Abstract
The environmental resistome has been recognized as the origin and reservoir of antibiotic resistance genes and considered to be dynamic and ever expanding. In this study, a targeted gene sequencing approach revealed that the polymorphic diversity of the aminoglycoside-inactivating enzyme AAC(6′)-Ib was ecological niche-specific. AAC(6′)-Ib-cr, previously known as a clinical variant, was prevalent in various soils and the intestines of chickens and humans, suggesting that this variant might not have arisen from adaptive mutations in the clinic but instead originated from the environment. Furthermore, ecologically dominant polymorphic variants of AAC(6′)-Ib were characterized and found to display different substrate specificities for quinolones and aminoglycosides, conferring the altered resistance spectra. Interestingly, a novel variant with the D179Y substitution showed an extended resistance spectrum to the recently developed fluoroquinolone gemifloxacin. Our results suggest that soil and animal microbiomes could be major reservoirs of antibiotic resistance; polymorphic diversity expands the antibiotic resistome in the environment, resulting in the potential emergence of novel resistance.
Introduction
The development of sequencing technologies and bioinformatic tools to analyze massive sequence data has accelerated studies on the antibiotic resistome and suggested that antibiotic resistance genes are ancient and ubiquitous in the natural environment (D'Costa et al., 2007;;D'Costa et al., 2007;; Finley et al., 2013; Martinez et al., 2015). Recent studies have focused on the vast diversity of the resistome and the dissemination mechanisms of antibiotic resistance genes from the environment to the clinic (D'Costa et al., 2007; Perry and Wright, 2013; Martinez et al., 2015). Polymorphisms of antibiotic-inactivating enzymes in the environment can contribute to the diversity of the resistome, as genetic polymorphisms can confer plasticity to those proteins and affect their substrate specificities, leading to changes in the resistance spectra (Cambray and Mazel, 2008; Walkiewicz et al., 2012; Galan et al., 2013), as shown for polymorphisms of β-lactamase from soil bacteria (Demaneche et al., 2008). Although shotgun metagenomics can be the most direct method to decipher the complexity of the environmental resistome, revealing polymorphisms of a specific gene in the environment is not easy to due to the limited sequencing depth. Recent attempts to analyze the ARG abundance in various metagenomic data showed that clinically relevant resistance genes were absent or detected in low abundance (Chen et al., 2013; Hu et al., 2013; Nesme et al., 2014). Targeted gene sequencing by high-throughput sequencing technologies can be a good approach to elucidate such polymorphic diversity of functional genes in depth (Iwai et al., 2010; Schmieder and Edwards, 2012; Lee et al., 2014; Zhou et al., 2015). In this study, aminoglycoside 6'-N-acetyltransferase (AAC(6′)-Ib) was chosen for a targeted gene sequencing analysis due to its clinical importance and suitability for interpreting functionally important polymorphisms (Robicsek et al., 2006; Cambray and Mazel, 2008; Maurice et al., 2008; Vetting et al., 2008; Ramirez et al., 2013). The clinically relevant variant AAC(6′)-Ib-cr, which possesses mutations in two amino acid residues (W102R and D179Y) of AAC(6′)-Ib, is known to confer structural plasticity to the protein required for adaptation to new antibiotics, resulting in the extension of resistance to fluoroquinolone, a different class of antibiotics (Robicsek et al., 2006; Maurice et al., 2008; Vetting et al., 2008). In the present study, we explored the diversity of AAC(6′)-Ib in various ecological niches to demonstrate whether such polymorphic diversity occurring in nature is correlated with function: prominent polymorphic variants of AAC(6′)-Ib were identified and their functional implications were studied to reveal the extended resistance spectra of novel AAC(6′)-Ib variants in the natural environment.
Materials and methods
Sampling and DNA extraction
Information on the sampling sites is provided in (Supplementary Table 1). The geographical location of the sampling sites and the population density of the nearest cities are displayed in Supplementary Figure 1. River samples were obtained from four sites of Han river and two sites of Hancheon river. Water samples were also collected from different types of wastewater treatment plants (urban, university and hospital). Soil samples were taken from six mountain and seven agricultural sites. Information on human fecal samples is given in Supplementary Table 2. For the chicken intestine samples, the luminal contents of the ileum and cecum from six broiler chickens (RSS 308) assessed in a previous study were used (Choi et al., 2014). Environmental DNA from water and soil samples, human fecal DNA, and chicken intestinal content DNA were extracted using FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA), QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA) and UltraClean Fecal DNA kit (MO BIO Laboratory, Carlsbad, CA, USA), respectively. The experimental procedures followed the manufacturer’s instructions.
Targeted gene sequencing
A strategy for targeted gene sequencing of AAC(6′)-Ib protein was described in Supplementary Figure 2. PCR primers and conditions for the amplification of the aac(6′)-Ib gene from environmental DNA followed the method of Park et al. (Park et al., 2006). The barcode primers are provided in Supplementary Table 3. Pfu polymerase (Bioneer Co., Daejeon, Korea) was employed for amplification. The obtained amplicon was sequenced using the 454 GS-FLX+ platform (Roche, Branford, CT, USA) at Chunlab (Seoul, Korea). The MOTHUR software platform (v.1.35.1) (Schloss et al., 2009) was used for allocating raw sequences into different samples according to barcode sequences and removing low-quality and shot-length sequence reads (<350 bp). Chimeric sequences were removed using the UCHIME software package (Edgar et al., 2011). Samples with an excessive number of sequence reads were sub-sampled to 3000 sequences per sample. Singleton sequences from each sample were discarded. All non-singleton sequences were aligned with the MAFFT software (v.7) (Katoh and Standley, 2013) and trimmed to defined start and end positions. The trimmed sequences were used for translation to obtain protein sequences. Sequencing errors in homopolymer regions were manually edited by comparison with the reference protein (DQ303918). Frame-shifted sequence reads caused by insertions or deletions and stop codon-containing sequence reads were discarded. After translation, nucleotide sequences that encode the same amino acid sequence were assigned to a unique protein sequence. The relative abundance of each protein sequence was determined by the number of sequence reads that were assigned to a unique protein sequence.
Sequence analysis
The adequacy of the sequencing depth was verified by monitoring the saturation of the rarefaction curve. The composition, abundance and phylogenetic distance of unique protein sequences at each sampling site were used for the principal coordinates analysis (PCoA) based on the weighted pairwise UniFrac method (Lozupone and Knight, 2005) implemented in the QIIME software (Caporaso et al., 2010). Shannon entropy values (H′) and the relative abundance of polymorphic amino acids at 133 residues in the AAC(6′)-Ib protein were calculated as previously described (Iwai et al., 2010). Statistical tests were performed using analysis of similarity (ANOSIM) (Clarke, 1993) and non-parametric multivariate analysis of variance (Adonis) (Anderson, 2001) implemented in ‘vegan’ R package (Dixon, 2003).
Construction of phylogenetic tree with relative abundance
The software package Beast (v.2.4.0) (Bouckaert et al., 2014) was used to construct a Bayesian maximum clade credibility phylogenetic tree for all unique AAC(6′)-Ib protein sequences. The Beauti software was employed to create an XML file to run Markov Chain Monte Carlo (MCMC) analyses for ten million states in Beast. The Bayesian analysis was performed using the Yule model and a strict clock model with default parameters. Samples of trees and parameters were collected every 1000 steps after discarding a burn-in of 10%. The log file obtained from the run was subjected to the Tracer program (v.1.6.0) to visualize convergence. A maximum clade credibility tree was generated by TreeAnnotator (v.2.4.0) from the output of the MCMC run after the initial trees were discarded (50%) as burn-in. The type information and the relative abundance of branches in the tree were added using Interactive Tree of Life (iTOL v.3) (Letunic and Bork, 2016).
Construction of AAC(6′)-Ib variants for susceptibility testing and protein expression
The major variants used for the functional analyses were generated by site-directed mutagenesis (Stratagene, La Jolla, CA, USA). A gene encoding AAC(6′)-Ib-cr from the plasmid pAC3 (KM204147) of Aeromonas sp. strain C3 was used to produce the variant genes. Variants IbL, RD, RG and WY were constructed with specific primers according to the manufacturer’s instructions (Supplementary Table 3) and the mutated sequences were confirmed by sequencing analysis (Solgent, Daejeon, Korea). To analyze their contribution to resistance in E. coli, all variants were cloned into the pBR322 and pUC18 plasmids under the control of the natural promoter of AAC(6′)-Ib-cr from pAC3 (Supplementary Table 3). Briefly, the promoter region was cloned into the pBR322 plasmid, and the variant genes were then inserted downstream of the promoter to produce pBR322 constructs. For the pUC18 constructs, the variant genes together with the promoter region were amplified with Pro_F and aac6_R2 primers by PCR and inserted into the pUC18 plasmid (Supplementary Table 3). All variant genes were cloned into the pET-28a plasmid (Novagen, Madison, WI, USA) for heterologous expression and purification of the variant proteins (Supplementary Table 3). Protein expression and purification were performed according to the manufacturer’s instructions (Novagen).
N-Acetyltransferase activity assay
For ciprofloxacin, 2 μg of purified protein, 0.5 mm acetyl-CoA, 0.3 mm ciprofloxacin (Sigma-Aldrich, St. Louis, MO, USA) and 1 mm magnesium chloride were mixed in 200 μl of reaction buffer (50 mm Tris-HCl, pH 8.0). For gemifloxacin, 20 μg of purified protein, 0.5 mm acetyl-CoA, 0.2 mm gemifloxacin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and 1 mm magnesium chloride were mixed in 200 μl of reaction buffer (50 mm Tris-HCl, pH 8.0). The reaction was stopped by the addition of 5% (v/v) of a 2 m hydrochloride solution. The chemical structure of N-acetylated gemifloxacin was determined by LC–MS/MS and proton nuclear magnetic resonance (1H-NMR) analyses. An LTQ Velos mass spectrometer (Thermo Scientific, Waltham, MA, USA) and Accela PDA detector (Thermo Scientific) were used for the identification of N-acetylated fluoroquinolones. The chemical structures of the metabolites were predicted through a comparison of the full-scan and product-ion mass spectra. 1H-NMR analysis was performed using a 600 MHz NMR spectrometer (VNS, Varian Inc., Walnut Creek, CA, USA) and deuterated acetonitrile was used as a solvent. The obtained spectra were analyzed by MestReNova software v.8.1.0 (Mestrelab Research, Spain) according to those from the previous studies (Al-Hadiya and Mahmoud, 2011; Kim et al., 2017). The N-acetylation of fluoroquinolone was analyzed by HPLC. The analysis was performed using an Atlantis dC-18 column (4.6 mm × 250 mm; Waters Corp., Milford, MA, USA) and the Varian ProStar HPLC (Varian Inc., Walnut Creek, CA, USA) system set at 280 nm with a diode-array detector. The mobile phase consisted of a linear gradient from 10% to 95% acetonitrile containing 0.1% formic acid at a flow rate of 1 ml/min. For kanamycin, 0.9-3.5 μg of purified protein, 0.1 mm acetyl-CoA, 0.005% (w/v) kanamycin sulfate (Sigma-Aldrich), and 1 mm 5,5′-dithio-bis-(2,2)-nitroenzoic acid (Sigma-Aldrich) were mixed in 3 ml of reaction buffer (89 mM 4-morpholineethanesulfonic acid buffer, pH 5.7) (Benveniste and Davies, 1971). The increase in A412 was measured. All reactions were performed in duplicate or triplicate and statistical analysis was performed by Student’s t-test. Enzyme units were defined as the amount of enzyme that catalyzed the conversion of 1 nmol of antibiotic substrate to its N-acetylated form per minute at 37 °C.
Antibiotic susceptibility test
The disk-diffusion and micro-broth dilution assays were performed according to protocols based on the CLSI guide (Wiegand et al., 2008). M9 minimal and Mueller Hinton media (Difco Laboratories) were used for the disk-diffusion and broth dilution assays, respectively. pBR322 clones were employed for the broth dilution assay for ciprofloxacin and kanamycin. pUC18 clones were used for the disk-diffusion assay against ciprofloxacin, gemifloxacin and kanamycin and for the broth dilution assay against gemifloxacin.
Nucleotide sequence accession numbers
The nucleotide sequences described in this study have been deposited in the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) with Bioproject accession no. PRJNA360018.
Results
Targeted gene sequencing for AAC(6′)-Ib
Forty-one samples were collected from a variety of ecological niches in South Korea, including rivers, wastewater treatment plants, mountain and agricultural soils, chicken intestines and human feces, and subjected to targeted gene sequencing (Supplementary Figure 1; Supplementary Tables 1 and 2). A partial fragment (482 bp) of the gene (600 bp) was amplified and sequenced using the 454 GS-FLX+ platform. The obtained non-singleton nucleotide sequences (68 667 sequence reads) were processed to produce well-curated protein sequences for diversity analyses. The processed sequences (133 amino acids) covered the complete acetyltransferase domain from residues 54 to 186 of the reference protein (DQ303918). A total of 460 unique protein sequences consisting of 44 026 sequence reads were finally obtained and regarded as environmental variants that represented the diversity of AAC(6′)-Ib (Supplementary Table 4).
Ecological niche-specific diversity and abundance of AAC(6′)-Ib
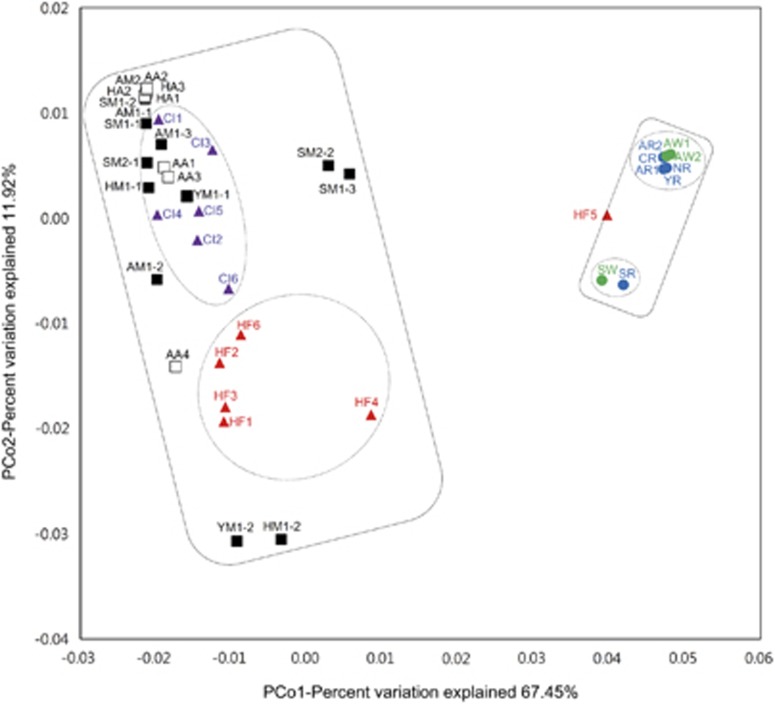
PCoA of weighted UniFrac distances for the AAC(6′)-Ib protein sequence data showed that the AAC(6′)-Ib communities clustered into two distinctive groups according to their ecological niches: (1) soil and intestine and (2) water samples (Figure 1). The water samples were further sub-categorized not by sample source (river and wastewater treatment plants) but instead by geographical location (for example, the metropolitan city Seoul and other rural areas; Figure 1; Supplementary Table 1). The soil samples showed relatively high diversity and the intestine samples were further sub-clustered by their origin (human and poultry). These results suggest that the polymorphic diversity of a single resistance gene can also be determined by ecology, as shown in previous studies on the close correlation between the resistome and ecology (Gibson et al., 2015).
Figure 1.
Principal coordinates analysis of weighted UniFrac distances based on 460 AAC(6′)-Ib protein sequences in various environmental samples. All samples are displayed as follows: river (blue circle), wastewater treatment plants (green circle), mountain soil (black square), agricultural soil (white square), chicken intestine (purple triangle) and human feces (red triangle). The main and sub-categories with statistically significant differences (Supplementary Table 5) are indicated by solid and dashed lines, respectively.
Polymorphisms, abundance and phylogeny of AAC(6′)-Ib variants in various ecological niches
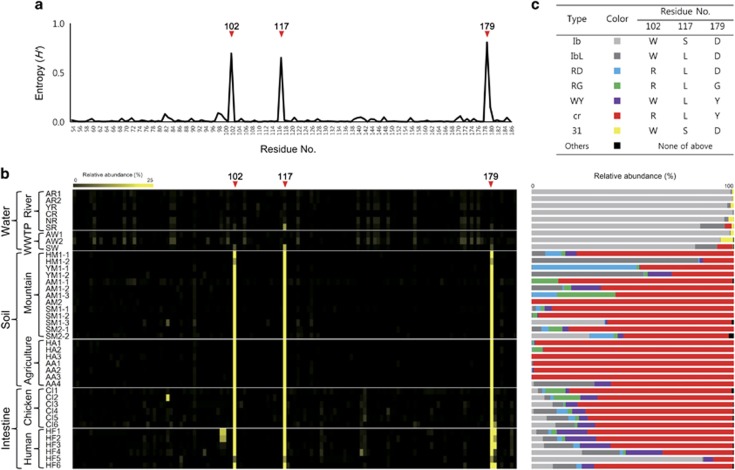
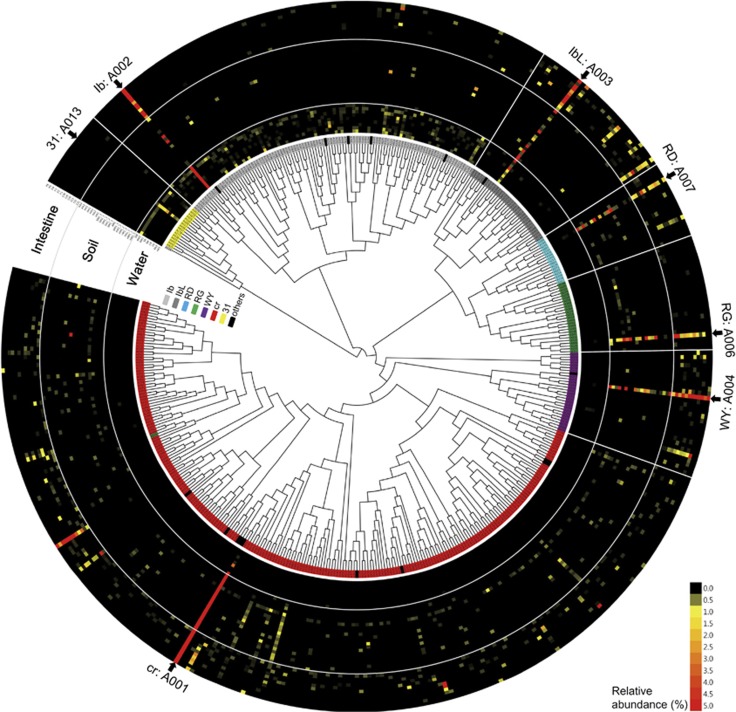
As shown through Shannon entropy analysis of all AAC(6′)-Ib sequences (Figure 2a), most of the amino acid residues were conserved, with the exception of three residues: 102, 117 and 179. When the degree of polymorphism at each AAC(6′)-Ib residue was expressed as a heatmap for the different ecological niches (Figure 2b), these three residues were greatly altered in the soil and intestine samples. Similar patterns of polymorphism in these residues were also shown in the 1395 AAC(6′)-Ib sequences reported in the NCBI GenBank database (Supplementary Figure 3). Remarkably, these residues were the mutation sites in the clinically important variant AAC(6′)-Ib-cr, which was prevalent in the soil and intestinal environments (Figure 2c). Other variants harboring different combinations of amino acid substitutions at these residues were also present in these environments (Figure 2c). Among the 460 unique protein sequences, seven polymorphism-based sequences identified due to polymorphisms at these residues accounted for 84% of the total sequence reads (94.5% in water, 93.9% in soil and 69.2% in intestine; Table 1). Out of 460 unique protein sequences, 444 sequences were assigned to one of seven types, including AAC(6′)-31 type, which was also detected in the amplicon sequencing (Mendes et al., 2007), and the following six polymorphic types of AAC(6′)-Ib: Ib (wild-type AAC(6′)-Ib), IbL (S117L), RD (W102R and S117L), RG (W102R, S117L and D179G), WY (S117L and D179Y) and cr (W102R, S117L and D179Y) (Figure 2c; Table 1). Only 16 unique protein sequences consisting of 0.2% of total sequence reads were not assigned to one of the seven polymorphism-based sequence types and these sequences were designated as ‘others’ in Figures 2c and 3. The composition of the seven major types in each sample was also ecology-specific (Figure 2c), as shown in the PCoA results. The wild-type Ib was dominant in all water samples, whereas the cr type was dominant in the soil and intestine samples. The RD, RG and WY types were mainly detected in the soil and intestine samples, whereas type 31 was found only in the water samples. A Bayesian maximum clade credibility phylogenetic tree of all AAC(6′)-Ib variants showed the diversity and abundance of entire sequences in the different ecological niches (Figure 3). The tree also indicated that 460 unique protein sequences formed distinctive sequence-based lineages congruent with the seven major polymorphism-based sequence types, which represented almost all sequences. Furthermore, PCoA results of weighted Unifrac distances based on seven major types of AAC(6′)-Ib (Supplementary Figure 4) also showed a similar coordination among the different ecosystems to those based on 460 unique protein sequences (Figure 1). The most abundant variant sequences representing each type are shown in the heatmap (arrows in Figure 3), and their relative abundances in the water, soil and intestine samples are presented in Table 1.
Figure 2.
Polymorphism of AAC(6′)-Ib and the relative abundance of representative types in various ecological niches. (a) Shannon entropy at 133 amino acid residues of AAC(6′)-Ib among the entire sequences obtained from all samples. (b) Heatmap of the degree of polymorphism at 133 amino acid residues of AAC(6′)-Ib in each sample. (c) Major types of AAC(6′)-Ib and their relative abundance in each sample. Others (black) indicate sequences that were not assigned to the seven major types.
Table 1. Representative variant sequences of seven major types and their characteristics obtained in this study.
| Representative variant sequence | Type |
Relative abundancea
(%) |
Identityb (%) |
Residue |
Resistance phenotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| Water | Soil | Intestine | 102 | 117 | 179 | ||||
| A002 | Ib | 88.44 | 1.90 | 11.12 | 100.0 | W | S | D | AGR |
| A003 | IbL | 3.53 | 7.20 | 4.35 | 99.2 | W | L | D | AGR |
| A007 | RD | 0.03 | 6.07 | 1.68 | 98.5 | R | L | D | AGR CipR |
| A006 | RG | 0.01 | 4.51 | 3.64 | 97.7 | R | L | G | AGR CipR |
| A004 | WY | 0.01 | 1.99 | 7.05 | 98.5 | W | L | Y | AGR CipR GemR c |
| A001 | cr | 1.34 | 72.28 | 41.35 | 97.7 | R | L | Y | AGR CipR |
| A013 | 31 | 1.09 | 0.00 | 0.00 | 86.5 | W | S | D | AGR |
| Total | 94.49 | 93.94 | 69.20 | ||||||
Abbreviations: AG, aminoglycoside; Cip, ciprofloxacin; Gem, gemifloxacin; R, resistance.
Relative abundance indicates percentage of each sequence in different samples.
Sequence identity with the representative variant sequence (A002) of the wild type (Ib).
GemR, low-level resistance.
Figure 3.
Phylogenetic diversity of the AAC(6′)-Ib variants and their relative abundance in various ecological niches. Bayesian maximum clade credibility phylogeny and abundance profiles of 460 unique protein sequences of AAC(6′)-Ib were presented for 41 environmental samples. Each phylogenetic lineage is assigned to one of seven major types of AAC(6′)-Ib and is colored differently. Others (black) indicate sequences that were not assigned to the seven major types. Representative variant sequences of each type (Table 1) are indicated with arrows.
N-Acetylation activity and altered resistance of the AAC(6′)-Ib variants
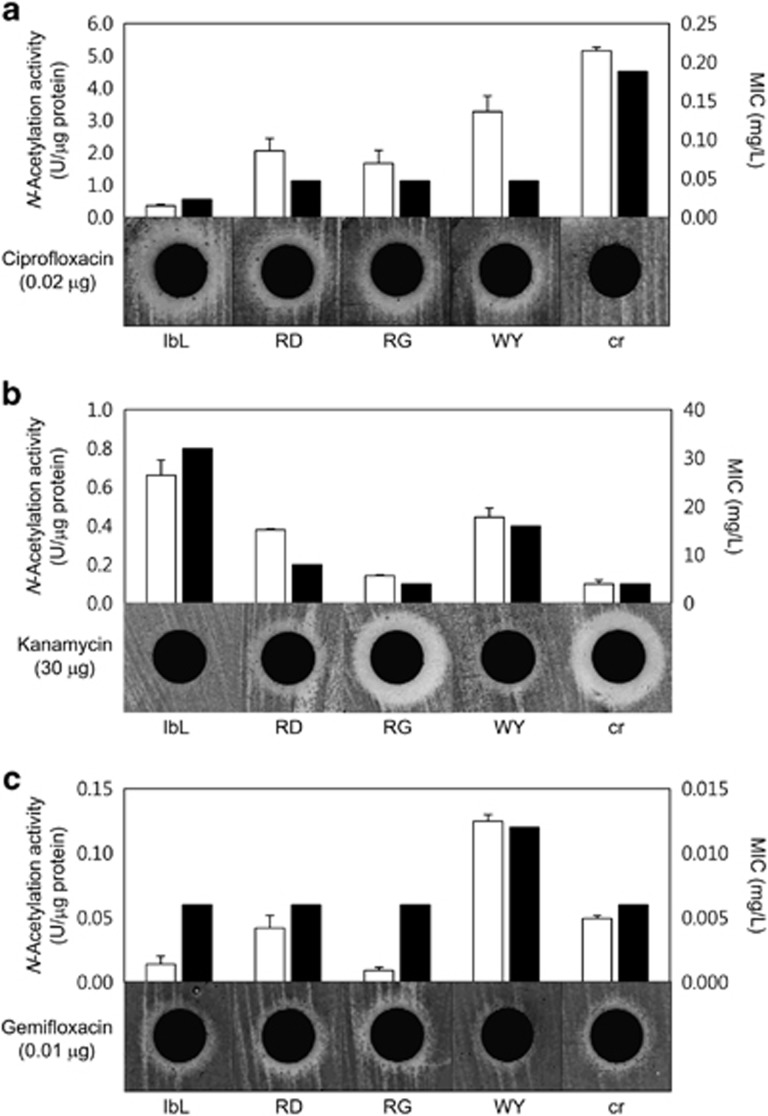
Polymorphisms of TEM β-lactamase have previously been implicated in causing the extended spectrum of antibiotic resistance (Salverda et al., 2010). A total of 5411 protein sequences obtained from the NCBI GenBank database exhibited variations in critical residues (Supplementary Figure 5), and these variations were closely related to their extended resistance spectra (Salverda et al., 2010). In the case of AAC(6′)-Ib, the cr variant was previously shown to have extended substrate specificity conferring resistance to both kanamycin and ciprofloxacin (Robicsek et al., 2006). Considering that the present study also revealed such polymorphism, it is possible that environmental variants of AAC(6′)-Ib might be related to their functionality. Among the seven major types, the IbL (control), RD, RG, WY and cr variants were selected for functional characterization because these variants are all identical except for the polymorphic residues 102 and 179. Corresponding recombinant proteins constructed by site-directed mutagenesis were purified, and their N-acetylation activity against ciprofloxacin was analyzed. The cr variant displayed the highest N-acetylation activity against ciprofloxacin (Figure 4a), and other variants such as RD, RG and WY also showed activities higher than IbL, which is known to exhibit a basal level of activity (Robicsek et al., 2006). The ciprofloxacin susceptibility of E. coli cells harboring these variant genes was tested through the disk-diffusion assay and the broth dilution method. As previously reported (Robicsek et al., 2006), the cr variant showed prominent resistance to ciprofloxacin compared with IbL, which was tested as the wild type (Figure 4a). Additionally, the RD, RG and WY variants displayed increased resistance to ciprofloxacin, although this resistance was low compared with that of the cr variant (Figure 4a), suggesting that the increased enzyme activity of these variants conferred the resistant phenotype (Table 1). The D179Y mutation in the WY variant was proposed to be important for fluoroquinolone N-acetylation activity, whereas the W102R mutation in the RD variant was not considered critical (Maurice et al., 2008; Vetting et al., 2008). Interestingly, when the RD and WY variants acquired ciprofloxacin resistance, they displayed less reduction in kanamycin N-acetylation activity and resistance than the cr variant, which exhibited a significant reduction as a trade-off (Figure 4b).
Figure 4.
N-Acetylation activities of the AAC(6′)-Ib variants and the related resistance against (a) ciprofloxacin, (b) kanamycin and (c) gemifloxacin. The N-acetylation activities of the purified variant enzymes are indicated with white bars. Statistical results (t-test) were provided in Supplementary Table 6. The altered resistance of E. coli DH5α harboring the variant genes is shown based on minimal inhibitory concentration values (black bars) and images of the disk-diffusion assay results.
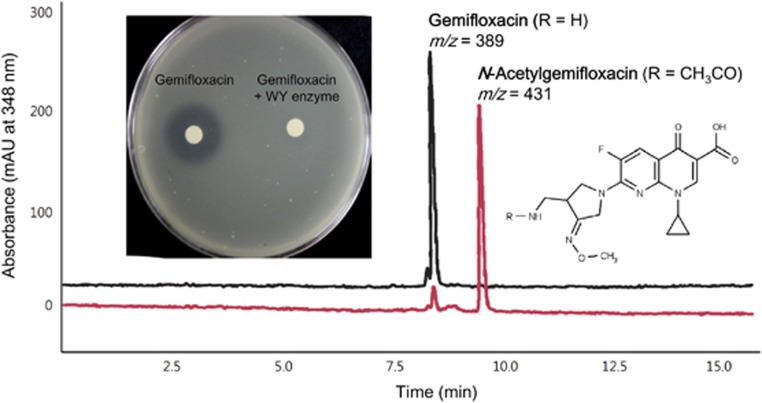
To further evaluate the extended resistance spectra of the AAC(6′)-Ib variants, various antibiotics containing primary or secondary amine moieties were also tested for N-acetylation activity. The 4th-generation fluoroquinolone gemifloxacin, against which the cr variant was previously known not to be resistant (Robicsek et al., 2006), was found to be N-acetylated by the WY variant (Figure 4c; Supplementary Tables 7 and 8). Although the gemifloxacin N-acetylation activity of the WY variant was much lower than the ciprofloxacin N-acetylation activity of the cr variant, the activity was sufficient to confer increased resistance to gemifloxacin compared with the other variants (Figure 4c; Table 1). Inactivation of gemifloxacin through N-acetylation was demonstrated using the disk-diffusion assay (Figure 5). Other fluoroquinolones that were recently developed, including tosufloxacin, clinafloxacin, moxifloxacin and zabofloxacin, were also tested with the WY variant; only zabofloxacin was proven to be a substrate of the WY variant (data not shown), suggesting that the aminoalkyl and methoxyimino groups at the pryrrolidine ring are important for binding at the catalytic site. Homology modeling based on protein-ligand binding suggested that the D179Y mutation was critical for recruiting and stacking the quinolone ring into the binding pocket via π-interactions as previously demonstrated (Maurice et al., 2008; Vetting et al., 2008), but the W102R mutation in the cr variant could result in a potential steric clash with the methoxy group of gemifloxacin (data not shown).
Figure 5.
N-Acetylation of gemifloxacin by the WY variant resulting in the loss of antimicrobial activity. The antimicrobial activities of gemifloxacin and N-acetylgemifloxacin were tested via disk-diffusion assays using authentic gemifloxacin and a reaction mixture of gemifloxacin and the WY variant enzyme against E. coli DH5α. The complete conversion of gemifloxacin was confirmed by HPLC analysis.
Discussion
The use of targeted gene sequencing approach has been emphasized to reveal the vast diversity of antibiotic resistance genes in the environment (Schmieder and Edwards, 2012). The results of our study showed that the polymorphic diversity of AAC(6′)-Ib was a natural phenomenon in the environment and that certain dominant polymorphisms were ecological niche-specific. The prevalence of the cr type as a dominant polymorphism in soil where the antibiotic selection pressure was absent suggested that the clinically relevant variant was a naturally-occurring polymorphic form. Additionally, the soil and intestine samples showed close relationships in their polymorphic diversity and the compositions of their major types (Figures 2c and 3). These results coincide with previous findings regarding interconnections between the resistomes of the human gastrointestinal tract, soil and clinical pathogens (Benveniste and Davies, 1973; Forsberg et al., 2012; Woolhouse et al., 2015). Comparative genomics analysis also revealed an inter-link of antibiotic resistance between humans and livestock (Mather et al., 2013; Ward et al., 2014). Therefore, the soil, animal and human microbiomes are considered major reservoirs of antibiotic resistance (D'Costa et al., 2007; O'Toole, 2014; Woolhouse et al., 2015). The cr variant was also found in hospital wastewater (SW) and Han river samples (SR) downstream of metropolitan Seoul but not in other water samples from much less populated regions (Figures 1 and 2c; Supplementary Figure 1). These results suggest that anthropogenic activity may influence the emergence of certain antibiotic resistance genes. In previous studies, it was proposed that steady increase in the clinical use of ciprofloxacin during the 1990s has generated selection pressure for the cr variant (Park et al., 2006; Robicsek et al., 2006; Jacoby et al., 2009). Moreover, several reports have shown that human activity is closely associated with the antibiotic resistome (Perry and Wright, 2014; Stalder et al., 2014; Li et al., 2015).
While the cr variant was previously reported to inactive ciprofloxacin and aminoglycosides (Robicsek et al., 2006), other types of polymorphic variants were functionally characterized in this study. A novel N-acetylation activity of the WY variant against gemifloxacin was discovered and the resulting resistance was demonstrated. According to previous reports (Baquero, 2001; Andersson and Hughes, 2014; Martinez et al., 2015), this low-level resistance in the environment should not be overlooked because it could be ‘a gateway to clinical resistance’, as previously shown for the cr variant (Robicsek et al., 2006; Hawkey and Jones, 2009). Furthermore, the altered resistance spectra of environmental variants may confer advantages under certain environmental conditions in various ecological niches. Indeed, we observed different enzyme activities of AAC(6′)-Ib variants under different pH levels and magnesium ion concentrations (Supplementary Figure 6).
Since these variants were discovered only in metagenomic sequences, attempts to isolate environmental bacteria harboring these variant sequences were conducted using four mountain soil samples tested for targeted gene sequencing. As a result, the cr variant was found to be a dominant type in soil isolates harboring the aac(6′)-Ib gene; other major variants, such as RD, RG and WY, were also detected (data not shown). Resistance to such a novel antibiotic in environmental bacteria may lead to the emergence of novel resistance in the clinic. Indeed, some major variants, such as the RD and WY variants, have recently been reported in clinical and animal isolates and have been found to be associated with mobile genetic elements (Supplementary Table 9; Moura et al., 2012; Deng et al., 2014; Vredenburg et al., 2014; Vaz-Moreira et al., 2016), suggesting that they may have already been disseminated to the clinic as a potential source of resistance. Since 460 variant sequences identified in this study were mostly not reported in the database, other novel polymorphic variants related to resistance could still be present. For example, the substitution G180S in the cr variant was prevalent in the intestine samples (8.4% of the total sequence reads of the intestine samples; Figures 2b and 3).
In addition to the previous discovery in polymorphic mutations conferring resistance to two different classes of antibiotics in the clinical isolate of the cr variant (Robicsek et al., 2006), our study demonstrated the altered resistance spectra of other polymorphic variants, including the novel gemifloxacin resistance of the WY variant. More extensive analysis of other environmental variants to determine their functionality will lead to the identification of a wider spectrum of multi-drug resistance and novel resistance mechanisms. In conclusion, our results suggested that polymorphisms occurring in nature could alter the substrate specificity of antibiotic-inactivating enzymes and therefore determine resistance spectra, ultimately expanding the antibiotic resistome throughout the environment.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF-2013R1A1A2012270), Ministry of Education, Science & Technology and also funded by the Korea Ministry of Environment (MOE) as ‘the Environmental Health Action Program (2016001350004)’.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Al-Hadiya BM, Mahmoud AM.2011. Gemifloxacin. Profiles Drug Subst Excip Relat Methodol 36: 151–168. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. 2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology 26: 32–46. [Google Scholar]
- Andersson DI, Hughes D. 2014). Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol 12: 465–478. [DOI] [PubMed] [Google Scholar]
- Baquero F. 2001). Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat 4: 93–105. [DOI] [PubMed] [Google Scholar]
- Benveniste R, Davies J. 1971). Enzymatic acetylation of aminoglycoside antibiotics by Escherichia coli carrying an R factor. Biochemistry 10: 1787–1796. [DOI] [PubMed] [Google Scholar]
- Benveniste R, Davies J. 1973). Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci USA 70: 2276–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D et al. 2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray G, Mazel D. 2008). Synonymous genes explore different evolutionary landscapes. PLoS Genet 4: e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. 2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Yang Y, Liang X, Yu K, Zhang T, Li X. 2013). Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ Sci Technol 47: 12753–12760. [DOI] [PubMed] [Google Scholar]
- Choi JH, Kim GB, Cha CJ. 2014). Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult Sci 93: 1942–1950. [DOI] [PubMed] [Google Scholar]
- Clarke KR. 1993). Non-parametric multivariate analyses of changes in community structure. Austral J Ecol 18: 117–143. [Google Scholar]
- D'Costa VM, Griffiths E, Wright GD. 2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol 10: 481–489. [DOI] [PubMed] [Google Scholar]
- D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C et al. 2011). Antibiotic resistance is ancient. Nature 477: 457–461. [DOI] [PubMed] [Google Scholar]
- Demaneche S, Sanguin H, Pote J, Navarro E, Bernillon D, Mavingui P et al. 2008). Antibiotic-resistant soil bacteria in transgenic plant fields. Proc Natl Acad Sci USA 105: 3957–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YT, Wu YL, Tan AP, Huang YP, Jiang L, Xue HJ et al. 2014). Analysis of antimicrobial resistance genes in Aeromonas spp. isolated from cultured freshwater animals in China. Microb Drug Resist 20: 350–356. [DOI] [PubMed] [Google Scholar]
- Dixon P. 2003). VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley RL, Collignon P, Larsson DG, McEwen SA, Li XZ, Gaze WH et al. 2013). The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57: 704–710. [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. 2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JC, Gonzalez-Candelas F, Rolain JM, Canton R. 2013). Antibiotics as selectors and accelerators of diversity in the mechanisms of resistance: from the resistome to genetic plasticity in the β-lactamases world. Front Microbiol 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MK, Forsberg KJ, Dantas G. 2015). Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 9: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey PM, Jones AM. 2009). The changing epidemiology of resistance. J Antimicrob Chemother 64(Suppl 1): i3–i10. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N et al. 2013). Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun 4: 2151. [DOI] [PubMed] [Google Scholar]
- Iwai S, Chai B, Sul WJ, Cole JR, Hashsham SA, Tiedje JM. 2010). Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J 4: 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA, Gacharna N, Black TA, Miller GH, Hooper DC. 2009). Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob Agents Chemother 53: 1665–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Thawng CN, Lee SH, Cha CJ. 2017). Unique features of Aeromonas plasmid pAC3 and expression of the plasmid-mediated quinolone resistance genes. mSphere 2: e00203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cantarel B, Henrissat B, Gevers D, Birren BW, Huttenhower C et al. 2014). Gene-targeted metagenomic analysis of glucan-branching enzyme gene profiles among human and animal fecal microbiota. ISME J 8: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yang Y, Ma L, Ju F, Guo F, Tiedje JM et al. 2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J 9: 2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. 2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Coque TM, Baquero F. 2015). What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 13: 116–123. [DOI] [PubMed] [Google Scholar]
- Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, Harris SR et al. 2013). Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341: 1514–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice F, Broutin I, Podglajen I, Benas P, Collatz E, Dardel F. 2008). Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep 9: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Castanheira M, Toleman MA, Sader HS, Jones RN, Walsh TR. 2007). Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac(6')-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob Agents Chemother 51: 2611–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Pereira C, Henriques I, Correia A. 2012). Novel gene cassettes and integrons in antibiotic-resistant bacteria isolated from urban wastewaters. Res Microbiol 163: 92–100. [DOI] [PubMed] [Google Scholar]
- Nesme J, Cecillon S, Delmont TO, Monier JM, Vogel TM, Simonet P. 2014). Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol 24: 1096–1100. [DOI] [PubMed] [Google Scholar]
- O'Toole DK. 2014). The natural environment may be the most important source of antibiotic resistance genes. MBio 5: e01285–e01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006). Prevalence in the United States of aac(6')-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50: 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Wright GD. 2013). The antibiotic resistance ‘mobilome’: searching for the link between environment and clinic. Front Microbiol 4: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Wright GD. 2014). Forces shaping the antibiotic resistome. Bioessays 36: 1179–1184. [DOI] [PubMed] [Google Scholar]
- Ramirez MS, Nikolaidis N, Tolmasky ME. 2013). Rise and dissemination of aminoglycoside resistance: the aac(6')-Ib paradigm. Front Microbiol 4: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH et al. 2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12: 83–88. [DOI] [PubMed] [Google Scholar]
- Salverda ML, De Visser JA, Barlow M. 2010). Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev 34: 1015–1036. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. 2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2012). Insights into antibiotic resistance through metagenomic approaches. Future Microbiol 7: 73–89. [DOI] [PubMed] [Google Scholar]
- Stalder T, Barraud O, Jove T, Casellas M, Gaschet M, Dagot C et al. 2014). Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J 8: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Moreira I, Varela AR, Pereira TV, Fochat RC, Manaia CM. 2016). Multidrug resistance in quinolone-resistant Gram-negative bacteria isolated from hospital effluent and the municipal wastewater treatment plant. Microb Drug Resist 22: 155–163. [DOI] [PubMed] [Google Scholar]
- Vetting MW, Park CH, Hegde SS, Jacoby GA, Hooper DC, Blanchard JS. 2008). Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6')-Ib and its bifunctional, fluoroquinolone-active AAC(6')-Ib-cr variant. Biochemistry 47: 9825–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredenburg J, Varela AR, Hasan B, Bertilsson S, Olsen B, Narciso-da-Rocha C et al. 2014). Quinolone-resistant Escherichia coli isolated from birds of prey in Portugal are genetically distinct from those isolated from water environments and gulls in Portugal, Spain and Sweden. Environ Microbiol 16: 995–1004. [DOI] [PubMed] [Google Scholar]
- Walkiewicz K, Benitez Cardenas AS, Sun C, Bacorn C, Saxer G, Shamoo Y. 2012). Small changes in enzyme function can lead to surprisingly large fitness effects during adaptive evolution of antibiotic resistance. Proc Natl Acad Sci USA 109: 21408–21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MJ, Gibbons CL, McAdam PR, van Bunnik BA, Girvan EK, Edwards GF et al. 2014). Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl Environ Microbiol 80: 7275–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I, Hilpert K, Hancock RE. 2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3: 163–175. [DOI] [PubMed] [Google Scholar]
- Woolhouse M, Ward M, van Bunnik B, Farrar J. 2015). Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci 370: 20140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. 2015). High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. MBio 6: e02288–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.