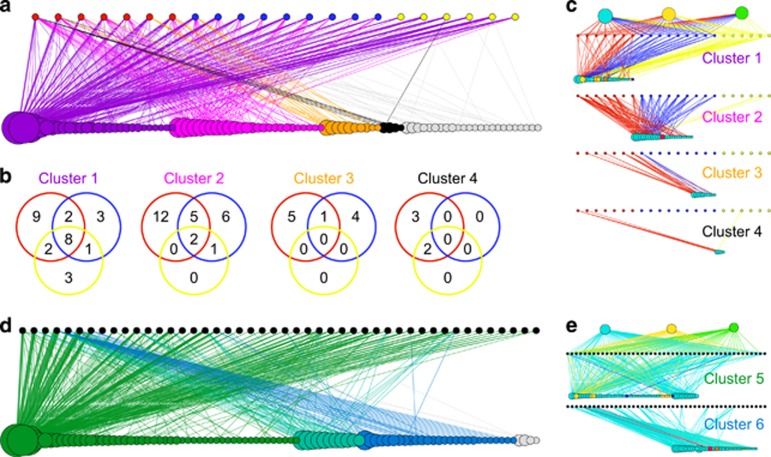
The structure of host–symbiont diversity was examined using bipartite networks, which map the occurrence of reefs or hosts to symbiont OTUs (Figures 3a and d). Our analysis showed that the abundant Symbiodinium OTUs seem to represent a resource shared across the Arabian Seas, which were accompanied by many endemic, regional OTUs supporting the argument of the existence of different selective environmental pressures between these regions (Ziegler et al., 2017) (Figure 3b). The largest Symbiodinium co-occurrence cluster showed a high degree of mixing between reefs and regions and the three smaller clusters captured more specialized communities that are connected by (that is, share) single OTUs between regions (Figure 3b). Symbiodinium phylogenetic diversity increased with cluster size, with the smallest clusters (3 and 4) being comprised only of members of clade C, and clusters 1 and 2 of clade A, B, D, F, and G in addition to clade C, respectively (Figure 3c). The remaining OTUs (n=24), not identified as belonging to co-occurrence clusters, were mostly characterized by occurrences in a single reef or host. The host–symbiont networks revealed that coral taxa have very few extreme specialists, in terms of ‘private’ host OTUs, or generalists, in terms of having consistently many OTUs (Figure 3d). Symbiodinium in turn has many specialists, that is, OTUs that appeared only in one or a few coral host genera, which were those rare OTUs composing the tail of the rank-abundance plot (Figure 1a), and a few extreme generalists that were found at a high prevalence across different corals and regions and are those with the highest rank abundance in the rank-abundance plot (Figure 3d). Similar to the reef-based clusters, the host taxon-based co-occurrence clusters were comprised of a phylogenetically mixed Symbiodinium community, dominated by clade C (Figure 3e).
Figure 3.
Coral host–Symbiodinium bipartite networks over reefs (a) and host genera (d) from three regions across the Arabian Seas and bipartite networks for significant OTU co-occurrence clusters over reefs (c) and hosts (e). Top nodes represent reefs over three sampling regions for (a, c) (red=Red Sea, blue=Gulf of Oman, yellow=Persian/Arabian Gulf), and the number of shared OTUs per region and cluster is illustrated with Venn diagrams (b). Top nodes in (d, e) represent host taxa; all bottom nodes represent Symbiodinium OTUs. In (a, d) OTUs are ordered by cluster (color codes as in Figure 2) and by abundance (indicated by relative size) and in (c, e) OTUs are ordered by abundance (size) with the three most abundant OTU arranged at the top of the graph for clarity and colored by clade (Symbiodinium clade A=green, B=dark orange, C=cyan, D=yellow, F=blue, G=red). Lines represent observed associations. Corals have very few extreme specialists or generalists and Symbiodinium have many specialists and a few extreme generalists.
Host–Symbiodinium community robustness to perturbations
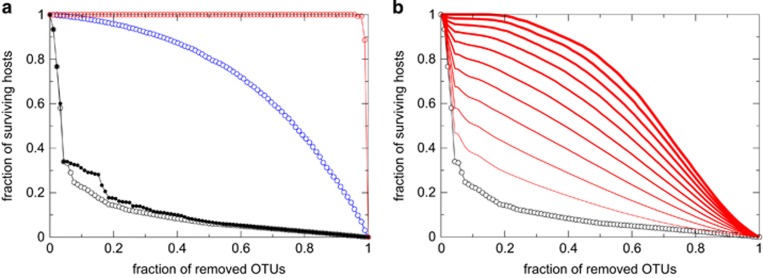
We assessed the robustness of the coral system against environmental perturbation by measuring the size of the largest connected component of the bipartite network as OTUs were removed (Figure 4), implying OTUs to differ in their resistance to stress. As OTUs were removed from the bipartite network, the number of hosts represented in the network decreased at rates that were dependent on the removal strategy of the OTUs. The decrease in the number of remaining connected hosts at rates faster than the linear loss expected by random removal occurred when highly connected OTUs, that is, those that associate with many hosts (that is, with high degrees), were removed first. This suggests that host–symbiont assemblages rely disproportionally on common Symbiodinium taxa and that these common taxa confer resistance to coral systems against random loss of OTUs from the community (Figure 4a). Our modeling results are based on the assumption of fixed roles of symbiont community members (Lee et al., 2016). However, a host–symbiont community composed of few, common Symbiodinium taxa would be rendered highly vulnerable to impacts (for example, disease, environmental extremes) leading to catastrophic ‘mortality’ of the dominant OTUs. Indeed, recent evidence suggests that rare Symbiodinium members may replace dominant ones with shifting environmental conditions, therefore providing resilience to host–symbiont communities (Boulotte et al., 2016; Hume et al., 2016). To address possible scenarios of emerging rare members, we introduced a probability (p) in an adaptive robustness analysis that an alternative OTU could take over the prevalence and abundance of a removed OTU. Increasing the replacement probability p increased the number of surviving hosts, defined as hosts that retain at least one OTU (Figure 4b). For a replacement probability of 10%, a removal of 10% of OTUs led to a doubling in the number of surviving samples (Figure 4b). Consistently, the OTUs responsible for the replacement belonged to the rare proportion of OTUs. The OTUs involved in >50% of the replacements had a prevalence of 1, that is, they only appeared in one sample. The average increase of prevalence for this set was almost 2 (1.96), representing a signature of spreading of rare OTUs upon loss of the dominant OTUs.
Figure 4.
Robustness of the coral–Symbiodinium community, measured as the fraction of remaining samples, to removal of Symbiodinium OTUs; (a) removal strategies depicted: random order (blue), according to their degree (that is, presence in coral samples; white), or according to their abundance (black). Removing components in increasing order of degree (red) shows a more stable scenario compared to removal in random order. When components with the highest degree are removed first (white), the robustness of the system quickly decreases and falls below linear loss of species, indicating that common Symbiodinium OTUs contribute overproportionally to maintaining host–symbiont assemblages. (b) Allowing for adaptation by introducing the probability that a removed OTU can be replaced by co-occurring OTUs increases the fraction of surviving samples. The probability that an existing OTU replaces the removed OTU increases from 0.1 to 1.0 at intervals of 0.1 from bottom to top (red lines).
Discussion
The finding of a rare and diverse Symbiodinium community within coral hosts parallels the existence of a rare bacterial biosphere (sensu Sogin et al., 2006). This cryptic and diverse community structure of Symbiodinium within corals was only uncovered recently through the application of NGS approaches. Further, we found this community to be characterized by a rank-abundance pattern following a power-law relationship that matches those reported for planktonic bacterial assemblages (cf. Figure 2 in Pedrós-Alió, 2012). A key question is whether the rare biosphere in general, and that of coral–algal symbionts in particular, have a functional role in the ecosystem (Lee et al., 2016).
Several lines of evidence suggest that the uneven Symbiodinium community structure, composed of few abundant and many rare members, may potentially present a so-far overlooked aspect of coral holobiont functioning. First, the prevalent and dominant components of the symbiont community cannot explain the breadth of responses of corals to stress or their resilience to locally restricted environmental perturbations, precisely because the dominant components are shared across wide environmental gradients and across reefs with contrasting status. This further points to the differentiating elements of the assemblage, composed of the rare symbionts, as the components that may underpin such diversity in response. Interactions between members of the bacterial symbiont community can support holobiont functions and traits that cannot be supplied by single taxa in isolation, for example, synergistic effects between pairs of bacterial taxa increase fungal resistance in the freshwater polyp Hydra compared to polyps colonized by single bacterial taxa (Fraune et al., 2015). Similarly, the functional roles of Symbiodinium assemblages are presumably not carried out by individual OTUs but by assemblages of OTUs, as suggested by the existence of distinct co-occurrence networks involving rare components of the symbiont pool revealed by our analysis. This is in accordance with patterns of co-occurrence of diverse micro- and macroorganisms, which suggests that non-random community assembly supporting ecosystem function and stability may be a general feature of all domains of life (McCann, 2000; Horner-Devine et al., 2007).
Second, rare bacterial community members are shown to fulfill essential functions while at low abundance. They are often more active than abundant members as revealed by rRNA:rDNA ratios and respiration rates (Dimitriu et al., 2010; Campbell et al., 2011). Rare microbes sustain a vast functional gene pool and can indirectly enhance functionality of abundant microbes (reviewed in Jousset et al., 2017). For example, for coral-associated bacterial communities it was suggested that two widely distributed, but low abundant bacterial strains may contribute large proportions of defined metabolic processes within the network of coral host–bacterial functions (Ainsworth et al., 2015). Furthermore, functional diversity of rare species is well documented in assemblages of macroorganisms such as rainforest trees, reef and stream fishes, or birds (Mouillot et al., 2013; Leitão et al., 2016). Symbiodinium also represent a highly diverse group that was described on the level of differences between species and between strains within a species (Suggett et al., 2015; Parkinson et al., 2016). Given the large (functional) diversity of Symbiodinium and the similarity of Symbiodinium community structure to that of bacterial communities (Pedrós-Alió, 2006, 2012), we suggest that similar assumptions can be made regarding the repertoire and contribution of the rare Symbiodinium biosphere to community function, and further research should probe the functions provided by the rare Symbiodinium biosphere to the coral holobiont while at low abundance.
Further, the network co-occurrence analysis suggests the potential for the rare Symbiodinium community to play a role in conferring resistance to the coral host by replacing dominant Symbiodinium types lost under environmental stress. The emergence of a regionally cryptic Symbiodinium species as prevalent in the heat-selected corals of the Persian/Arabian Gulf (Hume et al., 2016) supports the notion that a phylogenetically diverse and rare algal biosphere may serve as a source of genomic innovation and standing genetic diversity providing the raw material for adaptation to changing environmental conditions (Sogin et al., 2006). As such, an otherwise rare component can become the dominant member and represents a source of adaptation to the coral host–symbiont system (Hume et al., 2016); albeit this reorganization may sometimes come with a trade-off in general ecological performance, such as decreased coral growth rates (Pettay et al., 2015).
The extensive data set used in our study entails samples with mixed communities of Symbiodinium that are comprised of combinations of various clade C symbionts, combinations of clade C and clade D symbionts, or clade A with clade C or clade D symbionts in varying abundances. The patterns in these data largely correspond to previously reported switching or shuffling scenarios for clade C to clade D dominance (Jones et al., 2008; Silverstein et al., 2015; Boulotte et al., 2016) or vice versa (Jones et al., 2008), between clade C types (Boulotte et al., 2016), and between clade A and C (Hume et al., 2015). In addition, some coral species, such as Pocillopora verrucosa and Porites lutea (both abundant in this data set) associate with a range of different symbionts throughout reef locations in the Red Sea (Sawall et al., 2015; Ziegler et al., 2015). Thus, they may at least potentially be able to undergo switching or shuffling events under stress scenarios. Despite the large gap of knowledge under which environmental conditions shuffling and switching may occur, which coral species can change their symbionts, or whether this is a selective mechanism, our modeling approach helps exploring the role of rare Symbiodinium OTUs in host–Symbiodinium community dynamics, thereby providing directions for future research.
The role that Symbiodinium background diversity plays in the robustness of the host–symbiont community assembly is a fundamental, but yet unresolved question. Previous models suggested high robustness of highly uneven host–symbiont communities (Fabina et al., 2013), but these conclusions were drawn without the exclusion of intragenomic ITS2 sequence variants that are prone to confound diversity estimates. In our study, we aimed to remove redundancy from intragenomic diversity by applying an OTU-based clustering approach. Consequently, our results suggest a rather low robustness of a static system when dominant Symbiodinium components are lost, while rare members remain unchanged and cannot become dominant. We integrated recent studies that suggest that rare Symbiodinium community members can become dominant over short time scales following stress events (Boulotte et al., 2016) and during long-term environmental adaptation (Hume et al., 2016) by conducting an adaptive robustness analysis (that is, allowing co-occurring endosymbionts to replace removed members). Comparing the results from both modeling approaches revealed a significant increase in the stability of the host–symbiont community when rare members can become dominant and replace the lost components. Thus, our model highlights a potential role for rare symbionts upon environmental change. It should be noted, however, that although our findings support a potential role of Symbiodinium community composition for holobiont functioning at large and for background symbionts as mediating components that buffer the community against stressors in particular, detailed and rigorous experiments should be conducted to confirm this notion.
Conclusions
Our results reveal a highly structured community assembly in coral host–Symbiodinium associations and suggest functional importance of the Symbiodinium community as the sum of all of its members, not only of the most dominant members. The host–Symbiodinium association is shaped by a few dominant symbionts as well as a rare, diverse community characterized by robust scaling power laws that escaped previous analyses based on reduced sample size and/or sequencing depth. The existence of distinct co-occurrence networks involving rare members of the symbiont pool suggests that the functional role of Symbiodinium in the holobiont is not determined by a single individual taxon, but rather by assemblages of symbionts. Intriguingly, the inclusion of rare microbial members in robustness analyses resulted in a significant stability increase of the host–symbiont community and thus further highlights a potential role that rare symbionts may have in corals threatened by environmental change. Given that host–Symbiodinium association plays a central role in coral holobiont response to perturbation and resilience, future studies should focus on experimental elucidation of the specific functions contributed to the holobiont by rare Symbiodinium members and the synergistic effects arising from their interactions.
Data accessibility
Sequence data used in this publication can be accessed in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA306572.
Acknowledgments
Research reported in this publication was supported by competitive research funding from King Abdullah University of Science and Technology (KAUST), Office of Sponsored Research (OSR) under Red Sea Research Center CCF Award No. FCC/1/1973-22-01 and baseline research funds to CRV. We thank the three anonymous referees for their comments that helped improving this manuscript.
Author contributions
Conceptualization, data interpretation and writing-review and editing: MZ, VME, CMD, and CRV; model testing: VME; writing-original draft: MZ, VME, and CRV; visualization: MZ and VME; funding acquisition: CMD and CRV.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abrego D, Ulstrup KE, Willis BL, van Oppen MJ. (2008). Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc Biol Sci 275: 2273–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth T, Krause L, Bridge T, Torda G, Raina J-B, Zakrzewski M et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9: 2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albergante L, Blow JJ, Newman TJ. (2014). Buffered qualitative stability explains the robustness and evolvability of transcriptional networks. eLife 3: e02863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif C, Daniels C, Bayer T, Banguera-Hinestroza E, Barbrook A, Howe CJ et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol Ecol 23: 4418–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans R, van Oppen MJH. (2006). The role of zooxanthellae in the thermal tolerance of corals: a 'nugget of hope' for coral reefs in an era of climate change. Proc Biol Sci 273: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulotte NM, Dalton SJ, Carroll AG, Harrison PL, Putnam HM, Peplow LM et al. (2016). Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 10: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddemeier RW, Fautin DG. (1993). Coral bleaching as an adaptive mechanism—a testable hypothesis. Bioscience 43: 320–326. [Google Scholar]
- Cadotte MW, Dinnage R, Tilman D. (2012). Phylogenetic diversity promotes ecosystem stability. Ecology 93: S223–S233. [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauset A, Shalizi CR, Newman MEJ. (2009). Power-law distributions in empirical data. SIAM Rev 51: 661–703. [Google Scholar]
- Cooper TF, Ulstrup KE, Dandan SS, Heyward AJ, Kühl M, Muirhead A et al. (2011). Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc Biol Sci 278: 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debroas D, Hugoni M, Domaizon I. (2015). Evidence for an active rare biosphere within freshwater protists community. Mol Ecol 24: 1236–1247. [DOI] [PubMed] [Google Scholar]
- Dimitriu PA, Lee D, Grayston SJ. (2010). An evaluation of the functional significance of peat microorganisms using a reciprocal transplant approach. Soil Biol Biochem 42: 65–71. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabina NS, Putnam HM, Franklin EC, Stat M, Gates RD. (2013). Symbiotic specificity, association patterns, and function determine community responses to global changes: defining critical research areas for coral-Symbiodinium symbioses. Global Change Biol 19: 3306–3316. [DOI] [PubMed] [Google Scholar]
- Fraune S, Anton-Erxleben F, Augustin R, Franzenburg S, Knop M, Schroder K et al. (2015). Bacteria-bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J 9: 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, Gomez E et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318: 1737–1742. [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O, Cai R, Poloczanska ES, Brewer PG, Sundby S, Hilmi K et al. (2014). The ocean. In: Barros VR, Field CB, Dokken DJ, Mastrandrea MD, Mach KJ, Bilir TE, et al. (eds). Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press: Cambridge, UK and New York, NY, USA, pp 1655–1731.
- Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA et al. (2007). A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Kerry JT, Álvarez-Noriega M, Álvarez-Romero JG, Anderson KD, Baird AH et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543: 373–377. [DOI] [PubMed] [Google Scholar]
- Hume BC, D'Angelo C, Smith EG, Stevens JR, Burt J, Wiedenmann J. (2015). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world's hottest sea, the Persian/Arabian Gulf. Sci Rep 5: 8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume BCC, Voolstra CR, Arif C, D’Angelo C, Burt JA, Eyal G et al. (2016). Ancestral genetic diversity associated with the rapid spread of stress-tolerant coral symbionts in response to Holocene climate change. Proc Natl Acad Sci USA 113: 4416–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives AR, Carpenter SR. (2007). Stability and diversity of ecosystems. Science 317: 58–62. [DOI] [PubMed] [Google Scholar]
- Jones AM, Berkelmans R, van Oppen MJ, Mieog JC, Sinclair W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc Biol Sci 275: 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A, Bienhold C, Chatzinotas A, Gallien L, Gobet A, Kurm V et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J 11: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh M. (2003). Foraging adaptation and the relationship between food-web complexity and stability. Science 299: 1388–1391. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Parkinson JE, Reimer JD. (2012). A genetics-based description of Symbiodinium minutumsp. nov. and S. psygmophilumsp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J Phycol 48: 1380–1391. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Wham DC, Pettay DT, Parkinson JE, Keshavmurthy S, Chen CA. (2014). Ecologically differentiated stress-tolerant endosymbionts in the dinoflagellate genus Symbiodinium (Dinophyceae) Clade D are different species. Phycologia 53: 305–319. [Google Scholar]
- Lancichinetti A, Radicchi F, Ramasco JJ, Fortunato S. (2011). Finding statistically significant communities in networks. PLoS One 6: e18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Jeong HJ, Jang SH, Lee SY, Kang NS, Lee KH et al. (2016). Most low-abundance 'background' Symbiodinium spp. are transitory and have minimal functional significance for symbiotic corals. Microb Ecol 71: 771–783. [DOI] [PubMed] [Google Scholar]
- Leitão RP, Zuanon J, Villéger S, Williams SE, Baraloto C, Fortunel C et al. (2016). Rare species contribute disproportionately to the functional structure of species assemblages. Proc R Soc B 283: 20160084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KS. (2000). The diversity-stability debate. Nature 405: 228–233. [DOI] [PubMed] [Google Scholar]
- Mieog JC, van Oppen MJH, Cantin NE, Stam WT, Olsen JL. (2007). Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26: 449–457. [Google Scholar]
- Mieog JC, VANO MJ, Berkelmans R, Stam WT, Olsen JL. (2009). Quantification of algal endosymbionts (Symbiodinium in coral tissue using real-time PCR. Mol Ecol Resour 9: 74–82. [DOI] [PubMed] [Google Scholar]
- Mouillot D, Bellwood DR, Baraloto C, Chave J, Galzin R, Harmelin-Vivien M et al. (2013). Rare species support vulnerable functions in high-diversity ecosystems. PLoS Biol 11: e1001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Baumgarten S, Michell CT, Baums IB, LaJeunesse TC, Voolstra CR. (2016). Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol Evol 8: 665–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós-Alió C. (2006). Marine microbial diversity: can it be determined? Trends Microbiol 14: 257–263. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. (2012). The rare bacterial biosphere. Ann Rev Mar Sci 4: 449–466. [DOI] [PubMed] [Google Scholar]
- Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC. (2015). Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci USA 112: 7513–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley KM, Davies SW, Kenkel CD, Willis BL, Matz MV, Bay LK. (2014). Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS One 9: e94297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaka-Kudla ML. (1997) Global biodiversity of coral reefs: a comparison with rainforests. In: Reaka-Kudla ML, Wilson DE (eds). Biodiversity II: Understanding and Protecting our Biological Resouces. Joseph Henry Press: Washington DC, USA, pp 83–108. [Google Scholar]
- Roberts CM, McClean CJ, Veron JEN, Hawkins JP, Allen GR, McAllister DE et al. (2002). Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 295: 1280–1284. [DOI] [PubMed] [Google Scholar]
- Saichev AI, Malevergne Y, Sornette D. (2009) Theory of Zipf's Law and Beyond, Vol. 632. Springer Science & Business Media: Berlin/Heidelberg, Germany. [Google Scholar]
- Sawall Y, Al-Sofyani A, Hohn S, Banguera-Hinestroza E, Voolstra CR, Wahl M. (2015). Extensive phenotypic plasticity of a Red Sea coral over a strong latitudinal temperature gradient suggests limited acclimatization potential to warming. Sci Rep 5: 8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DA, Trench RK. (1980). Genetic variation in Symbiodinium (=Gymnodiniummicroadriaticum Freudenthal, and specificity in its symbiosis with marine invertebrates. I. Isoenzyme and soluble protein patterns of axenic cultures of Symbiodinium microadriaticum. Proc R Soc Lond B Biol 207: 405–427. [Google Scholar]
- Silverstein RN, Cunning R, Baker AC. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob Change Biol 21: 236–249. [DOI] [PubMed] [Google Scholar]
- Simkin MV, Roychowdhury VP. (2011). Re-inventing Willis. Phys Rep 502: 1–35. [Google Scholar]
- Skopina MY, Vasileva AA, Pershina EV, Pinevich AV. (2016). Diversity at low abundance: the phenomenon of the rare bacterial biosphere. Microbiology 85: 272–282. [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored 'rare biosphere'. Proc Natl Acad Sci USA 103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggett DJ, Goyen S, Evenhuis C, Szabó M, Pettay DT, Warner ME et al. (2015). Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol 208: 370–381. [DOI] [PubMed] [Google Scholar]
- Thomas L, Kendrick GA, Kennington WJ, Richards ZT, Stat M. (2014). Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol Ecol 23: 3113–3126. [DOI] [PubMed] [Google Scholar]
- Thornhill DJ, LaJeunesse TC, Santos SR. (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation, pseudogenes, and PCR artifacts confound biodiversity estimates. Mol Ecol 16: 5326–5340. [DOI] [PubMed] [Google Scholar]
- Whitacre JM, Bender A. (2010). Networked buffering: a basic mechanism for distributed robustness in complex adaptive systems. Theor Biol Med Model 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SP, Fisher PL, van Oppen MJ, Davy SK. (2015). Intra-genomic variation in symbiotic dinoflagellates: recent divergence or recombination between lineages? BMC Evol Biol 15: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yule GU. (1925). A mathematical theory of evolution, based on the conclusions of Dr J. C. Willis, F.R.S. Philos Trans R Soc Lond B 213: 21–87. [Google Scholar]
- Ziegler M, Roder C, Büchel C, Voolstra CR. (2015). Niche acclimatization in Red Sea corals is dependent on flexibility of host-symbiont association. Mar Ecol Prog Ser 533: 149–161. [Google Scholar]
- Ziegler M, Arif C, Burt JA, Dobretsov S, Roder C, LaJeunesse TC et al. (2017). Biogeography and molecular diversity of coral symbionts in the genus Symbiodinium around the Arabian Peninsula. J Biogeogr 44: 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data used in this publication can be accessed in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA306572.