Abstract
Arbuscular mycorrhizal fungi (AMF; phylum Gomeromycota) associate with plants forming one of the most successful microbe–plant associations. The fungi promote plant diversity and have a potentially important role in global agriculture. Plant growth depends on both inter- and intra-specific variation in AMF. It was recently reported that an unusually large number of AMF taxa have an intercontinental distribution, suggesting long-distance gene flow for many AMF species, facilitated by either long-distance natural dispersal mechanisms or human-assisted dispersal. However, the intercontinental distribution of AMF species has been questioned because the use of very low-resolution markers may be unsuitable to detect genetic differences among geographically separated AMF, as seen with some other fungi. This has been untestable because of the lack of population genomic data, with high resolution, for any AMF taxa. Here we use phylogenetics and population genomics to test for intra-specific variation in Rhizophagus irregularis, an AMF species for which genome sequence information already exists. We used ddRAD sequencing to obtain thousands of markers distributed across the genomes of 81 R. irregularis isolates and related species. Based on 6 888 variable positions, we observed significant genetic divergence into four main genetic groups within R. irregularis, highlighting that previous studies have not captured underlying genetic variation. Despite considerable genetic divergence, surprisingly, the variation could not be explained by geographical origin, thus also supporting the hypothesis for at least one AMF species of widely dispersed AMF genotypes at an intercontinental scale. Such information is crucial for understanding AMF ecology, and how these fungi can be used in an environmentally safe way in distant locations.
Introduction
In 1934, Baas-Becking (1934) was the first to propose a worldwide distribution of microorganisms, saying that ‘Everything is everywhere, the environment selects’. A recent study of the global distribution of arbuscular mycorrhizal fungi (AMF), comprising the phylum Glomeromycota, suggests that the statement by Baas-Becking might be true for this phylum. Using virtual taxa (VT; Opik et al., 2010), discriminated by a 520 bp region of the 18 S (small sub-unit or SSU) rRNA gene, 93% of all AMF taxa were found to be present on more than one continent and 34% of species occurred on all continents except Antarctica (Davison et al., 2015). Global distribution of one-third of a phylum in eukaryotes is particularly unusual, especially in organisms with a low dispersal rates (Tedersoo et al., 2014). Records of cosmopolitan species, excluding marine (Finlay, 2002) and invasive or pest species (Margaritopoulos et al., 2009), are rare. Birds have great dispersal capabilities, but even so, only six out of 10 000 species have a cosmopolitan distribution. Even those six species exhibit distinct genetic substructure (Monti et al., 2015).
Several findings in fungal population genetics suggest that, for some species, a cosmopolitan distribution reflects hidden endemism (Taylor et al., 2006). Some species first thought to be cosmopolitan are actually comprised of cryptic species with distinct geographical ranges such as the Basidiomycete Schizophyllum commune (James et al., 1999), Aspergilllus fumigatus and six other cited Ascomycete species (Pringle et al., 2005).
Morton (1990) hypothesized that AMF speciation occurred prior to the breakup of Pangea and that fungal lineages co-evolved with the emergence of plant species thereafter, thus, resulting in global distribution. Alternatively, Davison et al. (2015) favoured a hypothesis based on a more recent VT-molecular clock phylogeny that such widespread distribution was the result of a recent dispersal of spores, mediated by strong storms, as well as previously underestimated human activities. In either case, interpretations may be misleading if there is an underestimation of true species and an overestimation of species with an intercontinental distribution. The use of such short SSU sequences and a 97% similarity threshold to construct VT taxa (Davison et al., 2015) might not be informative enough to resolve significant AMF clades (Bruns and Taylor, 2016; Schlaeppi et al., 2016). Their argument is based on the premise that two taxa that share a 97% similarity in their SSUs may have diverged genetically many millions of years ago into different species. Davison et al. (2015) justified the use of VT because they are ‘phylogenetically defined sequence groups that exhibit a taxonomic resolution similar to that of morphological species’. However, the absence of clear morphological distinctions and indistinguishable life history traits between Rhizophagus intraradices and Rhizophagus irregularis, together with highly supported divergence based on rDNA sequences (Stockinger et al., 2009), provides compelling evidence of cryptic speciation in the Rhizophagus clade. Similarly, other species may be genetically divergent but are morphologically similar (Rosendahl et al., 2009). This is more likely to occur in small eukaryotes with limited phenotypic space for evolution of new traits but with a less constrained capacity for genotypic evolution (Taylor et al., 2000). Discovery of these cryptic taxa requires the sampling of globally distributed AMF taxa and the application of population genomics tools because a large number of markers distributed across the genome would give a finer resolution suitable to detect genetic differences among populations (Bruns and Taylor, 2016; Opik et al., 2016).
A high throughput sequencing technique, double digested restriction-site associated DNA sequencing (ddRAD-seq) is a reliable way of obtaining a large number of genome-wide markers (Parchman et al., 2012; Peterson et al., 2012). This overcomes bias created by use of single gene sequences and low similarity thresholds (for example, the SSU rRNA gene). This approach was previously applied to R. irregularis, an AMF species that has a putative global distribution and can be cultured under in vitro conditions. In vitro culturing allows the extraction of DNA that is free of contaminant microorganisms or plant DNA. This technique was previously shown to be reliable for Fusarium (Talas and McDonald, 2015) and in a study measuring inter- and intra- isolate variation in a population of R. irregularis (Wyss et al., 2016). This multi-locus approach can provide a much finer resolution of the divergence between different R. irregularis isolates, which in turn can provide data to resolve whether this species has evolved under strong or relaxed geographical constraints.
Describing the genetic diversity of AMF species, and especially the genetic diversity of R. irregularis at a wide geographical scale, is fundamental to understanding biogeographic patterns. However, it is not only important for resolving biogeographic questions but also has strong implications for interpretation of ecological, agronomic, and environmental studies. First, AMF are important symbiotic partners that interact with many plant species. Approximately 300–1600 predicted AMF taxa form symbioses with over 200 000 plant species (van der Heijden et al., 2015). The fungi have the capacity to efficiently absorb nutrients, particularly phosphate, from the soil and give them to the plant (Smith and Read, 2008) and richness of AMF taxa promotes plant diversity (van der Heijden et al., 1998). Most ecological studies have not considered the role of intra-specific AMF variation even though intra-specific differences in AMF can have larger effects on P uptake and variation in plant growth than inter-specific differences (Munkvold et al., 2004; Mensah et al., 2015; Rodriguez and Sanders, 2015). Thus, measurements of genetic variation within AMF species could be highly ecologically relevant. Second, genetic variability in R. irregularis population causes significant variation in plant growth (Koch et al., 2006). The potential of using naturally existing genetic variation in this species to develop more efficient strains to increase plant growth has been demonstrated by Angelard et al. (2010) where five-fold differences in rice growth could be achieved due to genetic variation in R. irregularis strains. Defining the total genetic variation in this AMF species is important for future programmes using the genetic variation in this fungus for more efficient inoculants (Sanders, 2010). Third, in vitro grown R. irregularis significantly increases yields of the globally important crop cassava (Ceballos et al., 2013). R. irregularis is potentially a good candidate species for large-scale inoculation of tropical crops because it can be mass-produced in contaminant-free in vitro conditions and because it appears to have a global distribution. However, before introducing high growth-promoting R. irregularis isolates from one location to another, it is important to establish with population genomics techniques the risk of introducing genetically novel isolates into a new environment with the potential to become invasive. If R. irregularis has a very wide geographical distribution of very genetically similar isolates then the risk of introducing exotic genetic material is low. If there is strong geographically determined genetic structure among R. irregularis populations then such a risk is much higher (Rodriguez and Sanders, 2015).
In this study, 61 isolates identified morphologically and genetically as R. irregularis were obtained from various locations across Europe, North Africa, Middle East and North America and were propagated in vitro and sequenced with ddRAD-seq. To the 61 isolates, data from 20 other isolates, that had been sequenced previously with the same ddRAD-seq protocol by Wyss et al. (2016), were added. We addressed three central questions: (i) Is the low level of endemism reported by Davison et al. (2015) applicable to R. irregularis and related species when using high resolution genome-wide markers that are capable of showing within population genetic variation? (ii) Is there significant genetic diversification among continents? (iii) Is there evidence that some quantitative traits vary in accordance with the genetic variation observed in this fungal species?
Materials and methods
Fungal isolates and culturing
R. irregularis (known previously as Glomus intraradices, Glomus irregulare or more recently as Rhizoglomus irregulare) were obtained from collections and biobanks such as BEG (http://www.i-beg.eu), GINCO (http://www.mycorrhiza.be/ginco-bel/) and INVAM (https://invam.wvu.edu/), from small enterprises: Symbiom (Czech Republic; https://www.symbiom.cz/en/ and INOQ (Germany), and from research groups (IRTA: http://www.irta.cat, TERI: http://www.teriin.org, or were present in our group at the University of Lausanne. For more details on nomenclature, see Supplementary Note 1. Sixty-one isolates were collected (Supplementary Table 1). The collection included the R. irregularis holotype, DAOM197198, cultured in vitro in two laboratories (Switzerland: DAOM197198-CH and Czech Republic: DAOM197198-CZ), the holotype of R. intraradices, FL208 grown in vitro from pot culture material provided by INVAM and R. proliferus that was used as an outgroup for some analyses. All isolates in this study were cultured in vitro with Ri T-DNA transformed carrot roots in a two-compartment culture system (St-Arnaud et al., 1996) to obtain fungal spores free from host plant DNA or other microorganisms. Some isolates were received as soil samples and were transferred to in vitro single spore cultures (Supporting Information, Supplementary Note 2).
DNA extraction, amplification and sequencing of the phosphate transporter and the SSU
Three-month-old sporulating in vitro cultures were used for DNA extraction. The medium in compartments containing only spores and hyphae was dissolved in 500 ml of citrate buffer (0.0062 m of citric acid anhydrous and 0.0028 m of sodium citrate tribasic dihydrate) for 1 h with a magnetic stirrer. Spores and hyphae were collected by filtering in citrate buffer through a sieve with 32 μm openings and rinsed with ddH20. Spores and hyphae were frozen in liquid nitrogen and ground with a sterile pestle. DNA was extracted with the DNeasy Plant Mini Kit (QIAGEN Inc., Venlo, Netherlands) following the manufacturer’s instructions. Species identity of each isolate was verified by sequencing the Phosphate Transporter Gene (PTG), which is considered a phylogenetically informative marker by Sokolski et al. (2011).
The primers AML2 and NS31, previously used by Davison et al. (2015), were used to amplify the 18 S (SSU) rRNA gene of all isolates. This was done to observe whether this marker could reveal genetic differences among the isolates used in this study and to compare those differences with the genetic variation revealed with ddRAD-seq data. The amplified PCR product was cloned with a StrataClone PCR Cloning Kit (Agilent Technologies, Santa Clara, CA, USA) following manufacturer protocols and one to two clones were sequenced. For more information of PTG and SSU amplification, sequencing and sequence cleaning see Supplementary Note 3.
ddRAD sequencing
Double-digest restriction-site associated DNA sequencing was carried out on DNA of the 61 isolates with three to five independent biological replicates per isolate (Wyss et al., 2016; Supplementary Note 4). The sampling design is given in Supplementary Note 5. The ddRAD-seq protocol was performed on 233 DNA samples. Between 1 and 25 ng μl−1 of genomic DNA was digested with the EcoRI and MseI enzymes (NEB). Between 36 and 90 samples were pooled into each of four libraries with independent barcode for each sample. Library quality was verified on a Fragment Analyser (Advanced Analytical Technologies, Inc., Ankeny, IA, USA). Each library was then sequenced at the Lausanne Genomics Technologies Facility (GTF) on an Illumina HiSeq sequencer in one lane to give 100 bp paired-end reads.
In silico workflow
An additional ddRAD-seq data set containing 20 isolates of R. irregularis, with 3 biological replicates of each isolate, was included in the analyses (Wyss et al., 2016), thus increasing the total number of isolates analysed to 81 (and the number of samples analysed to 299). Read quality was controlled with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The computations were performed at the Vital-IT (http://www.vital-it.ch) centre for high-performance computing of the Swiss Institute of Bioinformatics (SIB). After Illumina adaptor and low quality read trimming and demultiplexing (Supplementary Note 6), the reads were mapped to the DAOM197198 R. irregularis genome assembly N6 with the Novoalign software (Novocraft-Technologies, 2014). We chose this assembly, because it was the most complete single nucleus genome assembly of this fungus to date (Lin et al., 2014). FreeBayes v1.0.2 (Garrison and Marth, 2012) software was used to call single-nucleotide polymorphisms (SNPs), insertions and deletions (indels) and multiple nucleotide polymorphisms (MNPs). The –p option was set up to 10, assuming a potential number of alleles up to 10 and a minimum frequency of each allele of 10%. VCFfilter from the VCFLib library (Garrison and Marth, 2012) was used to call positions with a phred quality >30. Markers falling in coding and non-coding regions were defined based on a prediction of coding regions with GeneMark-ES (Ter-hovhannisyan et al., 2008; Wyss et al., 2016). Positions identified as repeated elements in the genome, using RepeatModeler Open-1-0 (Smit and Hubley, 2008) and RepeatMasker Open-3.0 (Smit et al., 1996), were removed from subsequent analyses (Wyss et al., 2016). An in silico double digestion EcoRI-MseI in the original genome allowed identification of predicted fragments and all fragments that potentially mapped two or more times to the DAOM197198 genome. We did not consider samples with less than 450 000 reads after demultiplexing, and samples with less than 300 000 reads uniquely mapping to the N6 assembly for further analyses. This decreased the number of samples to 262 (Supplementary Table 2). The four libraries, RAD7, RAD8, RAD10B and RAD12 are available at NCBI (bioproject accession number PRJNA357003).
SNP data sets
Three data sets were generated and analysed. All the positions included in these data sets were shared among all isolates, so that no missing data was present for any isolate or replicate. These positions were located in coding and non-coding regions and contained at least 10 reads of coverage. The first data set included all 262 samples and included 498 mono-allelic and poly-allelic positions in total. This data set allowed us to conduct analyses to study genetic variation among all isolates and all species included in this study. A second data set included only one replicate of each isolate, where the replicate chosen to represent each isolate was the one with the highest number of uniquely mapped reads and the highest number of loci covered. This allowed us to conduct more precise analysis on the genetic differences among all isolates using a larger number of polymorphic sites than that available in data set 1. Using this selection, a total of 2491 mono-allelic positions were concatenated for 68 isolates. In this data set, one isolate out of the 14 identified using the PTG sequences as R. proliferus was used as an outgroup. A third data set comprised a single replicate of each isolate identified in the PTG tree as R. irregularis. By considering polymorphic positions that were only shared among all isolates identified as R. irregularis, a database with a larger number of polymorphic positions could be generated, allowing us to conduct analyses with a high resolution to identify and quantify genetic differences within the species R. irregularis. In this data set, 6888 mono-allelic positions were concatenated. A summary of the three data sets is found in Table 1 and information about the samples used in each of the three data sets is found in Supplementary Table 2.
Table 1. The three data sets used for the phylogenetic analysis and population structure analysis.
| No. of samples | No. of isolates | No. of sites | Taxa included in data set | |
|---|---|---|---|---|
| Data set 1 | 265 | 81 | 489 | R. intraradices, R. irregularis, Rhizophagus sp. LPA8-CH3, R. proliferus |
| Data set 2 | 68 | 68 | 2491 | R. intraradices, R. irregularis, Rhizophagus sp. LPA8-CH3, R. proliferus (as outgroup) |
| Data set 3 | 59 | 59 | 6888 | R. irregularis |
Phylogenetic analyses and 18 S rRNA clustering
Phylogenetic analysis of the PTG is described in Supplementary Note 3. After sequence cleaning, SSU cloned sequences of each isolate were clustered at 97, 98 and 99% similarity thresholds with the script pick_otus and the uclust clustering algorithm in the QIIME environment (Caporaso et al., 2010; Edgar, 2010). Each clone was also blasted against the MaarjAM database (Opik et al., 2010) and the best VT hit was retained. The sequences used for the PTG phylogeny comprised 71 from this study and 25 from Sokolski et al. (2011). All PTG sequences used to reconstruct this phylogeny have been deposited in NCBI GenBank as accession numbers KY348541—KY348610 and KY436236. The SSU sequences were deposited in the NCBI GenBank with accession numbers KY436237-KY436352.
Scalar distances were calculated according to the equation 2 of Wyss et al. (2016). These distances were computed on the first ddRAD-seq dataset and were analysed with the package ape, version 3.5 (Paradis et al., 2004) in the R software 3.3.2. A dendrogram was built using the function hclust and plot.phylo (Wyss et al., 2016). A bootstrapping method described by Wyss et al. (2016), was applied to calculate support values. The second and third data sets were analysed with Mr Bayes (Huelsenbeck and Ronquist, 2001), implementing Markov chain Monte Carlo (MCMC) probability analysis. A mixed model was set up with the command lset nst=mixed rates=gamma. The MCMC chains were run for 1 000 000 generations with the reversible jump MCMC (RJ-MCMC) procedure, avoiding the selection of only one model of substitution rate. The chains were run until the standard deviation (s.d.) of split frequencies reached 0.01. All PSRF+ values reached a value of 1.
Population structure, species and clade delimitation
A constrained correspondence analysis (CCA) built with the function cca from the package vegan, version 2.4-1 (Dixon, 2003) was applied to the first data set and based on a matrix of scalar distances among the 262 samples and the 498 positions. We did this to examine how all the different isolates used in this study would cluster.
BP&P (Yang and Rannala, 2010, version 3.3) and STRUCTURE (Pritchard et al., 2000, version), two programmes based on the Bayesian algorithm, were used in order to define species and genotype boundaries. First, the 2491 positions of the second data set were concatenated and tested under the multispecies coalescent (MSC) model (Yang, 2002; Rannala and Yang, 2003). Using this model, species were delimited using a user-specified guide tree (Yang and Rannala, 2010; Rannala and Yang, 2013). The BPP software only evaluates the models that can be generated by collapsing nodes on the guide tree using the reversible jumps algorithm. We have added this text into the manuscript. The specific guide tree used is based on the phylogeny generated in Mr Bayes, as follows:
(R. proliferus, (R. intraradices, (Rhizophagus sp. LPA8-CH3, ((R. irregularis Gp3, R. irregularis Gp4), ((R. irregularis Gp1A, R. irregularis Gp1B), R. irregularis Gp2))))),
where Rhizophagus sp. LPA8-CH3 formed a distinct sister clade to R. irregularis comprising two isolates, R. intraradices is a known species, and R. irregularis groups Gp1A, Gp1B, Gp2, Gp3, Gp4 are distinct genetic groups within the phylogenetic analysis. Three models were run with varying ancestral population size (θ) and root age (τ0) following the protocol of Leaché and Fujita (2010) (Supplementary Note 7).
Population structure in the third data set was analysed using an admixture ancestry model with the programme STRUCTURE, without prior knowledge of location and with correlated allele frequencies between populations. STRUCTURE (Pritchard et al., 2000) is a programme suitable for defining species groups (K) even when the Hardy-Weinberg equilibrium is not respected and is also robust for analysis of population structure with data originating from organisms with low recombination rates (Falush et al., 2003, 2007). The MCMC chains were run for 100 000 generations, with 10 000 generations of burn-in. Each run from K1 to K7 was replicated 10 times. The delta K (Evanno et al., 2005) was computed using STRUCTURE HARVESTER (Earl and vonHoldt, 2012) in order to define the most likely number of K present in the sample. In order to concatenate all the replicates for each K, CLUMPP was used (Jakobsson and Rosenberg, 2007). Graphical output of cluster assignment as barplots and pie charts for mapping each isolate to their geographical origin was performed in R.
Finally, an analysis of molecular variance (AMOVA) was performed on the data using Continent as a factor with the highest number of isolates (that is, Europe and North America), with 1000 permutations, based on scalar genetic distances (pegas package version 0.9, Paradis, 2010). A Mantel test with the package ape (Paradis et al., 2004, version 3.5) and with 1000 permutation was also performed to test for isolation by distance, by correlating a geographical distance matrix and the matrix of phylogenetic scalar distance based on data set 3 derived from ddRAD-seq.
A previous study (Croll et al., 2008) analysed the genetic structure among 29 R. irregularis isolates with a reduced set of markers comprising ten microsatellites, two mitochondrial LSU gene introns and one nuclear gene intron. Those 29 isolates were also present in this study, allowing a comparison of the results. We produced a matrix of phylogenetic scalar distance, based on the ddRAD-seq data set, but this time with the 29 isolates. This matrix was then compared with a Mantel test to a matrix of phylogenetic Jaccard distance based on 13 markers (Croll et al., 2008). This analysis combined with the other analyses indicated that the ddRAD-seq data were robust (Supplementary Note 8).
Measurement of hyphal density
Previous studies have shown differences in hyphal density among some of the isolates used in this study. We wanted to see whether hyphal density patterns of R. irregularis varied in accordance with the genetic differences observed among R. irregularis isolates. After sequencing and analyzing all ddRAD-seq of each isolate, nine isolates spread across the major branches of the phylogeny were cultured for 3 months in vitro with five replicates. There were only three replicates of one of the isolates, DAOM197198-CZ. Following 3 months of growth, each plate was photographed using a camera attached to a stereomicroscope in order estimate hyphal density (Supplementary Note 9).
Results
Phylogeny based on phosphate transporter gene and SSU gene clustering
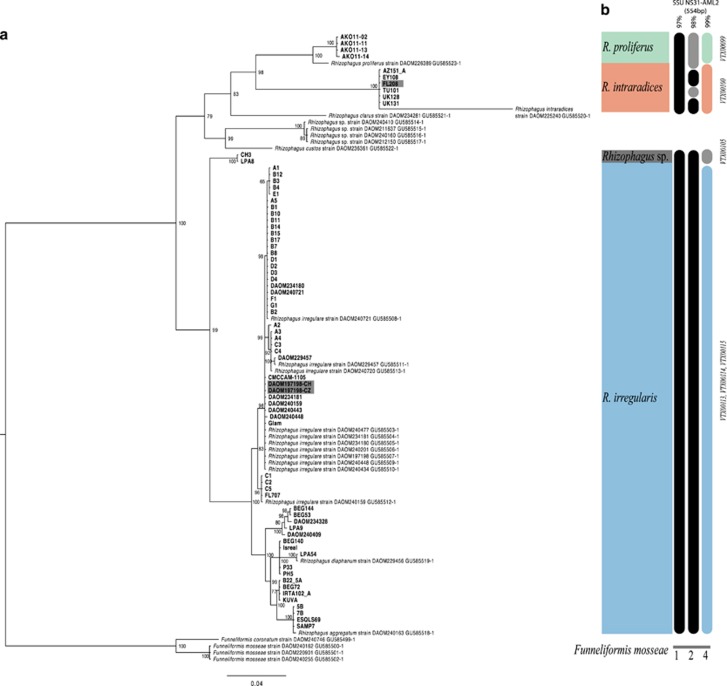
Sequences of the PTG ranged between 800 and 1191 bp in length, with 282 SNPs. Previously published sequences (Sokolski et al., 2011) and sequences of the reference isolates FL208 and DAOM197198, combined with the other PTG sequences from this study, resolved three Rhizophagus species; R. proliferus, R. intraradices and R. irregularis (Figure 1a). Twenty-two different haplotypes were identified with a nucleotide diversity (PiT) of 0.0806. In the R. irregularis clade, 13 haplotypes were detected with a nucleotide diversity of 0.0158 and 49 segregating sites. As expected, sequences of the reference strain DAOM197198 clustered within the R. irregularis clade. A highly supported sister clade to R. irregularis was designated as a distinct putative species Rhizophagus sp. LPA8-CH3 based on the two isolates comprising this group. The isolate FL208, considered as the holotype of R. intraradices (Stockinger et al., 2009), clustered with five other isolates and with a previously published sequence of R. intraradices.
Figure 1.
Phylogeny constructed using phosphate transporter gene (PTG) sequences (a) and schematic clustering of the SSU (b). Additional sequences from Sokolski et al. (2011). Sequences of Funneliformis mosseae were used to root the tree. Holotypes of R. intraradices (FL208) and R. irregularis (DAOM197198) shown in dark grey. R. proliferus (green), R. intraradices (red), R. irregularis (blue) and Rhizophagus sp. LPA8-CH3 (dark grey). Numbers at nodes represent bootstrap support for consensus tree bipartitions. SSU sequences for all isolates clustered at 97, 98 and 99% and corresponding virtual taxa number (VT) of the first hit after blast on MaarjAM database.
Sequences of the same SSU region studied by Davison et al. (2015) of 554 bp, revealed that at the 97% similarity threshold the different Rhizophagus species used in this study could not be discriminated from each other (Figure 1b; Supplementary Table 3). Only at the 99% similarity threshold, could the 3 species of Rhizophagus plus the putative species be distinguished from each other (Figure 1b). At the 99% similarity threshold, R. proliferus, R. intraradices and the putative Rhizophagus sp. LPA8-CH3, each occurred in one separate VT (as seen in the MaarjAM database). R. irregularis isolates occurred in three different VT.
Analysis of ddRAD-seq data
Each of the four ddRAD-seq libraries generated more than 300 million reads (Supplementary Table 4). Barcodes to de-multiplex the samples in each library are listed in Supplementary Table 2.
Sequencing details for each replicate of each isolate are reported in Supplementary Table 2. The ddRAD sequencing on DNA from 81 isolates resulted in 509 563 positions covered with a mean of 12 282 SNPs and 1936 indels per isolate when compared with the reference genome assembly. Not all of these positions had sufficient depth of coverage across all isolates in all samples. Among R. intraradices isolates, the number of positions in non-repeated coding and non-coding regions, covered by at least 10 reads, ranged from 23 668 in UK131-1 to 80 460 in UK128-1. Among R. proliferus isolates, the number of positions ranged from 60 795 in AKO11-11-2 to 77 186 in AKO11-13-1. Among R. irregularis isolates, positions ranged from 73 030 in ESQLS69-1 to 361 950 in DAOM240448-5.
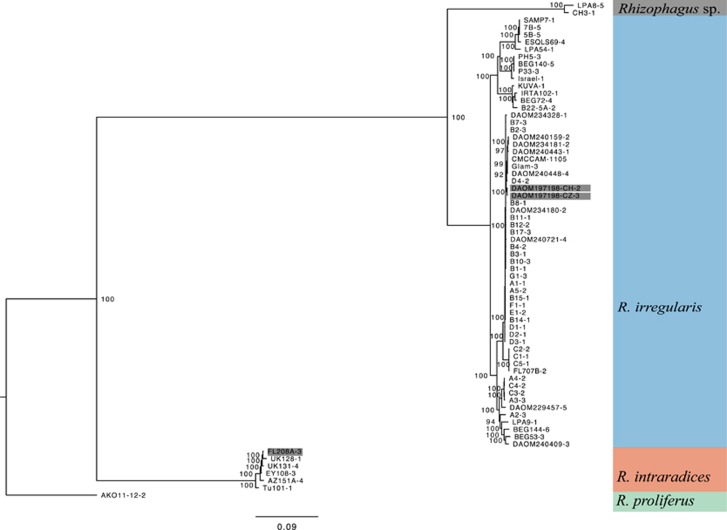
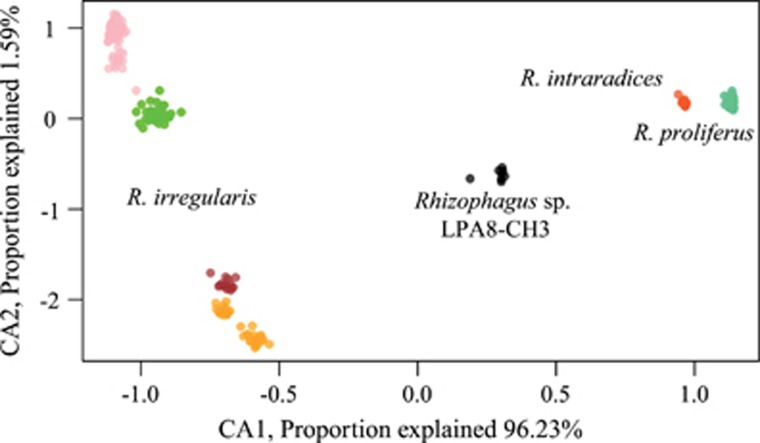
Based on the phylogenies of both ddRAD-seq data sets 1 and 2, and a CCA built with the first data set (Supplementary Figure 1; Figures 2 and 3), similar clade structure to that found with the PTG phylogeny was obtained: R. proliferus (n=14), R. intraradices (n=6) and R. irregularis (n=59). These data provided additional support for recognising Rhizophagus sp. LPA8-CH3 (n=2) as being genetically distinct from R. irregularis and R. intraradices. Four mains branches within the R. irregularis tree as well as four main groups of isolates within R. irregularis were clearly recognised in the CCA.
Figure 2.
Phylogenetic tree based on a concatenation of 2491 SNPs across the genome (data set 2) of three species of Rhizophagus and Rhizophagus sp. LPA8-CH3 and 68 isolates. Only one replicate per isolate was used. Names are composed of the isolate name followed by the replicate number (short name in Supplementary Table 2). Numbers at nodes represent bootstrap support for consensus tree bipartitions. Colour coding follows that of Figure 1.
Figure 3.
Constrained correspondence analysis (CCA) built on a scalar distance matrix computed among all samples and replicates (n=262) and based on 489 shared mono-allelic or poly-allelic positions. The four AMF species are well separated along the X-axis. Colour coding follows that of previous figures. In this figure the colour for R. irregularis is split into four main groups (pink, green, brown, orange).
Phylogeography and population genetic structure
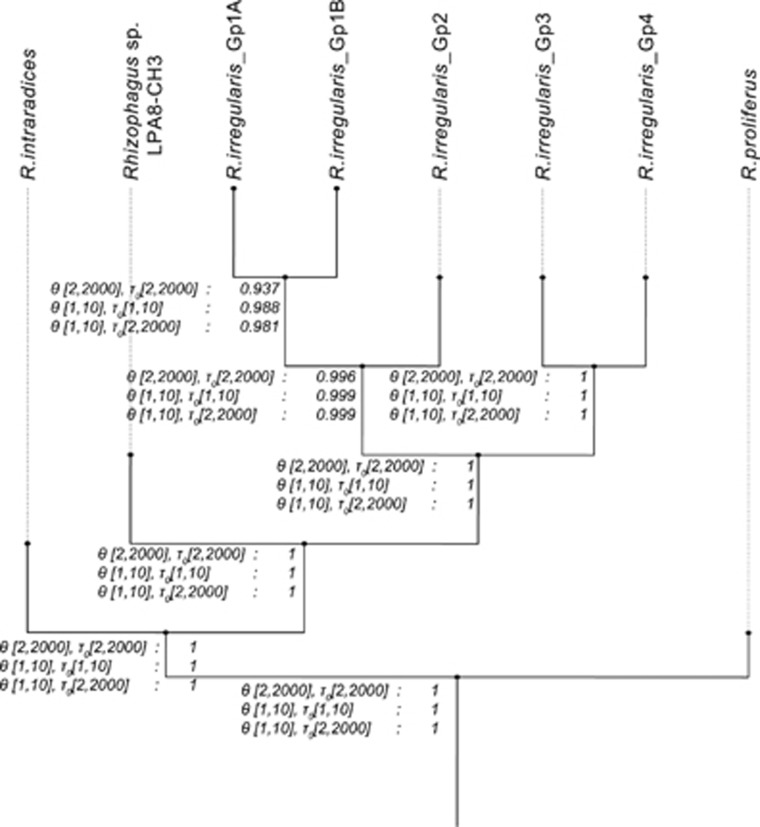
Rhizophagus irregularis and R. intraradices isolates co-occurred in Europe, the Middle East, North Africa and Northern America (Supplementary Figure 2). We analysed the speciation probability among Rhizophagus species as well as within R. irregularis. The MSC model implemented with data set 2 using BP&P showed that in three models, the three distinct species were highly supported: R. irregularis, R. intraradices and R. proliferus and one putative species: Rhizophagus sp. LPA8-CH3 (Figure 4). A probability of >0.95 would normally be considered likely to represent a speciation event. In this analysis we also found significant divergence between groups Gp3 and Gp4 and between groups Gp1 and Gp2 within R. irregularis. Two of the three tests exceeded the >0.95 probability threshold for genetic differentiation between Gp1A and Gp1B.
Figure 4.
Bayesian species delimitation tree (based on data set 2). Marginal probabilities of speciation for three models with variable population size (θ) and divergence time (τ0) are presented at each node.
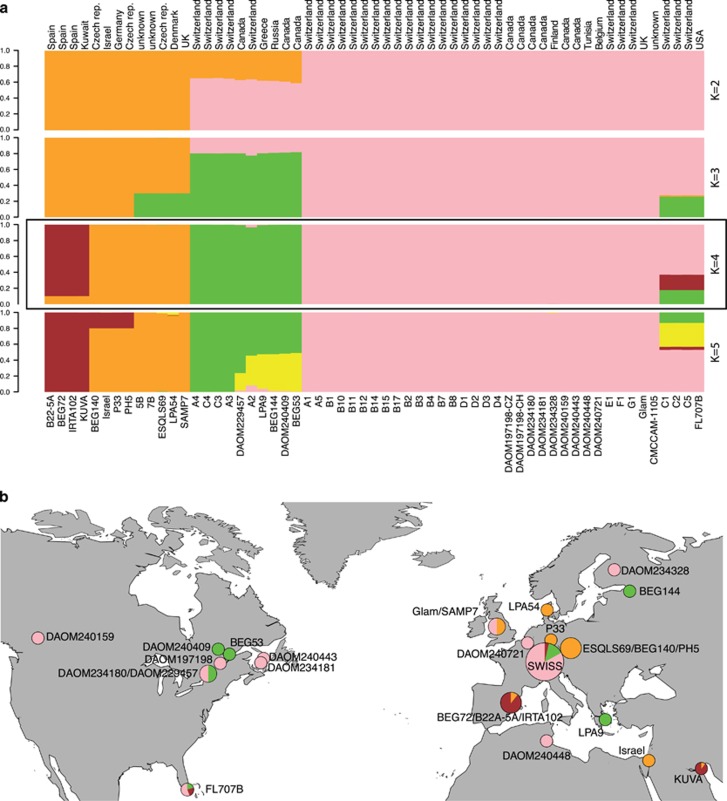
The analysis using STRUCTURE and the Delta K on data set 3 (59 R. irregularis isolates) suggested the possibility of either two or four divergent clades within the sampled R. irregularis isolates (Figure 5a), with Delta K values supporting a slightly higher probability of four groups than two (Supplementary Figure 3). BP&P and STRUCTURE analyses collectively suggested that the R. irregularis isolates were divided into at least four well-defined genetic groups.
Figure 5.
Assignment to 4 genetic groups with STRUCTURE and distribution map of genetic groups. (a) Vertical bars represent one R. irregularis isolate of and colours represent the assignment to different genetic groups (K). The delta K value for K=4 was slightly higher than for K=2. (b) Colour coding corresponds to results obtained with STRUCTURE, based on data set 3. Several isolates are of unknown geographical origin and are not presented here. The name of each isolate is written near its respective pie chart, except for the 29 Swiss isolates that are represented as the Swiss population.
The distribution of the divergent genetic groups of R. irregularis did not follow a geographical pattern (Figure 5b). Indeed, the Mantel test used to test for isolation by distance and an AMOVA using genetic distance and Continents as factor (America=9, Europe=37) did not reveal significant genetic differences (AMOVA: df=1, P=0.395) between the populations of the two different continents or any significant isolation by distance (z-statistic: 112835, P-value: 0.518). Moreover, isolates of each of the four genetic groups could be found in highly distant locations. For example, R. irregularis group Gp4 occurred in several geographically distant places in north America (Florida and Canada), in Europe from Switzerland to Finland and in North Africa (Tunisia). Similar patterns were evident in the other genetic groups. Two strains (DAOM234181 and DAOM240159) appeared to be the same clonal haplotype and originated from two locations separated by almost 4000 km in Canada. Isolates belonging to the two close genetic groups Gp3 and Gp4 coexisted in the same soil in Switzerland and both groups also co-occurred in geographically close sampling sites in Canada.
The population structure of 29 R. irregularis isolates measured using 13 markers (Croll et al., 2008) reflected that observed with the large number of SNPs identified in this study. The Jaccard distances among isolates measured with microsatellite data were significantly correlated with the scalar distance calculated on 2100 positions from ddRAD-seq data (Supplementary Figure 4, Mantel statistic r: 0.9624, P-value<0.001).
Extraradical hyphal density
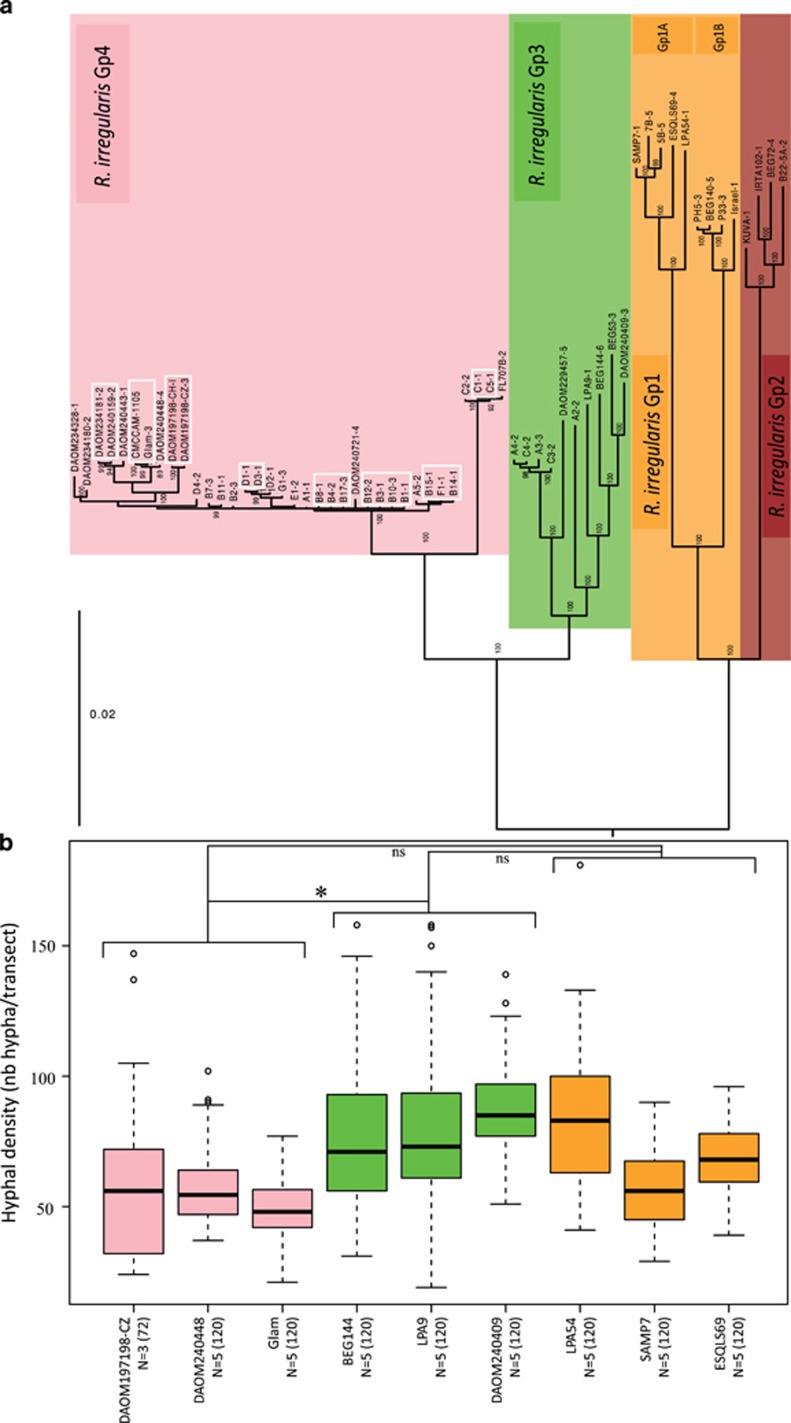
Data set 3 provided the finest resolution to discriminate between R. irregularis isolates and showed, based on 6888 SNPs, that genetic variation within each of the four main genetic groups also existed (Figure 6a). Significant differences in extraradical hyphal density among the nine chosen R. irregularis isolates, and among three genetic groups (Gp1, Gp3 and Gp4), was found (Figure 6b; lmer; Genetic groups: dF=2, F value=5.94, P=0.035*). Isolates in the genetic group Gp3 produced a significantly higher density of extraradical hyphae than those in Gp4. Other comparisons between genetic groups were non-significant (Gp3–4, P=0.01* Gp1–4, P=0.09; Gp1–3, P=0.18).
Figure 6.
Phylogeny of the four genetic groups and hyphal density of 9 R. irregularis isolates from three genetic groups. Colour coding follows that in previous figures and corresponds to the four genetic groups. (a) Phylogeny based on 6 888 concatenated SNPs across the R. irregularis genome constructed using data from 59 isolates (data set 3). The white squares represent clonal haplotypes. (b) Extraradical hyphal density of R. irregularis isolates from three genetic groups. Genetic group 2 (Gp2) was not included. On the x-axis, N represents the number of plates and the number in parentheses represents the number of transects where the hyphae were counted. The significance among the three genetic groups and obtained with the mixed model is indicated above the boxplots (ns: non-significant, *P-value<0.05).
Discussion
Describing the genetic diversity of AMF species, and especially that of the broadly distributed R. irregularis group, is fundamental to understanding their biogeographic patterns. Data confirmed that both R. irregularis and R. intraradices are widely distributed geographically across at least two continents (North America and Europe). Moreover, the four cryptic genomic forms within the species R. irregularis, identified by thousands of SNP markers obtained in this study, also have a broad geographic distribution. These data collectively support the hypothesis of Davison et al. (2015) that endemism is low in some Glomeromycota, not only at the 97% level of clustering resolution using VT and at the species level, but also amongst significantly diverged genomic forms within a species.
We confirmed with both the PTG and ddRAD-seq loci that the two reference isolates FL208 and DAOM197198 (holotypes of R. intraradices and R. irregularis, respectively) indeed belonged to two distinct clades and that a number of the other isolates in this study also clustered into these two clades. The four distinct genetic groups identified by variation in thousands of genome-wide SNPs among R. irregularis isolates were not fully resolved by the PTG and SSU phylogenies. This result stresses the need for higher resolution and larger number of markers for understanding AMF biogeography.
While the four genetically defined R. irregularis groups cannot be fully ranked at the species level in this study, the data indicate that, at most, a negligible amount of gene flow occurs among the groups, even though they coexist in the same soil. Thus, rather than referring to these as different cryptic species, we define the genetically different groups as cryptic genomics forms of the fungus, meaning that they are genetically distinct, but would likely not be distinguishable by morphological studies or using single gene markers. Anastomosis has been observed in the laboratory at very low frequency between isolates in groups Gp3 and Gp4 originating from the same location (Croll et al., 2009). However, if this were common in nature, then such defined genetic groups would not be expected in the data sets generated in this study because intermediates should occur.
The four genetic groups were found across large geographical distances. The most striking examples were genetic groups Gp3 and Gp4, which occur at several different localities in Europe, Northern Africa, Canada and the USA. Some isolates from the same genetic group occurred more than 8000 km apart. The four genetic groups described here were detected because of the high resolution obtained by the large number of ddRAD-seq markers. Thus, Bruns and Taylor (2016) were correct that low-resolution markers such as the one used by Davison et al. (2015) would have failed to detect such genetic groups and would have assigned them to only one VT. However, regardless of the level of resolution informed by the SNPs or the 18 S (SSU) rRNA, the same pattern of low endemism observed by Davison et al. (2015) was evident for R. irregularis. This suggests, as proposed by another study (Rosendahl et al., 2009), that at least the AMF species studied in detail so far, from the species level down to the intra-specific level, have a wide distribution attributable either to high dispersal via anthropogenic activities (Davison et al., 2015) or by slower dispersal over millions of years (Morton, 1990). Since many of the fungi in this study were isolated from agricultural fields, human agriculture may well be responsible for the dispersal of this species. We do not, however, consider it likely that the wide distribution of some of the groups is due to the application of exotic inoculum containing Rhizophagus irregularis because the more widespread use of such inocula is actually very recent and many of the donated cultures were isolated from the soil long before the use of commercial inocula.
The considerable genomic variability described here for a large set of isolates has not previously been reported for R. irregularis. Given that genetically different AMF isolates can cause large differences in plant growth (Munkvold et al., 2004; Koch et al., 2006), it would now be important to identify if variation in plant growth during the symbiosis with these different forms is greater among or within R. irregularis genetic groups. In this present study, we found evidence that the extraradical hyphal density, a phenotypic trait that is known to impact the phosphate acquisition capacity of the fungus and benefit to the plant (Jakobsen et al., 1992), differed significantly among genetic groups. Identifying whether the within species genetic and phenotypic variability in these fungi has consequences for plant ecology would be an important step to understand the link between fungal communities and plant communities as well as an important step for using AMF in agriculture for increasing crop yields.
The focus on the biogeography of R. irregularis in this study has broad agronomic implications. Genetically different R. irregularis applied to rice (Angelard et al., 2010) resulted in differential rice growth. Significant yield increases in cassava in the field have been achieved by applying in vitro produced R. irregularis (Ceballos et al., 2013). R. irregularis is a potentially strong candidate species for large-scale inoculation of tropical crops because it can be mass produced in contaminant-free in vitro conditions and because of its global distribution. The occurrence of R.irregularis genotypes distributed across diverse environments suggests broad adaptability to different soil types.
The introduction of non-native exotic strains with a strong geographically determined genetic structure among populations could be perceived as a risk for potential invasiveness (Rodriguez and Sanders, 2015; Schlaeppi et al., 2016). However, that concern is alleviated for R. irregularis, given that distribution of genetically similar isolates is widespread. For example, if isolates Gp3 and Gp4 from Switzerland were introduced in a field sites in Canada, the presence of these genotypes there negate any classification as exotic. We propose that a population genomic study of R. irregularis from tropical soils should be undertaken to verify if similar patterns occur.
Attention then would focus on the design of multi-isolate commercial inocula, where relatedness of isolates growing on the same plant strongly impacts plant biomass (Roger et al., 2013). The variation found in the 81 isolates of this study provides a bank of genetic diversity that can be utilised to test for crop compatibility and symbiotic efficiency and effectiveness. The variability described in this study has the advantage that each isolate is referenced and stored as in vitro pure culture and could be at any time mass-produced and used for breeding programmes (Rodriguez and Sanders, 2015), ecological experiments (Koch et al., 2006) and agronomic trials (Ceballos et al., 2013).
This study is, to our knowledge, the first to show that almost-clonal isolates of R. irregularis occurred in highly distant localities up to 4000 km apart. It also confirms the findings of Davison et al. (2015) of low endemism and similar genomic forms on multiple continents. At the same time, our results also show that, as Bruns and Taylor (2016) suggested, considerable genetic divergence can indeed be hidden by the use of low-resolution markers. The presence of different genetic groups of R. irregularis across the globe should be now investigated by isolating and genotyping isolates from agronomic and natural ecosystems from other continents such as Australia, Africa, Asia and South America.
Acknowledgments
We thank Aleš Látr and Miroslav Vosatka at Symbiom, Dirk Redecker, Amelia Campubri, Yolande Dalpé, Alok Adholeya and Louis Mercy for providing AMF isolates. We thank Jeremy Bonvin, Marie Strehler, Laura Alvarez Lorenzana, Jérôme Wassef, Jérôme Goudet, Lucas Villard and Nicolas Salamin for their help and three anonymous reviewers for their constructive comments. We thank Keith Harshman at the Lausanne University Genomic Technologies Facility. All bioinformatics computations were performed at the Vital-IT (http://www.vital-it.ch) Center for High Performance Computing of the Swiss Institute of Bioinformatics. This study was funded by the Swiss National Science Foundation (Grant number: 31003A_162549 to IRS).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Angelard C, Colard A, Niculita-Hirzel H, Croll D, Sanders IR. (2010). Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr Biol 20: 1216–1221. [DOI] [PubMed] [Google Scholar]
- Baas Becking LGM. (1934) Geobiologie of Inleiding Tot de Milieukunde. W.P. Van Stockum & Zoon (in Dutch): The Hague, The Netherlands. [Google Scholar]
- Bruns TD, Taylor JW. (2016). Comment on 'Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism'. Science 351: 826–b. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos I, Ruiz M, Fernandez C, Pena R, Rodriguez A, Sanders IR. (2013). The in vitro mass-produced model mycorrhizal fungus, Rhizophagus irregularis, significantly increases yields of the globally important food security crop cassava. Plos One 8e70633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Wille L, Gamper HA, Mathimaran N, Lammers PJ, Corradi N et al. (2008). Genetic diversity and host plant preferences revealed by simple sequence repeat and mitochondrial markers in a population of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 178: 672–687. [DOI] [PubMed] [Google Scholar]
- Croll D, Giovannetti M, Koch AM, Sbrana C, Ehinger M, Lammers PJ et al. (2009). Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 181: 924–937. [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Ouml, pik M, Adholeya A, Ainsaar L et al. (2015). Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- Dixon P. (2003). VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- Earl DA, Vonholdt BM. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359–361. [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. (2002). Global dispersal of free-living microbial eukaryote species. Science 296: 1061–1063. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907 [q-bio.GN].
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jakobsen I, Abbott LK, Robson AD. (1992). External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. 1. Spread of hyphae and phosphorus flow into roots. New Phytol 120: 371–380. [Google Scholar]
- Jakobsson M, Rosenberg NA. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- James TY, Porter D, Hamrick JL, Vilgalys R. (1999). Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. Evolution 53: 1665–1677. [DOI] [PubMed] [Google Scholar]
- Koch AM, Croll D, Sanders IR. (2006). Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol Lett 9: 103–110. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK. (2010). Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proc R Soc B Biol Sci 277: 3071–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Limpens E, Zhang ZH, Ivanov S, Saunders DGO, Mu DS et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. Plos Genet 10: e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritopoulos JT, Kasprowicz L, Malloch GL, Fenton B. (2009). Tracking the global dispersal of a cosmopolitan insect pest, the peach potato aphid. BMC Ecol 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensah JA, Koch AM, Antunes PM, Kiers ET, Hart M, Bucking H. (2015). High functional diversity within species of arbuscular mycorrhizal fungi is associated with differences in phosphate and nitrogen uptake and fungal phosphate metabolism. Mycorrhiza 25: 533–546. [DOI] [PubMed] [Google Scholar]
- Monti F, Duriez O, Arnal V, Dominici JM, Sforzi A, Fusani L et al. (2015). Being cosmopolitan: evolutionary history and phylogeography of a specialized raptor, the Osprey Pandion haliaetus. BMC Evolution Biol 15: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JB. (1990). Species and clones of arbuscular mycorrhizal fungi (Glomales, zygomycetes) - their role in macroevolutionary and microevolutionary processes. Mycotaxon 37: 493–515. [Google Scholar]
- Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I. (2004). High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol 164: 357–364. [DOI] [PubMed] [Google Scholar]
- Novocraft-Technologies.. (2014). Novoalign. Available from http://www.novocraft.com.
- Opik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM et al. (2010). The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188: 223–241. [DOI] [PubMed] [Google Scholar]
- Opik M, Davison J, Moora M, Partel M, Zobel M. (2016). Response to comment on 'Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism'. Science 351: 826–c. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Paradis E. (2010). pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26: 419–420. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle CA. (2012). Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol 21: 2991–3005. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. (2012). Double Digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. Plos One 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW. (2005). Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59: 1886–1899. [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang ZH. (2003). Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang ZH. (2013). Improved reversible jump algorithms for bayesian species delimitation. Genetics 194: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Sanders IR. (2015). The role of community and population ecology in applying mycorrhizal fungi for improved food security. ISME J 9: 1053–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger A, Colard A, Angelard C, Sanders IR. (2013). Relatedness among arbuscular mycorrhizal fungi drives plant growth and intraspecific fungal coexistence. ISME J 7: 2137–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl S, McGee P, Morton JB. (2009). Lack of global population genetic differentiation in the arbuscular mycorrhizal fungus Glomus mosseae suggests a recent range expansion which may have coincided with the spread of agriculture. Mol Ecol 18: 4316–4329. [DOI] [PubMed] [Google Scholar]
- Sanders IR. (2010). 'Designer' mycorrhizas? Using natural genetic variation in AM fungi to increase plant growth. ISME J 4: 1081–1083. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K, Bender SF, Mascher F, Russo G, Patrignani A, Camenzind T et al. (2016). High-resolution community profiling of arbuscular mycorrhizal fungi. New Phytol 212: 780–791. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R. (2008). RepeatModeler Open-1.0 http://www.repeatmasker.org.
- Smit AFA, Hubley R, Green P. (1996), RepeatMasker Open-3.0 http://www.repeatmasker.org.
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis 3rd ed. Elsevier Ltd.: London, UK. [Google Scholar]
- Sokolski S, Dalpe Y, Piche Y. (2011). Phosphate transporter genes as reliable gene markers for the identification and discrimination of arbuscular mycorrhizal fungi in the genus Glomus. Appl Environ Microbiol 77: 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA. (1996). Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res 100: 328–332. [Google Scholar]
- Stockinger H, Walker C, Schussler A. (2009). 'Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol 183: 1176–1187. [DOI] [PubMed] [Google Scholar]
- Talas F, McDonald BA. (2015). Genome-wide analysis of Fusarium graminearum field populations reveals hotspots of recombination. BMC Genomics 16: 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. (2006). Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc B Biol Sci 361: 1947–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R et al. (2014). Global diversity and geography of soil fungi. Science 346: 1078. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72. [Google Scholar]
- van der Heijden MGA, Martin FM, Selosse MA, Sanders IR. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205: 1406–1423. [DOI] [PubMed] [Google Scholar]
- Wyss T, Masclaux FG, Rosikiewicz P, Pagni M, Sanders IR. (2016). Population genomics reveals that within-fungus polymorphism is common and maintained in populations of the mycorrhizal fungus Rhizophagus irregularis. ISME J 10: 2514–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. (2002). Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162: 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH, Rannala B. (2010). Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci USA 107: 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.