Abstract
Vertebrate (cellular retinaldehyde-binding protein) and Drosophila (prolonged depolarization afterpotential is not apparent [PINTA]) proteins with a CRAL-TRIO domain transport retinal-based chromophores that bind to opsin proteins and are necessary for phototransduction. The CRAL-TRIO domain gene family is composed of genes that encode proteins with a common N-terminal structural domain. Although there is an expansion of this gene family in Lepidoptera, there is no lepidopteran ortholog of pinta. Further, the function of these genes in lepidopterans has not yet been established. Here, we explored the molecular evolution and expression of CRAL-TRIO domain genes in the butterfly Heliconius melpomene in order to identify a member of this gene family as a candidate chromophore transporter. We generated and searched a four tissue transcriptome and searched a reference genome for CRAL-TRIO domain genes. We expanded an insect CRAL-TRIO domain gene phylogeny to include H. melpomene and used 18 genomes from 4 subspecies to assess copy number variation. A transcriptome-wide differential expression analysis comparing four tissue types identified a CRAL-TRIO domain gene, Hme CTD31, upregulated in heads suggesting a potential role in vision for this CRAL-TRIO domain gene. RT-PCR and immunohistochemistry confirmed that Hme CTD31 and its protein product are expressed in the retina, specifically in primary and secondary pigment cells and in tracheal cells. Sequencing of eye protein extracts that fluoresce in the ultraviolet identified Hme CTD31 as a possible chromophore binding protein. Although we found several recent duplications and numerous copy number variants in CRAL-TRIO domain genes, we identified a single copy pinta paralog that likely binds the chromophore in butterflies.
Keywords: phototransduction, chromophore, retinol-binding proteins, tandem duplication, copy number variations
Introduction
Phenotypic differences between organisms may be driven by small nucleotide changes in protein coding or regulatory regions, or by whole gene or genome duplications (Stern 2000; Hersh and Carroll 2005; Demuth et al. 2006). Gene duplications in particular are hypothesized to be an important mechanism for evolutionary change because these events give rise to new material for novelties and may facilitate the emergence of new genes (Ohno 1970; Long et al. 2003). Often, gene duplications result in pseudogenization. However, there are at least two mechanisms by which duplicated genes can remain functional regardless of redundancy: 1) in neofunctionalization, a duplicated gene develops a new function different from the ancestral gene and 2) in subfunctionalization the two paralogs each have part of the function of an ancestral gene (Lynch and Conery 2000; Long et al. 2003; Zhang 2003). Gene duplications and rearrangements have resulted in large gene families. Genes are classified as part of a gene family when they share common sequence motifs and sometimes may have related general functions (Henikoff et al. 1997).
Lineage-specific gene family expansions are hypothesized to be a mechanism by which eukaryotic species can adapt and diversify (Lespinet et al. 2002). In support of this, studies in mammals suggest that changes to the size of large gene families are likely arising through lineage specific gene loss or gain rather than by changes in gene number at branch sites (Demuth et al. 2006). Gene families that are subject to expansions or reductions have a wide variety of functions, including immunity and sensory perception (Cooper et al. 2007; Dopman and Hartl 2007; Conrad et al. 2010). Chemosensory genes in particular have been widely studied in a number of species and have been found to vary in copy number between and within species (Nei et al. 2008; Nozawa and Nei 2008). Copy number variation (CNV) is a DNA segment 1 kb or longer whose copy number varies between individuals, as a result of recent gene duplications or deletions (Stranger et al. 2007). Insects have been studied for CNV by focusing on gene families with lineage-specific duplications; these genes are candidates for CNVs (Zhang 2003). As an example, the butterfly Heliconius melpomene and the pea aphid Acyrthosiphon pisum both have lineage-specific gene expansions and CNV of olfactory and gustatory receptors correlated with host plant specialization (Briscoe et al. 2013; Duvaux et al. 2015).
The CRAL-TRIO domain gene family is another family that is evolving by lineage-specific duplication in insects and has undergone an expansion in Lepidoptera (moths and butterflies; Smith and Briscoe 2015). Lepidoptera thus have almost twice as many CRAL-TRIO domain genes relative to other insects (Smith and Briscoe 2015). The lineage-specific duplications of this gene family make it a candidate to study for CNV (Zhang 2003). Furthermore, the specific functions of the members of this family remain unknown, with one or two exceptions. The CRAL-TRIO domain is an N-terminal structural region, ∼170 amino acids long, common to several proteins that bind and transport tocopherols (Panagabko et al. 2003; Sigrist et al. 2012). The CRAL-TRIO domain gene family includes a cellular retinaldehyde-binding protein (CRALBP) which is essential to vertebrate vision due to its function in chromophore transport (Wu et al. 2006). The visual pigment chromophore is derived from vitamin-A. In photoreceptor cells, opsin proteins bind a chromophore molecule (in humans 11-cis-retinal and in butterflies 11-cis-3-hydroxyretinal) to form rhodopsin. Rhodopsin initiates the phototransduction cascade when photon absorption changes the chromophore configuration from 11-cis to all-trans (von Lintig et al. 2010). In humans, mutations to CRAL-TRIO domain genes result in a variety of retinal and neurological diseases (Maw et al. 1997; Bomar 2003; Min et al. 2003). Mutations in RLBP1, the gene encoding CRALBP in humans, results in retinitis pigmentosa (Maw et al. 1997) and mutations in a gene encoding αTTP results in ataxia with vitamin E deficiency (AVED) (Min et al. 2003). Moreover, mutations in human Atcay, a CRAL-TRIO domain containing gene, are associated with Cayman ataxia, and a mouse homolog of atcay causes ataxia and dystonia in jittery mice (Bomar 2003).
In insect genomes, CRAL-TRIO domain genes are numerous, however, their function remains largely unexplored except for prolonged depolarization afterpotential is not apparent (pinta). PINTA in Drosophila is a CRAL-TRIO domain protein belonging to the SEC14 superfamily that, similar to CRALBP, shuttles the chromophore from retinal pigment cells to photoreceptor cells (Wang and Montell 2005). PINTA protein is required for the biosynthesis of rhodopsin. Drosophila with mutated pinta genes have low expression of Rh1, the protein component of the light-sensitive rhodopsin found in R1-6 photoreceptor cells (Wang and Montell 2005). Similarly, another member of the SEC14 superfamily squid RALBP functions in retinal binding in cephalopods (Ozaki et al. 1994; Speiser et al. 2014). Although there is an expansion of CRAL-TRIO domain genes in Lepidoptera, no pinta ortholog has been found in this group. The functions of CRALBP and PINTA suggest that a distinct CRAL-TRIO domain protein might be serving an essential role in lepidopteran visual systems by transporting the chromophore.
Presently, most of our knowledge about photoreceptor determination, phototransduction, and chromophore transport comes from studies in Drosophila. However, a recent analysis of 80 vision genes in the Manduca sexta genome (Kanost et al. 2016) found that at least four gene families involved in photoreceptor differentiation pathways have undergone lepidopteran-specific gene duplications including corkscrew, embryonic lethal/abnormal vision, rhabdomeric opsins, and genes encoding CRAL-TRIO domain containing proteins. Since CRAL-TRIO domain genes have undergone an expansion in Lepidoptera and their functions in other organisms suggest a role in vision, it is worth exploring the function of these genes in a butterfly species. Heliconius melpomene provides a good system in which to investigate the evolution of CRAL-TRIO domain genes due to the availability of a reference genome and a growing collection of resequenced genomes (Martin et al. 2013; Davey et al. 2016). In addition, we have generated RNA-Seq data from Heliconius melpomene tissues with which to investigate the expression of the CRAL-TRIO domain genes.
Here, we aim to 1) characterize the molecular evolution of the CRAL-TRIO domain gene family and to 2) identify a candidate gene for chromophore transport in butterflies. We used RNA-Seq data from H. melpomene head, antennae, legs, and mouth parts to make a de novo transcriptome assembly from which to identify CRAL-TRIO domain gene transcripts. We also investigated the reference genome to search for any CRAL-TRIO domain genes that may be found in the genome but not expressed in the tissues we sampled. We found support for the expansion of the CRAL-TRIO domain gene family in butterflies by identifying 43 CRAL-TRIO domain genes in the H. melpomene genome comparable with the 42 found in Manduca sexta (Smith and Briscoe 2015). We also investigated 18 resequenced H. melpomene genomes (Martin et al. 2013) for structural variation (specifically CNV) and found that 32 of the 43 genes in the reference genome had either a large duplication or deletion in at least one of the resequenced genomes. Further, to identify a CRAL-TRIO domain gene functioning in vision, we did a differential expression analysis between tissue types and found one CRAL-TRIO domain gene (Hme CTD31) that is upregulated in head tissue. RT-PCR and immunohistochemistry shows that Hme CTD31 is expressed in the compound eye and not the brain, and Hme CTD31 is localized to the retinal pigment and trachea cells making it a candidate chromophore binding protein. We used mass spectrometry to sequence eye proteins associated with an ultraviolet fluorescing pigment and found a match for our CRAL-TRIO domain protein Hme CTD31. These various lines of evidence suggest that we have found a CRAL-TRIO domain gene that binds the chromophore in butterflies.
Materials and Methods
CRAL-TRIO Domain Gene Phylogeny
A phylogeny (Smith and Briscoe 2015) was expanded by adding homologs of the CRAL-TRIO domain gene family found in H. melpomene. Smith and Briscoe identified CRAL-TRIO domain genes from the genomes of Manduca sexta, Danaus plexippus, Drosophila melanogaster, Anopheles gambiae, Apis mellifera, Tribolium castaneum, and Bombyx mori (Smith and Briscoe 2015). To expand this repertoire, we used BLAST + (Basic Local Alignment Search Tool) (Camacho et al. 2009) to identify CRAL-TRIO domain gene homologs in a de novo transcriptome of H. melpomene rosina. These contig sequences were extracted and added to the alignment. Contig nucleotide sequences were translated and curated in MEGA by finding the correct reading frame from start to stop codon. Sequences with missing homologs were blasted against the H. melpomene melpomene reference genome v. 2 (Davey et al. 2016), from which additional sequences were recovered. Manual annotations of the genes not included in the transcriptome and not annotated in the reference genome were done by extracting the nucleotide sequence around the area where there was a BLAST hit to a CRAL-TRIO domain gene. The extracted nucleotides were annotated and translated in AUGUSTUS (Stanke and Morgenstern 2005) and aligned to a BLAST output of the genome to correct the sequence. 215 amino acid sequences were aligned using MUSCLE (Edgar 2004) with default settings, and this alignment was then modified manually. A Bayesian phylogenetic tree was made using MrBayes (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2011) with a BLOSUM62 (Henikoff and Henikoff 1992) model for 1,000,000 generations. The phylogeny was color coded using iTOL (Letunic and Bork 2016).
Structural Variation
To detect copy number variation (CNV) in these genes, we aligned reads for 18 resequenced H. melpomene genomes generated by Martin et al. (Martin et al. 2013), European Nucleotide Archive: ERP002440. Read mapping to the reference genome for four subspecies (six H. melpomene melpomene, four H. melpomene rosina, four H. melpomene amaryllis, and four H. melpomene aglaope) was performed using bwa (Li and Durbin 2009), and samtools was used to index and sort the files (Li et al. 2009). Pindel was used to examine mapping results to detect structural variation (Ye et al. 2009). Pindel looks for read pairs for which one read maps uniquely to the genome while the other read is unmapped to determine the structural breakpoint and direction of unmapped reads (Ye et al. 2009).
RNA Library Preparation
We extracted RNA from whole heads (excluding antennae and mouth parts) of three male and three female H. melpomene butterflies. We also extracted RNA from the head, antennae, legs, and mouth parts (lapial palps + proboscis) of one male and one female H. melpomene specimen to increase our biological replicates to n = 4. Butterflies were placed in −80 °C and stored until RNA extraction. RNA was extracted using TRIzol (Life Technologies, Grand Island, NY) and purified using a NucleoSpin RNA II kit (Macherey-Nagel, Bethlehem, PA). Purified RNA was quantified using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY) and quality checked using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). A TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA) was used to prepare sequencing libraries. Libraries with distinct adapter sequences were quantified, quality checked, normalized, and pooled according to their concentrations. Pooled libraries were run on a 2% agarose gel. A Geneclean III kit (MP Biomedical, Santa Ana, CA) was used to recover DNA from the gel (∼240–600 bp), and Agencourt AMPure XP (Beckman Coulter, Brea, CA) beads were used for a second purification. Sequencing was conducted at the UCI Genomics High-Throughput Facility using a HiSeq 2500 (Illumina, San Diego, CA), paired end 100-cycle sequence run.
Assembly and Read-Mapping
RNA-Sequencing data were obtained for three H. melpomene males and three female antennae, legs, and mouth parts from a previous RNA-Seq study (ArrayExpress: E-MTAB-1500) (Briscoe et al. 2013). We created eight new head libraries from four males and four females. In addition, we made a new antennae, legs, and mouth parts library for one H. melpomene male and one female. The raw sequencing data for the 14 new libraries were deposited in ArrayExpress archive under E-MTAB-6249 and E-MTAB-6342. All libraries were parsed using custom perl and python scripts. A de novo transcriptome assembly was constructed using Trinity (Grabherr et al. 2011; Haas et al. 2013) by including one library per tissue type (head, legs, antennae, mouthparts) for one male and one female, eight libraries total. We made a de novo assembly because the CRAL-TRIO domain genes were not all annotated in the genome and a transcriptome recovered more sequences that were complete. The reference transcriptome was deposited in Dryad under doi: 10.5061/dryad.857n9. Each sequenced library was then mapped back to the reference assembly using RSEM (Li and Dewey 2011) from which we extracted raw read count data, FPKM (Fragments Per Kilobase of exon per Million fragments mapped), and TPM (Transcripts Per Kilobase Million). FPKM was further normalized using NOISeq (Tarazona et al. 2011).
Since some of the CRAL-TRIO domain genes were not recovered in the transcriptome, we manually annotated these genes and read mapped each library as described earlier to the nucleotide sequences of the 43 genes. TPM expression was scaled to the values of whole-transcriptome analysis. We then used two-way ANOVAs to test if these genes varied by sex, tissue type, or sex and tissue type interaction.
edgeR
We performed differential gene expression analysis for all Trinity assembled contigs using edgeR (Robinson and Smyth 2007, 2008; Robinson et al. 2010; Mccarthy et al. 2012). To analyze genes differentially expressed by tissue type, we did pairwise comparisons of head versus antennae, head versus legs, and head versus mouth parts using a generalized linear model with terms for tissue, sex, the interaction of sex and tissue and included a term to correct for batch effects of sequencing on different lanes (∼batch + tissue + sex + sex×tissue). Each analysis included filtering to remove contigs expressed at <1 count per million (CPM) for at least four groups, and between sample normalization using a trimmed mean of the log expression ratios (TMM) (Robinson and Oshlack 2010). Contigs were considered significantly differentially expressed when the false discovery rate (FDR) was <0.05 and the log fold change (logFC) was >1. We did FDR corrections using the qvalue package and using a Bonferroni correction (Storey and Tibshirani 2003; Dabney and Storey 2013).
RT-PCR
To localize where in the head the candidate gene was expressed, we performed reverse transcription polymerase chain reaction (RT-PCR) using RNA from a single individual male and female H. melpomene antennae, retina, and brain tissue. Animals were sacrificed a day after eclosion by squeezing the thorax. Heads were dissected in petri dishes in Ringer’s solution, the retina and brain tissue were transferred to 1.7-ml microtubules on ice. Total RNA was extracted from these tissues using TRIzol (Life Technologies, Grand Island, NY) and quantified using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY). RNA was treated with DNAse I (Fisher Scientific, Pittsburgh, PA). Primers were designed using Primer3 (supplementary table S1, Supplementary Material online) (Koressaar and Remm 2007; Untergasser et al. 2012). Each 25 μl reaction had 2.5 μl Choice PCR buffer (Denville Scientific, South Plainfield, NJ), 2.5 μl dNTPs (2 mM), 0.5 μl Choice-Taq Blue (Denville Scientific, South Plainfield, NJ), 0.5 μl (1:20 diluted) SuperScript II Reverse Transcriptase (Life Technologies, Grand Island, NY), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), 18 μl H2O, and 1 μl RNA (12 μg/ml). The PCR reaction consisted of 42 °C for 30 min, 20 cycles of (95 °C for 30 s, 55 °C for 30 s, and 68 °C for 55 s), 68 °C for 7 min, 4 °C hold. We visualized amplification by running the PCR products on a 2% agarose gel.
Immunohistochemistry
An antibody against the peptide N-CLRPGKPTNYDELFGID-C of the Heliconius melpomene CTD31 was generated in chicken and immunoaffinity purified (New England Peptide, Gardner, MA). We also used a rabbit antibody against the nymphalid Limenitis astyanax LWRh opsin sequence (Frentiu et al. 2007) to label LWRh expressing cells in H. melpomene (McCulloch et al. 2016). Eyes were fixed, sucrose protected, cryosectioned, and immunolabeling was performed as described in McCulloch et al. (2016). Slides were placed in 100% ice-cold acetone for 5 min, then washed 3 × 10 min in 0.1 M Phosphate-buffered saline (PBS). Slides were then placed in 0.5% sodium dodecyl sulfate in 0.1 M PBS for 5 min. Each slide was blocked for 1 h at room temperature using 8% (v/v) normal goat serum, and 0.3% Triton X-100 in 0.1 M PBS. Slides were incubated with 1:75 chicken anti-CTD31 and 1:15 rabbit anti-LWRh antibodies in blocking solution overnight at 4 °C. Slides were washed 3 × 10 min in 0.1 M PBS and then incubated with 1:1,000 goat antichicken Alexafluor 488 and 1:500 goat antirabbit Alexafluor 555 secondary antibodies in blocking solution for 2 h at room temperature in the dark. Slides were washed once more 3 × 10 min in 0.1 M PBS in the dark. Slides were stored for imaging by coverslipping with Aqua Poly/Mount (Polysciences, Inc. Cat. # 18606). Image stacks were taken using a Zeiss LSM700 Confocal Microscope under 20× objective at the UC Irvine Optical Biology Core Facility. Maximum intensity projections and two-channel composites were generated using Fiji. Brightfield images were taken using untreated sections and were viewed with epifluorescence microscopy using a Zeiss Axioskop 2 under a 20× lens. Images were taken using a Canon PowerShot S5 and associated Canon software. Contrast and brightness were adjusted for clarity using Adobe Photoshop and Fiji.
Western Blot and Mass Spectrometry
Butterfly heads were removed and immediately placed at −80°C until they were shipped together with an aliquot of anti-CTD31 antibody to Zyagen (San Diego, CA) overnight on dry ice. Immunoblotting was performed by Zyagen. Proteins were extracted by mechanical homogenization in protein lysis buffer and estimated protein concentration using a BCA kit. Total protein was fractionated through two large gels (SDS–PAGE) at different concentrations (20, 40, 60, and 80 μg each gel). Protein from the two gels was then transferred to Polyvinylidene fluoride (PVDF) membranes. The membrane of the first gel was blocked with 5% milk in TBST for 2 h, then incubated with primary antibody anti-CTD31 at a concentration of 1:100 at 4 °C overnight. Membrane was washed 3 times in TBST then incubated with secondary antibody (antichicken-peroxidase antibody) from Jackson ImmunoResearch (West Grove, PA) at a concentration of 1:5,000 for 1 h. After several washes, membrane was incubated for 5 min with chemiluminescence substrate. Two major protein bands were observed ∼35 kDa.
To visualize which protein may be interacting with the chromophore, eight aliquots (50 μg each) of butterfly head proteins were fractionated through a large native gel by Zyagen (San Diego, CA). One lane was cut and visualized on UV light to locate bands that fluoresce. Gel pieces containing three protein bands were collected in 15-ml tubes and were shipped to UC Irvine. The samples were immediately transferred to a Proteomics & Mass Spectrometry Facility in the school of Biological Sciences (Irvine, CA) for nano LC-MS/MS mass spectrometry using an LTQ Velos Pro mass analyzer (Thermo-Fisher). The resulting peaklists were compared against our translated transcriptome along with a database of common contaminants using Mascot 2.5 to score (Matrix Science, Boston, Massachusetts). Mascot scores are the probability that the ion score of the experimental data match the ion scores of the database sequence; protein scores <67 are significant (P < 0.05).
Results and Discussion
Phylogeny and Chromosomal Location
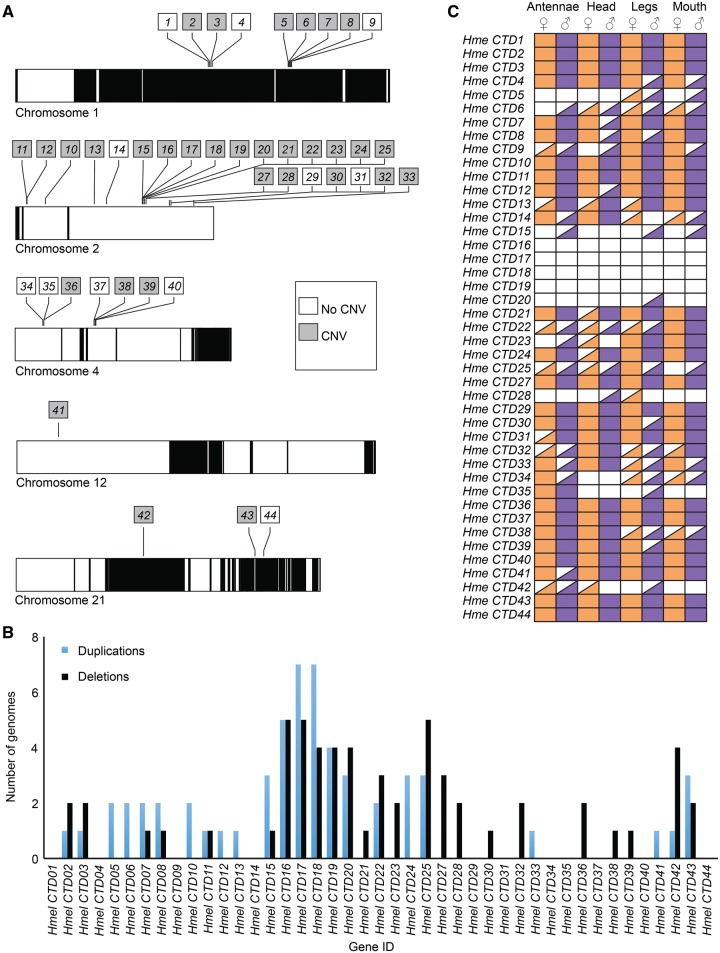
We identified a total of 43 CRAL-TRIO domain genes (Hme CTD) in the H. melpomene reference genome and 28 of them were recovered in a de novo assembly (supplementary table S2, Supplementary Material online). We found H. melpomene orthologs of most previously identified insect CRAL-TRIO domain genes (Smith and Briscoe 2015). We also discovered a recent duplication (Hme CTD38 and CTD39) since Heliconius shared a common ancestor with Danaus plexippus, and an expansion of CRAL-TRIO domain genes (Hme CTD16-20 and Hme CTD24-25; fig. 1). We refer to recent paralogs found in the reference genome as recent duplications; we refer to multiple duplications as an expansion, and genes with CNV are those that are duplicated or deleted in resequenced genomes compared with the reference. We named the H. melpomene CRAL-TRIO domain genes according to their location on scaffolds and since many genes are on similar scaffolds, we decided to map these genes on to chromosomes (fig. 2A). We found that all 43 genes were located on a total of 5 chromosomes and 23 of the genes were on a single chromosome, chromosome 2 (fig. 2A). Only one gene in this family (Hme CTD44) is intronless and likely arose through retrotransposition (Zhang 2003).
Fig. 1.
—Bayesian phylogeny of insect CRAL-TRIO domain proteins. Phylogeny includes sequences from Anopheles gambiae (green), Apis mellifera (gray), Bombyx mori (light blue), Drosophila melanogaster (black), Danaus plexippus (orange), Heliconius melpomene (red), Manduca sexta (purple), and Tribolium casteneum (dark blue). The Bayesian tree was found using MrBayes with a BLOSUM62 model of amino acid substitution. The Heliconius expansion as well as Drosophila pinta and Heliconius Hme CTD31 are indicated on the phylogeny with black lines and arrows.
Fig. 2.
—CRAL-TRIO domain gene location, copy number variation, and mRNA presence patterns. (A) CRAL-TRIO domain genes are located on five chromosomes, many in tandem. Alternating black and white chromosomal regions represent scaffolds. Shaded squares represent genes with copy number variation, duplicated and/or deleted in at least 1 of 18 resequenced genomes. (B) Number of genomes in which CRAL-TRIO domain genes are deleted (black) or duplicated (blue) in 18 Heliconius melpomene resequenced genomes. (C) mRNA presence patterns of CRAL-TRIO domain genes in H. melpomene male and female antennae, head, legs, and mouth parts. Filled square represents complete presence (>1 TPM for all replicates), half-filled square represents partial presence (>1 TPM for at least one replicate but not all four), and no fill represents lack of transcript mRNA.
New genes also arise by tandem duplication which themselves arise by unequal crossing over resulting in new gene copies adjacent to each other or by segmental duplications which can be dispersed throughout the genome and experience few recombination events (Jelesko et al. 1999; Baumgarten et al. 2003; Zhang 2003; Cannon et al. 2004). Most of the CRAL-TRIO domain genes were located in tandem suggesting that this gene family is the result of early segmental duplications and recent tandem duplications or early and recent tandem duplications with rearrangements in H. melpomene (fig. 2A). Moreover, areas of gene duplication can be hotspots for chromosomal rearrangement and might be enriched for copy number variation (CNV) (Sharp et al. 2005). In D. melanogaster, tandem duplications are significantly enriched near areas with CNVs (Dopman and Hartl 2007). The physical locations of CRAL-TRIO domain genes display arrays of tandem duplications making this gene family a good candidate for studying CNV (Redon et al. 2006).
Copy Number Variation
We used Pindel (Ye et al. 2009) to look for duplications and deletions 1 kb or larger (Stranger et al. 2007) of these CRAL-TRIO domain genes in resequenced genomes of four H. melpomene subspecies, H. melpomene melpomene, H. melpomene rosina, H. melpomene amaryllis, and H. melpomene aglaope (Martin et al. 2013). The average size of these genes including introns was 3,648 bp, coding sequences being ∼304 amino acids long. Nine genes (Hme CTD1-9) were located on chromosome 1; Hme CTD2, 3 and 5-8, had potential CNV in at least 1 of the 18 sampled genomes (fig. 2A). Hme CTD2-3 were duplicated in one H. melpomene aglaope individual and were deleted in two genomes (H. m. melpomene and H. m. aglaope; fig. 2B andsupplementary tables S3 and S4, Supplementary Material online). Hme CTD5-8 were duplicated in two genomes (H. m. melpomene and H. m. aglaope; fig. 2B andsupplementary table S3, Supplementary Material online), but Hme CTD7-8 were deleted in one H. m. amaryllis genome (supplementary table S4, Supplementary Material online).
Twenty-three genes (Hme CTD10-33) were located on chromosome 2 with more complex patterns of CNV. Hme CTD10-13, 15-20, 22, 24-25, and 33 were potentially duplicated in one or more resequenced genome (fig. 2B). Of these duplicates, Hme CTD11-13 and 33 were duplicated in one resequenced genome (supplementary table S3, Supplementary Material online). Hme CTD10 and 22 were duplicated in two genomes (supplementary table S3, Supplementary Material online). Hme CTD15, 20, 24 and 25 were duplicated in three genomes (supplementary table S3, Supplementary Material online). Hme CTD19 was duplicated in four genomes (supplementary table S3, Supplementary Material online). Hme CTD16 was duplicated in 5 genomes and 17-18 were duplicated in 7 of the 18 resequenced genomes (supplementary table S3, Supplementary Material online). Multiple CRAL-TRIO domain genes were also deleted in at least one resequenced genome: Hme CTD11, 15-23, 25, 27-28, 30 and 32 (fig. 2A). Of these, Hme CTD11, 15, 21 and 30 were deleted in one resequenced genome (fig. 2B andsupplementary table S4, Supplementary Material online). Hme CTD23, 28, and 32 were deleted in two genomes (fig. 2B andsupplementary table S4, Supplementary Material online). Hme CTD22 and 27 were deleted in three genomes (fig. 2B andsupplementary table S4, Supplementary Material online). Hme CTD18-20 were deleted in four genomes (fig. 2B andsupplementary table S4, Supplementary Material online). Hme CTD16-17 and 25 were deleted in five genomes (fig. 2B andsupplementary table S4, Supplementary Material online). One sequence identified in the de novo transcriptome, Hme CTD26, was excluded from analysis because the translation of the mRNA contig included stop codons and BLAST results suggested it was a chimeric sequence of Hme CTD24 and 25, most likely due to a Trinity misassembly. In some instances, duplications and deletions are large enough to change the presence or absence of a few genes in close proximity. Genes with the most duplications/deletions were duplicated/deleted in different subspecies; this shows that there is CNV between and within subspecies.
Seven genes (Hme CTD34-40) were found on chromosome 4; none were duplicated but CTD36 was deleted in one H. m. amaryllis and one H. m. aglaope, and CTD38-39 were both deleted in one H. m. melpomene genome (fig. 2B andsupplementary table S4, Supplementary Material online). One CRAL-TRIO domain gene (Hme CTD41) was located on chromosome 12, this gene was duplicated in one H. m. aglaope genome (fig. 2B andsupplementary table S3, Supplementary Material online). Lastly, Hme CTD42-44 were on chromosome 21; CTD41 was duplicated in one genome, CTD42 was duplicated in one genome and deleted in four genomes, CTD43 was duplicated in three and deleted in two genomes, and all resequenced genomes had one copy of CTD44 (fig. 2B andsupplementary tables S3 and S4, Supplementary Material online). To summarize, we found potential CNV in 32 of the 43 CRAL-TRIO domain genes. Intriguingly, we found no CNV in Hme CTD31, our candidate chromophore-binding protein (see below).
We refer to our findings of structural variation as “potential” duplications or deletions because the results were derived through bioinformatic inference which is subject to error (Emerson et al. 2008; Alkan et al. 2011). Pindel uses read mapping information in order to find paired reads in which one read maps to the reference and the other mate does not to identify break points and direction of unmapped reads (Ye et al. 2009). For a few large areas with a lot of potential structural variation, Pindel could not differentiate whether the break was due to a duplication or deletion. Although current CNV analyses are subject to error, finding replication of duplications or deletions in more than one resequenced genome as we found in some instances is evidence that these results are meaningful. We investigated the breakpoints for genes that were duplicated/deleted in multiple resequenced genomes and found that a majority of genes had similar breakpoints in at least two individuals (supplementary tables S3 and S4, Supplementary Material online). In addition, a different study investigated CNVs in H. melpomene rosina using three discovery methods and found support for duplications in the genome location of Hme CTD5-9 and CTD16-18 (Pinharanda et al. 2016, 2017). That study also used Pacific Biosciences (PacBio) long molecule sequencing of H. melpomene and H. cydno to validate the findings of CNVs on chromosome 2. They found support for CNV in Hme CTD10-12 using one discovery method and found many instances of CNVs in scaffold Hmel202006 using the three discovery methods (Pinharanda et al. 2016, 2017).
Twenty of our CRAL-TRIO domain genes were located on scaffold Hmel202006 including the genes within the H. melpomene expansion (Hme CTD16-20 and 24-25). We find the most CRAL-TRIO domain genes in tandem at a scaffold where our study and another found a large amount of CNV (Pinharanda et al. 2016, 2017). An interesting observation of CNV in this gene family was that all of the genes within the H. melpomene CRAL-TRIO expansion have potential CNV between individuals. In particular Hme CTD16-20 have potential CNV in the highest number of resequenced genomes (n = 9, 9, 8, 6, 5) relative to other CRAL-TRIO domain genes. These results suggest that this area in the genome could be a hotspot for structural variation potentially due to unequal crossing over because similar duplicates are located in tandem.
The adaptive significance of CNV is still under investigation. As mentioned previously, the number of chemosensory receptor genes present between and within animal species is variable (Nozawa and Nei 2008) and their distribution suggests CNV is the result of genomic drift that can lead to adaptive evolution (Nozawa and Nei 2008). In Drosophila melanogaster, duplications with functional sequences were found to be possibly beneficial (Dopman and Hartl 2007). CNV affects phenotypes through its direct influence on gene expression. In humans, CNV can lead to Mendelian and complex diseases by affecting gene dosage (Redon et al. 2006). The HapMap project found a substantial amount of CNV between humans, and an association analysis determined that most significant CNV-associations had a positive correlation between gene copy number and gene expression levels (Stranger et al. 2007). Several positively selected duplication and deletion events in D. melanogaster have also been linked to gene expression variation (Emerson et al. 2008; Schmidt et al. 2010; Catalán et al. 2016).
Studies in Drosophila suggest CNV persists due to positive selection on paralogs that have tissue-specific expression (Dopman and Hartl 2007). To determine expression patterns for CRAL-TRIO domain genes we looked at gene presence and absence in the head, antennae, legs, and mouth parts of male and female H. melpomene (n = 4/sex). Here, we refer to complete presence as having >1 TPM for all replicates, partial >1 TPM for at least one replicate but not all four, and absence as mRNA expression <1 TPM for all replicates (fig. 2C). Some genes varied in presence patterns between tissue types such as Hme CTD22, 28, and 38 (fig. 2C). Hme CTD4-9, 12-15, 20-21, 23-25, 30-35, 39, 41, and 42 had different presence patterns between sexes for one or more of the tissues examined. Although patterns of gene presence or absence (fig. 2C) provide an idea of which genes are expressed and where, absolute and differential expression needs to be analyzed to detect potential gene functions (see below).
CNV may be one contributor to the speciation of Heliconius, which has undergone a radiation in Central and South America (Kozak et al. 2015; Pinharanda et al. 2017). A recent study sought to identify adaptive CNV between two sympatric hybridizing species with distinct wing patterns, H. melpomene and H. cydno (Pinharanda et al. 2017). That study found four duplications with strong signals of divergent selection: these included an odorant binding protein, a serine protease, a regulator of the cell cycle and nitrogen compound metabolic processes, and one near the gene cortex which regulates wing color patterns (Nadeau et al. 2016; Pinharanda et al. 2017). The identification of an odorant binding protein supports the finding of Heliconius species having CNV of olfactory and gustatory receptor genes for putative host plant recognition in oviposition behavior (Briscoe et al. 2013). Divergent selection of a serine protease could be associated with Heliconius pollen feeding behavior (Smith et al. 2016). This raises the question as to what is the function of the CRAL-TRIO domain genes which have potential CNV between and within species.
Differential Expression Analysis
Members of the CRAL-TRIO domain protein family are believed to be involved in transporting hydrophobic molecules. In particular, a member of this gene family (pinta) transports the chromophore necessary for phototransduction in Drosophila, however we did not find an ortholog in Lepidoptera (fig. 1). To detect whether any of the CRAL-TRIO domain genes in H. melpomene might have this function, we did a differential expression analysis to identify CRAL-TRIO domain genes upregulated in head tissues (relative to antennae, legs, and mouth parts), potentially involved in vision. We built a reference transcriptome assembly consisting of 68,388 transcripts and 31,193 contigs with an N50 of 2,627. On an average ten million reads mapped to the transcriptome and each library averaged 79% read mapping (supplementary table S5, Supplementary Material online). The transcriptome was deposited in Dryad under data identifier doi:10.5061/dryad.857n9 and the raw RNA-Seq reads were deposited in ArrayExpress archive under accession E-MTAB-6249 and E-MTAB-6342.
Differential gene expression analysis comparing heads versus antennae yielded 4,868 Differentially Expressed (DE) contigs using qvalue and 1,173 using Bonferroni for false discovery rate correction (supplementary table S6, Supplementary Material online), 561 of these 1,173 contigs were upregulated in heads (table 1). Analysis of head versus legs mRNA gave 6,108 DE contigs using qvalue and 1,472 using Bonferroni (supplementary table S7, Supplementary Material online), of these contigs 928 were upregulated in heads. Heads versus mouth parts comparison yielded 6,176 DE contigs using qvalue and 1,486 using Bonferroni (supplementary table S8, Supplementary Material online), 914 of these were upregulated in heads (table 1).
Table 1.
Summary of Differentially Expressed (DE) and Upregulated Contigs
| qvalue | Bonferroni | Upregulated in Headsa | |
|---|---|---|---|
| Head vs. antennae | 4,868 | 1,173 | 561 |
| Head vs. legs | 6,108 | 1,472 | 928 |
| Head vs. mouth | 6,176 | 1,486 | 914 |
These contigs are upregulated in heads using a Bonferroni FDR correction.
CRAL-TRIO Domain Genes Expression
To find if any CRAL-TRIO domain genes were potentially upregulated in H. melpomene heads, we inspected our significantly DE gene list for CRAL-TRIO domain genes. By using the Bonferroni method to correct for multiple tests, only one CRAL-TRIO domain contig was upregulated in the head versus antennae comparisons, Hme CTD31 (table 2). This gene was also upregulated across comparisons when qvalue was used to correct for multiple tests. Hme CTD22 was upregulated in head versus antennae and head versus legs when using qvalue, but Hme CTD31 was the only contig upregulated across all comparisons. In addition, when we plotted the TPM for all genes across tissues, it became apparent that Hme CTD31 is very highly expressed in male and female heads (fig. 3 and supplementary figs. S1–S4, Supplementary Material online).
Table 2.
Head Expression Patterns of CRAL-TRIO Domain Contigs
| Qvalue |
Bonferroni |
|||||
|---|---|---|---|---|---|---|
| Gene ID | H vs. A | H vs. L | H vs. M | H vs. A | H vs. L | H vs. M |
| Hme CTD1 | Not DE | Not DE | Down | Not DE | Not DE | Not DE |
| Hme CTD2 | Not DE | Down | Down | Not DE | Not DE | Not DE |
| Hme CTD3 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD4 | Down | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD6 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD8 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD9 | Down | Down | Down | Not DE | Not DE | Not DE |
| Hme CTD10 | Down | Down | Down | Down | Down | Not DE |
| Hme CTD11 | Down | Not DE | Not DE | Down | Not DE | Not DE |
| Hme CTD12 | Down | Down | Down | Down | Down | Not DE |
| Hme CTD13 | Down | Down | Down | Down | Not DE | Down |
| Hme CTD14 | Down | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD15 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD21 | Down | Down | Down | Down | Down | Down |
| Hme CTD22 | Up | Up | Down | Not DE | Not DE | Not DE |
| Hme CTD29 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD30 | Down | Down | Not DE | Not DE | Not DE | Not DE |
| Hme CTD31 | Up | Up | Up | Up | Not DE | Not DE |
| Hme CTD32 | Not DE | Not DE | Not DE | Not DE | Not DE | Not DE |
| Hme CTD34 | Down | Down | Down | Down | Down | Not DE |
| Hme CTD35 | Down | Not DE | Not DE | Down | Not DE | Not DE |
| Hme CTD36 | Down | Down | Down | Down | Down | Down |
| Hme CTD37 | Down | Down | Down | Down | Down | Not DE |
| Hme CTD38 | Down | Not DE | Not DE | Down | Not DE | Not DE |
| Hme CTD39 | Down | Down | Not DE | Not DE | Not DE | Not DE |
| Hme CTD40 | Down | Not DE | Down | Not DE | Not DE | Not DE |
| Hme CTD41 | Down | Down | Down | Not DE | Not DE | Not DE |
| Hme CTD43 | Not DE | Not DE | Down | Not DE | Not DE | Down |
Not DE, not differentially expressed; Up, upregulated in heads; Down, downregulated in heads.
Fig. 3.
—Expression of CRAL-TRIO domain genes. (A) Transcripts per kilobase million (TPM) of CRAL-TRIO domain genes in female antennae, head, legs, and mouth parts. (B) TPM of CRAL-TRIO domain genes in male antennae, head, legs, and mouth parts.
To investigate patterns of gene expression in the rest of the CRAL-TRIO domain genes, we used two-way ANOVAs to test if these genes varied by sex, tissue type, or sex and tissue type interaction (supplementary table S9, Supplementary Material online). We found that most genes varied by tissue type, including Hme CTD1, 2, 4, 5, 7-14, 21-24, 27, 29, 31, 33-40, 43, and 44 (supplementary table S9 and figs. S5–S7, Supplementary Material online). Only two genes varied by sex Hme CTD7 and 8 (supplementary table S9 and figs. S5–S7, Supplementary Material online).
The ANOVA analysis and the genome-wide DE analysis showed that Hme CTD31 is a candidate pigment binding protein due to high expression in H. melpomene heads. The top NCBI blastp (protein to protein alignment) results for this gene are CRAL-TRIO domain containing protein and alpha-tocopherol transport protein. We found CRAL-TRIO domain genes that were upregulated in other tissues such as Hme CTD11 and CTD12 in the antennae. We do not know the specific function of these genes, but it is possible that they play a role in mediating the activation of other sensory receptors. Studies identifying chemosensory proteins have found some potential sensory receptors that are similar in sequence to opsins (Troemel et al. 1995). Opsins and some chemosensory receptors, such as olfactory, gustatory, and ionotropic receptors, belong to the rhodopsin-type superfamily of receptors but the groups vary in rate of molecular evolution. Opsins are more conserved between species, although gene duplications exist (see Sison-Mangus et al. 2008; Pohl et al. 2009; McCulloch et al. 2017), whereas olfactory, gustatory, and ionotropic receptors have duplicated extensively resulting in large gene families with a lot of copy number variation (Raible et al. 2006; van Schooten et al. 2016). Since these receptors have similar mechanisms of activation and similar functions in sensory perception, it is possible that the hydrophobic molecules with which they interact can be transported by proteins that are also similar to each other. In the cotton bollworm H. armigera four chemosensory proteins are expressed in both the eyes and proboscis; these proteins bind β-carotene and retinol (Zhu et al. 2016). That study demonstrates that proteins belonging to a family that responds to chemicals can have modified functions to have a role as a carrier for dietary carotenoids and visual processing in insects. Likewise, it is possible that Hme CTD11 and 12, upregulated in antennae, have functions in mediating olfaction through subfunctionalization.
Hme CTD31 Candidate Chromophore Transporter
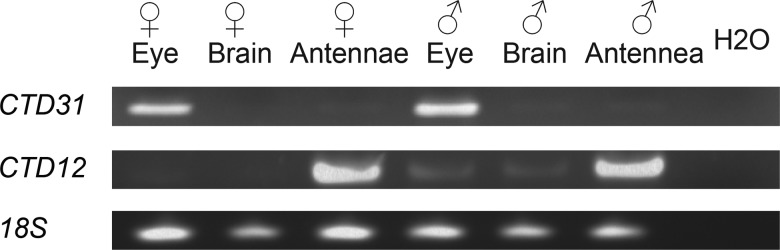
Hme CTD31 is a candidate gene to explore for functions in visual pigment transport due to its upregulation in heads. However, head libraries were generated using whole head mRNA, so we used reverse transcription PCR (RT-PCR) to dissect whether Hme CTD31 was expressed in the eye, brain, or both. We used the 18 S rRNA gene as a positive control for normalized mRNA presence. We also used Hme CTD12 and antennae tissue to validate TPM expression patterns. We expected to see Hme CTD31 expressed in the eye and brain but not in the antennae, and Hme CTD12 only expressed in the antennae. Hme CTD12 was only amplified in the antennae as expected (fig. 4). However, RT-PCR showed that Hme CTD31 was only expressed in male and female eyes and not in the brain or the antennae (fig. 4). Additional support for Hme CTD31 having a potential role in butterfly vision came from exploring the expression of CRAL-TRIO domain genes in heads of a different butterfly species, B. anynana (Macias-Muñoz et al. 2016) (accession numbers E-MTAB-3887 and doi: 10.5061/dryad.f98s6). We found that the B. anynana ortholog of Hme CTD31 is the most highly expressed CRAL-TRIO domain gene in Bicyclus butterfly heads (supplementary fig. S8, Supplementary Material online) further supporting that expression of this gene is important in the compound eye across butterfly species.
Fig. 4.
—Hme CTD31 RT-PCR. RT-PCR of Hme CTD31, Hme CTD12, and 18 S in female and male eye, brain, and antennae.
To localize where the Hme CTD31 protein is expressed in the H. melpomene eye, we designed an antibody against a unique peptide to perform immunohistochemistry. Our protein of interest has a predicted weight of ∼35 kDa, and an immunoblot of proteins extracted from whole head tissue using this antibody indicates it binds to a protein of the expected size (fig. 5A). We saw another band <35 kDa and that maybe the same protein but running through the gel differently due to phosphorylation of specific residues in the protein. Hme CTD31 has sites that are potentially phosphorylated with a probability score <0.75 at sites 7, 74, 109, 127, 175, 233, 275, and 28 (Blom et al. 1999).
Fig. 5.
—Hme CTD31 Western Blot. (A) Western blot using head tissue and Hme CTD31 antibody performed by Zyagen (San Diego CA). Arrow indicates expected band. (B) Butterfly head protein run on a native gel shows three bands that fluoresce under UV light. Arrows indicate the location of the upper, middle, and lower bands which were cut out and sequenced using mass spectrometry.
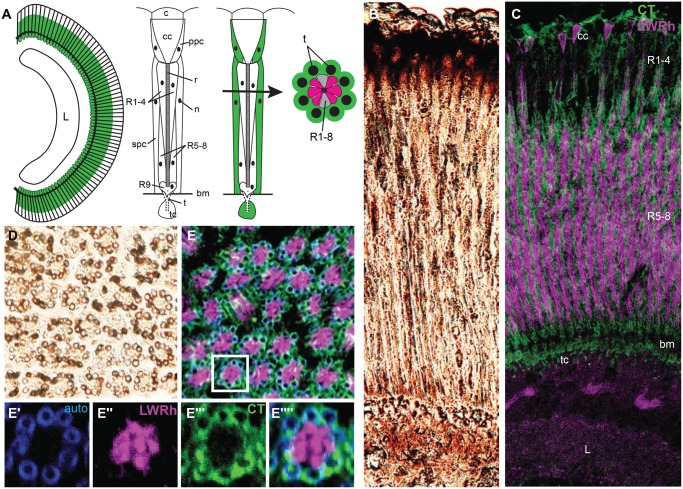
Next, to identify the cellular localization of the protein we examined longitudinal and transverse sections of the butterfly compound eye (fig. 6A). Each Heliconius ommatidium consists of a cornea, crystalline cone, and nine photoreceptor cells with a fused rhabdom and a tiered cell body arrangement. Primary pigment cells surround the crystalline cone and secondary pigment cells surround the photoreceptor cells. Brightfield images of a longitudinal section of the compound eye showed that there is pigment at the top of the ommatidia, around or within each ommatidium for its entire length, and below the basement membrane in tracheal cells (fig. 6B). A transverse image showed that the ommatidia are surrounded by eight tracheoles which have pigment along the tracheal walls (fig. 6D).
Fig. 6.
—Immunohistochemistry of Hme CTD31 in Heliconius melpomene eye and optic lobe. ( A) Drawing of a longitudinal view of a compound eye and lamina, and longitudinal and transverse sections of a single ommatidium. Green highlights where we find Hme CTD31 expression; L, lamina; c, cornea; cc, crystalline cone; ppc, primary pigment cells; r, rhabdom; R1-9 conventional Lepidoptera numbering of photoreceptor cells; n, cell nucleus; spc, secondary pigment cells; bm, basement membrane; t, trachea; and tc, tracheal cell. (B) Brightfield longitudinal section showing pigments in the H. melpomene retina. (C) Longitudinal section with Hme CTD31 and LW opsin staining; Hme CTD31 is in green and LW opsin is in magenta. (D) Brightfield image of a transverse section of a butterfly eye, pigment is seen in the structures surrounding the ommatidia. (E) Transverse view of a butterfly eye stained for LW and Hme CTD31. (E′) autofluorescence showing tracheoles surrounding an individual ommatidium. (E′′) LW opsin staining showing where the LW photoreceptor cells are. (E′′′) CTD31 staining showing where the CRAL-TRIO domain protein Hme CTD31 is expressed. (E′′′′) merged image of LWRh, CTD31, and autofluorescence.
We used polyclonal antibodies against Hme CTD31 and the long wavelength opsin (LWRh) to visualize where Hme CTD31 was expressed in relation to photoreceptor cells (McCulloch et al. 2016). We found that Hme CTD31 is found in the primary pigment cells, secondary pigment cells, and tracheal cells (fig. 6C). The tracheal cells project tracheoles up and around the ommatidia, and these structures autofluoresce under blue light (488 nm laser) due to the presence of chitin (fig. 6E) (Iwata et al. 2014). Hme CTD31 is also expressed in the cell bodies surrounding the tracheole walls (fig. 6E). Hme CTD31 immunohistochemical results were similar to those of a retinol binding protein in the family Papilionidae, Papilio retinol binding protein (RBP). Papilio RBP binds retinol and was found to be expressed in primary pigment cells, secondary pigment cells, and tracheal cells (Wakakuwa et al. 2004). However, Hme CTD31 is expressed in the lower two-thirds of the ommatidia, rather than along the entire length, whereas Papilio RBP is found in the entire length of the ommatidia. The difference in where Papilio RBP and Hme CTD31 are located in Papilio and Heliconius, respectively, might be due to the difference in ommatidium morphology. Papilio RBP also does not belong to the CRAL-TRIO domain gene family. However, an ortholog of the gene encoding Papilio RBP in H. melpomene (Hme comp30064) was upregulated in heads relative to other tissue types (supplementary tables S6–S8 and fig. S9, Supplementary Material online). It is possible that Hme CTD31 and Papilio RBP are both necessary to transport the retinal molecule in different configurations as in vertebrates (McBee et al. 2001). The study characterizing pinta suggested there might be additional proteins in the primary pigment cells that are required for biosynthesis of the chromophore (Wang and Montell 2005).
It is also possible that Hme CTD31 functions in binding filtering pigments. From the RT-PCR and immunohistochemistry alone, we cannot confirm to what molecule Hme CTD31 binds but its upregulation in heads and localization in the ommatidia suggest that this protein has a role in butterfly vision. To confirm whether Hme CTD31 binds a chromophore, proteins from butterfly heads were run on a native gel and examined under UV light (fig. 5B). In the swallowtail butterfly, Papilio RBP bound to the chromophore fluoresces under UV light (Wakakuwa et al. 2004). We found three fluorescing bands which were cut and sequenced using mass spectrometry. Our candidate protein Hme CTD 31 is one of the top 20 proteins matching peptide fragment fluorescing upper (consisting of insoluble material), middle, and lower bands as detected by mass spectrometry (table 3). This evidence further supports our hypothesis that Hme CTD31, a CRAL-TRIO domain containing protein, is binding the chromophore molecule in butterflies. Hme CTD31 likely transports the vitamin-A derived chromophore molecule similar to vertebrate CRALBP and Drosophila PINTA. Drosophila cralbp and pinta both belong to the CRAL-TRIO and SEC14 superfamilies yet PINTA is the one shown experimentally to bind retinal (Wang and Montell 2005). Similarly, RALBP also belongs to the SEC14 superfamily and also functions in retinal binding in cephalopods (Ozaki et al. 1994; Speiser et al. 2014). These previous findings and our results suggest a conserved role for at least some of the CRAL-TRIO domain proteins, even if the specific function in this pathway is undertaken by nonorthologous members of the expanded gene family.
Table 3.
Top 20 Proteins from Upper, Middle, and Lower Bands Detected by Mass Spectrometry Sorted by Upper Band Mascot Score
| Accession | Protein Family | Upper |
Middle |
Lower |
|||
|---|---|---|---|---|---|---|---|
| Mascot Score | Peptide Matches | Mascot Score | Peptide Matches | Mascot Score | Peptide Matches | ||
| comp33735_c0 | Rfabg | 5766 | 198 | 1699 | 67 | 45 | 2 |
| comp31078_c1 | Atpalpha | 2204 | 62 | 1557 | 41 | 561 | 16 |
| comp31397_c0 | betaTub56D | 1587 | 47 | 925 | 26 | 594 | 18 |
| comp27767_c0 | Vha68-2 | 1542 | 39 | 946 | 24 | 298 | 12 |
| comp32095_c0 | ATPsynbeta | 1488 | 41 | 855 | 25 | 719 | 25 |
| comp28890_c0 | Gapdh2 | 1204 | 33 | 820 | 26 | 368 | 8 |
| comp15204_c0 | kdn | 1187 | 31 | 787 | 24 | 660 | 17 |
| comp27239_c0 | CG1635 | 1051 | 29 | 692 | 18 | 127 | 2 |
| comp31202_c0 | alpha-Spec | 998 | 38 | 1262 | 51 | 39 | 3 |
| comp26414_c0 | PyK | 997 | 30 | 784 | 19 | 483 | 13 |
| comp31948_c0 | Pp2A-29B | 963 | 20 | 475 | 17 | 584 | 19 |
| comp29636_c0 | CG2663 | 947 | 43 | 514 | 24 | 494 | 24 |
| comp33018_c0 | TER94 | 890 | 29 | 534 | 15 | 227 | 10 |
| comp30615_c0 | nrv3 | 867 | 19 | 786 | 21 | 139 | 4 |
| comp14607_c0 | Pgi | 836 | 24 | 667 | 23 | 358 | 11 |
| comp29963_c1 | Gdh | 833 | 29 | 1103 | 40 | 677 | 27 |
| comp28746_c0 | blw | 822 | 27 | 588 | 19 | 57 | 3 |
| comp25729_c0 | Hsc70-4 | 817 | 32 | 2565 | 93 | 945 | 32 |
| comp29025_c0 | Mdh2 | 817 | 25 | 764 | 21 | 200 | 7 |
| comp31520_c0 | alphaTub84B | 741 | 26 | 609 | 16 | 262 | 10 |
| comp30064_c0 | CG10476 | 517 | 12 | 271 | 5 | 309 | 7 |
Note.—Mascot protein scores >67 are significant (P < 0.05). Bold indicates comp29636_c0, which corresponds to CTD31. comp30064 corresponds to the Papilio RBP homolog in Heliconius melpomene.
To summarize, we investigated a large gene family whose function in insects is only known for one gene in one species: pinta transports the chromophore molecule in Drosophila and is necessary for phototransduction. Although other members of the CRAL-TRIO domain gene family have undergone an expansion, we found no ortholog of the pinta gene in Lepidoptera. In H. melpomene, we found an expansion of genes in close proximity suggesting that CRAL-TRIO domain genes are evolving by tandem duplications. We also found copy number variation of CRAL-TRIO domain genes between individuals. Although the function of these genes is not known, we hypothesized that one or more of these genes could have a role in vision similar to pinta and we were able to identify one candidate gene upregulated in H. melpomene heads and two other genes upregulated in antennae. This gene, Hme CTD31, was found in eye mRNA and its protein product was localized to secondary and primary pigment cells and to a protein gel band that fluoresces under UV light. Interestingly, Hme CTD31 is a single copy gene across the 18 resequenced genomes we investigated, suggesting a critical function. We have thus identified a CRAL-TRIO domain containing gene that likely encodes a chromophore binding protein in butterflies, a paralogous member of the pinta gene family that is rapidly evolving in butterflies.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Ana Catalán, Gilbert Smith, Ali Mortazavi, Kevin Thornton, J. J. Emerson, and Francisco Ayala for advice on data handling, and Jennifer Briner for comments on the manuscript. We also thank James Baldwin-Brown and Mahul Chakraborty for bioinformatic assistance, and Junsung Im and Paul Gershon for technical assistance with the mass spectrometry. Nucleotide sequences for the CRAL-TRIO domain containing genes were submitted to GenBank, Accession: MG434596-MG434667. This work was supported in part from National Science Foundation grants (IOS-1257627 and IOS-1656260 to A.D.B.) and a National Science Foundation Graduate Research Fellowship to A.M.M.
Literature Cited
- Alkan C, Coe BP, Eichler EE.. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet. 12(5):363–376.http://dx.doi.org/10.1038/nrg2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G.. 2003. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165(1):309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S.. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 294(5):1351–1362. [DOI] [PubMed] [Google Scholar]
- Bomar JM. 2003. Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nat Genet. 35(3):264–269.http://dx.doi.org/10.1038/ng1255 [DOI] [PubMed] [Google Scholar]
- Briscoe AD, et al. 2013. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9(7):e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G.. 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán A, et al. 2016. An indel polymorphism in the MtnA 3’ untranslated region is associated with gene expression variation and local adaptation in Drosophila melanogaster. PLoS Genet. 12(4):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464(7289):704–712.http://dx.doi.org/10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Nickerson DA, Eichler EE.. 2007. Mutational and selective effects on copy-number variants in the human genome. Nat Genet. 39(7s):S22–S29. [DOI] [PubMed] [Google Scholar]
- Dabney A, Storey JD.. 2013. qvalue: Q-value estimation for false discovery rate control. R package version 1.36.0.
- Davey JW, et al. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 6(3):695–708.http://dx.doi.org/10.1534/g3.115.023655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW.. 2006. The evolution of mammalian gene families. PLoS One 1:e85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL.. 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 104(50):19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvaux L, et al. 2015. Dynamics of copy number variation in host races of the pea aphid. Mol Biol Evol. 32(1):63–80.http://dx.doi.org/10.1093/molbev/msu266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.http://dx.doi.org/10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M.. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320(5883):1629–1631. [DOI] [PubMed] [Google Scholar]
- Frentiu FD, et al. 2007. Adaptive evolution of color vision as seen through the eyes of butterflies. Proc Natl Acad Sci U S A. 104(Suppl 1):8634–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652.http://dx.doi.org/10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512.http://dx.doi.org/10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, et al. 1997. Gene families: the taxonomy of protein paralogs and chimeras. Science 278(5338):609–614.http://dx.doi.org/10.1126/science.278.5338.609 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG.. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 89(22):10915–10919.http://dx.doi.org/10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh BM, Carroll SB.. 2005. Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development 132(7):1567–1577.http://dx.doi.org/10.1242/dev.01737 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755.http://dx.doi.org/10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Iwata M, Ohno Y, Otaki JM.. 2014. Real-time in vivo imaging of butterfly wing development: revealing the cellular dynamics of the pupal wing tissue. PLoS One 9(2):e89500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesko JG, Harper R, Furuya M, Gruissem W.. 1999. Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 96(18):10302–10307.http://dx.doi.org/10.1073/pnas.96.18.10302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, et al. 2016. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem Mol Biol. 76:118–147.http://dx.doi.org/10.1016/j.ibmb.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M.. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291.http://dx.doi.org/10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- Kozak KM, et al. 2015. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 64(3):505–524.http://dx.doi.org/10.1093/sysbio/syv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L.. 2002. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12(7):1048–1059.http://dx.doi.org/10.1101/gr.174302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760.http://dx.doi.org/10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W.. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 4(11):865–875. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155.http://dx.doi.org/10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Macias-Muñoz A, Smith G, Monteiro A, Briscoe AD.. 2016. Transcriptome-wide differential gene expression in Bicyclus anynana butterflies: female vision-related genes are more plastic. Mol Biol Evol. 33(1):79–92. [DOI] [PubMed] [Google Scholar]
- Martin SH, et al. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828.http://dx.doi.org/10.1101/gr.159426.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maw MA, et al. 1997. Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat Genet. 17(2):198–200.http://dx.doi.org/10.1038/ng1097-198 [DOI] [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, Pepperberg DR.. 2001. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog Retin Eye Res. 20(4):469–529. [DOI] [PubMed] [Google Scholar]
- Mccarthy DJ, Chen Y, Smyth GK.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. 40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch KJ, et al. 2017. Sexual dimorphism and retinal mosaic diversification following the evolution of a violet receptor in butterflies. Mol Biol Evol. 34(9):2271–2284. [DOI] [PubMed]
- McCulloch KJ, Osorio D, Briscoe AD.. 2016. Sexual dimorphism in the compound eye of Heliconius erato: a nymphalid butterfly with at least five spectral classes of photoreceptor. J Exp Biol. 219(Pt 15):2377–2387. [DOI] [PubMed] [Google Scholar]
- Min KC, Kovall RA, Hendrickson WA.. 2003. Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency. Proc Natl Acad Sci U S A. 100(25):14713–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, et al. 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534(7605):106–110.http://dx.doi.org/10.1038/nature17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M.. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 9(12):951–963.http://dx.doi.org/10.1038/nrg2480 [DOI] [PubMed] [Google Scholar]
- Nozawa M, Nei M.. 2008. Genomic drift and copy number variation of chemosensory receptor genes in humans and mice. Cytogenet Genome Res. 123(1–4):263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer-Verlag. [Google Scholar]
- Ozaki K, et al. 1994. Molecular characterization and functional expression of squid retinal-binding protein. J Biol Chem. 269:3838–3845. [PubMed] [Google Scholar]
- Panagabko C, et al. 2003. Ligand specificity in the CRAL-TRIO protein family. Biochemistry 42(21):6467–6474.http://dx.doi.org/10.1021/bi034086v [DOI] [PubMed] [Google Scholar]
- Pinharanda A, Martin SH, Barker SL, Davey JW, Jiggins CD.. 2016. Data from: The comparative landscape of duplications in Heliconius melpomene and Heliconius cydno. Dryad Digit Repos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinharanda A, Martin SH, Barker SL, Davey JW, Jiggins CD.. 2017. The comparative landscape of duplications in Heliconius melpomene and Heliconius cydno. Heredity 118(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl N, Sison-Mangus MP, Yee EN, Liswi SW, Briscoe AD.. 2009. Impact of duplicate gene copies on phylogenetic analysis and divergence time estimates in butterflies. BMC Evol Biol. 9:99.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, et al. 2006. Opsins and clusters of sensory G-protein-coupled receptors in the sea urchin genome. Dev Biol. 300(1):461–475. [DOI] [PubMed] [Google Scholar]
- Redon R, et al. 2006. Global variation in copy number in the human genome. Nature 444(7118):444–454.http://dx.doi.org/10.1038/nature05329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, Smyth GK.. 2010. edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK.. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23(21):2881–2887.http://dx.doi.org/10.1093/bioinformatics/btm453 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK.. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9(2):321–332. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J, Teslenko M.. 2011. MrBayes version 3.2 manual: tutorials and model summaries. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574.http://dx.doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schmidt JM, et al. 2010. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6(6):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, et al. 2005. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 77(1):78–88.http://dx.doi.org/10.1086/431652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, et al. 2012. New and continuing developments at PROSITE. Nucleic Acids Res. 41(D1):344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison-Mangus MP, Briscoe AD, Zaccardi G, Knuttel H, Kelber A.. 2008. The lycaenid butterfly Polyommatus icarus uses a duplicated blue opsin to see green. J Exp Biol. 211(3):361–369. [DOI] [PubMed] [Google Scholar]
- Smith G, Briscoe AD.. 2015. Molecular evolution and expression of the CRAL_TRIO protein family in insects. Insect Biochem Mol Biol. 62:168–173.http://dx.doi.org/10.1016/j.ibmb.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Smith G, Macias-Muñoz A, Briscoe AD.. 2016. Gene duplication and gene expression changes play a role in the evolution of candidate pollen feeding genes in Heliconius butterflies. Genome Biol Evol. 8(8):2581–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser DI, et al. 2014. Using phylogenetically-informed annotation (PIA) to search for light-interacting genes in transcriptomes from non-model organisms. BMC Bioinformatics 15:350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B.. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Web Server):465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2000. Evolutionary developmental biology and the problem of variation. Evolution 54(4):1079–1091.http://dx.doi.org/10.1111/j.0014-3820.2000.tb00544.x [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, et al. 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315(5813):848–853.http://dx.doi.org/10.1126/science.1136678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A.. 2011. Differential expression in RNA-seq: a matter of depth. Genome Res. 21(12):2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI.. 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83(2):207–218. [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schooten B, Jiggins CD, Briscoe AD, Papa R.. 2016. Genome-wide analysis of ionotropic receptors provides insight into their evolution in Heliconius butterflies. BMC Genomics 17:254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Kiser PD, Golczak M, Palczewski K.. 2010. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci. 35(7):400–410.http://dx.doi.org/10.1016/j.tibs.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakakuwa M, Ozaki K, Arikawa K.. 2004. Immunohistochemical localization of Papilio RBP in the eye of butterflies. J Exp Biol. 207(Pt 9):1479–1486. [DOI] [PubMed] [Google Scholar]
- Wang T, Montell C.. 2005. Rhodopsin formation in Drosophila is dependent on the PINTA retinoid-binding protein. J Neurosci. 25(21):5187–5194.http://dx.doi.org/10.1523/JNEUROSCI.0995-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, et al. 2006. CRALBP ligand and protein interactions. Adv Exp Med Biol. 572:477–483. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z.. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25(21):2865–2871.http://dx.doi.org/10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18(6):292–298.http://dx.doi.org/10.1016/S0169-5347(03)00033-8 [Google Scholar]
- Zhu J, et al. 2016. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int J Biol Sci. 12(11):1394–1404.http://dx.doi.org/10.7150/ijbs.16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.