Abstract
Oxygen-dependent microbial oxidation of sulfur compounds leads to the acidification of natural waters. How acidophiles and their acidic habitats evolved, however, is largely unknown. Using 16S rRNA gene abundance and composition data from 72 hot springs in Yellowstone National Park, Wyoming, we show that hyperacidic (pH<3.0) hydrothermal ecosystems are dominated by a limited number of archaeal lineages with an inferred ability to respire O2. Phylogenomic analyses of 584 existing archaeal genomes revealed that hyperacidophiles evolved independently multiple times within the Archaea, each coincident with the emergence of the ability to respire O2, and that these events likely occurred in the recent evolutionary past. Comparative genomic analyses indicated that archaeal thermoacidophiles from independent lineages are enriched in similar protein-coding genes, consistent with convergent evolution aided by horizontal gene transfer. Because the generation of acidic environments and their successful habitation characteristically require O2, these results suggest that thermoacidophilic Archaea and the acidity of their habitats co-evolved after the evolution of oxygenic photosynthesis. Moreover, it is likely that dissolved O2 concentrations in thermal waters likely did not reach levels capable of sustaining aerobic thermoacidophiles and their acidifying activity until ~0.8 Ga, when present day atmospheric levels were reached, a time period that is supported by our estimation of divergence times for archaeal thermoacidophilic clades.
Introduction
Hyperacidic environments (pH<3) are pervasive landscape features, albeit isolated, and are commonly associated with volcanic systems or sulfide ore deposits. These environments define the lower pH limit for life, are major sources of economically important metals and have the potential to contribute to the pollution of natural waters (Johnson and Quatrini, 2016). As such, hyperacidic environments are of substantial interest to microbiologists, geochemists, geologists and planetary scientists, and have been subject to extensive research to understand the processes that contribute to their formation and the adaptations that allow for their habitation. Considerable research has been conducted on the role of microbial activity in the formation of hyperacidic waters (Johnson et al., 1993; Baker and Banfield, 2003). However, the events that led to the evolution of acidophiles and their role in the generation of acidic habitats are underexplored.
Two dominant processes are involved in the formation of hyperacidic natural waters: magmatic degassing that contributes hydrochloric, hydrofluoric and sulfuric acid (from disproportionation of sulfur dioxide and subsequent oxidation of hydrogen sulfide or elemental sulfur) and the oxidation of mineral sulfides that produces sulfuric acid (Nordstrom et al., 2000). Common to both processes are the oxidation of sulfur compounds, including free sulfide (H2S), mineral sulfides (for example, FeS2) or elemental sulfur (S0), and the concomitant production of sulfate and protons. High-potential oxidants capable of driving sulfide/S0 oxidation include nitrate (NO3−), ferric iron ions (FeIII) and O2 (Falkowski et al., 2008). Mechanisms of generating NO3− include lightning-catalyzed oxidation of atmospheric dinitrogen and precipitation of nitrogen oxides as NO3−, a process that has been decreasing in importance since 3.8 Ga (Navarro-Gonzalez et al., 2001), and biological oxidation of ammonia, which requires O2 (Godfrey and Falkowski, 2009). Likewise, the generation of FeIII requires either abiological photo-oxidation of ferrous iron (FeII; Braterman et al., 1983), which has been of negligible importance after the onset of an ozonated atmosphere following the Great Oxidation Event (GOE) ~2.5 Ga (Kim et al., 2013), or one of several biological oxidative processes (Emerson et al., 2010). Biological oxidation of FeII requires O2 or NO3− as an oxidant (Emerson et al., 2010) or FeII-oxidizing phototrophic organisms (Widdel et al., 1993). The involvement of phototrophic organisms in FeIII generation in acidic environments is of putative importance only in environments with temperatures <56 °C—the upper temperature limit for phototrophs in acidic (pH<4.0) environments (Boyd et al., 2012). Thus, the remaining and most likely oxidant capable of driving H2S/S0 oxidation and the development of hyperacidic hydrothermal ecosystems is atmospheric O2 rather than local O2, given temperature constraints on photosynthesis.
Previous studies indicate that abiotic oxidation of H2S with O2 can be extremely slow in aqueous solutions with pH⩽6 (Chen and Morris, 1972; Zhang and Millero, 1993; D’Imperio et al., 2008). Further, at elevated pH (that is, in marine waters) where abiotic oxidation of sulfide is quicker, rates can be several orders of magnitude slower than biotic oxidation (Luther et al., 2011), although biotic oxidation is not always necessary to explain observed rates (Millero, 2001). Further, the rate of abiotic oxidation of S0 with O2 is extremely slow (Nordstrom et al., 2005). Finally, the growth of several sulfur-oxidizing, aerobic thermoacidophiles results in the acidification of the cultivation medium (Shivvers and Brock, 1973; Giaveno et al., 2013). These observations, coupled with evidence for the widespread distribution of aerobic organisms capable of oxidizing both H2S and S0 in acidic hot springs (Zillig et al., 1994; Inskeep et al., 2013; Xie et al., 2014; Huber and Stetter, 2015) and the capability of sulfur-oxidizing organisms to acidify their environments, strongly suggest that biological activity is involved in the generation of many low-pH (<3.0) hydrothermal ecosystems (Nordstrom et al., 2005; Nordstrom et al., 2009; Johnson and Quatrini, 2016).
Here, we hypothesize that O2-dependent biological oxidation of sulfur compounds in hydrothermal ecosystems is dependent on a non-point source of O2 and that the widespread development of low-pH (<3.0) hot spring ecosystems could only have occurred after O2 began to accumulate in the atmosphere to levels capable of supporting and sustaining aerobic biological activity ~0.8 Ga. As such, we further hypothesized that the microorganisms that dominate and are presumptively responsible for forming acidic hot springs belong to recently evolved lineages that incorporate aerobic metabolism to meet energetic demands associated with their acidophilic lifestyles. To begin to assess these hypotheses, we measured microbial community abundances and composition among geochemically diverse hot springs in Yellowstone National Park (YNP), Wyoming, USA, to determine the abundance of taxonomic lineages found in thermal acidic springs. We then investigated the evolutionary history of thermoacidophiles using phylogenomic analyses of publicly available genomes from cultured and uncultured taxa. To better understand the adaptations that led to increased acid tolerance, we used comparative genomics to determine the protein complements that differentiate thermoacidophiles from other members of their higher-order lineages. Finally, we present these results in context of changes in atmospheric O2 over geologic time to evaluate the hypothesis that both thermoacidophiles and the habitats that they are putatively responsible for generating are recent evolutionary and geologic innovations, respectively.
Materials and methods
Spring temperature and pH were determined with a portable pH meter and temperature-compensated probe (WTW 3300i or 3110; WTW, Weilheim, Germany) that was calibrated daily with standardized buffer solutions. Hot spring sediments were sampled from hot springs and DNA was extracted, purified and quantified as previously described (Colman et al., 2016). Quantitative PCR quantification and bacterial/archaeal 16S rRNA gene amplification and sequencing via the 454 Titanium platform were also performed as previously described (Hamilton et al., 2013). Raw untrimmed 454 sequence data are deposited in the NCBI SRA database under SRA accession SRR3181942. Metadata for aerobic respiration capability were inferred from the nearest cultivated isolate (or genomic reference) and are provided in Supplementary Table S1. Statistical analyses of quantitative PCR data and community compositional differences in relation to geochemical parameters were conducted in the base R package v. 3.2.0 (R Core Team, 2015).
Available archaeal genomes were downloaded from the NCBI genome portal and cross-referenced against the JGI Integrated Microbial Genomes database to ensure that representatives available in either database were collected (Supplementary Table S2). Genome completeness of each downloaded genome was estimated based on the presence of 104 single-copy archaeal-specific phylogenetic marker genes using the Amphora2 software package (Wu and Scott, 2012). Only genomes with >50% of the phylogenetic marker genes present in the assembly were included to reduce bias from low-coverage genomes (n=584; median completion=99% 0.05 percentile completeness=67%). Phylogenetic analysis was conducted using a concatenated alignment of the phylogenetic marker genes, with individual alignment of each of the individual 104 gene data sets using Clustal Omega v. 1.2 (Sievers et al., 2011). The concatenated alignment was subjected to Maximum Likelihood (ML) analysis using RAxML v. 8.2.4 (Stamatakis, 2014), specifying the LG protein substitution model (PROTCATLG) and evaluation of node support with 100 ML bootstraps. pH optima and the ability to respire O2 (facultative aerobe or obligate aerobe) for cultivars, where available, were collected from published cultivation data of isolates from the same species (Supplementary Table S3). For genomes representing uncultured species or species without pH optima descriptions, environmental pH (uncultured species) or culture conditions (cultivars) were collected from published reports or culture collections, where available (Supplementary Table S3). Where pH ranges were only available, the mean of the range of pH values tested for each cultivar was used.
Proteins for all genomes were clustered into protein bins using CD-HIT v. 4.6.5 (Fu et al., 2012) at the 30% homology level after first clustering into 90% homology level bins, followed by a second clustering at the 60% homology level. Protein bin presence/absence was then normalized for each genome to account for unequal protein bins/genome using the ‘normalize.rows’ function of the R package ‘vegetarian’ (Charney and Record, 2012). The normalized presence/absence table was then used to construct NMDS plots using the ‘vegdist’ function for distance matrix calculation and the metaMDS ordination function in the R package ‘vegan’ (Oksanen et al., 2015). The ‘indval’ function of the labdsv R package (Roberts, 2015) was used to identify ‘indicator’ protein bins that were highly enriched in the group of interest relative to other lineages in their respective superphyla. The non-normalized presence/absence matrix of protein cluster bins among genomes was used in the indicator analyses. ‘Indicator values’ represent a measure of fidelity and frequency within specified groups relative to others. Phylogenetic groups were determined based on the placement of genomes within the marker-gene phylogeny and were referenced against published phylogenies. The nomenclature of the DPANN superphylum was used as in Castelle et al. (2015). Enriched protein cluster bins for Thermoplasmatales were identified as those with >0.87 indicator value, which allowed for enriched bins to be missing in 13% (1/8 total) of the Thermoplasmatales genomes. Enriched protein cluster bins for the Sulfolobales were only considered with >0.90 indicator values. Indicator analyses were conducted for the Thermoplasmatales and Sulfolobales separately while only considering other members of either the Euryarchaeota or TACK superphyla, respectively. Protein annotations were taken from the representative bin sequence (as determined in CD-HIT) and augmented by manual Conserved Domain Database searches of protein sequences. Protein bins that were highly enriched in the Thermoplasmatales and Sulfolobales were analyzed phylogenetically as described above. The nesting of the Thermoplasmatales within Sulfolobales/Crenarchaeota was assessed by visually inspecting the phylogenies and determining whether the Sulfolobales/Crenarchaeota were paraphyletic with respect to Thermoplasmatales protein placement.
To estimate the time of divergence for the Sulfolobales and Thermoplasmatales, a bacterial-rooted time tree was constructed using a subset (n=100) of the total archaeal taxa used in the above phylogenomic analyses. Archaeal taxa were selected to reduce data set size and computation time by including multiple representatives of the major taxonomic orders present in the full data set while removing taxa that were phylogenetically redundant. Three bacterial taxa were selected to represent an outgroup: Hydrogenobaculum sp. Y04AAS1 (IMG genome ID: 642555132), Thermocrinis ruber (IMG genome ID: 2512875013) and Thermotoga maritima MSB8 (IMG genome ID: 2519899531). Protein-coding genes for each bacterial genome were surveyed for homologs of the 104 archaeal housekeeping genes using Amphora2. The bacterial genomes consisted of 38.5%, 38.0% and 41.3% of the 104 archaeal housekeeping genes, respectively. Although the inclusion of genomes in the analysis that share <50% of the gene data set is not ideal, there are no other taxonomic groups that can serve as outgroups to the entirety of the archaeal domain, Bacteria have been used elsewhere to root whole-domain archaeal phylogenomic trees (Petitjean et al., 2015) and empirical analyses indicate that as little as 10–20% data inclusion in large protein concatenations are needed to reconstruct accurate phylogenies from whole genomes (Delsuc et al., 2005).
Individual housekeeping genes were then aligned using Clustal Omega, concatenated and subjected to ML analyses in RAxML, as in the archaeal-only analyses described above. Node support was assessed using resampling estimated log-likelihood bootstrapping (Minh et al., 2013) as implemented in RAxML. Divergence estimates for each node were then estimated using the Reltime method (Tamura et al., 2012) as implemented in MEGA7 (Kumar et al., 2016) using the LG substitution model and only using alignment positions with >50% coverage in the data set (28 407 positions total). The divergence of Archaea from Bacteria was calibrated with a maximum age estimate as 3.83 Ga, which corresponds to the earliest and perhaps most well-vetted date for evidence for potential life on Earth via isotopically light graphite inclusions in metasediments from Greenland (Mojzsis et al., 1996; McKeegan et al., 2007). A minimum age estimate of 3.46 Ga was used for the archaeal domain, which is the earliest evidence for isotopically light, microbially produced methane from methane-bearing inclusions within hydrothermal deposits from Australia (Ueno et al., 2006).
Results and discussion
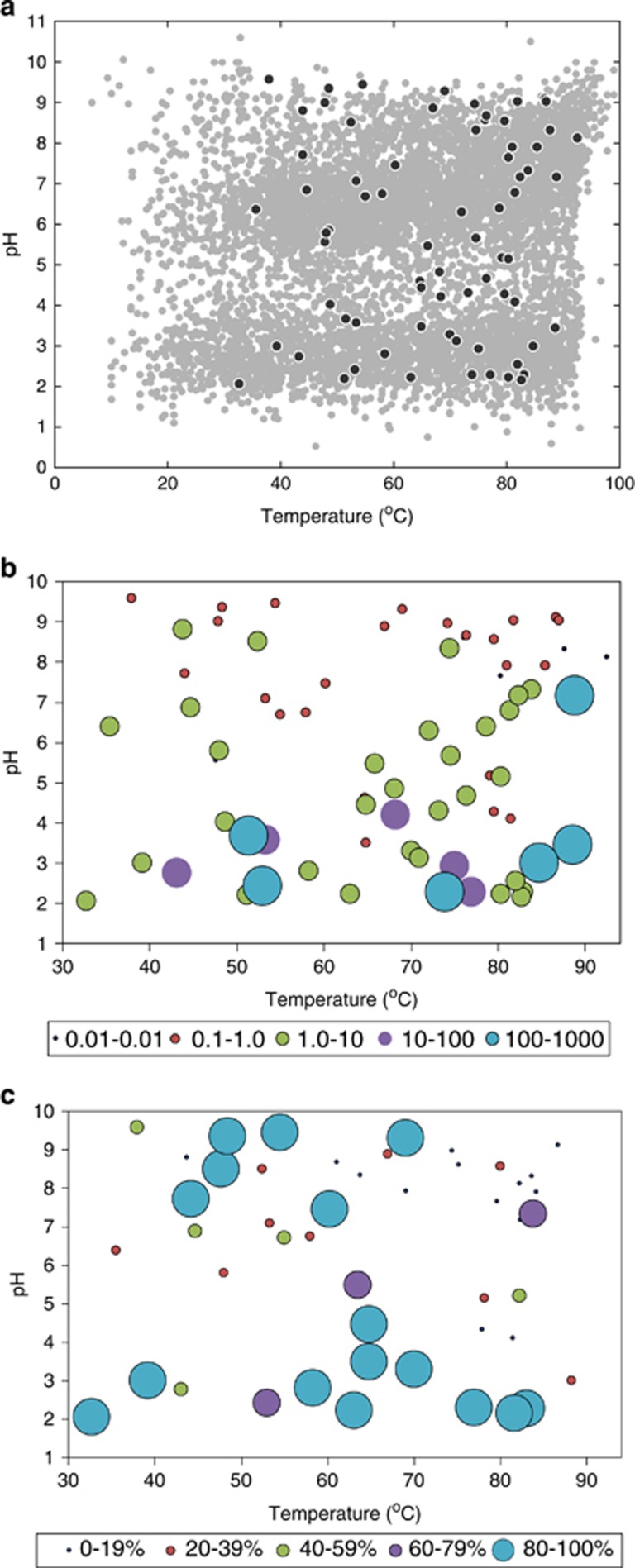
The diversity, distribution and inferred physiological characteristics of microbial populations inhabiting 72 representative hot spring environments in YNP that span a pH range of 2.1–9.6 and a temperature range of 32.7–92.5 °C were determined (Figure 1a and Supplementary Table S4). Quantitative PCR of archaeal and bacterial 16 S rRNA genes (as proxies for population sizes) revealed a transition toward archaeal dominance in low-pH springs (Figure 1b). Regression analyses of the log-transformed ratio of archaeal to bacterial 16S rRNA gene copies indicated a highly significant and inverse relationship with pH (β=−0.22, adjusted R2=0.29, P =6.42 x 10−7; Supplementary Figure S1) but no significant correlation with temperature (β=0.00, adjusted R2=−0.01, P=0.83). A multiple linear regression model incorporating both pH and temperature reiterated that pH was primarily responsible for the observed population ratios, whereas temperature was of negligible influence (data not shown). It should, however, be noted that many of the springs sampled here (57/72) have temperatures >50 °C and thus the lack of an apparent effect of temperature on archaeal dominance is likely related to the generally limited sampling of low-temperature hot springs. Analysis of variance tests of archaeal/bacterial 16S rRNA gene copy quantitative PCR ratios in relation to pH while parsing samples into low/high-pH groups at thresholds ranging from pH 3.0 to 7.0 indicated that a pH 4.0 threshold best segregated the data and supports the observation that archaeal dominance is pronounced in springs with pH<4.0 (Figure 1b, Supplementary Table S5 and Supplementary Figure S2).
Figure 1.
Temperature and pH of hot springs that were sampled and the dominance of aerobic Archaea in acidic hot springs. (a) Temperature and pH for spring samples (black circles; n=72) are overlaid on temperature and pH of thermal features downloaded from the Yellowstone National Park Research Coordination Network database (gray circles; n=7693). (b) The ratio of archaeal:bacterial 16S rRNA genes from 72 YNP hot springs plotted as a function of spring temperature and pH. Legend below x axis provides size and color for corresponding ratio magnitude. (c) The percentage of the archaeal community inferred to be capable of aerobic respiration, based on physiological inference to the closest related cultivar, nearest cultivated isolate or genome in springs that yielded 16S rRNA gene sequence.
Although the number of 16S rRNA gene operons present in a genome can vary and thus complicate the estimation of population sizes via quantification of their copy numbers (Acinas et al., 2004), 16S rRNA gene copy numbers in archaeal genomes are generally much lower than that of Bacteria (Acinas et al., 2004). Moreover, a survey of 16S rRNA gene operon copy numbers within the rrnDB operon database (Stoddard et al., 2015) revealed that the archaeal orders Sulfolobales, Desulfurococcales and Thermoproteales (which are among the predominant taxa reported in these springs; discussed below; Supplementary Table S1) do not contain multiple 16S rRNA gene copies, whereas the predominant bacterial orders (Aquificales, Thermales, Firmicutes and Proteobacteria) all consist of genera with multiple 16S rRNA gene operons. Consequently, our estimates of bacterial population sizes via quantification of 16S rRNA genes are likely to overestimate true population sizes. Thus, qualitatively, our estimates of archaeal predominance in low-pH springs (as archaeal/bacterial population ratios) are likely to underestimate true community dominance. Archaeal population dominance over Bacteria has also been observed in an acidic (pH ~2.5) New Zealand hot spring when temperatures exceed ~65 °C (Ward et al., 2017) and in the hottest and most acidic of 20 YNP hot spring metagenomes (Inskeep et al., 2013). The major transition toward archaeal dominance in springs with pH<4.0 is consistent with cultivar-based inferences from globally distributed environments, which suggest that Archaea have an ecological advantage over Bacteria in acidic thermal habitats (Valentine, 2007).
Pyrotag sequencing of archaeal 16S rRNA genes recovered from the springs analyzed here and physiological inference based on taxonomic identities suggest that the transition to archaeal dominance in acidic hot spring ecosystems is accompanied by an increased ability to integrate O2 respiration into energy metabolism (Figure 1c). Linear regression analyses indicated a significantly negative correlation between the percent of the archaeal community inferred to be aerobic and pH (β=−9.15, adjusted R2=0.36, P=1.82 × 10−5), but no significant correlation with temperature (β=−0.53, adjusted R2=0.05, P=0.09). Parsing samples into low/high-pH groups at thresholds from pH 3.0 to 7.0 indicated that pH 4.0 best segregated the communities based on inferred aerobic capacity and supports the observation that communities in springs with pH<4.0 are dominated by Archaea with aerobic respiratory physiologies (Supplementary Table S6 and Supplementary Figure S3). This trend is driven by an increased prevalence of sequences with affiliation to organisms in the Thermoplasmatales and Crenarchaeal-associated lineages in the lowest pH springs, the majority of which are capable of using O2 in energy metabolism (Supplementary Table S1). In contrast to lower-temperature acidic environments (that is, acid mine drainages; Tyson et al., 2004), Bacteria did not represent significant fractions of the acidophilic microbial communities analyzed here. Although acidophilic Bacteria can inhabit high-temperature acidic springs (primarily aerobic Hydrogenobaculum spp. of the Aquificales order), they are not dominant where temperatures exceed ~75 °C (Supplementary Table S1; Inskeep et al., 2013). These results are consistent with the ubiquitous distribution and dominance of largely aerobic Thermoplasmatales and Crenarchaeota (particularly the Sulfolobales order) in high-temperature acidic hot springs in YNP and elsewhere (Inskeep et al., 2013; Xie et al., 2014; Ward et al., 2017).
It has been previously suggested that Archaea dominate the hottest and most acidic environments because of key adaptations that allow them to cope with chronic energy stress (Valentine, 2007). Our data suggest an alternative, but not mutually exclusive, explanation for archaeal dominance in acidic geothermal systems. Here, the development of acidic thermal waters is driven largely by microbially mediated O2-dependent oxidation of S0 or H2S (Nordstrom et al., 2005; Nordstrom et al., 2009) by members of acidophilic lineages such as the Sulfolobales, due in part to meet energetic demands associated with habitation of these environments. It follows that the dominance of these lineages in acidic environments may result from the modification of their local environment (that is, niche) in the form of sulfuric acid production, thereby excluding less well-adapted populations in a process of geological–biological feedbacks like what has been described as niche construction (Odling-Smee et al., 1996). The process of niche construction allows for the modification of an environment by the activity of an organism, with the modified environment favoring the fitness or selectivity of the modifying organism and their progeny. In the case of extant thermoacidophiles, it is possible that the biological production of acidity may have resulted in the development of slightly more acidic, high-temperature niche space that promoted the radiation of those organisms responsible for acidification, and their respective lineages. Over time and through successive generations, this process could have manifested in divergence of acidophilic lineages away from neutrophilic ancestors in a unidirectional manner.
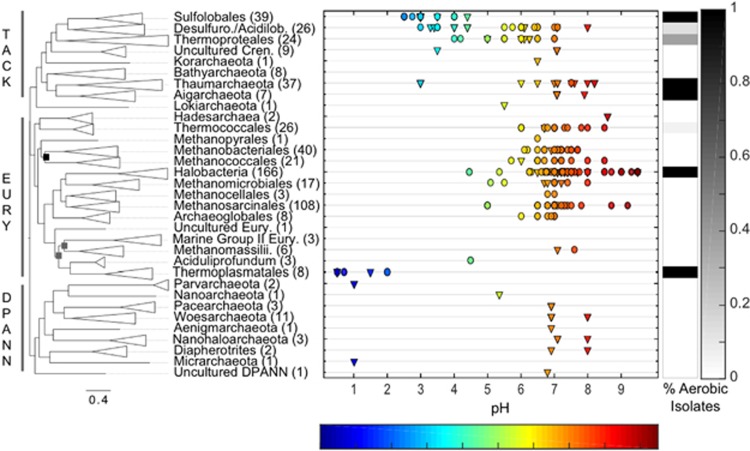
To examine further the role of O2 in the evolution of acidophilic Archaea, we compiled publicly available archaeal genomes (Supplementary Table S2), corresponding pH optima (or culture conditions, for cultivars) or environmental pH (for uncultured organisms), and O2 usage data (for cultivars). Phylogenomic reconstructions using single-copy phylogenetic marker genes reveal that acidophily (pH optima/environmental pH⩽3) is present in multiple archaeal lineages and is particularly enriched in the Thermoplasmatales and Sulfolobales (Figure 2). Notably, all acidophilic lineages, including the Thermoplasmatales and Sulfolobales, are nested among higher-order neutrophilic and alkaliphilic lineages. This indicates that acidophily is a derived physiological trait that has evolved independently and multiple times from neutrophilic or moderately acidophilic ancestors. Although environmental surveys have detected additional uncultivated Archaea (for example, Micrarchaeota, Parvarchaeota, Thaumarchaeota-related and Geoarchaeota) in acidic environments (Baker et al., 2010; Kozubal et al., 2013; Beam et al., 2014), they are all nested among largely uncultured lineages that are also prevalent in circumneutral or alkaline environments, suggesting that acidophily is a derived trait in these lineages as well. In addition, devolution of acidophilic lineages to moderately acidophilic or neutrophilic sublineages is not observed in our phylogenomic reconstruction, suggesting that evolution toward an increased acidophily has largely been a unidirectional evolutionary process. Together, these observations suggest relatively recent, multiple origins of acidophilic Archaea (Figure 2).
Figure 2.
Phylogenetic placement of archaeal acidophiles, pH optima and corresponding O2 usage profiles. The Maximum Likelihood tree was constructed using a concatenation of between 53 and 104 phylogenetic marker genes for 584 archaeal genomes (median n=103; 0.05 percentile n=70). Order-level (or above) lineages are collapsed in the tree. All nodes shown exhibited bootstraps >95% except where black boxes (>70%) or gray boxes (< 70%) are shown. Scale bar shows expected number of substitutions per site. pH optima (scatterplot) and O2 usage data (black/white heatmap on right, as a % of isolates that are aerobic) are given for each lineage where cultivars are available. pH optima for cultivars are shown as circles, whereas environmental pH of reconstructed genomes or cultivation media pH (non-optima) are shown as triangles.
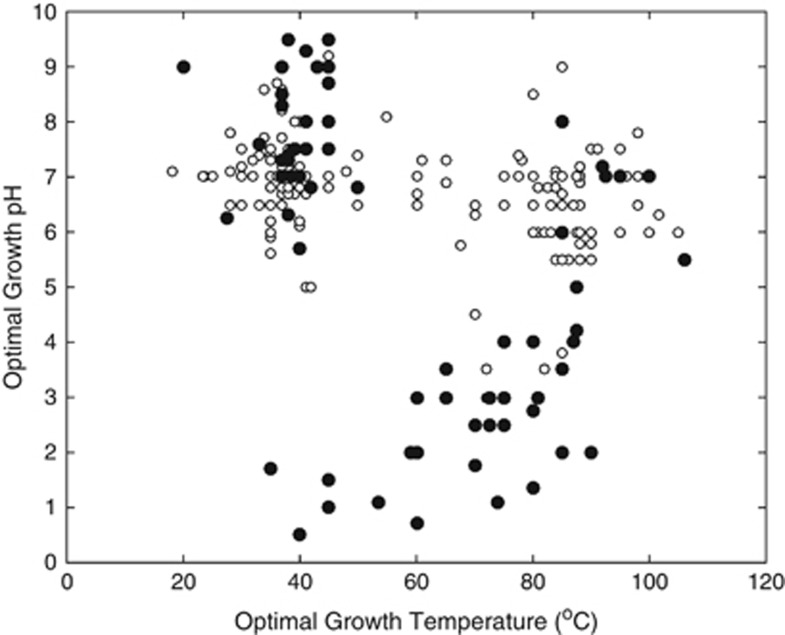
Mapping of O2 use among available cultivars on the archaeal phylogeny reveals predominance in the Sulfolobales and Thermoplasmatales, pointing to the importance of high-energy-yielding metabolisms in adapting to and inhabiting acidic environments (Figure 2). Indeed, analysis of temperature and pH optima along with O2 usage of available archaeal cultivars reveal that taxa with growth optima of pH 3.0 or less consistently exhibit the ability to respire O2 (Figure 3). As oxygenic phototrophs are excluded from acidic hot springs in YNP with temperatures >56 °C (Boyd et al., 2012), the O2 that supports thermoacidophiles and most likely supported their ancestors would have only been available through diffusion from atmospheric sources (or as dissolved O2 transported via groundwater). Although all characterized acidophilic bacteria isolated to date also exhibit aerobic metabolic capacity, only one sulfur-oxidizing genus within the Aquificae (Hydrogenobaculum) and another methanotrophic genus (Methylacidiphilum) within the Verrucomicrobia are known to grow optimally at ⩾60 °C, but neither are known to grow optimally above 65 °C (Dopson, 2016). This suggests that Bacteria are of marginal importance in the formation of high-temperature acidic environments today and, by extension, the geologic past.
Figure 3.
Optimal growth pH, optimal incubation temperature and O2 usage for archaeal cultivars. Each point represents a single archaeal cultivar (N=255) that is colored based on the ability (aerobe; black circles; n=76) or inability (anaerobe; white circles; n=179) to incorporate O2 into their metabolism.
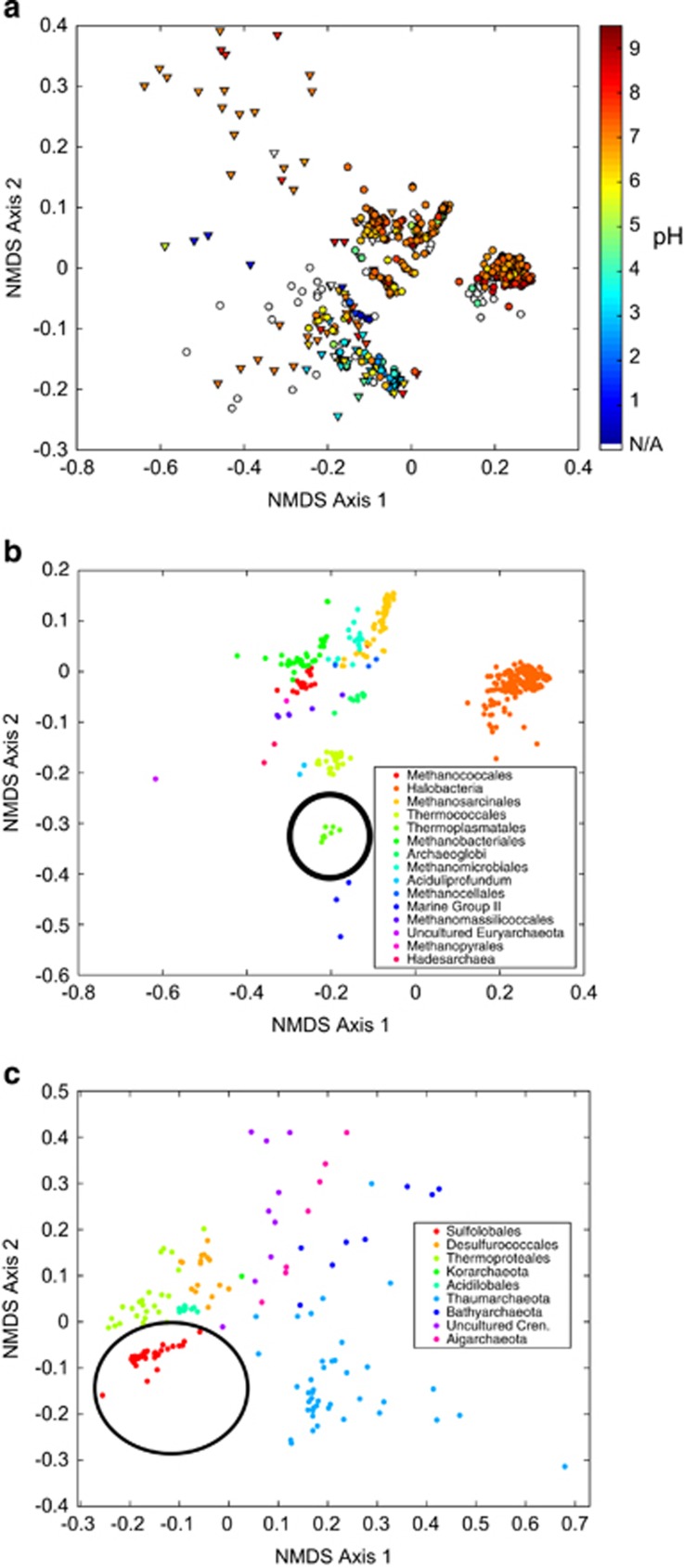
Comparative genomics was used to identify assemblages of protein-encoding genes that may contribute to successful habitation of hyperacidic habitats. Ordination of a dissimilatory matrix of protein homolog distances encoded among archaeal genomes revealed clustering of acidophile genomes (Figure 4a and Supplementary Figure S4). This indicates that the proteins encoded in acidophile genomes are similar despite belonging to disparate phylogenetic lineages. In addition, the proteins encoded by Sulfolobales and Thermoplasmatales genomes are, themselves, distinct from other groups of the TACK and Euryarchaeota superphyla, respectively (Figures 4b and c). These observations suggest that phylogenetically distinct taxa may have converged on similar protein-coding gene complements during their diversification into acidic habitats.
Figure 4.
Similarity in protein-coding genes among archaeal genomes. (a) Nonmetric multidimensional scaling (NMDS) plot of protein-coding gene bins among all archaeal genomes. Symbols are as in Figure 2: cultivar genomes are shown as circles and reconstructed genomes from environmental samples are shown as triangles. Symbol color refers to pH optima or environmental pH according to the scale on the right. (b) NMDS plots including only Euryarchaeota genomes and (c) TACK superphylum genomes. Thermoplasmatales and Sulfolobales are indicated with black circles in b, c, respectively. Each point represents a genome, and points are colored according to taxonomic orders as given by the legends on the right. Axes represent relative positioning of genomes to one another, such that points closer together are more similar, and points farther apart are less similar.
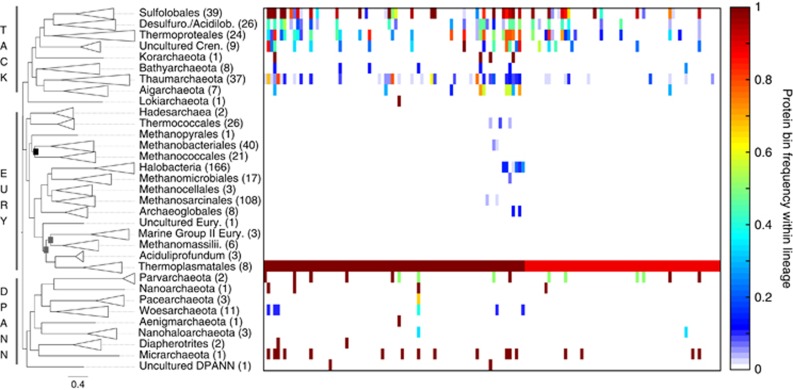
To identify those protein complements that contributed to the differentiation of acidophiles from their higher-order lineages, protein bins were identified that demarcated the Thermoplasmatales and Sulfolobales from the Euryarchaeota and TACK groups, respectively. Predicted membrane-associated permeases or transporters comprised ~25% of the protein bins that distinguished the Thermoplasmatales from other euryarchaeotes (Figure 5 and Supplementary Table S7). Of the 138 Thermoplasmatales-enriched bins, 32 were also present in >50% of Sulfolobales genomes (Figure 5 and Supplementary Table S7), which is consistent with observations that genomes from the Thermoplasmatales species Picrophilus torridus and Thermoplasma acidiphilum share significant homology with Sulfolobus genomes (Ruepp et al., 2000; Futterer et al., 2004). Many of the shared proteins are permease- or membrane transport-related, including amino acid and solute transporters/permeases (Supplementary Table S7), which supports the assertion that these protein functions are essential for intracellular pH homeostasis and thus, habitation of acidic environments (Baker-Austin and Dopson, 2007). In contrast, the enrichment of Thermoplasmatales-like proteins in Sulfolobales members is not observed (Supplementary Figure S5). These results suggest that the differentiation of Thermoplasmatales from the Euryarchaeota may be due, in part, to the acquisition of genes from a Sulfolobales-like ancestor, but that the converse is not supported.
Figure 5.
Heatmap showing the distribution of Thermoplasmatales-enriched protein-coding genes among non-Euryarchaeota genomes. The Maximum Likelihood tree is the same as shown in Figure 2. Protein bin distribution is only shown for those protein-coding genes with >87% frequency within the Thermoplasmatales lineage (n=138), and which most differentiate the Thermoplasmatales from other Euryarchaeaota based on ‘indicator’ values. Annotations for each protein bin are given in Supplementary Table S7.
Phylogenetic analysis of the Thermoplasmatales-enriched bins indicates a nesting of Thermoplasmatales homologs within crenarchaeal outgroups in most (65.6%) of the protein phylogenies (data not shown). These data support the hypothesis that the Thermoplasmatales, which are found in lower-temperature springs when compared with Sulfolobales, diverged from other Euryarchaeota due, at least in part, to horizontal acquisition of genes from an acidophilic crenarchaeal-like ancestor. This scenario is consistent with the hypothesis that aerobic sulfur-oxidizing Sulfolobales are responsible for the formation of high-temperature acidic niche space (for example, >65 °C), which may then facilitate the generation of lower-temperature acidic niche space for later diverging, less thermophilic acidophile lineages, such as the Thermoplasmatales. The downstream physical location of Thermoplasmatales (lower-temperature transects) relative to the upstream physical location of the Sulfolobales (higher-temperature transects) in hydrothermal systems may have contributed to the apparent unidirectional transfer of genes from the Sulfolobales to the Thermoplasmatales. Extensive horizontal gene transfer (HGT; at the inter- and intra-specific levels) has been documented in acidophiles (Whitaker et al., 2005; Simmons et al., 2008; Schonknecht et al., 2013) and may be a general mechanism by which lineages acquire traits that allow for successful adaptation to acidophilic environments.
Experimental evolution of Sulfolobus solfataricus toward increased acidophily supports the hypothesis that adaptation to a progressively more acidic environment necessitates increased metabolic energy yield, in part, to meet the increased biosynthetic demands associated with oxidative stress (McCarthy et al., 2016). Serial passage of S. solfataricus over 3 years into progressively more acidic culture conditions resulted in a substantial decrease in the minimum pH that supported growth (pH 0.60 for evolved strain; pH 1.64 for parental strain). Transcriptomic analyses of the derived and parental strains showed that increased acid tolerance in derived strains was accompanied with increased expression of catabolic functions (for example, TCA cycle genes) and anabolic functions (membrane biosynthesis genes). Comparative analysis of the genome of the derived and parental strain, however, revealed only several point mutations in membrane-related and transporter-associated genes (McCarthy et al., 2016), suggesting that evolved acidophilic strains differed primarily at the level of gene regulation rather than at the level of acquisition of new traits or trait variants. Adaptation toward increased acidophily in the natural environment, however, might be expected to be influenced more by HGT rather than by single point mutations. For example, in open natural systems it is possible that environmental DNA from lysed cells, exogenous sources and viruses—the latter of which are common in acidic hydrothermal environments (Bolduc et al., 2015; Gudbergsdottir et al., 2016) and in cultures of known acidophiles (Hochstein et al., 2016)—may accelerate HGT and diversification toward more acidic habitats. Indeed, comparative genomic analyses performed here and elsewhere (Ruepp et al., 2000; Futterer et al., 2004; Schonknecht et al., 2013) underscore the importance of HGT in the evolution of acidophilic lineages.
These collective observations prompt the intriguing question as to when thermoacidophiles and their acidic habitats emerged. Aerobic organisms within the Sulfolobales, such as S. solfataricus, grow optimally at O2 concentrations between ~1.5% and 24% v/v corresponding to 0.87 and 13.9 mM O2 at 80 °C, respectively, whereas no growth is observed in the absence of O2 (Simon et al., 2009). Oxygenation of Earth’s atmosphere did not begin until after the evolution of oxygenic photosynthesis, and oxygen concentrations did not reach appreciable levels until after the GOE between ~2.4 and 2.1 Ga. The timing of the GOE is widely supported by paleosol evidence indicating a substantial change in readily oxidized, redox-sensitive compounds such as pyrite (FeS2) and uraninite (UO2) in addition to the disappearance of mass-independent sulfur-isotope fractionations from the sedimentary rock record after this time (Farquhar et al., 2000; Canfield, 2005). However, the availability of atmospheric oxygen following the GOE has only recently come into focus.
Accepted models have bounded atmospheric O2 somewhere between ~1% and 40% of present atmospheric levels during the mid-Proterozoic (1.8–0.8 Ga) based on sulfur-isotope evidence for anoxic deep oceans and paleosol chemical profiles, and particularly those of Fe3+ (Rye and Holland, 1998; Canfield, 1998; Canfield, 2005; Lyons et al., 2014). Emerging data arising from chromium (Cr) isotopic records (documenting decreased Cr redox cycling, which is a highly oxygen-sensitive process) indicate that atmospheric oxygen levels likely decreased after the GOE to levels that did not exceed >~0.1% present atmospheric level during the mid-Proterozoic (Frei et al., 2009; Planavsky et al., 2014; Lyons et al., 2014; Cole et al., 2016). A final increase in atmospheric oxygen to near present atmospheric level during the late Proterozoic (0.8–0.55 Ga) is evinced by multiple lines of geochemical observations including carbon isotopic analyses indicating high organic carbon burial rates in ocean sediments (Knoll et al., 1986), and increased Cr redox cycling (Frei et al., 2009; Cole et al., 2016) among other observations (Canfield, 2005; Holland, 2006; Lyons et al., 2014). Thus, given the above evidence, the nonlinear decrease in O2 solubility with increasing water temperature (Shock et al., 2010), and the necessity for a non-point source of oxygen availability in high-temperature acidic springs (discussed above), it follows that aerobic thermoacidophiles did not likely emerge until at the earliest, the most recent rise of atmospheric O2 to present atmospheric level in the latter period of the mid-Proterozoic ~0.8 Ga.
An alternative hypothesis is that aerobic thermoacidophiles emerged before or during the GOE but lost niche space during the intervening ~1.5 billion years when atmospheric O2 plummeted and were confined to refugia, only to re-emerge and radiate ~0.8 Ga. Others have hypothesized that the spike in the abundance of Cr in sedimentary iron deposits dated to the GOE or just after the GOE, along with a lack of Cr redox cycling, could be explained by the release of reduced Cr (III) from acid weathering of a global terrestrial pyrite reservoir. It was hypothesized that this weathering was driven by the activity of low-temperature aerobic pyrite-oxidizing acidophiles that could hypothetically have been present during this time frame (Konhauser et al., 2011). The above evidence for extremely low mid-Proterozoic oxygen levels and the return of Cr abundances to pre-GOE levels in multiple sedimentary records during the mid-Proterozoic (Frei et al., 2009; Konhauser et al., 2011; Cole et al., 2016) potentially supports the aerobic acidophile niche-collapse hypothesis.
To assess whether one of the two above hypotheses is supported via molecular dating of the archaeal phylogeny, we constructed a bacterial-rooted time tree using a subset of archaeal taxa (n=100) from our phylogenomic data set. Using maximum and minimum age estimates for the divergence of all Archaea as 3.83 Ga (the age of the earliest potential evidence for life based on isotopically light graphite inclusions; Mojzsis et al., 1996; McKeegan et al., 2007) and 3.46 Ga (the earliest isotopic evidence for microbially produced methane; Ueno et al., 2006), the Most Recent Common Ancestor (MRCA) of the Sulfolobales was estimated to diverge at 1.053 Ga (95% confidence interval (CI): 1.601–0.656 Ga), whereas the divergence date of the MRCA of the Thermoplasmatales was estimated as 0.843 Ga (95% CI: 1.301–0.505 Ga; Supplementary Figure S6). These dates are broadly consistent with earlier estimates using smaller genomic data sets (Battistuzzi et al., 2004) and provide further support that aerobic thermoacidophiles radiated late in Earth history and are coincident with estimates for the timing of the mid-Proterozoic atmospheric rise in oxygen at ~0.8 Ga. Moreover, the estimated radiation of the Sulfolobales before that of the Thermoplasmatales provides additional support for the hypothesis that the Thermoplasmatales may have adapted to thermoacidophilic lifestyles, in part, because of HGT from Sulfolobales-like ancestors that predominate at higher-temperature hydrothermal settings (that is, HGT occurring down temperature gradients, rather than up gradient). However, it should be noted that dating phylogenies, and particularly those of Archaea, is problematic because of the lack of available calibration points for lineages. Thus, divergence estimates that are based on a single calibration point should be treated tentatively. Intriguingly, previous analyses have provided evidence for the divergence of chemotrophic sulfur-oxidizing Proteobacteria involved in the oxidation of sulfides in marine sediments at a similar time frame (0.64–1.05 Ga; Canfield and Teske, 1996). The late Proterozoic divergence of these mesophiles is coincident with the late Proterozoic rise in oxygen and large-scale changes in the isotopic fractionation of sedimentary sulfides that can be best explained by the emergence of an oxidative sulfur cycle driven by sulfide and sulfur-oxidizing non-photosynthetic Bacteria (Canfield and Teske, 1996). Whether the consilience between our date estimates for the origin of sulfur-oxidizing thermoacidophilic Archaea and those for mesophilic marine sulfide and sulfur-oxidizing Bacteria are coincidental, or rather, are linked via HGT between Archaea and Bacteria is a topic that is of interest for better understanding the origins of both sulfur-oxidizing microbial lineages and their role in the sulfur cycle.
Despite the suggestion that low-temperature mesophiles may have been involved in the generation of acidic environments following the GOE (Konhauser et al., 2011), the likelihood that discretely distributed, highly oxygenated and high-temperature refugia would have existed throughout the mid-Proterozoic that could continuously support aerobic thermoacidophiles certainly is low. Thus, our data support the hypothesis for a Neoproterozoic origin for archaeal thermoacidophiles. Taking into account the role of biology in accelerating the kinetics of reactions that lead to the acidification of hydrothermal waters, we contend that it is unlikely that thermal acidic ecosystems, such as those found in the YNP geothermal system, were widespread before the emergence of aerobic sulfur-oxidizing thermoacidophiles. Rather, these observations more likely suggest a recent emergence of thermoacidophiles and their habitats, which may have arisen and evolved in concert through a process of geological–biological feedbacks over the past ~0.8 Ga. Finally, confirmation of the emergence of thermoacidophilic Archaea during the mid to late Proterozoic, either through geological data or biological data, would provide a much-needed calibration point for archaeal phylogenies, allowing for more accurate molecular clock simulations to be conducted.
Conclusions
Acidic hydrothermal ecosystems in YNP are dominated by Archaea, which is consistent with data from other globally distributed hydrothermal systems (Xie et al., 2014; Ward et al., 2017). Physiological inference suggests that the transition to archaeal-dominated acidic hydrothermal habitats is accompanied by an increase in the ability to integrate O2 into respiration, likely due, in part, to meet the bioenergetic demands associated with mitigating oxidative stress (McCarthy et al., 2016). This observation, coupled to data indicating that the generation of acidic environments typically requires the O2-dependent oxidation of sulfur compounds, suggests that thermoacidophilic Archaea and the acidity of their habitats co-evolved after the emergence of oxygenic photosynthesis. Further, we hypothesize that these events may have taken place through a series of geological–biological feedbacks in a process that is similar to what has been described as niche construction (Odling-Smee et al., 1996). Although the process of niche construction may be little explored in microbial ecology, it is likely a widespread phenomenon in microbial evolution considering how quickly microorganisms can influence the geochemical composition of their local environments, the apparent ease by which they can acquire new traits via HGT, and their relatively fast rates of reproduction.
Phylogenomic analyses support these conclusions and suggest that the evolution of acidophily in Archaea has occurred independently in divergent lineages relatively recently and was potentially aided by HGT. Taken together, these results expand our understanding of the ecological prevalence of Archaea in hot springs and provide physiological and evolutionary context for their current dominance in these environments. Moreover, these observations point toward a hypothesis of biological modification of natural environments in shaping the diversification of an organism’s progeny (that is, niche construction), local co-inhabitants or downstream inhabitants (that is, facilitation). Further laboratory experimentation of aerobic thermoacidophiles is needed to assess how adaptation toward increased acidophily affects ecological fitness in the adapted and ancestral pH niches as well as the potential impacts on incipient species divergence.
Acknowledgments
This work was supported by a NASA Exobiology and Evolutionary Biology grant (NNX13AI11G) to ESB, NSF grant EAR-1529963 to ELS and a Montana Space Grant Consortium grant to DRC and ESB. The NASA Astrobiology Institute is supported by grant NNA15BB02A (to ELS and ESB). We thank Christie Hendrix and Stacey Gunther at YNP for research permitting. We would also like to thank Paul G Falkowski and D Kirk Nordstrom for thoughtful comments on a previous version of the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. (2004). Divergence and redundancy of 16 S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Banfield JF. (2003). Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44: 139–152. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD et al. (2010). Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA 107: 8806–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C, Dopson M. (2007). Life in acid: pH homeostasis in acidophiles. Trends Microbiol 15: 165–171. [DOI] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. (2004). A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol 4: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam JP, Jay ZJ, Kozubal MA, Inskeep WP. (2014). Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J 8: 938–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc B, Wirth JF, Mazurie A, Young MJ. (2015). Viral assemblage composition in Yellowstone acidic hot springs assessed by network analysis. ISME J 9: 2162–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Fecteau KM, Havig JR, Shock EL, Peters JW. (2012). Modeling the habitat range of phototrophs in Yellowstone National Park: toward the development of a comprehensive fitness landscape. Front Microbiol 3: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braterman PS, Cairns-Smith AG, Sloper RW. (1983). Photo-oxidation of hydrated Fe2+ - significance for banded iron formations. Nature 303: 163–164. [Google Scholar]
- Canfield DE, Teske A. (1996). Late Proterozoic rise in atmospheric oxygen concentration inferred from phylogenetic and sulphur-isotope studies. Nature 382: 127–132. [DOI] [PubMed] [Google Scholar]
- Canfield DE. (1998). A new model for Proterozoic ocean chemistry. Nature 396: 450–453. [Google Scholar]
- Canfield DE. (2005). The early history of atmospheric oxygen: homage to Robert M. Garrels. Annu Rev Earth Planet Sci 33: 1–36. [Google Scholar]
- Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ et al. (2015). Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25: 690–701. [DOI] [PubMed] [Google Scholar]
- Charney N, Record S. (2012). vegetarian: jost diversity measures for community data, version 2.1..
- Chen KY, Morris JC. (1972). Kinetics of oxidation of aqueous sulfide by O2. Environ Sci Technol 6: 529. [Google Scholar]
- Cole DB, Reinhard CT, Wang X, Gueguen B, Halverson GP, Gibson T et al. (2016). A shale-hosted Cr isotope record of low atmospheric oxygen during the Proterozoic. Geology 44: 555–558. [Google Scholar]
- Colman DR, Feyhl-Buska J, Robinson KJ, Fecteau KM, Xu H, Shock EL et al. (2016). Ecological differentiation in planktonic and sediment-associated chemotropic microbial populations in Yellowstone hot springs. FEMS Microbiol Ecol 92: 1–13. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkamnn H, Philippe H. (2005). Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6: 361–375. [DOI] [PubMed] [Google Scholar]
- D'Imperio S, Lehr CR, Oduro H, Druschel G, Kuhl M, McDermott TR. (2008). Relative importance of H2 and H2S as energy sources for primary production in geothermal springs. Appl Environ Microbiol 74: 5802–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopson M. (2016) Physiological and phylogenetic diversity of acidophilic bacteria. In:Quatrini R, Johnson DB (eds). Acidophiles. Caister Academic Press: Norfolk, UK. [Google Scholar]
- Emerson D, Fleming EJ, McBeth JM. (2010). Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64: 561–583. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320: 1034–1039. [DOI] [PubMed] [Google Scholar]
- Farquhar J, Bao H, Thiemens M. (2000). Atmospheric influence of earth’s earliest sulfur cycle. Science 289: 756–758. [DOI] [PubMed] [Google Scholar]
- Frei R, Gaucher C, Poulton SW, Canfield DE. (2009). Fluctuations in Precambrian atmospheric oxygenation recorded by chromium isotopes. Nature 461: 250–254. [DOI] [PubMed] [Google Scholar]
- Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer O, Angelov A, Liesegang H, Gottschalk G, Schleper C, Schepers B et al. (2004). Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc Natl Acad Sci USA 101: 9091–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaveno MA, Urbieta MS, Ulloa JR, Toril EG, Donati ER. (2013). Physiologic versatility and growth flexibility as the main characteristics of a novel thermoacidophilic Acidianus strain isolated from Copahue geothermal area in Argentina. Microb Ecol 65: 336–346. [DOI] [PubMed] [Google Scholar]
- Godfrey LV, Falkowski PG. (2009). The cycling and redox state of nitrogen in the Archaean ocean. Nat Geosci 2: 725–729. [Google Scholar]
- Gudbergsdottir SR, Menzel P, Krogh A, Young M, Peng X. (2016). Novel viral genomes identified from six metagenomes reveal wide distribution of archaeal viruses and high viral diversity in terrestrial hot springs. Environ Microbiol 18: 863–874. [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Peters JW, Skidmore ML, Boyd ES. (2013). Molecular evidence for an active endogenous microbiome beneath glacial ice. ISME J 7: 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein RA, Amenabar MJ, Munson-McGee JH, Boyd ES, Young MJ. (2016). Acidianus tailed spindle virus: a new archaeal large tailed spindle virus discovered by culture-independent methods. J Virol 90: 3458–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland HD. (2006). The oxygenation of the atmosphere and oceans. Phil Trans R Soc B 361: 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Stetter KO. (2015) Sulfolobales. Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons: Hoboken, NJ, USA. [Google Scholar]
- Inskeep WP, Jay ZJ, Tringe SG, Herrgard MJ, Rusch DB, et alCommittee YNPMPS. (2013). The YNP Metagenome Project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front Microbiol 4: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Ghauri MA, Mcginness S. (1993). Biogeochemical cycling of iron and sulfur in leaching environments. FEMS Microbiol Rev 11: 63–70. [Google Scholar]
- Johnson DB, Quatrini R. (2016) Acidophile microbiology in space and time. In: Quatrini R, Johnson DB (eds). Acidophiles. Caister Academic Press: Norfolk, UK. [Google Scholar]
- Kim JD, Yee N, Nanda V, Falkowski PG. (2013). Anoxic photochemical oxidation of siderite generates molecular hydrogen and iron oxides. Proc Natl Acad Sci USA 110: 10073–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH, Hayes JM, Kaufman AJ, Swett K, Lambert IB. (1986). Secular variation in carbon isotope ratios from Upper Proterozoic successions of Svalbard and east Greenland. Nature 321: 832–838. [DOI] [PubMed] [Google Scholar]
- Konhauser KO, Lalonde SV, Planavsky NJ, Pecoits E, Lyons TW, Mojzsis SJ et al. (2011). Aerobic bacterial pyrite oxidation and acid rock drainage during the Great Oxidation Event. Nature 478: 369–373. [DOI] [PubMed] [Google Scholar]
- Kozubal MA, Romine M, Jennings R, Jay ZJ, Tringe SG, Rusch DB et al. (2013). Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J 7: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther GW III, Findlay AJ, Macdonald DJ, Owings SM, Hanson TE, Beinart RA et al. (2011). Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons TW, Reinhard CT, Planavsky NJ. (2014). The rise of oxygen in Earth's early ocean and atmosphere. Nature 506: 307–315. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Johnson T, Pavlik BJ, Payne S, Schackwitz W, Martin J et al. (2016). Expanding the limits of thermoacidophily in the archaeon Sulfolobus solfataricus by adaptive evolution. Appl Environ Microb 82: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KD, Kudryavtsev AB, Schopf JW. (2007). Raman and ion microscopic imagery of graphitic inclusions in apatite from older than 3830 Ma Akilia supracrustal rocks, west Greenland. Geology 35: 591–594. [Google Scholar]
- Millero FJ. (2001) The Physical Chemistry of Natural Waters. Vol. 2. Wiley-Interscience: NJ, USA. [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojzsis S, Arrhenius G, McKeggan KD, Harrison TM, Nutman AP, Friend CRL. (1996). Evidence of life on Earth before 3,800 million years ago. Nature 384: 55–59. [DOI] [PubMed] [Google Scholar]
- Navarro-Gonzalez R, McKay CP, Mvondo DN. (2001). A possible nitrogen crisis for Archaean life due to reduced nitrogen fixation by lightning. Nature 412: 61–64. [DOI] [PubMed] [Google Scholar]
- Nordstrom DK, Alpers CN, Ptacek CJ, Blowes DW. (2000). Negative pH and extremely acidic mine waters from Iron Mountain, California. Environ Sci Technol 34: 254–258. [Google Scholar]
- Nordstrom DK, Ball JW, McCleskey RB. (2005) Ground water to surface water: chemistry of thermal outflows in Yellowstone National Park. In: Inskeep W, Mcdermott T (eds). Geothermal Biology and Geochemistry in Yellowstone National Park. Montana State University Publications: Bozeman, MT, USA. [Google Scholar]
- Nordstrom DK, McCleskey RB, Ball JW. (2009). Sulfur geochemistry of hydrothermal waters in Yellowstone National Park: IV acid-sulfate waters. Appl Geochem 24: 191–207. [Google Scholar]
- Odling-Smee FJ, Laland KN, Feldman MW. (1996). Niche construction. Am Nat 147: 641–648. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015). vegan: Community Ecology Package, 2.3 edn (https://cran.r-project.org/web/packages/vegan/).
- Petitjean C, Deschamps P, Lopez-Garcia P, Moreira D. (2015). Rooting the domain Archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biol Evol 7: 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planavsky NJ, Reinhard CT, Wang X, Thomson D, McGoldrick P, Rainbird RH et al. (2014). Low Mid-Proterozoic atmospheric oxygen levels and the delayed rise of animals. Science 346: 635–638. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2015) R: A language and environment for statistical computing version 3.2.0. R Foundation for Statistical Computing: Vienna, Austria, http://www.R-project.org/. [Google Scholar]
- Roberts DW. (2015). labdsv: Ordination and multivariate analysis for ecology, R package version 1.7 (http://ecology.msu.montana.edu/labdsv/R).
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW et al. (2000). The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407: 508–513. [DOI] [PubMed] [Google Scholar]
- Rye R, Holland HD. (1998). Paleosols and the evolution of atmospheric oxygen: a critical review. Am J Sci 298: 621–672. [DOI] [PubMed] [Google Scholar]
- Schonknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M et al. (2013). Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339: 1207–1210. [DOI] [PubMed] [Google Scholar]
- Shivvers DW, Brock TD. (1973). Oxidation of elemental sulfur by Sulfolobus acidocaldarius. J Bacteriol 114: 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock EL, Holland M, Meyer-Dombard D, Amend JP, Osburn GR, Fischer TP. (2010). Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochim Cosmochim Acta 74: 4005–4043. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, Dibartolo G, Denef VJ, Goltsman DS, Thelen MP, Banfield JF. (2008). Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol 6: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G, Walther J, Zabeti N, Combet-Blanc Y, Auria R, van der Oost J et al. (2009). Effect of O2 concentrations on Sulfolobus solfataricus P2. FEMS Microbiol Lett 299: 255–260. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. (2015). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43: D593–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. (2012). Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci USA 109: 19333–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. (2006). Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440: 516–519. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Valentine DL. (2007). Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol 5: 316–323. [DOI] [PubMed] [Google Scholar]
- Ward L, Taylor MW, Power JF, Scott BJ, McDonald IR, Stott MB. (2017). Microbial community dynamics in Inferno Crater Lake, a thermally fluctuating geothermal spring. ISME J 11: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. (2005). Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol 22: 2354–2361. [DOI] [PubMed] [Google Scholar]
- Widdel F, Schnell S, Heising S, Ehrenreich A, Assmus B, Schink B. (1993). Ferrous iron oxidation by anoxygenic phototrophic bacteria. Nature 362: 834–836. [Google Scholar]
- Wu M, Scott AJ. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28: 1033–1034. [DOI] [PubMed] [Google Scholar]
- Xie W, Zhang CL, Wang J, Chen Y, Zhu Y, de la Torre JR et al. (2014). Distribution of ether lipids and composition of the archaeal community in terrestrial geothermal springs: impact of environmental variables. Environ Microbiol 17: 1600–1614. [DOI] [PubMed] [Google Scholar]
- Zhang J, Millero FJ. (1993). The products from the oxidation of H2S in seawater. Geochim Cosmochim Acta 57: 1705–1718. [Google Scholar]
- Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J et al. (1994). Screening for Sulfolobales, their plasmids and their viruses in icelandic solfataras. Syst Appl Microbiol 16: 609–628. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.