Abstract
Objective
Mediator 1 (MED1) interacts with transcription factors to regulate transcriptional machinery. The role of MED1 in macrophage biology and the relevant disease state remains to be investigated.
Approach and Results
To study the molecular mechanism by which MED1 regulates the M1/M2 phenotype switch of macrophage and the effect on atherosclerosis, we generated MED1/apolipoprotein E (ApoE) double-deficient (MED1ΔMac/ApoE−/−) mice and found that atherosclerosis was greater in MED1ΔMac/ApoE−/− mice than MED1fl/fl/ApoE−/− littermates. The gene expression of M1 markers was increased and that of M2 markers decreased in both aortic wall and peritoneal macrophages from MED1ΔMac/ApoE−/− mice, whereas MED1 overexpression rectified the changes in M1/M2 expression. Moreover, low-density lipoprotein receptor-deficient mice received bone marrow from MED1ΔMac mice showed greater atherosclerosis. Mechanistically, MED1 ablation decreased the binding of peroxisome proliferator-activated receptor γ (PPARγ) and enrichment of H3K4me1 and H3K27ac to upstream region of M2 marker genes. Furthermore, interleukin 4 induction of PPARγ and MED1 increased the binding of PPARγ or MED1 to the PPAR response elements of M2 marker genes.
Conclusions
Our data suggest that MED1 is required for the PPARγ-mediated M2 phenotype switch, with M2 marker genes induced but M1 marker genes suppressed. MED1 in macrophages has an anti-atherosclerotic role via PPARγ-regulated transactivation.
Keywords: MED1, atherosclerosis, macrophage polarization
Subject Codes: Atherosclerosis, Inflammation, Genetically Altered and Transgenic Models
Introduction
Mediator, containing multiple subunits, transduces regulatory information from enhancers to promoters to facilitate transcription factor binding and RNA polymerase II recruitment to the promoter of target genes.1, 2 Mediator subunit 1 (MED1) often co-localizes or co-occupies with cell-type-specific master transcription factors and active histone markers at the enhancer/promoter region of the target genes.3 In mammalian cells, MED1 was originally identified as a peroxisome proliferator-activated receptor (PPAR)-binding protein, which increases the transcriptional activity of PPARγ.4 Subsequently, MED1 was found to also bind to the thyroid hormone receptor-associated protein (TRAP) complex and function as a coactivator for several other nuclear receptors, including retinoic acid receptor, retinoid×receptor (RXR), vitamin-D-receptor, farnesoid X receptor (FXR), estrogen receptor, and glucocorticoid receptor.4–6 Furthermore, MED1 regulates several transcription regulators, including p53, five GATA family members, p300, PPARγ coactivator-1 α, and CCAAT/enhancer-binding protein β (C/EBPβ).7–9 By interacting with a myriad nuclear receptors and transcriptional regulators, MED1 plays an important role in transcriptional regulation.10, 11 In terms of biological functions at the tissue level, MED1 deficiency in mouse embryonic fibroblasts impaired PPARγ-stimulated adipogenesis and PPARγ-regulated gene expression.12 Liver-specific MED1 knockout in mice inhibited hepatic steatosis induced by a glucocorticoid receptor agonist or PPARγ overexpression.13, 14 Mice with muscle-specific MED1 knockout showed enhanced insulin sensitivity, improved glucose tolerance and resistance to high-fat diet-induced obesity.15 In addition, cardiomyocyte-specific ablation of MED1 in mice results in lethality due to dilated cardiomyopathy-related ventricular dilation and heart failure, suggesting that MED1 may play a role in cardiovascular diseases.16 Overall, MED1 may have pivotal but distinct roles in various pathophysiological states depending on the tissue.
The innate immune response and lipid retention in monocytes/macrophages is imperative in the initiation, progression, and exacerbation of atherosclerosis.17, 18 Tissue macrophages have immense plasticity and can be classified into the classically activated pro-inflammatory M1 macrophages and alternatively activated anti-inflammatory M2 macrophages.19 M1 macrophages show an elevated level of pro-inflammatory cytokines, including interleukin 1-β (IL-1β), tumor necrosis factor α (TNF α), and monocyte chemoattractant protein 1 (MCP-1). In contrast, anti-inflammatory M2 macrophages have various functions, including regulation of immunity, maintenance of tolerance and tissue repair/wound healing.20
PPARγ activation primes human monocytes into alternative M2 macrophages; thus, PPARγ transactivates anti-inflammatory genes such as IL-10 via its PPAR-responsive element (PPRE).21, 22 PPARγ activation by 15d-PGJ2, a synthetic PPARγ ligand, upregulates several M2 markers such as arginase 1 (Arg1).23 Additionally, PPARγ suppresses the expression of pro-inflammatory genes by interacting with pro-inflammatory transcription factors such as AP-1, NFAT, and NF-κB.24, 25 In the context of atherosclerosis, apolipoprotein E (ApoE)-deficient (ApoE−/−) mice receiving troglitazone, a PPARγ agonist, showed decreased atherosclerosis.26 Furthermore, PPARγ deletion in macrophages increased atherosclerosis in mice with a low-density lipoprotein receptor (LDLR)-null (LDLR−/−) background.27
Because MED1 positively regulates PPARγ, which is anti-atherosclerotic in macrophages, we hypothesized that MED1 is also involved in the development of atherosclerosis. For this purpose, we generated a mouse line with MED1 ablation specifically in monocytes/macrophages (MED1ΔMac). Atherosclerosis was significantly enhanced in MED1ΔMac/ApoE−/− double knockout mice or LDLR−/− mice receiving MED1ΔMac bone marrow. Furthermore, MED1ΔMac macrophages showed a profound inflammatory state, as evidenced by the phenotype transition from M2 to M1. At the molecular level, MED1 was crucial for PPARγ acting on the PPRE in the upstream region of M2 marker genes. Thus, targeting MED1 in macrophages may become a novel strategy for treating atherosclerosis.
Materials and methods
Materials and methods are available in the online-only Data Supplement.
Results
MED1 deficiency in macrophages increases atherosclerosis
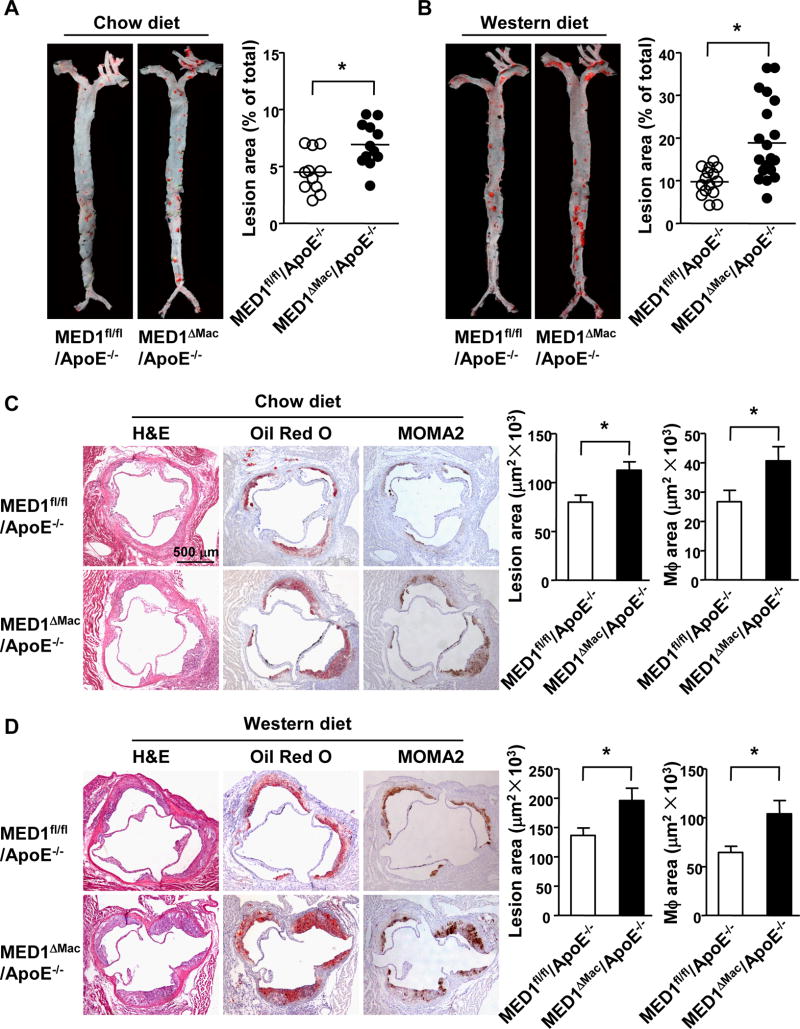
To investigate the role of MED1 in macrophages in atherosclerosis, we developed macrophage-specific MED1 knockout (MED1ΔMac) mice (Supplemental Figure I), and then introduced MED1ΔMac or floxed MED1 allele (MED1fl/fl)28 into mice with an ApoE-null background to generate MED1ΔMac/ApoE−/− and MED1fl/fl/ApoE−/− mouse lines. The 2 groups of mice, both starting at age of 8 weeks, were fed normal chow or a Western diet for 12 weeks, and then killed for assessing atherosclerosis. En face Oil Red O staining revealed significantly greater atherosclerotic lesion area in the aortic tree of MED1ΔMac/ApoE−/− than MED1fl/fl/ApoE−/− littermates under both the chow and Western diet [4.5± 0.5% vs 6.9 ± 0.6% (Figure 1A)] and [9.7 ± 0.8% vs 18.8 ± 2.0% (Figure 1B)], for an increase of 54% and 93%, respectively. Atherosclerosis was also increased greatly in the aortic root of MED1ΔMac/ApoE−/− mice as compared with MED1fl/fl/ApoE−/− littermates fed a chow or Western diet (79.8 ± 7.1×103 vs 112.6 ± 8.4×103 µm2 and 136.2 ± 13.0×103 vs 195.8 ± 21.1×103 µm2) (Figure 1C–D). In sections of aortic roots stained with MOMA2 antibody recognizing monocytes/macrophages, MOMA2-positive areas were greater in MED1ΔMac/ApoE−/− than MED1fl/fl/ApoE−/− mice fed a chow or Western diet (26.7±3.8×103 vs 40.7± 4.8×103 µm2 and 64.5±6.3×103 vs 103.9±13.7×103 µm2) (Figure 1C–D). Furthermore, TUNEL staining showed increased apoptosis in the aortic roots of MED1ΔMac/ApoE−/− mice fed Western diet (Supplemental Figure II). However, the lipid profile was comparable between the 2 groups of mice under either diet (Supplemental Tables I and II). These results suggest that MED1 silencing in a monocytic lineage is sufficient to exacerbate both spontaneous and diet-induced atherosclerosis.
Figure 1. MED1 deficiency in monocytes/macrophages promotes atherosclerosis in mice.
Eight-week old MED1fl/fl/ApoE−/− male mice and age-matched MED1ΔMac/ApoE−/− male littermates were fed a chow (n=11 and n=12, respectively) or Western diet (n=15 and n=21, respectively) for 12 weeks. (A, B) En face Oil Red O staining of aortic specimens. (C, D) Cross sections of aortic roots from mice stained with hematoxylin and eosin (H&E), Oil Red O (atherosclerosis), or MOMA2 (macrophage) antibody (n =8–12 mice per group). Data are mean±SEM. * p<0.05.
MED1 deficiency promotes macrophage M1 polarization
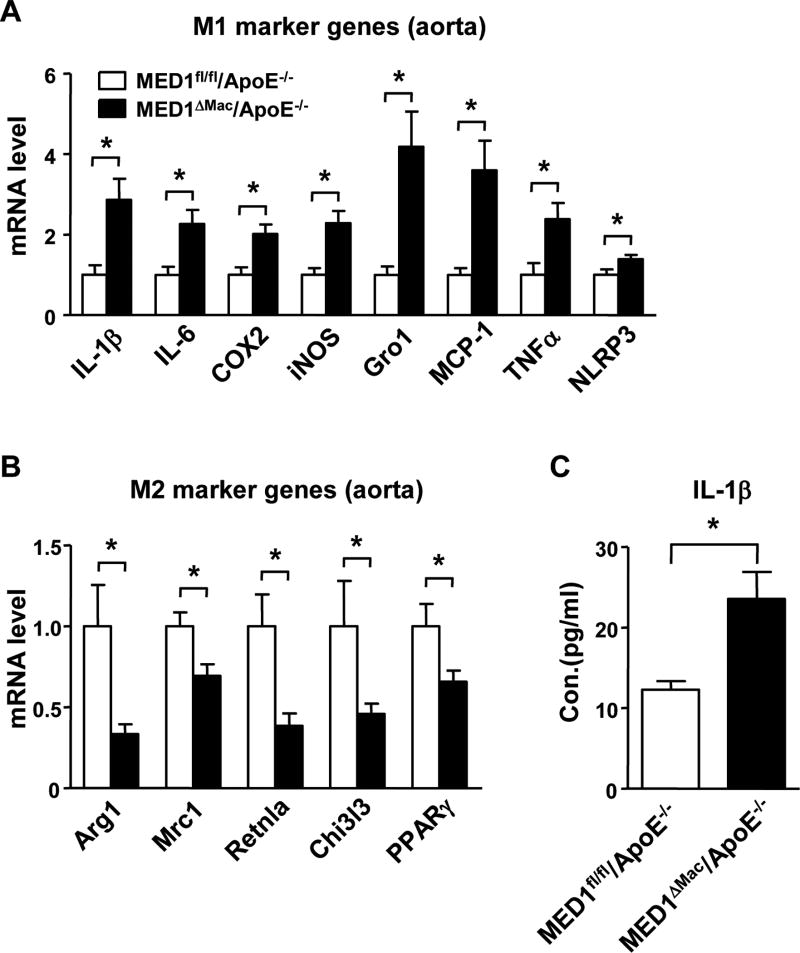
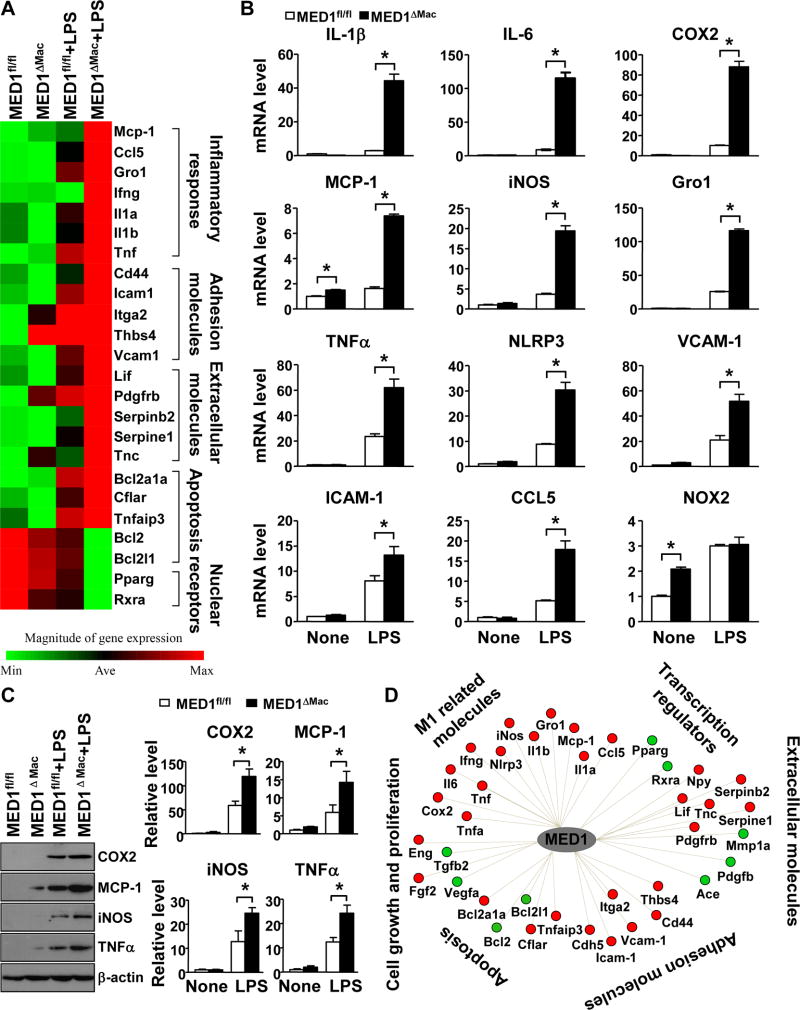
Given the increased MOMA2-positive areas in atheroprone areas of MED1ΔMac/ApoE−/− mice, which indicate increased residence of monocytes/macrophages in the vessel wall, we next sought to determine whether MED1 deficiency in monocytes/macrophages promotes the pro-inflammatory M1 phenotype in the vessel wall. Analysis of tissue extracts from aortic specimens showed that the expression of M1 marker genes, including IL-1β, IL-6, cyclooxygenase 2 (COX2), inducible nitric oxide synthase (iNOS), chmeokine (C-X-C motif) ligand 1 (i.e., Gro1), chemokine (C-C motif) ligand 2 (MCP-1), and TNFα, was significantly greater in the aortic tissue of MED1ΔMac/ApoE−/− mice than MED1fl/fl/ApoE−/− littermates (Figure 2A). In addition, the level of NLR family pyrin domain-containing 3 (NLRP3) was increased, so NLRP3 inflammasome might be induced in the aorta of MED1ΔMac/ApoE−/− mice. In contrast, the level of M2 marker genes, including Arg1, mannose receptor C type 1 (Mrc1), resistin-like α (Retnla/Fizz1), chitinase 3-like 3 (Chi3l3) as well as PPARγ, was lower in aortic tissue of MED1ΔMac/ApoE−/− than MED1fl/fl/ApoE−/− mice (Figure 2B). Of note, the circulatory level of IL-1β was greatly elevated in MED1ΔMac/ApoE−/− mice (Figure 2C), which was consistent with the NLRP3 induction in the arterial wall. In addition, we performed immunostaining on sections of aortic roots of MED1ΔMac/ApoE−/− and MED1fl/fl/ApoE−/− control mice on Western diet for 4 weeks (Supplemental Figure III). M1-like macrophages (MOMA2+iNOS+) were increased in the early stage of atherosclerosis in MED1ΔMac/ApoE−/− mice (Supplemental Figure III). Therefore, the increased M1/M2 polarization in macrophages deficient in MED1 may drive atherosclerosis initiation and progression.
Figure 2. MED1 deficiency in monocyte/macrophages aggravates inflammatory response in the aorta.
(A) RT-qPCR analysis of the mRNA level of M1 marker genes (IL-1β, IL-6, COX2, iNOS, and Gro1), MCP-1, TNFα, and NLRP3 and (B) M2 marker genes (Arg1, Mrc1, Retnla, Chi3l3, and PPARγ) in aortas of MED1fl/fl/ApoE−/− mice and their MED1ΔMac/ApoE−/− littermates (n=8 in each group) fed a Western diet for 12 weeks. (C) ELISA of the plasma level of IL-1β in MED1fl/fl/ApoE−/− and MED1ΔMac/ApoE−/− mice (n=17 and n=22, respectively). Data are mean±SEM from 3 independent experiments. In (A, B), the levels of mRNA are compared with those in MED1fl/fl/ApoE−/− mice set to 1. *p< 0.05.
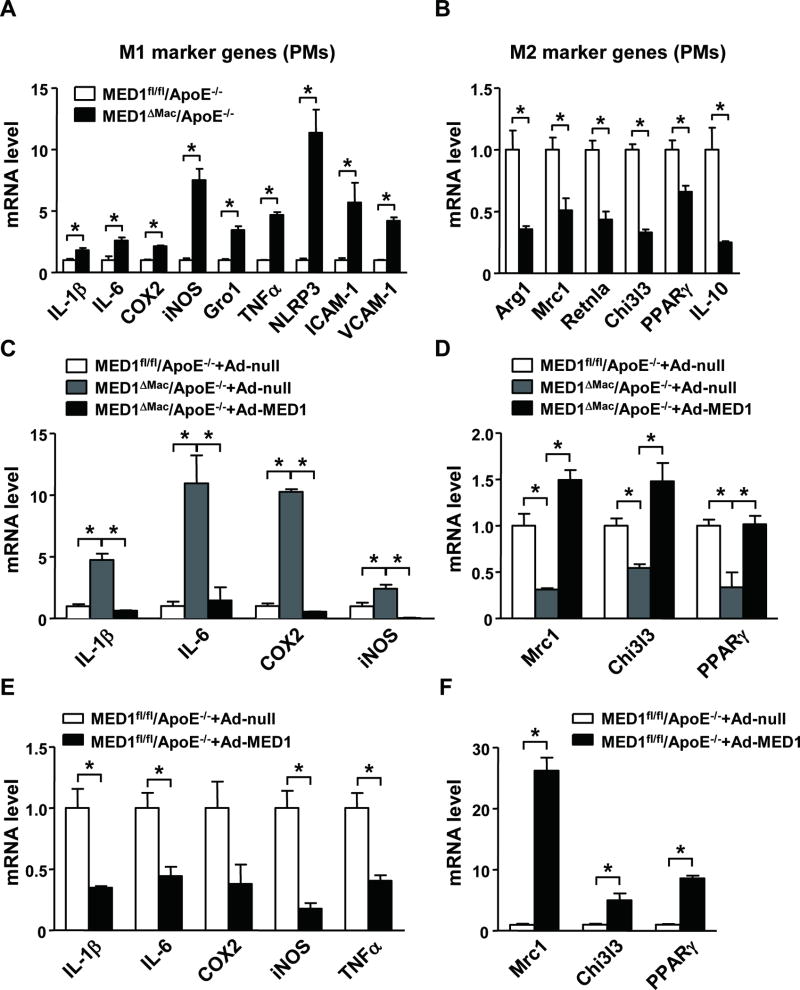
To examine whether the M1 polarization found in the aorta of MED1ΔMac/ApoE−/− mice was attributed to the imbalanced expression of pro- versus anti-inflammatory genes in MED1-deficient macrophages, we compared the expression profile of M1 and M2 marker genes in peritoneal macrophages (PMs) isolated from the 2 lines of mice. Relative to PMs from MED1fl/fl/ApoE−/− mice, those from MED1ΔMac/ApoE−/− mice showed augmented mRNA expression of IL-1β, IL-6, COX2, iNOS, Gro1, TNFα, NLRP3, vascular cell adhesion molecular 1 (VCAM-1), and intercellular adhesion molecule 1 (ICAM-1) (Figure 3A). However, the mRNA expression of M2 marker genes (e.g., Arg1, Mrc1, Retnla, and Chi3l3), PPARγ, and IL-10 was significantly reduced in MED1ΔMac/ApoE−/− PMs (Figure 3B). We further performed a rescue experiment by complementarily expressing MED1 in MED1ΔMac/ApoE−/− PMs. Adenovirus-mediated MED1 expression decreased the levels of pro-inflammatory genes, including IL-1β, IL-6, COX2, and iNOS, whose levels were comparable to that in MED1fl/fl/ApoE−/− PMs transfected with Ad-null (Figure 3C). In contrast, complementary expression of MED1 reversed the level of Mrc1, Chi3l3, and PPARγ M2 marker genes (Figure 3D). To test whether MED1 was sufficient for suppressing M1 marker genes, we conducted a gain-of-function experiment by overexpressing MED1 in MED1fl/fl/ApoE−/− PMs. Compared with Ad-null overexpression, Ad-MED1 overexpression decreased the mRNA levels of IL-1β, IL-6, COX2, iNOS, and TNFα in wild-type PMs (Figure 3E) but increased the expression of Mrc1, Chi3l3, and PPARγ M2 marker genes (Figure 3F).
Figure 3. MED1 deficiency promotes macrophage M1 polarization.
(A, B) Peritoneal macrophages were isolated from MED1fl/fl/ApoE−/− mice and MED1ΔMac/ApoE−/− littermates (n=6 in each group). RT-qPCR analysis of the mRNA level of M1 and M2 marker genes in pooled macrophages. (C, D) Peritoneal macrophages pooled from 6 MED1ΔMac/ApoE−/−mice or 6 MED1fl/fl/ApoE−/− littermates were infected with Ad-null (50 multiplicity of infection [MOI]) or Ad-MED1 (50 MOI). (E, F) RT-qPCR analysis of mRNA levels of (E) IL-1β, IL-6, COX2, iNOS, TNFα and (F) Mrc1, Chi3l3, and PPARγ in MED1fl/fl/ApoE−/− macrophages infected with Ad-null or Ad-MED1. In (A–D), the levels of mRNA were compared to those in MED1fl/fl/ApoE−/− macrophages set to 1. In E and F, the Ad-null infected levels were set to 1. Data are mean±SEM from 3 independent experiments. *p< 0.01.
MED1 deficiency in macrophages increases atherosclerosis in LDLR−/− mice
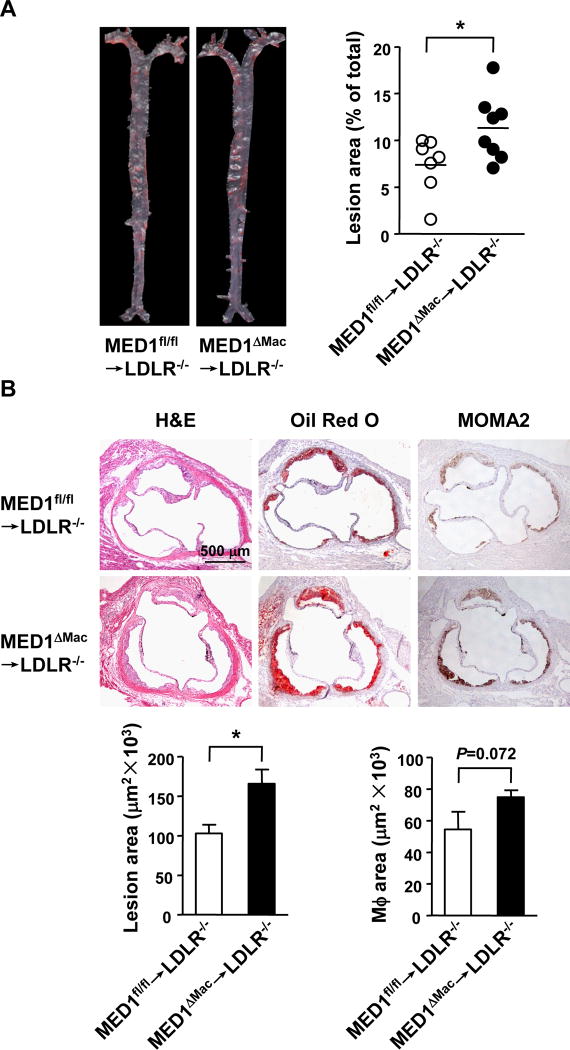
Given that the monocytes/macrophages residing in the arterial wall emanate from bone marrow-derived monocytic cells and indeed PMs from MED1ΔMac/ApoE−/− mice showed a phenotype shift from M2 to M1, we used bone marrow transplantation to further explore the role of MED1 in atherosclerosis. The use of LDLR−/−, but not ApoE−/−, mice was because ApoE-sufficient donors can reduce the atherosclerotic phenotype in ApoE-deficient recipients.29 LDLR−/− mice were lethally irradiated, and then received bone marrow from MED1ΔMac or MED1fl/fl donors. After a chow diet for 6 weeks, then a Western diet for 12 weeks, the 2 groups of mice were killed for assessing atherosclerosis. LDLR−/− mice receiving MED1ΔMac bone barrow (MED1ΔMac →LDLR−/−) showed a 54% increase in lesion size as compared with controls receiving MED1fl/fl bone marrow (MED1fl/fl→LDLR−/−) (11.3 ±1.2% vs 7.4 ±1.1%) (Figure 4A). As well, the aortic root of MED1ΔMac →LDLR−/− mice showed a 61% increase in lesion size (165.4±18.1×103 vs 102.7±11.1×103 µm2) and 38% increase in MOMA2-positive areas [54.4±11.1×103 vs 74.9±4.4 ×103 µm2 (Figure 4B)]. With comparable lipid profiles in the 2 groups of animals (Supplemental Table III), MED1 deficiency in bone marrow-derived macrophages resulted in increased atherosclerosis in LDLR−/− mice. Collectively, Figures 1–4 suggest that an imbalance between pro-inflammatory M1 and anti-inflammatory M2 gene expression caused by macrophage-specific MED1 silencing is associated with spontaneous and accelerated atherosclerosis in ApoE−/− and LDLR−/− mice.
Figure 4. MED1 deficiency in macrophages increases atherosclerosis in LDLR−/− mice.
Bone marrow from MED1fl/fl or MED1ΔMac donor mice were transplanted to 8-week-old and lethally irradiated LDLR−/− male mice [MED1fl/fl→LDLR−/− (n=7) and MED1ΔMac→LDLR−/− (n=8)]. After a 6-week chow diet then 12-week Western diet, recipient mice were killed. Atherosclerotic lesion areas in the aorta tree (A) and aortic roots (B) were analyzed and data are presented as mean±SEM. * p<0.05.
MED1 deficiency in macrophages potentiates innate immune stimulation
With results from animal experiments in Figures 1–4 suggesting the atheroprotective role of MED1 in macrophages, we then investigated the involved mechanism. Firstly, we performed quantitative RT-PCR (qPCR) analysis for MED1 expression in PMs after stimulation with LPS, an innate immune stimulation that is well known to induce M1 polarization. The expression of MED1 and PPARγ was strongly repressed by LPS treatment (Supplemental Figure IV).30 Next, the differential gene expression in MED1fl/fl and MED1ΔMac PMs stimulated with or without LPS was analyzed using mouse atherosclerosis PCR array. The heat map in Figure 5A reveals that the expression of 20 genes involved in innate immune response and M1 polarization was significantly higher in MED1ΔMac than MED1fl/fl macrophages after LPS treatment. Upregulated genes in MED1ΔMac macrophages were the pro-inflammatory genes MCP-1, chemokine (C-C motif) ligand 5 (CCL5), Gro1, IFNγ, IL-1α, IL-1β, and TNFα; adhesion molecules CD44, ICAM-1, integrin subunit α2 (i.e., ITGA2), thrombospondin 4 (i.e., THBS4), and VCAM-1; apoptosis-related genes BCL2-related protein A1α (BCL2A1α), caspase-8 (i.e., CASP8) and FADD-like apoptosis regulator (i.e., CFLAR), and TNFα-induced protein 3 (TNFαIP3, also known as A20); and extracellular molecules, including leukemia inhibitory factor LIF, PDGFB, serpin family B member 2 (SERPINB2), serpin family E member 1 (SERPINE1), and tenascin C (TNC). Conversely, downregulated genes were PPARγ, RXRα, and BCL2, and Bcl-2-like 1 (BCL2L1) anti-apoptosis genes. Using qPCR, we further validated the mRNA expression of M1 marker genes in MED1fl/fl and MED1ΔMac macrophages challenged with LPS. Levels of M1 markers, including IL-1β, IL-6, COX2, MCP-1, iNOS, Gro1, TNFα, NLRP3, VCAM-1, ICAM-1, and CCL5, were confirmed to be higher in LPS-treated MED1ΔMac than MED1fl/fl macrophages (Figure 5B). The protein levels of these inflammatory markers, such as COX2, MCP-1, iNOS, and TNFα were consistently increased in MED1ΔMac macrophages treated with LPS (Figure 5C). Cytoscape software was then used to construct an MED1-related gene regulatory network that consolidated the results in Figure 5A and B. Such in silico analysis shown in Figure 5D demonstrated that MED1 deficiency downregulated PPARγ and concurrently upregulated molecules facilitating a pro-inflammatory and dysregulated redox state, which potentiated the phenotype switch from M2 to M1 macrophages. The pro-inflammatory nature of MED1ΔMac PMs is also supported by the nuclear portion of p65 NF-κB being slightly increased in MED1ΔMac PMs and the C/EBPβ mRNA level increased in LPS-stimulated MED1ΔMac PMs (Supplemental Figure V).
Figure 5. MED1 deficiency in macrophages potentiates an inflammatory response.
Peritoneal macrophages isolated from MED1fl/fl and MED1ΔMac mice were treated with or without LPS (50 ng/ml) for 6 hr. (A) Heat map from microarray assay shows up- and downregulation of selected atherosclerosis-related genes. (B) qPCR analysis of the mRNA level of the indicated genes. All experiments were repeated 3 times (n=6 per group). * p<0.05. (C) Western blot analysis of the expression of COX2, MCP-1, iNOS, and TNFα. β-actin was used as loading control. Samples were pooled from 6 animals in each of the indicated groups. * p<0.05. (D) Cytoscape reconstruction of the MED1-related gene regulatory network summarizing results in A and B. Red and green circles represent up- and downregulated genes with MED1 deficiency in macrophages.
PPARγ induction of M2 marker genes depends on MED1
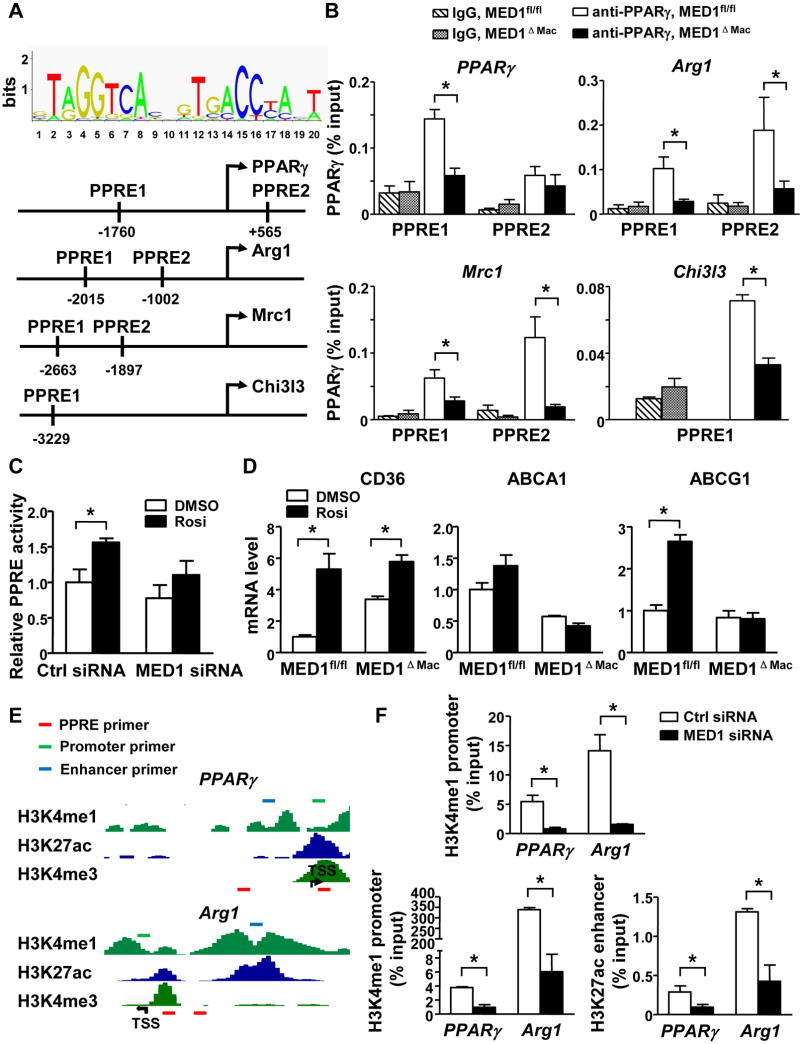
Because PPARγ is required for the maturation of M2 macrophages and MED1 is essential for PPARγ-regulated transactivation,12, 13, 31 we then studied how MED1 is involved in the PPARγ-mediated M2 phenotype switch. We first used ChIP assay to examine the effect of MED1 on the binding of PPARγ to the PPRE in the promoter region of M2 marker genes including PPARγ, Arg1, Mrc1, and Chi3l3 (Figure 6A). PGC1α lacking the PPRE in the promoter was used as positive control (Supplemental Figure VI). MED1 deficiency in MED1ΔMac macrophages significantly decreased the binding of PPARγ to the PPRE in the promoter region of PPARγ, Arg1, Mrc1 and Chi3l3 (Figure 6B). Consistently, MED1 knockdown significantly decreased the binding of PPARγ to the PPRE in the promoter region of PPARγ, Arg1, Mrc1 and Chi3l3 (Supplemental Figures VII and VIII).
Figure 6. MED1 mediates PPARγ-induced M2 genes.
(A) Bioinformatics analysis of PPREs in the upstream region of M2 marker genes (PPARγ, Arg1, Mrc1, and Chi3l3). (B) PMs were isolated from MED1fl/fl and MED1ΔMac mice, and then nuclear extracts were obtained. ChIP assay was performed with the use of anti-PPARγ to detect PPARγ enrichment at the upstream region of the PPARγ, Arg1, Mrc1, and Chi3l3 gene. IgG was used as the isotype control. The primer sets used in qPCR were sequences adjacent to the predicted PPREs. Bar graphs represent the binding of PPARγ as % of input. (C) RAW264.7 cells were transfected with PPRE-TK-luc reporter constructs together with control or MED1 siRNA. The transfected cells were then stimulated with PPARγ agonist rosiglitazone (Rosi) (10 mM) for 24 hr. Luciferase activity was measured and normalized to that of β-gal. (D) qPCR was performed to detect the mRNA level of CD36, ABCA1 and ABCG1 in MED1fl/fl and MED1ΔMac macrophages treated with Rosi. Graphs represent mean±SEM from 6 mice per group. * p<0.05. (E) The epigenetic landscapes of the upstream region of mouse PPARγ and Arg1 [obtained from http://epgg-test.wustl.edu/d/mm9/ENCFF001JYI.bigWig,(H3K4me1), http://epgg-test.wustl.edu/d/mm9/ENCFF001JYV.bigWig,(H3K27ac) and http://epgg-test.wustl.edu/d/mm9/ENCFF001JYO.bigWig (H3K4me3)]. The red, green, and blue boxes represent the locations of primers detecting the respective PPRE, promoter, and enhancer. (F) RAW264.7 cells were transfected with control or MED1 siRNA. ChIP assay of the respective enrichment of H3K4me1 and H3K27ac on the promoter and enhancer; H3K27ac on the enhancer of the PPARγ and Arg1 gene. Data represent as % of input and are mean±SEM from 3 independent experiments * p<0.05.
We then investigated whether the agonist-enhanced PPARγ activity depends on MED1 by transfecting RAW264.7 cells with PPRE-TK-luc reporter constructs together with control or MED1 siRNA. The transfected cells were then stimulated with PPARγ agonist rosiglitazone. As shown in Figure 6C, rosiglitazone treatment significantly induced the PPRE-driven transcriptional activity in RAW264.7 cells transfected with control siRNA. However, this induction was attenuated in cells with MED1 knocked down. In line with this result, rosiglitazone treatment increased the expression of the PPARγ-regulated genes, including CD36, ABCA1, and ABCG1 in MED1fl/fl, when compared with MED1ΔMac macrophages (Figure 6D).
Transcriptional activation is associated with a euchromatin state in the promoter and/or enhancer regions. Indeed, macrophage-unique PPARγ binding sites in the genome coincide with the active histone marker H3K9ac.32 Accordingly, we used ChIP-qPCR assay to investigate whether MED1 regulates the expression of M2 genes by modulating the chromatin states at the promoter and/or enhancer region of the M2 marker genes. Nuclear extracts were immunoprecipitated first with antibodies for H3K4me1 (recognizing both active enhancers and promoters) and H3K27ac (recognizing active enhancers). With PPARγ and Arg1 as representative M2 marker genes, we profiled the promoter and enhancer regions of these genes defined in ENCODE data (Figure 6E). Primer sets were then designed for qPCR assessment of the enrichment of H3K4me1 and H3K27ac in these upstream regions. Compared to control RAW264.7 cells, MED1 knockdown significantly reduced the enrichment of H3K4me1 at both promoter and enhancer regions, whereas H3K27ac was less abundant in the enhancer region of PPARγ and Arg1 (Figure 6F).
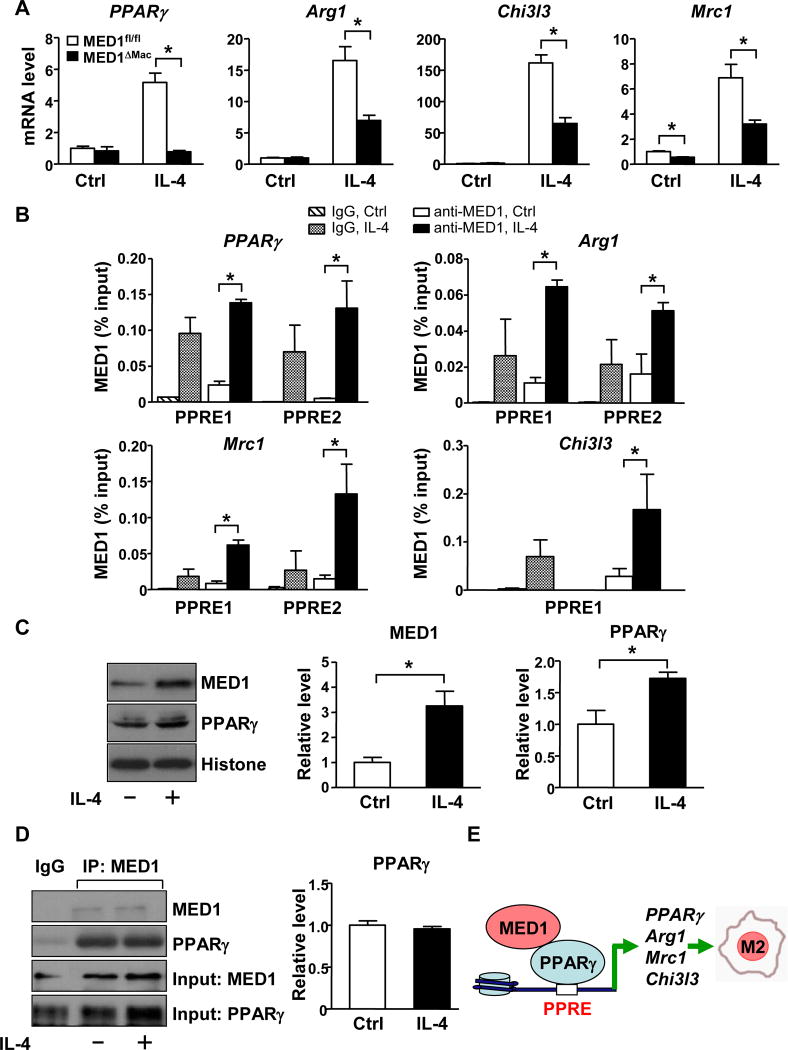
IL-4 induces activation of alternative M2 macrophages with attendant increase in the expression of PPARγ and other prototypical target genes that characterize the M2 phenotype.20 To further explore the role of MED1 in PPARγ-dependent M2 transition, we compared the mRNA level of M2 marker genes in MED1fl/fl and MED1ΔMac macrophages with or without IL-4 stimulation. At the basal level (without IL-4 treatment), the expression of M2 markers, including PPARγ, Arg1, and Chi3l3, was comparable between MED1ΔMac and MED1fl/fl macrophages, whereas Mrc1 expression was lower in MED1ΔMac macrophages (Figure 7A). With IL-4 treatment, the expression of these M2 markers was strongly induced in MED1fl/fl macrophages. However, such IL-4 induction of M2 genes was significantly attenuated in MED1ΔMac macrophages (Figure 7A). The decreased expression of M2 markers was also recapitulated in RAW264.7 cells transfected with MED1 siRNA (Supplemental Figure IX).
Figure 7. IL-4 induction of M2 marker genes is MED1-dependent.
Peritoneal macrophages isolated from MED1fl/fl and MED1ΔMac mice (A) and RAW264.7 cells (B–D) were treated with or without IL-4 (10 ng/ml) for 16 hr. (A) RT-qPCR of the mRNA level of PPARγ, Arg1, Chi3l3, and Mrc1 compared with MED1fl/fl without IL-4 set to 1. (B) ChIP assay was performed to detect the enrichment of MED1 at the predicted PPREs of PPARγ, Arg1, Chi3l3, and Mrc1. IgG was used as the isotype control. Bar graphs represent the binding of MED1 as % of input. (C) Western blot analysis of the protein level of MED1 and PPARγ. Histone bands indicate the loading control. (D) MED1 was immunoprecipitated, then immunoblotted with anti-MED1 or anti-PPARγ antibody. In (A–D), data are mean±SEM from at least three independent experiments, * p<0.05. The bar graphs represent the comparison with those without IL-4 treatment set to 1. (E) A graphic presentation of the mechanism by which MED1 in macrophages is atheroprotective through its regulation of macrophage polarization.
Because MED1 was essential for the euchromatin state of the upstream region of the M2 genes such as PPARγ and Arg1 (Figure 6D), we next explored the effect of IL-4 on MED1 binding to the PPREs in PPARγ, Arg1, Chi3l3, and Mrc1. IL-4 treatment indeed greatly increased the enrichment of MED1 to the PPREs of these genes (Figure 7B). Of note, with IL-4 treatment, the level of PPARγ and MED1 increased in RAW264.7 cells (Figure 7C). Coimmunoprecipitation revealed that MED1 interacted with PPARγ to a similar extent with or without IL-4 treatment (Figure 7D). However, MED1 had little, if any, interaction with either p65 NF-kB or C/EBPβ with or without IL-4 treatment (Supplemental Figure X). Thus, IL-4 induction of the M2 marker genes would be caused by the increased amount of MED1/PPARγ that transactivated the target genes.
Discussion
The principal finding of this study is that MED1 in macrophages is essential for an atheroprotective phenotype, which is mainly supported by the increased atherosclerosis in ApoE−/− or LDLR−/− mice with MED1 deficiency in bone marrow-derived macrophages. These results reveal a new function for MED1 in macrophages in addition to its roles in lipogenesis in liver and insulin sensitivity in muscle. The underlying mechanism is that MED1, functioning as a transcriptional coactivator, is essential for the PPARγ-mediated M2 polarization. Previous study by Odegaard et al. indicated that PPARγ preferentially binds to the PPRE located at the promoter region of M2 marker genes (e.g., Arg1).31 Our results further demonstrate that such transactivation of PPARγ on M2 marker genes depends on MED1 serving as a coactivator (summarized in Figure 7E).
Given that PPARγ is a master transcription factor for the monocytic lineage, MED1 may be an integrated part of the PPARγ transcriptional machinery in macrophages. However, MED1 also participates in the transcriptional activation regulated by several pro-inflammatory transcription factors such as NF-κB and C/EBPβ.9, 33 Although MED1 in macrophages facilitates PPARγ-mediated M2 gene expression, this positive effect seems to be omitted in regulating pro-inflammatory genes, including M1 markers that are regulated by NF-κB and C/EBPβ. Regarding possible mechanism, data presented in Supplemental Figure X demonstrated that MED1 had little interaction with either p65 NF-κB or C/EBPβ in RAW264.7 cells under basal condition. Moreover, neither p65 NF-κB nor C/EBPβ increased their interaction with MED1 during the IL-4-induced M2 polarization. Consistently, the reciprocal ChIP experiments revealed no detectable binding of p65 NF-κB to the upstream region of M1 marker genes including COX2, iNOS, and TNFα in RAW264.7 cells treated with IL-4 or infected with Ad-MED1 (data not shown). Thus, MED1 repression of the M1 genes may not involve a direct binding of NF-κB or C/EBPβ.
MED1, acting as a super-enhancer, may change the euchromatin status of M2 genes via histone modification and enhancer activity, which also depends on the expression level or activity of lineage-dependent transcription factors such as PPARγ in macrophages.34 Although MED1 is ubiquitously expressed, tissue-related functions depend on the MED1-enhanced expression of genes regulated by tissue-related master transcription factors.3 Hence, MED1 interacting with PPARγ to maintain an optimal level of M2 genes is essential to maintain macrophage homeostasis. Conversely, MED1 deficiency in macrophages would lead to impaired maturation of PPARγ-driven M2 macrophages. Data presented in Figure 7C showed that M2 stimuli (e.g., IL-4) increased the level of MED1 and PPARγ in RAW264.7 cells. On the other hand, M1 stimuli (e.g., LPS) suppressed the expression of MED1 and PPARγ in macrophages (Supplemental Figure IV).30 Thus, it is reasonable to infer that the abundance of the MED1/PPARγ complex contributes to M1 versus M2 polarization. In terms of atherosclerosis, an optimal level of MED1 in monocytes/macrophages would be athero-protective. This postulation is supported by an increase in the M1-like macrophages in the early stage of atherosclerosis in MED1ΔMac/ApoE−/− mice (Supplemental Figure III).
MED1 also seems to regulate signal-dependent transcription factor (SDTF)-dependent expression.35 In accordance with the concept of SDTFs, PPARγ induced by IL-4 (Figure 7E) can be considered SDTF-dependent regulation during the M2 polarization. In the context of epigenetic regulation, the IL-4 induction of PPARγ level seems to increase PPARγ binding to the PPRE at the upstream region of various M2 genes.20 Thus, the enhanced atherosclerosis in MED1ΔMac/ApoE−/− mice (Figure 1) would be due mainly to the impaired MED1/PPARγ regulation of M2 polarization in monocytes/macrophages. Intriguingly, MED1 deficiency in mouse liver causes hypercholesterolemia (Borensztajn and Reddy, unpublished results) and therefore MED1 would regulate cholesterol metabolism in hepatocytes, which in turn affects the lipoprotein profile in the circulation. In the current study, lipid profiles were comparable between MED1ΔMac/ApoE−/− mice and their MED1fl/fl/ApoE−/− littermates under a chow diet or atherogenic Western diet (Supplemental Tables I and II), so MED1 in macrophages may not alter lipoprotein metabolism in the circulation. Although increased cholesterol content is implicated in macrophage M1 polarization,36 cellular cholesterol level in MED1fl/fl and MED1ΔMac PMs with or without oxLDL stimulation seemed to be comparable (Supplemental Figure XI). Given the technical limitations in our analysis of cellular cholesterol content, a contribution of cellular cholesterol and lipid raft cholesterol to changes in macrophage signaling and M1/M2 phenotypic change should not be excluded.
Besides PPARγ, MED1 can interact with many other nuclear receptors, some of which have been shown to exert anti-inflammatory effects in macrophages.37 Using bioinformatics, we found that the promoter regions of the M2 genes (e.g., PPARγ, Arg1, Mrc1 and Chi3l3) do include the putative binding sites for RXRα, RARα, RORα, FXR, and HNF4α (Supplemental Figure XIIA). Similar approach has been used to infer the binding sites of NF-κB, C/EBPβ, RXRα, RARα, RORα, FXR, and HNF4α in the promoters of M1 marker genes (e.g., COX2, iNOS and TNF) (Supplemental Figure XIIB). This in silico analysis suggests that these nuclear receptors may also be involved in the anti-inflammatory effect exerted by MED1 in macrophages.
Super-enhancers were originally defined by the differential level of MED1 enrichment at multiple loci in the genome.3 MED1 interacting with BRD4, an epigenetic reader that recognizes acetylated lysines, is involved in the inflammatory response in myeloma tumor cells.38 Inhibition of BRD4 by the thienotriazolodiazepine JQ1 reduces the MED1 enrichment, especially in the super enhancer regions.38 JQ1 also attenuates TNFα-induced NF-κB activation in endothelial cells, thereby decreasing atherosclerosis.39 This observation may seem contradictory to our finding that MED1 deficiency was pro-M1, or pro-atherosclerotic, whereas suppression of MED1 binding to enhancers by JQ1 is anti-atherosclerotic.3, 39 A multitude of explanations may rationalize such paradox. First, our experimental approaches involved tissue-specific MED1 ablation in macrophages, whereas in the previous study, BRD4 inhibition decreased MED1 super enhancer binding in endothelial cells. These differential results in macrophages versus endothelial cells agree with the notion that MED1-modulated gene expression is tissue-dependent. Nonetheless, the paradox is that neither study can justify why only a certain set of genes can be transcriptionally activated, given that MED1 is ubiquitously important in the Mediator complex. Thus, pro- and anti-inflammatory stimulating cues combined with tissue-specific signals may cause a unique recruitment of pioneer transcription factors, co-activators, and/or co-suppressors to the MED1-associated Mediator complex so that a set of genes are selectively activated. An alternative explanation is that distinct signals may result in conformational changes of MED1, which in turn affect the genomic loci of MED1 super enhancer binding that may direct long-distance chromatin remodeling and ensuing gene expression.
In summary, our data strongly support that MED1 in macrophages is anti-atherosclerotic because of PPARγ-dependent M2 polarization. However, the detailed molecular insights of the selective induction of M2 marker genes require further investigation to interpret the epigenetic landscapes of MED1 interactions at the whole genome level with functionally opposing stimuli.
Supplementary Material
Highlights.
MED1 deficiency in macrophages results in the increase of atherosclerosis in ApoE−/− and LDLR−/− mice.
MED1 regulates the M1/M2 phenotype switch of macrophage.
MED1 facilitates the binding of PPARγ to the PPRE in the promoter region of M2 marker genes, such as Arg1, Mrc1 and Chi3l3.
Acknowledgments
We thank Laboratory Animal Center for their technical support and Ting Lei for technical assistance.
Sources of Funding
This work was supported by grants from the National Natural Science Foundation of China (81200207, 81070250, 81670452, 81270349) (L.B., E.L., J.Y.S.), Natural Science Foundation of Shaanxi Province (2014JQ4138) (L.B.), a Public Service Platform Grant from Shaanxi Province (2014FWPT-07) (E.L.), and US National Institutes of Health grant HL108735 (J.Y.S.).
Abbreviations
- MED1
Mediator 1
- PPARγ
Peroxisome proliferator-activated receptor γ
- ApoE
Apolipoprotein E
- C/EBPβ
CCAAT/enhancer-binding protein β
- TNFα
Tumor necrosis factor α
- MCP-1
Monocyte chemoattractant protein 1
- PPRE
PPAR-responsive element
- LDLR
Low-density lipoprotein receptor
- Arg1
Arginase 1
- Mrc1
Mannose receptor C type 1
- NLRP3
NLR family pyrin domain-containing 3
- COX2
Cyclooxygenase 2
- iNOS
Inducible nitric oxide synthase
- Retnla
Resistin-like molecule α
- VCAM-1
Vascular cell adhesion molecular 1
- ICAM-1
Intercellular adhesion molecule 1
- LPS
Lipopolysaccharide
Footnotes
Disclosures
None.
References
- 1.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Roeder RG. The Mediator subunit MED1/TRAP220 is required for optimal glucocorticoid receptor-mediated transcription activation. Nucleic Acids Res. 2007;35:6161–6169. doi: 10.1093/nar/gkm661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK. Defects of the heart, eye, megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 8.Drane P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15:3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Gade P, Nallar SC, Raha A, Roy SK, Karra S, Reddy JK, Reddy SP, Kalvakolanu DV. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-beta-driven transcription in response to interferon-gamma. J Biol Chem. 2008;283:13077–13086. doi: 10.1074/jbc.M800604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Viswakarma N, Jia Y, Bai L, Vluggens A, Borensztajn J, Xu J, Reddy JK. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010:2010. doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 13.Bai L, Jia Y, Viswakarma N, Huang J, Vluggens A, Wolins NE, Jafari N, Rao MS, Borensztajn J, Yang G, Reddy JK. Transcription coactivator mediator subunit MED1 Is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia Y, Viswakarma N, Fu T, Yu S, Rao MS, Borensztajn J, Reddy JK. Conditional ablation of mediator subunit MED1 (MED1/PPARBP) gene in mouse liver attenuates glucocorticoid receptor agonist dexamethasone-induced hepatic steatosis. Gene Expr. 2009;14:291–306. doi: 10.3727/105221609788681213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci U S A. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia Y, Chang HC, Schipma MJ, et al. Cardiomyocyte-Specific Ablation of Med1 Subunit of the Mediator Complex Causes Lethal Dilated Cardiomyopathy in Mice. PLoS One. 2016;11:e0160755. doi: 10.1371/journal.pone.0160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li AC, Glass CK. The macrophage foam cell as a target for therapeutic intervention. Nat Med. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- 18.Libby P, Hansson GK. Inflammation and immunity in diseases of the arterial tree: players and layers. Circ Res. 2015;116:307–311. doi: 10.1161/CIRCRESAHA.116.301313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouhlel MA, Derudas B, Rigamonti E, Dievart R, Brozek J, Haulon S, Zawadzki C, Jude B, Torpier G, Marx N, Staels B, Chinetti-Gbaguidi G. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6:137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Thompson PW, Bayliffe AI, Warren AP, Lamb JR. Interleukin-10 is upregulated by nanomolar rosiglitazone treatment of mature dendritic cells and human CD4+ T cells. Cytokine. 2007;39:184–191. doi: 10.1016/j.cyto.2007.07.191. [DOI] [PubMed] [Google Scholar]
- 23.Penas F, Mirkin GA, Vera M, Cevey A, Gonzalez CD, Gomez MI, Sales ME, Goren NB. Treatment in vitro with PPARalpha and PPARgamma ligands drives M1-to-M2 polarization of macrophages from T. cruzi-infected mice. Biochim Biophys Acta. 2015;1852:893–904. doi: 10.1016/j.bbadis.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Wang N, Verna L, Chen NG, Chen J, Li H, Forman BM, Stemerman MB. Constitutive activation of peroxisome proliferator-activated receptor-gamma suppresses pro-inflammatory adhesion molecules in human vascular endothelial cells. J Biol Chem. 2002;277:34176–34181. doi: 10.1074/jbc.M203436200. [DOI] [PubMed] [Google Scholar]
- 25.Yang XY, Wang LH, Chen T, Hodge DR, Resau JH, DaSilva L, Farrar WL. Activation of human T lymphocytes is inhibited by peroxisome proliferator-activated receptor gamma (PPARgamma) agonists. PPARgamma co-association with transcription factor NFAT. J Biol Chem. 2000;275:4541–4544. doi: 10.1074/jbc.275.7.4541. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Ishibashi S, Perrey S, et al. Troglitazone inhibits atherosclerosis in apolipoprotein E-knockout mice: pleiotropic effects on CD36 expression and HDL. Arterioscler Thromb Vasc Biol. 2001;21:372–377. doi: 10.1161/01.atv.21.3.372. [DOI] [PubMed] [Google Scholar]
- 27.Babaev VR, Yancey PG, Ryzhov SV, Kon V, Breyer MD, Magnuson MA, Fazio S, Linton MF. Conditional knockout of macrophage PPARgamma increases atherosclerosis in C57BL/6 and low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005;25:1647–1653. doi: 10.1161/01.ATV.0000173413.31789.1a. [DOI] [PubMed] [Google Scholar]
- 28.Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, Le Roith D, Chambon P, Gonzalez FJ, Reddy JK. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 29.Linton MF, Atkinson JB, Fazio S. Prevention of atherosclerosis in apolipoprotein E-deficient mice by bone marrow transplantation. Science. 1995;267:1034–1037. doi: 10.1126/science.7863332. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Sun ZW, Zhao WT, Wang Z, Wu MJ, Pan YY, Yan H, Zhu JH. CD97/ADGRE5 Inhibits LPS Induced NF-kappa B Activation through PPAR-gamma Upregulation in Macrophages. Mediators of Inflammation. 2016 doi: 10.1155/2016/1605948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao W, Breese E, Bowers A, Hoggatt J, Pelus LM, Broxmeyer HE, Goebl M, Harrington MA. SIMPL enhancement of tumor necrosis factor-alpha dependent p65-MED1 complex formation is required for mammalian hematopoietic stem and progenitor cell function. PLoS One. 2013;8:e61123. doi: 10.1371/journal.pone.0061123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang DX, Glass CK. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol. 2013;51:T37–50. doi: 10.1530/JME-13-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamson S, Leitinger N. Phenotypic modulation of macrophages in response to plaque lipids. Current Opinion in Lipidology. 2011;22:335–342. doi: 10.1097/MOL.0b013e32834a97e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nature Reviews Immunology. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 38.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown JD, Lin CY, Duan Q, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.